Abstract
Emerging technologies are bringing single-cell genome sequencing into the mainstream; this field has already yielded insights into the genetic architecture and variability between cells that highlight the dynamic nature of the genome.
The idea of analyzing genomes at the single-cell level is quite old: the banded structure of polytene chromosomes was first reported in images of single cells from the salivary glands of insects in 1882 (ref. 1). In 1935, Calvin Bridges published a map of the Drosophila genome based on such images, which enabled the identification of large-scale genomic rearrangements that distinguished different individuals, lines and species2. More recently, there has been sustained effort to apply the polymerase chain reaction and other biochemical amplification technologies to single cells. Notable results include the analysis of recombination hot-spot usage in single sperm cells two decades ago3 as well as the routine analysis of single cells from embryos for preimplantation genetic diagnoses4. Given the century-long history of this field, it is quite reasonable to ask, why has there been a sudden recent flood of attention on single cells?
We argue that the answer has to do with phenomenal recent advances in the ability to analyze detailed sequence information from single cells. These are defined by a confluence of three factors: advances in technology that enable effective whole genome and transcriptome amplification, the relentless improvement in DNA-sequencing instruments with ever higher throughput and lower costs, and the invention of technologies for single-cell manipulation such as microfluidics and fluorescence-activated cell sorting. The last 5 years have seen a burst of papers from labs around the world that have developed expertise in single-cell gene expression and genome analysis, and commercial vendors have played a crucial role in helping expand access to these technologies. Single-cell genome analysis is now influencing areas as diverse as microbial ecology, cancer, prenatal genetic diagnosis and the study of human genome structure and variation (reviewed in refs. 5 and 6). In this Commentary, we will focus on recent highlights and our best guesses of where the field might be going next.
Single-cell sequencing of single-celled life
Microbial ecology is an ideal arena for single-cell genomics because the vast majority of microbes—99% of species, by most estimates—cannot currently be cultured. Uncultured species exist as biological ‘dark matter’, as they can only be observed indirectly by methods such as marker-gene sequence surveys. Although metagenomic approaches can help establish gene inventories from such complex environments, the fundamental link between organism and gene is lost. It is only through single-cell genomic approaches that one can understand the connections between a unicellular organism's identity and the functional capabilities provided by its genome. A consequence of this is that an enormous fraction of genetic and evolutionary diversity on earth is not fully accounted for in current genomic databases.
The first uncultured microorganism to be analyzed with single-cell sequencing was a bacterium that lives on human tooth plaque7. In recent years there have been more than a dozen publications on single-cell genomes from yet-uncultured microbes, and we believe that this trend is going to increase exponentially as technologies continue to improve. As these data accumulate, we may see the discovery of previously unknown microbial functions and metabolites, the identification of many new species that relate to human health in both positive ways (such as through the microbiome) and negative ways (as newly recognized pathogens), and even major revisions to the structure of the ‘tree of life’ and insights into the evolutionary relationship between eukaryotes, bacteria and archaea.
The diversity of morphology, physiology and genotypes to explore among microbes also creates technical challenges for single-cell analysis. Numerous sample-specific considerations come into play when choosing reaction formats and chemistries5. For instance, microbes often require stringent lysis conditions, and the need to match conditions to different microbe types may complicate protocols. Because DNA purification is not commonly applied before amplification, the amplification reagents must also be compatible with the lysis reagents and contents of the lysed cells. Complex lysis and amplification protocols are well-suited to microwell plates and integrated microfluidic devices where protocol steps can be automated; interestingly, the performance of biochemical amplifiers improves as reaction volumes are shrunk to nanoliters in microfluidic formats8. Simple protocols may be well suited to reverse-emulsion liquid-droplet systems, where thousands of individual microreactors can be quickly produced and processed. Nearly all of the microbial single-cell sequencing results to date have used one particular whole-genome amplification chemistry: multiple displacement amplification (MDA), which is an isothermal amplification scheme that uses random primers and that is based on the strand-displacement ability of Φ29 DNA polymerase9.
Human haplotypes
Human-genome analysis has rapidly progressed from determining the reference sequence for the ‘average’ human genome to prolific sequencing of personal genomes, and it may seem surprising that single-cell approaches have anything more to contribute. However, some aspects of the human genome have been very challenging to determine using conventional techniques. For example, all of us have two genomes within each of our cells—one from our mother and another from our father—and the location of sequence variation in each haploid genome can have a significant effect on gene expression, protein function and disease.
The best-known example of this is variation in human leukocyte antigen (HLA) genes, whose haplotype is important to understand for bone marrow transplants, but it applies just as well to compound heterozygous mutations—two mutations at a single locus that may be harmless when they reside in the same haplotype, but deleterious when distributed between the maternal and paternal allele. Current techniques have not been able to resolve these differences, known as haplotype determination, at a genome-wide level with any degree of precision. The best conventional methods for haplotype determination require additional sequencing within a family pedigree, mainly from parents. Clearly, this is not a practical approach in most clinical situations.
Single-cell chromosome isolation enabled the first genome-wide haplotype measurement, with haplotype phase determined across the lengths of entire chromosomes10. This work was quickly followed by related single-cell sequencing approaches using small numbers of cells11, or in the case of males, large numbers of individual sperm cells12. We expect that further application of these techniques and long-read sequencing technologies that determine haplotypes of genomic segments will accelerate analysis of refractory parts of the human genome. The HLA region, which is the most polymorphic part of the human genome and intimately involved in the immune system and many aspects of human health, is a particularly interesting target, but up to now it has been sequenced in only a limited number of individuals owing to its haplotype complexity.
Another area touched by single-cell genomics is the analysis of recombination patterns across human diversity. Recombination is the cutting and pasting of large blocks of the maternally and paternally inherited chromosomes to create entirely new genomes in sperm and egg cells, and it is a major contributor to genetic diversity in the human species. It is known that recombination does not happen with uniform probability across the genome; rather, certain ‘hot spots’ experience frequent recombination. One of the earliest contributions of single-cell genomics was to show that there is differential hot-spot usage between individuals: some spots may be hot for one person but not another13. More recently, single-cell approaches have enabled the measurement of genome-wide recombination patterns and mutation rates in individual sperm cells, enabling the first studies of genome-wide hot-spot behavior within individuals12,14. We expect that further genomic analyses of single sperm cells will enable the study of recombination mutants (for example, in individuals carrying rare alleles of PRDM9) as well as the potential diagnosis of those with meiotic dysfunction related to sterility and infertility.
Somatic variation
The value of sequencing individual human genomes is increasingly being recognized, yet a personal genome is actually an average of cellular genomes in the body, which also vary. Genomic differences in some cell types are well characterized and have been appreciated for decades; these include B cells of the immune system, which make such a strong commitment to expressing a particular unique antibody that they irreversibly reprogram that gene in their genome. Germ cells also differ through the well-characterized process of meiosis and recombination, as discussed above. Less well understood but still important are the gradual accumulation of mutations through errors in cell division and the movement of mobile genetic elements15.
This gradual accumulation of errors is associated both with aging in general and with cancer in particular, so it is not surprising that these areas will become important for single-cell genomic applications. To date, single-cell methods have been used to directly measure the de novo mutation rate in human sperm14 as well as immortalized human cell lines16. They have also been used to determine the order of mutations that occur in normal hematopoietic stem cells before their transformation to acute myelogenous leukemia17, to understand the lineage structure of leukemia tumors18 and to estimate the clonal structure of breast tumors19. Mosaic variation is also known to exist in adult neural tissue and has been impli-cated in neurodegenerive disorders such as Alzheimer's disease20. Recently, single-cell genomic techniques have been used to detect megabase-scale copy number varia-tion in a large fraction of induced pluripo-tent stem cell–derived neurons and normal postmortem brain cells using single-cell MDA and genomic analysis21. Similarly, single-cell MDA and PCR-based whole-genome amplification were used to show how retrotransposition of an L1 element is a potential driver of somatic mosaicism in the brain, and further, how a variant present in a third of cells or less can drive serious brain disease (hemimegalenceph-aly)22. A complementary method—fluo-rescence in situ hybridization—was used to show how the proportion of aneuploid neurons increases in aging mice23. This is a fascinating area, as there are varying degrees of evidence that mosaic somatic variation is functional in development24, is present in normal mature neural tissue21, may explain variability among ‘normal’ neural phenotypes, can cause neurological disease22, may contribute to psychiatric dis-ease and increases with aging23.
When to go single-cell
When does it make economic sense to invest in sequencing single-cell genomes? Tumor genomes are highly heterogeneous and accrue mutations at widely varied rates, so they may be obvious targets for single-cell sequencing. While bulk tumor sequencing does not allow the unambiguous deconvolution of component clonal populations, it does point to genomic loci with sequence heterogeneity where targeted single-cell sequencing can reveal further detail. Such a staged approach greatly reduces sequencing costs, increasing the number of single cells that can be sequenced for a given tumor.
It is not clear that it will ever become cost effective to sequence entire genomes from large numbers of single cells from a given tumor, but most of the same benefits can come from analyzing important subsets of the genome or using shallow sequencing to measure lower-resolution copy number changes in these cells, in a manner quite reminiscent of that of Bridges and his Drosophila genomes from 80 years ago! An intriguing potential alternative to the staged approach is to use a single-step method in which exome sequences from many single cells are directly measured, thereby allowing the bulk tumor exome to be ‘calculated’ while also revealing the true clonal diversity within the tumor; this approach can be comparable in cost to whole-tumor sequencing.
Sequence before life
Single-cell sequencing is sometimes the only alternative for probing rare or unique cells. Preimplantation genetic diagnosis (PGD) is a procedure used for couples trying to conceive a child through in vitro fertilization: a single cell is extracted from the embryo before implantation and the contents of this cell's genome are analyzed. Although meta-analyses of older clinical trials found that PGD is not an effective way to screen for genetic disease25, more modern procedures have had better success in randomly controlled trials and have shown that live birth rates can nearly double26. The application of genome-wide analysis methods, such as array comparative genomic hybridization, in this area opens up the possibility of higher-resolution measurements of the embryo genome before implantation27. We expect that higher-resolution genomic techniques will soon be applied to PGD, enabling the routine calling of structural variation and even point mutations in individual embryos. These data in turn will be used to make more careful assessments about which embryos are most likely to lead to successful pregnancies.
Future of the technology
The cost of sequencing will hopefully continue to decline. The past decade has also been a fertile one for the development of biochemical DNA amplifiers, and there are now several choices for single-cell experiments5. No single amplifier has emerged as the overall winner, and we would be surprised if that becomes the case. It is very difficult to point to a ‘best’ amplifier, since there are several different performance parameters to consider. In particular, one would like to understand the following as they relate to the specific application, sample type and reaction format: convenience (isothermal or thermocycled; single step or multiple reagent steps), cost (commercial or homemade), fidelity (off-target and contaminant amplification, uniformity or bias in amplification, coverage, error rates, artifacts such as chimeras) and gain (amount of amplification required).
Furthermore, comparing different amplification chemistries on a statistically relevant sample of single cells is a significant undertaking, and requires care to avoid confounding effects such as reaction volume, gain, reaction format, lysis conditions, contamination, sample-specific differences and random cell-to-cell variability. Such well-controlled comparisons are needed in order to identify the best matches among sample types, applications, amplification chemistries, reaction formats and sequencing approaches.
Finally, there is a continuing need for innovative technologies that automate single-cell isolation and genome amplification. The current state of the art operates at the level of hundreds of cells, and one can use commercially available cell sorters for the isolation steps, pipetting robots for cell lysis and amplification, or microfluidic devices that combine both steps in a fully automated and integrated fashion28. Automation and miniaturization are important to single-cell sequencing because sufficient sampling is important to fully characterize genomic diversity in the bulk material. We expect quite a bit of creativity in the development of arrayed and serialized microfluidic and microfabricated approaches. These will increase the throughput and decrease the cost of steps required for single-cell sequencing by orders of magnitude and will enable microbial and targeted human cell studies on thousands of cells in a single experiment. We believe that it is only a matter of time until large projects are launched which will systematically characterize the genomes and transcriptomes of hundreds of thousands of single cells.
Single-cell genomic analysis represents a suite of rapidly developing technologies that touch on a wide variety of fundamental and applied problems in the life sciences. We look forward to the continuing impact of single-cell sequencing as amplification chemistries and reaction formats diversify, and as the community innovates ways of applying this technology to extract information from biological systems.
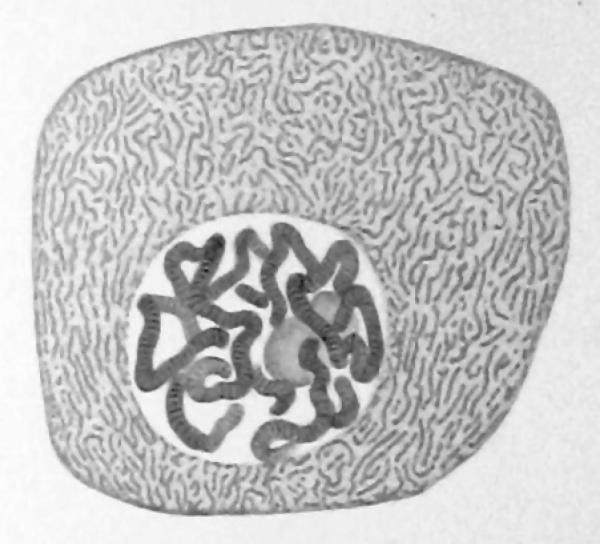
A single-cell genome image of polytene chromosomes from the 1882 monograph by Flemming (ref. 1).
Footnotes
COMPETING FINANCIAL INTERESTS
The authors declare competing financial interests: details are available in the online version of the paper (doi:10.1038/nmeth.2783).
Contributor Information
Paul C Blainey, Department of Biological Engineering, Massachusetts Institute of Technology, and Broad Institute of MIT and Harvard, Cambridge, Massachusetts, USA.
Stephen R Quake, Departments of Applied Physics and Bioengineering, Stanford University and Howard Hughes Medical Institute, Stanford, California, USA..
References
- 1.Flemming W. Zellsubstanz, Kern und Zelltheilung. Vogel; 1882. [Google Scholar]
- 2.Bridges CB. J. Hered. 1935;26:60–64. [Google Scholar]
- 3.Hubert R, MacDonald M, Gusella J, Arnheim N. Nat. Genet. 1994;7:420–424. doi: 10.1038/ng0794-420. [DOI] [PubMed] [Google Scholar]
- 4.Handyside AH, et al. N. Engl. J. Med. 1992;327:905–909. doi: 10.1056/NEJM199209243271301. [DOI] [PubMed] [Google Scholar]
- 5.Blainey PC. FEMS Microbiol. Rev. 2013;37:407–427. doi: 10.1111/1574-6976.12015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Shapiro E, Biezuner T, Linnarsson S. Nat. Rev. Genet. 2013;14:618–630. doi: 10.1038/nrg3542. [DOI] [PubMed] [Google Scholar]
- 7.Marcy Y, et al. Proc. Natl. Acad. Sci. USA. 2007;104:11889–11894. doi: 10.1073/pnas.0704662104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Marcy Y, et al. PLoS Genet. 2007;3:1702–1708. doi: 10.1371/journal.pgen.0030155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Dean FB, Nelson JR, Giesler TL, Lasken RS. Genome Res. 2001;11:1095–1099. doi: 10.1101/gr.180501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fan HC, Wang J, Potanina A, Quake SR. Nat. Biotechnol. 2011;29:51–57. doi: 10.1038/nbt.1739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Peters BA, et al. Nature. 2012;487:190–195. doi: 10.1038/nature11236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Lu S, et al. Science. 2012;338:1627–1630. doi: 10.1126/science.1229112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Arnheim N, Calabrese P, Tiemann-Boege I. Annu. Rev. Genet. 2007;41:369–399. doi: 10.1146/annurev.genet.41.110306.130301. [DOI] [PubMed] [Google Scholar]
- 14.Wang J, Fan HC, Behr B, Quake SR. Cell. 2012;150:402–412. doi: 10.1016/j.cell.2012.06.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lynch M. Trends Genet. 2010;26:345–352. doi: 10.1016/j.tig.2010.05.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zong C, Lu S, Chapman AR, Xie XS. Science338. 2012:1622–1626. doi: 10.1126/science.1229164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Jan M, et al. Sci. Transl. Med. 2012;4:149ra118. doi: 10.1126/scitranslmed.3004315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Shlush LI, et al. Blood. 2012;120:603–612. doi: 10.1182/blood-2011-10-388629. [DOI] [PubMed] [Google Scholar]
- 19.Navin N, et al. Nature. 2011;472:90–94. doi: 10.1038/nature09807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Arendt T. Mol. Neurobiol. 2012;46:125–135. doi: 10.1007/s12035-012-8262-0. [DOI] [PubMed] [Google Scholar]
- 21.McConnell MJ, et al. Science. 2013;342:632–637. doi: 10.1126/science.1243472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Evrony GD, et al. Cell. 2012;151:483–496. doi: 10.1016/j.cell.2012.09.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gundry M, Li W, Maqbool SB, Vijg J. Nucleic Acids Res. 2012;40:2032–2040. doi: 10.1093/nar/gkr949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Martin SL. Nature. 2009;460:1087–1088. doi: 10.1038/4601087a. [DOI] [PubMed] [Google Scholar]
- 25.Mastenbroek S, Twisk M, van der Veen F, Repping S. Hum. Reprod. Update. 2011;17:454–466. doi: 10.1093/humupd/dmr003. [DOI] [PubMed] [Google Scholar]
- 26.Rubio C, et al. Fertil. Steril. 2013;99:1400–1407. doi: 10.1016/j.fertnstert.2012.11.041. [DOI] [PubMed] [Google Scholar]
- 27.Yang Z, et al. Mol. Cytogenet. 2012;5:24. doi: 10.1186/1755-8166-5-24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Landry ZC, Giovanonni SJ, Quake SR, Blainey PC. Methods Enzymol. 2013;531:61–90. doi: 10.1016/B978-0-12-407863-5.00004-6. [DOI] [PMC free article] [PubMed] [Google Scholar]