Summary
Kidney allograft status is currently characterized using the invasive percutaneous needle core biopsy procedure. The procedure has become safer over the years, but challenges and complications still exist including sampling error, inter-observer variability, bleeding, arteriovenous fistula, graft loss, and even death. Because the most common type of acute rejection is distinguished by inflammatory cells exiting the intravascular compartment and gaining access to the renal tubular space, we reasoned that a kidney allograft may function as an in vivo flow cytometer and sort cells involved in rejection into urine. To test this idea, we developed quantitative polymerase chain reaction (PCR) assays for absolute quantification of mRNA and pre-amplification protocols to overcome the low RNA yield from urine. Here, we review our single center urinary cell mRNA profiling studies that led to the multicenter Clinical Trials in Organ Transplantation (CTOT-04) study and the discovery and validation of a 3-gene signature of 18S rRNA-normalized measures of CD3ε mRNA and IP-10 mRNA and 18S rRNA that is diagnostic and predictive of acute cellular rejection in the kidney allograft. We also review our development of a 4-gene signature of mRNAs for vimentin, NKCC2, E-cadherin, and 18S rRNA diagnostic of interstitial fibrosis/tubular atrophy (IF/TA).
Keywords: urinary cell mRNA profiling, acute cellular rejection, kidney transplantation, interstitial fibrosis, PCR
Introduction
Each year, approximately 100,000 patients compete for the 11,000 deceased donor kidneys available for transplantation in the US and at least 5,100 deaths occur among those on the kidney transplant wait list (1). A significant contributor to the burgeoning wait list is the return of patients with a failed graft to the list – 15% of patients on the wait list in 2011 were listed for repeat transplant. The prevention of such graft failures would improve long-term graft survival rates as well as mitigate the existing organ shortage crisis.
The causes of kidney allograft failure are multiple, with acute rejection (AR) being a major contributor to graft loss. The incidence of AR in the United States is reported to be 11.6% for deceased donor and 10.0% for live donor recipients (1). The incidence is higher in studies that include surveillance biopsies. The incidence was 15.2% in the first year of transplantation in the recent Mayo study of 797 kidney graft recipients that included surveillance biopsies (2).
The most common type of AR is T-cell-mediated acute cellular rejection (ACR) (2–5). For example, among the 121 AR biopsies in the Mayo study, 76 were classified as ACR, 42 as borderline, and only 3 as antibody-mediated rejection (AMR) (2). Moreover, borderline changes, clinical ACR, or subclinical ACR in the first year of transplantation, detected in either a for-cause biopsy or a surveillance biopsy were all associated with a significant increase in the incidence of interstitial fibrosis and inflammation in kidney allografts, transplant glomerulopathy, and development of class II donor specific antibodies, and reduced graft survival [HR=3.07, 95% confidence interval (CI), 1.92–4.94, P<0.0001]. In a landmark study of 93,934 kidney transplantations performed in the United States between 1988 and 1996, Hariharan et al. (6) found that clinical acute rejection in the first year of transplantation negatively impacted long-term survival of kidney allografts; whereas the projected half-life for deceased donor grafts in those without clinical acute rejection increased from 8.8 years for transplants performed in 1988 to 17.9 years for transplants performed in 1995, the projected half-life increased from 7.0 years in 1988 to only 8.8 years in 1995 for those with an episode of clinical acute rejection. In a recent analysis of 48,179 patients transplanted between 2000 and 2007 in US, an episode of AR in the first year of transplantation was associated with an adjusted relative risk for all-cause graft loss as high as 5.5 (7).
ACR is a risk factor for chronic rejection, manifested as interstitial fibrosis and tubular atrophy (IF/TA) with or without concurrent inflammation in the allograft, and with progressive loss of graft function. It has also been documented that interstitial inflammation and tubulitis, the histological hallmarks of ACR, detected in the surveillance biopsies performed in patients with stable graft function (8) or observed in for-cause biopsies in patients with chronic graft dysfunction, are harbingers of graft loss (9). Importantly, treatment of intragraft inflammation, identified by surveillance biopsies, is associated with better graft histology and function (10, 11).
The invasive biopsy procedure to diagnose AR or IF/TA
AR and IF/TA are both diagnosed with the use of a percutaneous needle core biopsy procedure. While the procedure has become safer over the years, challenges and complications still exist including sampling error, inter-observer variability, bleeding, arteriovenous fistula, graft loss, and even death (12–14). The financial costs of the procedure are substantial, reported to be approximately $3,000 (15). With an estimated incidence of 0.4 biopsies/patient during the first year of transplantation, about 7,000 biopsies at a cost of $21 million are estimated to have been performed during the first year of transplantation in the 17,671 patients who received a kidney graft in 2011 (16, 17).
Development of urinary cell mRNA profiling as a noninvasive diagnostic tool
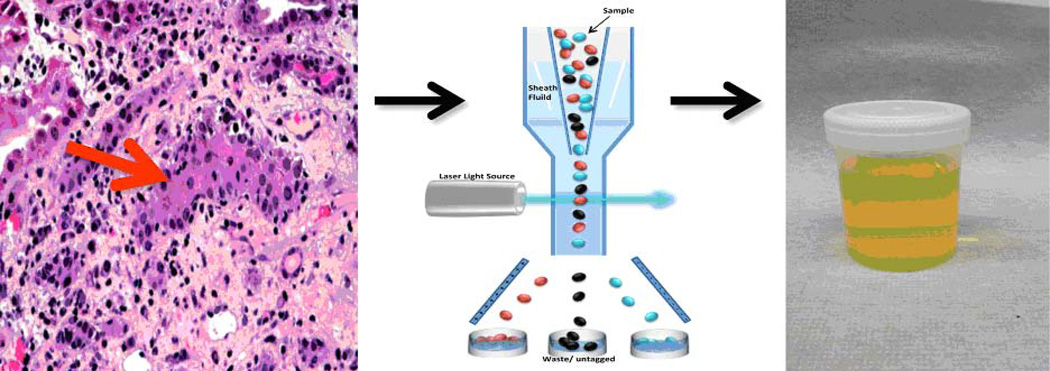
ACR is distinguished by interstitial inflammation and tubulitis with graft infiltrating cells gaining entry into the tubular space. We therefore reasoned that the allograft may function as an ‘in vivo flow cytometer’ that sorts cells involved in rejection into the urine (Fig. 1). We formulated the hypothesis that mRNA profiling of urinary cells would offer a noninvasive means of diagnosing ACR and initiated urinary cell mRNA profiling studies to investigate this concept. Because of the inherent difficulties in isolating RNA from urinary cells, we developed a pre-amplification step that enables measurement of multiple mRNA species from very small quantities of total RNA (18). We also developed an efficient method for absolute quantification of transcript numbers in the PCR assays (19). Armed with these innovations, we conducted single center studies followed by multicenter clinical trials.
Fig. 1. Conceptualization of the kidney allograft as an in-vivo flow cytometer.
Interstitial inflammation and tubilitis are the histological hallmarks of ACR in the kidney allograft (left panel). Since ACR involves infiltration of T lymphocytes and additional cells into the tubular space, the kidney allograft is conceptualized to function as an in vivo flow cytometer (middle panel) facilitating the entry of graft destructive/protective T lymphocytes and other immune cells and graft parenchymal cells into the urinary space. In this formulation, urinary cell mRNA profiling reflects intragraft events deterministic of allograft status.
Herein, we summarize our single center studies of urinary cell mRNA profiling that provided the scientific underpinning for the multicenter CTOT-04 study and the primary data from the CTOT-04 study. We also review our successful development of a urinary cell based gene signature for the noninvasive diagnosis of IF/TA, the most common feature of a failing kidney allograft.
Single Center Studies that Provided the Scientific Background for the CTOT-04 Study
In our first urinary cell mRNA profiling study, we examined whether levels of mRNA encoding cytotoxic attack molecules – perforin and granzyme B – are informative of human kidney allograft status. In the granule exocytosis pathway of cell-mediated cytotoxicity, a pathway implicated in acute cellular rejection, perforin, a pore forming protein, and granzyme B, an inducer of apoptosis, collaborate and contribute to targeted cell death. With the use of novel competitors, we found that the levels of mRNA for perforin in urinary cells predicted ACR with a sensitivity of 83% and a specificity of 83%, while granzyme B mRNA levels predicted ACR with a sensitivity of 79% and a specificity of 64% when the data analysis was restricted to patients with a biopsy-confirmed diagnosis of ACR (20).
Serine proteinase inhibitor-9 (PI-9) is an endogenous antagonist of granzyme B. We investigated whether urinary cell levels of PI-9 are diagnostic of kidney allograft status. In a fashion similar to perforin and granzyme B, levels of PI-9 were associated with kidney biopsy diagnosis and urinary cell levels of PI-9 predicted ACR with a sensitivity of 76% and a specificity of 79% (21).
It is well documented that intraepithelial homing of T cells is contingent upon the cell surface protein CD103. Since ACR is exemplified by tubulitis with intraepithelial homing of alloreactive T cells, we reasoned that urinary cell levels of CD103 would be diagnostic of ACR in the kidney allograft. This research hypothesis was validated by our finding that urinary cell levels of CD103 are diagnostic of ACR with a sensitivity of 59% and a specificity of 75% (22).
Chemokines and their receptors play a primary role in the trafficking of cells to inflammatory sites. We hypothesized that urinary cell levels of IP-10 and its receptor CXCR3 would help capture intragraft inflammation. Our research hypothesis was also based on elegant pre-clinical experimental studies implicating IP-10 and CXCR3 in inflammatory diseases including allograft rejection (23, 24). Our findings that AR is predicted with a sensitivity of 100% and a specificity of 78% using urinary cell levels of IP-10 mRNA and with a sensitivity of 63% and a specificity of 83% using urinary cell levels of CXCR3 mRNA validated our research hypothesis (25).
Multicenter CTOT-04 Study
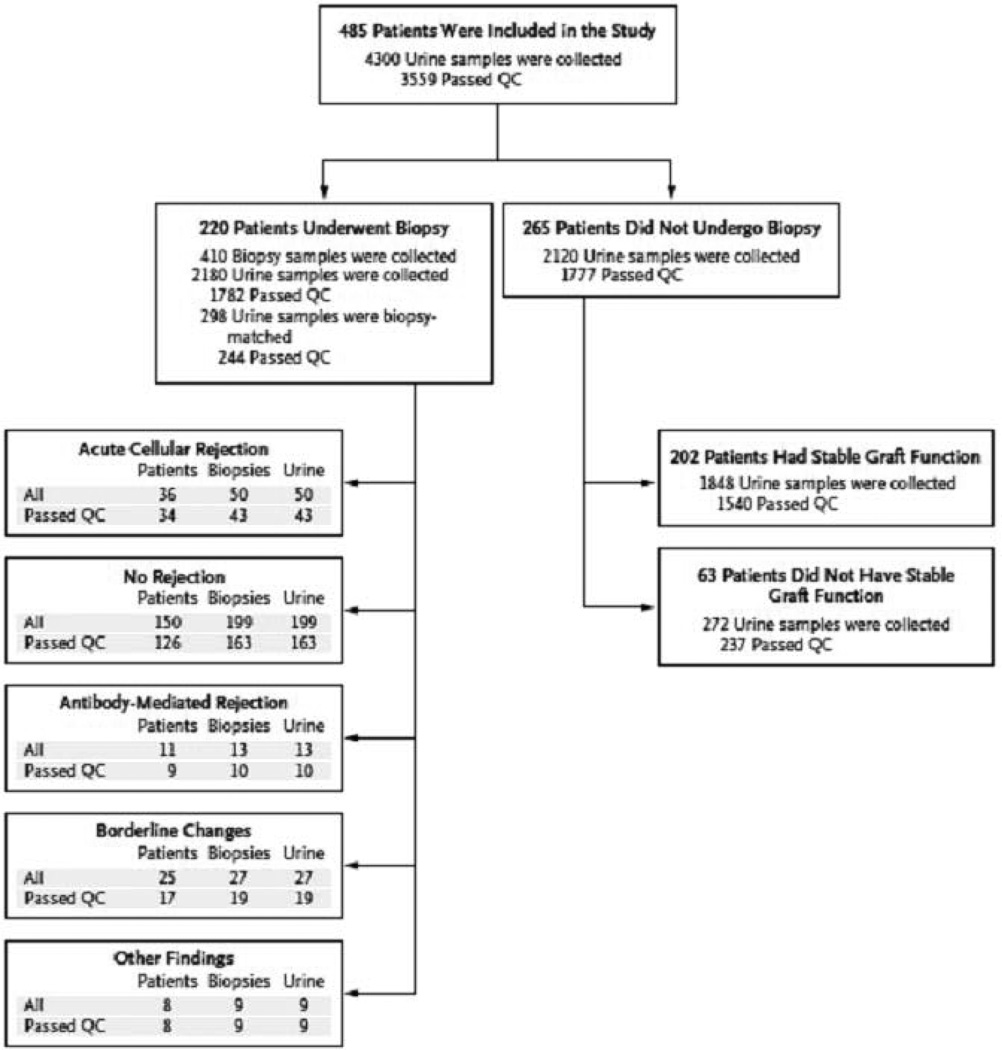
The CTOT-04 study was a prospective observational multicenter study with an initial target enrollment of 450 kidney transplant recipients from 5 clinical sites (3). The enrollment target was increased to 495 in view of dropouts and a total of 497 patients with end stage renal disease (ESRD) were enrolled. Five patients enrolled in CTOT-04 did not receive a kidney graft, and 7 of the 492 transplanted patients did not provide any urine specimens. Thus, our urinary cell mRNA profiling study had 485 patients. These 485 patients provided 4300 urine specimens for urinary cell mRNA profiling and 220 of 485 patients underwent at least one renal allograft biopsy (Fig. 2). We designed CTOT-04 as an observational study with each of the 5 participating sites being able to use site-specific immunosuppression and infection protocols. The rationale for this study design included the consideration that the urine mRNA profiling results should be generalizable to the kidney transplant population and not be restricted by the immunosuppressive regimen used in any one clinic.
Fig. 2. Patients, biopsy results, and urine samples.
A total of 4300 urine samples were collected from 485 kidney allograft patients for urinary-cell messenger RNA (mRNA) profiling on days 3, 7, 15, and 30, in months 2, 3, 4, 5, 6, 9, and 12 after transplantation, and at the time of any kidney-allograft biopsy and 2 weeks thereafter. Of the 4300 urine specimens, 3559 were classified as passing quality control (QC) and 741 were classified as not passing. A total of 220 patients underwent 410 kidney-allograft biopsies, and 265 did not undergo biopsy. The numbers of patients with biopsy-matched urine samples (urine samples that were collected from 3 days before to 1 day after biopsy and that passed QC) are shown for patients with acute cellular rejection (defined as Banff grade IA or higher), for those without any rejection features in the biopsy sample, for those with acute antibody-mediated rejection, for those with borderline changes, and for those with other biopsy findings. The number of patients listed under different diagnostic categories exceeds the 220 patients who underwent biopsy because several patients had multiple biopsies with different diagnoses. Among the 265 patients who did not undergo biopsy, 202 met the criteria for stable graft function, of whom 201 had urine samples that passed QC (from Suthanthiran et al. N Engl J Med 2013, with permission).
Urine samples (approximately 50 ml) were collected sequentially with the pre-specified schedule for collection being post-transplant days 3, 7, 15, and 30 and months 2, 3, 4, 5, 6, 9, and 12, and at the time of any renal allograft biopsy, prior to treatment and 2 weeks after biopsy (Fig. 2). All five centers followed an identical protocol with respect to urine sample collection. Urine cell pellets were prepared at the clinical sites with the use of a standard protocol for urine cell sedimentation, stored at −80°C and shipped in batches to the Gene Expression Monitoring (GEM) Core at Weill Cornell Medical College, New York, NY (WCMC).
The CTOT-04 study investigated the following hypotheses: (i) urinary cell mRNA profile at the time of a diagnostic biopsy predicts biopsy-based diagnosis of rejection and (ii) urinary cell mRNA profile is predictive of acute allograft rejection in the near future. We pre-specified seven mRNAs – perforin, granzyme B, proteinase inhibitor-9 (PI-9), CD103, CD3 epsilon chain (CD3ε), interferon inducible protein-10 (IP-10), and CXCR3 – for measurement and these members of the mRNA panel were selected on the basis of our single center studies associating their urinary cell levels with kidney allograft status (18, 20–22, 25). Two additional transcripts were also measured: mRNA encoding transforming growth factor-β1 (TGF-β1) and 18S ribosomal RNA (18S rRNA). These transcripts were included as quality control parameters (vide infra).
CTOT-04 study: methodology
We isolated total RNA from urinary cell pellets as described (3). The quantity and purity of the RNA were measured using the NanoDrop® ND-1000 UV-Vis spectrophotometer. We qualified a urine sample as quality control passed if in that urine sample, the 18S rRNA copy number was greater than or equal to 5×107 and its TGF-β1 mRNA copy number was greater than or equal to 100 copies per one microgram of RNA. With these parameters, 3559 (83%) of the 4300 urine samples were classified as quality control passed. The median quantity of total RNA isolated from the 3559 samples that passed quality control parameters was 560 ng (interquartile range: 333 ng to 974 ng) and the median purity of total RNA isolated for these samples was 1.80 (interquartile range: 1.67 to 1.89, perfect ratio: 2.0). We reverse transcribed the total RNA to cDNA using the TaqMan reverse transcription kit on the same day the total RNA was isolated and the total RNA was adjusted to one microgram in 100 microliters prior to reverse transcription. This adjustment normalized for the cell abundance in individual urinary cell pellets.
We designed gene specific oligonucleotide primers and TaqMan fluorogenic probes for the measurement of levels of mRNAs pre-specified in the CTOT-04 Protocol: CD3ε, perforin, granzyme B, IP-10, CXCR3, CD103, TGF-β1, PI-9, and 18S rRNA. The sequence and location of the gene specific oligonucleotide primer pairs and TaqMan probes used in the CTOT-04 study are provided in Table 1.
Table 1. Oligonucleotide Primers and TaqMan Probes used to Measure Levels of mRNA in the CTOT-04 Study.
The sequences and the locations of the oligonucleotide primers and probes that we designed and validated for the measurement of mRNA levels and 18S ribosomal RNA (rRNA) in the urine are shown. The fluorogenic TaqMan probes were labeled with 6-carboxy-fluorscein (FAM) at the 5’ end and with 6-carboxy-tetramethylrodamine (TAMRA) or minor groove binder (MGB) at the 3’ end. FAM functioned as the reported dye and TAMRA or MGB as the quencher.
| Type of mRNA |
GenBank Accession Number |
Sequence | Location |
|---|---|---|---|
| CD3-epsilon | NM_000733 | Sense: 5'-AAGAAATGGGTGGTATTACACAGACA-3' | 131–156 |
| Antisense: 5'-TGCCATAGTATTTCAGATCCAGGAT-3' | 233–209 | ||
| Probe: 5'-FAM-CCATCTCTGGAACCACAGTAATATTGACATGCC-TAMRA-3' | 170–202 | ||
| Perforin | M28393 | Sense: 5'-GGACCAGTACAGCTTCAGCACTG-3' | 492–514 |
| Antisense: 5'-GCCCTCTTGAAGTCAGGGTG-3' | 587–568 | ||
| Probe: 5'-FAM-TGCCGCTTCTACAGTTTCCATGTGGTACAC-TAMRA-3' | 526–555 | ||
| Granzyme B | J04071 | Sense: 5'-GCGAATCTGACTTACGCCATTATT-3' | 534–557 |
| Antisense: 5'-CAAGAGGGCCTCCAGAGTCC-3' | 638–619 | ||
| Probe: 5’-FAM-CCCACGCACAACTCAATGGTACTGTCG-TAMRA-3’ | 559–585 | ||
| IP-10 | NM_001565.1 | Sense: 5'-TGTCCACGTGTTGAGATCATTG-3' | 235–256 |
| Antisense: 5'-GGCCTTCGATTCTGGATTCA-3' | 309–290 | ||
| Probe: 5’-FAM TACAATGAAAAAGAAGGGTGAGAA-MGB-3’ | 258–281 | ||
| CXCR3 | NM_001504 | Sense: 5'-ACCCAGCAGCCAGAGCAC-3' | 41–58 |
| Antisense: 5'-CAACCTCGGCGTCATTTAGC-3' | 117–98 | ||
| Probe: 5'-FAM-CTTGGTGGTCACTCACCTCAAGGACCAT-TAMRA-3' | 69–96 | ||
| CD103 | XM_008508 | Sense: 5'-CGTGCTCAGCTCCCTTCTG-3' | 211–229 |
| Antisense: 5'-CCTGGTGTCCTCTTGGTTCTG-3' | 297–277 | ||
| Probe: 5’-FAM-ACCAAGACCCCAGCACCAACCATACCT-TAMRA-3’ | 231–257 | ||
| TGF-β1 | NM_000660 | Sense: 5'-GCGTGCTAATGGTGGAAACC-3' | 1170–1189 |
| Antisense: 5'-CGGAGCTCTGATGTGTTGAAGA-3' | 1263–1242 | ||
| Probe: 5’-FAM-ACAACGAAATCTATGACAAGTTCAAGCAGAGTACACA- TAMRA-3’ |
1191–1227 | ||
| PI-9 | NM_004155 | Sense: 5'-'TCAACACCTGGGTCTCAAAAAA-3' | 508–529 |
| Antisense: 5'- CAGCCTGGTTTCTGCATCAA-3' | 590–571 | ||
| Probe: 5’-FAM-AGCTACCCGGCAACAACTCTTCAATTTTACCT-TAMRA −3’ | 536–567 | ||
| 18S rRNA | K03432 | Sense: 5'-GCCCGAAGCGTTTACTTTGA-3' | 929–948 |
| Antisense: 5'-TCCATTATTCCTAGCTGCGGTATC-3' | 1009–986 | ||
| Probe: 5’-FAM-AAAGCAGGCCCGAGCCGCC-TAMRA-3’ | 965–983 |
Our laboratory has developed a pre-amplification enhanced real time quantitative PCR assay for absolute quantification of mRNA copy number (18). The PCR assay involves two steps. In the first pre-amplification step, the mRNAs to be measured are pre-amplified using sequence specific primer pairs without the TaqMan probe. The primary purpose of the pre-amplification step is to facilitate measurement of multiple mRNAs from a very small aliquot of cDNA. For example, whereas 5 microliters of cDNA is needed in the standard PCR assay to measure a single mRNA (duplicate wells with 2.5 microliters per well), the pre-amplification step and subsequent dilution of cDNA allows almost 50 different mRNA species to be measured from the same 5 microliters of cDNA. In the second absolute quantification step, the mRNA of interest is quantified with the use of a customized amplicon to construct a reference/standard curve. In the CTOT-04 study, we used an ABI Prism 7500/7900HT Fast detection system for the absolute quantification of mRNAs and 18S rRNA.
Statistical analysis
Details of statistical analysis including normalization, fitting the model, bootstrap model selection, internal validation of the fitted model, and calculation of the longitudinal perspective and retrospective trajectories have been reported (3). The validation statistics and plots were computed using Harrell’s R package “RMS, version 2.12.2” (http://cran.r-project.org/web/packages/rms/rms.pdf). All other analyses were performed using SAS, version 9.3 (Cary, NC).
CTOT-04 study: results
Urinary cell levels of mRNAs are diagnostic of ACR
We investigated the hypothesis that urinary cell levels of mRNA are diagnostic of Banff ACR. We tested this by measuring the 8 pre-specified mRNAs in 43 urine samples matched to 43 ACR biopsies from 34 patients and comparing the levels to those found in 163 samples matched to 163 biopsies without rejection (No Rejection Biopsy group, 126 patients) and 1540 samples from the Stable (no Biopsy) group (201 patients with urine specimens). Our primary goal was to examine whether the urine sample would serve as a surrogate for the invasive biopsy procedure. Thus, we used matched urine samples in those who underwent a biopsy. In those who did not undergo a biopsy (Stable group), all QC-passed samples were analyzed.
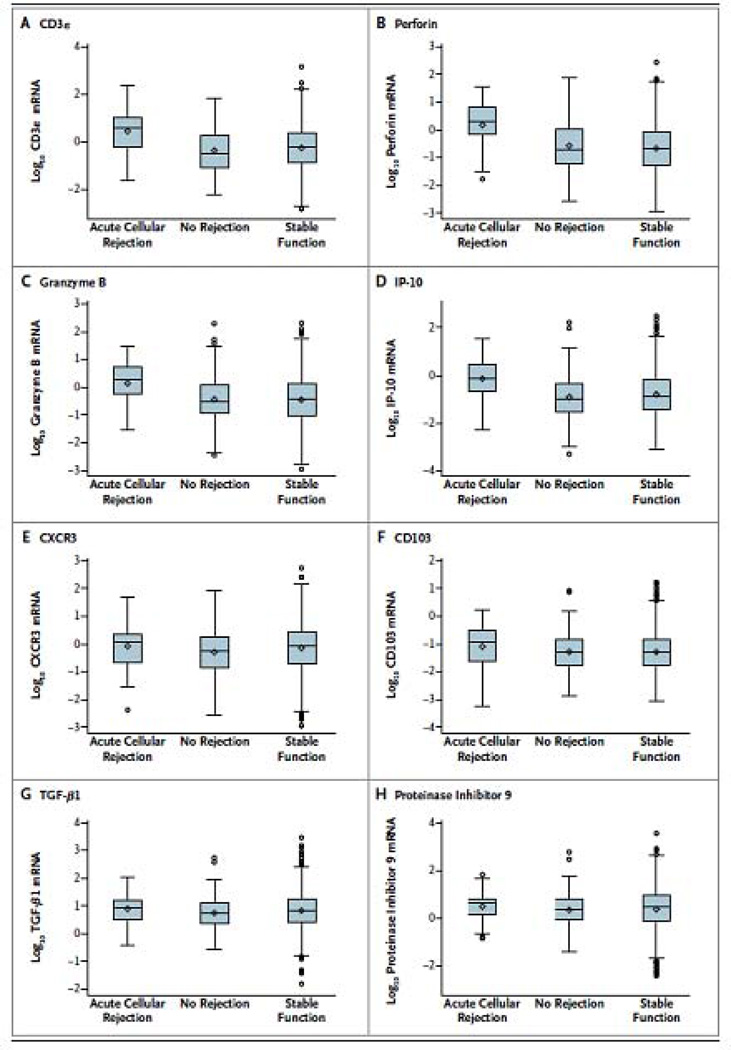
In accord with the research hypothesis, 18S-normalized levels of mRNA for CD3ε, perforin, granzyme B, and IP-10 in urinary cells differed significantly among the ACR, No Rejection Biopsy, and Stable (no biopsy) groups (P<0.0001 for all 4 mRNAs by Kruskal-Wallis test for 3-group comparison) (Fig. 3). Pairwise group comparisons (Mann-Whitney test) showed that the levels of mRNA for CD3ε, perforin, granzyme B, and IP-10 in the ACR group were higher than the levels in the No Rejection Biopsy group (P<0.0001 for all 4 mRNAs) and the Stable (no biopsy) group (P<0.0001 for all 4 mRNAs).
Fig. 3. Levels of mRNA in urinary cells.
Box-and-whisker plots show the log10-transformed ratios of mRNA copies per microgram of total RNA to 18S ribosomal RNA (rRNA) copies (X10−6) per microgram of total RNA for CD3ε, perforin, granzyme B, interferon-inducible protein 10 (IP-10), CXCR3, CD103, transforming growth factor β1 (TGF-β1), and proteinase inhibitor 9 in 43 urine samples matched to 43 biopsy specimens (from 34 patients) showing acute cellular rejection, 163 urine samples matched to 163 biopsy specimens (from 126 patients) showing no rejection, and 1540 longitudinally collected urine samples from 201 patients with stable graft function who did not undergo biopsy. The horizontal line within each box represents the median, the bottom and top of each box represent the 25th and 75th percentile values, and the thin vertical bars extend to the 10th and 90th percentile values; the diamond indicates the mean, and circles indicate outliers. The mRNA levels of CD3ε, perforin, granzyme B, and IP-10 differed significantly among the three groups (P<0.001 for all comparisons), but not the levels of CXCR3 (P = 0.06), CD103 (P = 0.13), TGF-β1 (P = 0.11), and proteinase inhibitor 9 (P = 0.38). P values are based on the Kruskal–Wallis test, with the log10-transformed, 18S-normalized mRNA levels treated as the dependent variable. Pairwise group comparisons by means of the Mann–Whitney test showed that the 18S-normalized mRNA levels for CD3ε, perforin, granzyme B, and IP-10 in patients with acute cellular rejection were significantly higher than the levels in those with specimens showing no rejection (P<0.001 for each mRNA) and in those with stable graft function (P<0.001 for each mRNA) (from Suthanthiran et al. N Engl J Med 2013, with permission).
Table 2A shows that urinary cell levels of 18S rRNA are higher in the ACR biopsy group than the levels in the No Rejection Biopsy group or the Stable (no biopsy) group (P=0.0002), and are higher in the ACR group than the Borderline biopsy group (P=0.04). Table 2A also shows that 18s rRNA normalized levels of mRNA for CXCR3 (P=0.06), CD103 (P=0.13), and PI-9 (P=0.38) are not associated with ACR in contrast to our single center studies showing that these mRNAs are associated with ACR (21, 22, 25). The reason for these mRNAs not being associated with ACR in the CTOT-04 study may include: (i) inclusion in the single center studies of patients without biopsy confirmation of allograft status whereas every patient used to develop the diagnostic signature in CTOT-04 had undergone an allograft biopsy, and especially (ii) analysis of levels of mRNA for association with ACR without 18S rRNA normalization in the single center studies versus analysis of levels of mRNA for association with ACR after 18S rRNA normalization in the CTOT-04 study. The second point is critical since non-normalized urinary cell levels of mRNA for CD103 (P<0.0001), CXCR3 (P<0.0001) and PI-9 (P=0.002) were all significantly associated with ACR in the CTOT-04 study (Table 2B), consistent with our previous publications (21, 22, 25).
Table 2.
| A. 18S rRNA Normalized, Log10 - Transformed Levels of mRNA in Urinary Cells. Levels of mRNA were measured with the use of preamplification enhanced real time quantitative PCR assays using gene specific oligonucleotide pairs and TaqMan probes. Median Copy Number (lower, upper quartiles) of each mRNA measure, normalized by 18s rRNA and log10-transformed are shown. Matched (urine collected minus 3 days to plus 1 day of biopsy) and quality control passed urine samples were included for those who underwent a biopsy, whereas all quality control passed urine samples collected longitudinally were included for the Stable (no biopsy) group. | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Type of mRNA |
Acute Cellular Rejection (N=43 samples 34 patients) |
No Rejection Group (N=163 samples 126 patients) |
Stable (No Biopsy) Group (N=1540 samples 201 patients) |
P Valuea | Borderline (N=19 samples 17 patients) |
Antibody Mediated Rej (N=10 samples 9 patients) |
Other (N=9 samples 8 patients) |
P Valueb ACR Vs. |
P-Valueb No Reject Vs. |
P Valueb Stable Vs. |
|
| CD3ε | 0.601 (−0.233, 0.995) |
−0.504 (−1.074, 0.247) |
−0.236 (−0.891, 0.341) |
<0.0001 | −0.024 (−0.703, 0.305) |
0.022 (−0.306, 0.385) |
−0.052 (−0.541, 0.429) |
No rejection Stable Borderline |
<0.0001 <0.0001 0.04 |
0.11 0.24 |
0.38 |
| Perforin | 0.289 (−0.160, 0.817) |
−0.689 (−1.215, 0.028) |
−0.688 (−1.279, −0.056) |
<0.0001 | −0.327 (−0.612, 0.087) |
−0.560 (−0.933, −0.164) |
−0.107 (−0.654, 0.287) |
No rejection Stable Borderline |
<0.0001 <0.0001 0.01 |
0.30 0.15 |
0.07 |
| Granzyme B | 0.261 (−0.222, 0.721) |
−0.523 (−0.910, 0.078) |
−0.430 (−1.039, 0.140) |
<0.0001 | −0.424 (−1.168, 0.368) |
−0.063 (−1.072, 0.380) |
0.354 (−0.662, 0.611) |
No rejection Stable Borderline |
<0.0001 <0.0001 0.01 |
0.80 0.95 |
0.94 |
| IP-10 | −0.114 (−0.655, 0.489) |
−1.029 (−1.534, −0.362) |
−0.874 (−1.424, −0.186) |
<0.0001 | −0.903 (−1.205, −0.531) |
−0.986 (−1.399, 0.150) |
0.314 (−0.028, 0.948) |
No rejection Stable Borderline |
<0.0001 <0.0001 0.002 |
0.09 0.61 |
0.91 |
| CXCR3 | 0.082 (−0.680, 0.338) |
−0.241 (−0.872, 0.259) |
−0.055 (−0.718, 0.455) |
0.06 | −0.458 (−0.932, 0.229) |
−0.419 (−1.186, 0.074) |
−0.289 (−1.340, 0.006) |
No rejection Stable Borderline |
0.10 0.66 0.21 |
0.02 0.66 |
0.21 |
| CD103 | −0.929 (−1.619, −0.517) |
−1.298 (−1.748, −0.830) |
−1.308 (−1.779, −0.848) |
0.13 | −1.145 (−1.948, −0.483) |
−1.184 (−2.040, −1.014) |
−1.369 (−1.534, −1.018) |
No rejection Stable Borderline |
0.05 0.05 0.59 |
0.96 0.70 |
0.72 |
| TGF-β1 | 0.928 (0.499, 1.181) |
0.756 (0.356, 1.113) |
0.837 (0.424, 1.242) |
0.11 | 0.584 (0.372, 1.341) |
1.088 (1.004, 1.288) |
0.709 (0.139, 0.830) |
No rejection Stable Borderline |
0.08 0.51 0.16 |
0.05 0.55 |
0.28 |
| PI-9 | 0.604 (0.155, 0.782) |
0.383 (−0.105, 0.816) |
0.445 (−0.137, 0.962) |
0.37 | 0.239 (−0.104, 0.681) |
0.787 (0.714, 0.984) |
0.438 (0.332, 0.907) |
No rejection Stable Borderline |
0.14 0.42 0.18 |
0.28 0.75 |
0.49 |
| 18S rRNA | 3.365 (2.798, 3.740) |
2.857 (2.427, 3.276) |
2.951 (2.491, 3.339) |
0.0002 | 2.994 (2.370, 3.442) |
3.145 (2.919, 3.221) |
3.310 (2.791, 3.326) |
No rejection Stable Borderline |
0.0001 0.0001 0.04 |
0.16 0.47 |
0.83 |
| B. Absolute Levels of mRNA in Urinary Cells. Median absolute copy number (lower, upper quartiles) of each mRNA measure, without 18S rRNA normalization and without log-transformation and median absolute copy number (lower, upper quartiles) of 18S rRNA (×10−6) without log-transformation are shown. Matched (urine collected minus 3 days to plus 1 day of biopsy) and quality control passed urine samples were included for those who underwent a biopsy, whereas all quality control passed urine samples collected longitudinally were included for the Stable (no biopsy) group. | |||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|
| Type of mRNA |
Acute Cellular Rejection (N=43 samples 34 patients) |
No Rejection Group (N=163 samples 126 patients) |
Stable (No Biopsy) Group (N=1540 samples 201 patients) |
P Valuea | Borderline (N=19 samples 17 patients) |
Antibody Mediated Rej (N=10 samples 9 patients) |
Other (N=9 samples 8 patients) |
P Valueb ACR Vs. |
P-Valueb No Reject Vs. |
P Valueb Stable Vs. |
|
| CD3ε | 8826 (2253, 21790) |
356 (13, 1812) |
497 (102, 2431) |
<0.0001 | 944 (81, 4388) |
943 (240, 2138) |
3192 (536, 4713) |
No rejection Stable Borderline |
<0.0001 <0.0001 0.001 |
0.06 0.23 |
0.51 |
| Perforin | 4119 (1093, 11148) |
223 (13, 1208) |
201 (13, 985) |
<0.0001 | 398 (192, 1564) |
273 (153, 1646) |
1595 (470, 2900) |
No rejection Stable Borderline |
<0.0001 <0.0001 0.0003 |
0.72 0.20 |
0.13 |
| Granzyme B | 3304 (743, 11571) |
337 (64, 1332) |
342 (63, 1663) |
<0.0001 | 614 (13, 1944) |
1123 (60, 2578) |
4605 (110, 10574) |
No rejection Stable Borderline |
<0.0001 <0.0001 0.0009 |
0.60 0.72 |
0.87 |
| IP-10 | 1445 (236, 7362) |
75 (13, 382) |
130 (13, 683) |
<0.0001 | 196 (13, 798) |
76 (13, 2104) |
4203 (859, 15162) |
No rejection Stable Borderline |
<0.0001 <0.0001 0.0003 |
0.02 0.43 |
0.94 |
| CXCR3 | 2122 (618, 6348) |
393 (123, 1616) |
770 (180, 2899) |
<0.0001 | 325 (60, 1782) |
203 (67, 2472) |
664 (68, 1837) |
No rejection Stable Borderline |
<0.0001 0.001 0.01 |
0.003 0.78 |
0.22 |
| CD103 | 234 (63, 596) |
13 (13, 135) |
13 (13, 162) |
<0.0001 | 13 (13, 272) |
13 (13, 161) |
59 (13, 236) |
No rejection Stable Borderline |
<0.0001 <0.0001 0.04 |
0.35 0.35 |
0.56 |
| TGF-β1 | 15142 (5187, 57855) |
4589 (896, 14668) |
5838 (1544, 21690) |
0.0001 | 5022 (992, 22670) |
15439 (8379, 28928) |
3169 (1510, 14065) |
No rejection Stable Borderline |
<0.0001 0.0002 0.01 |
0.05 0.72 |
0.73 |
| PI-9 | 9782 (1680, 28890) |
1756 (335, 9000) |
2153 (409, 11798) |
0.002 | 2180 (765, 9355) |
7527 (4389, 14564) |
5586 (1329, 6358) |
No rejection Stable Borderline |
0.0002 0.001 0.05 |
0.22 0.64 |
0.99 |
| 18S rRNA | 2320 (628, 5500) |
720 (267, 1866) |
894 (310, 2180) |
0.0002 | 985 (234, 2770) |
1398 (830, 1664) |
2040 (619, 2120) |
No rejection Stable Borderline |
0.0001 0.0001 0.04 |
0.16 0.47 |
0.83 |
P values calculated using Kruskal-Wallis test of no differences among the Acute Cellular Rejection biopsy group, No Rejection biopsy group and Stable (no biopsy) group.
P values calculated using Mann-Whitney test of no difference between two groups.
Development of a 3-gene diagnostic signature
A 3-gene model of 18S-normalized CD3ε mRNA, 18S-normalized IP-10 mRNA, and 18S rRNA (all logged) was the best-fitting parsimonious model, yielding the following diagnostic signature:
−6.1487 + 0.8534 log10(CD3ε/18S) + 0.6376 log10(IP-10/18S) + 1.6464 log10(18S)
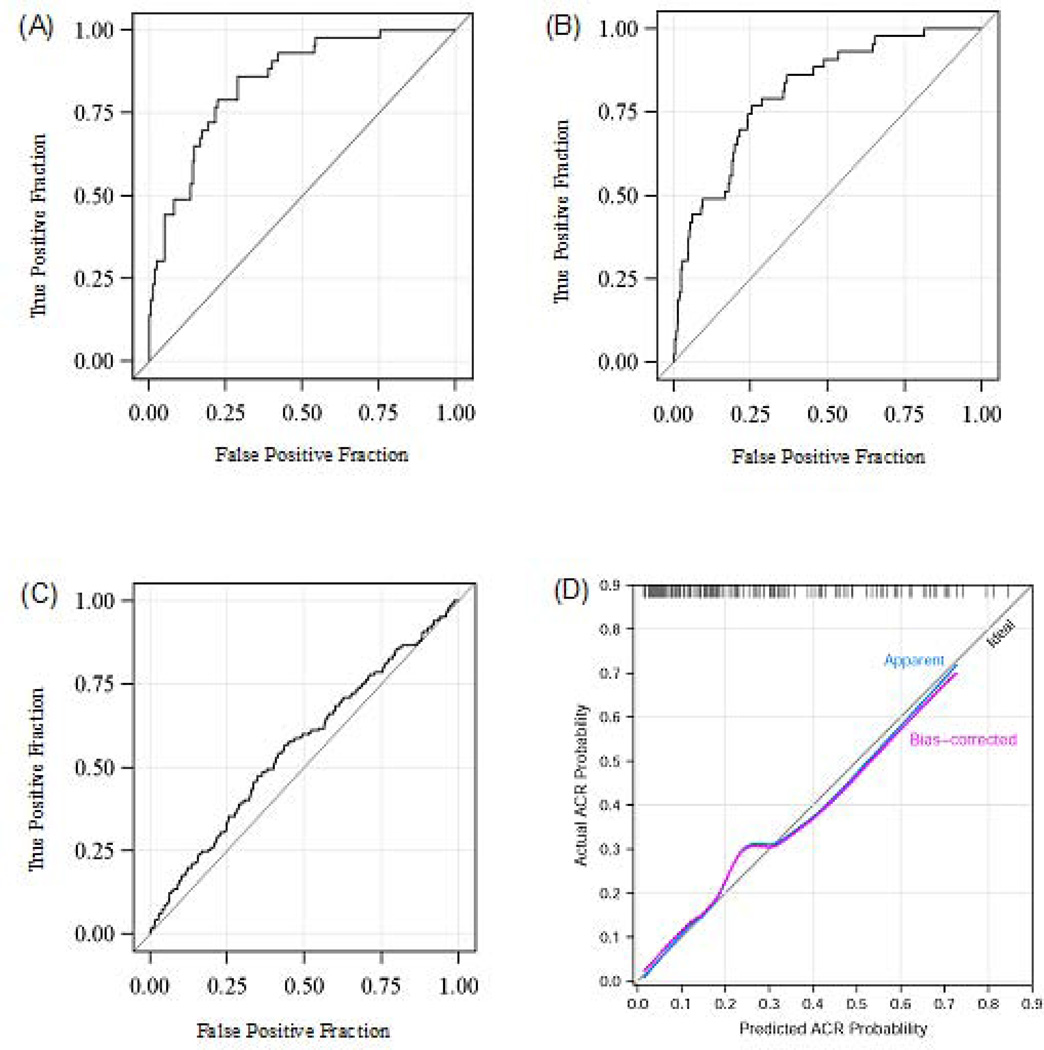
ROC curve analysis showed that this 3-gene signature yields an AUC (area under the curve) of 0.85 (95%CI 0.78 to 0.91, P<0.0001). Using the cutpoint of −1.213, which maximizes Youden’s index (26), this diagnostic signature had 79% sensitivity and 78% specificity to distinguish ACR biopsies from No Rejection biopsies (Fig. 4A). The Hosmer-Lemeshow test (27) indicated an excellent fit of this model to the data (χ2 = 4.84, df=8, p=0.77). The 3-gene signature also discriminated the ACR biopsy group from the Stable (no biopsy) group (Fig. 4B). The signature marginally distinguished the No Rejection biopsy group from the Stable (no biopsy) group (Fig. 4C).
Fig. 4. Receiver-operating-characteristic curves and calibration curve for the diagnostic signature.
The fraction of true positive results (sensitivity) and false positive results (1-specificity) for the diagnostic signature (calculated from 18S-normalized CD3ε mRNA, 18S-normalized IP-10 mRNA and 18S rRNA, all logged) as biomarker of ACR are shown in Panels A, B, and C, and the calibration plot based on bootstrap validation is shown in panel D. The area under the curve (AUC) was 0.85 (95 percent confidence interval, 0.78–0.91) for ACR vs. No Rejection Biopsy group (Panel A), 0.81 (95 percent confidence interval, 0.75–0.87) for ACR vs. Stable (no biopsy) group (Panel B) and 0.56 (95 percent confidence interval, 0.51–0.61) for No Rejection Biopsy group vs. Stable (no biopsy) group (Panel C). Adding 18S-normalized perforin to the best-fitting model provided no additional improvement, largely because perforin was very highly correlated with CD3ε. Alternative combinations of mRNA for perforin and IP-10 or perforin and CD3ε also predicted ACR accurately; the AUC for the combination of 18S-normalized perforin mRNA, 18S-normalized IP-10 mRNA and 18S rRNA was 0.84 (95% confidence Interval 0.78–0.90; P<0.0001), and this combination discriminated ACR biopsies with a sensitivity of 77% and a specificity of 76%. The AUC for the combination of 18S-normalized-CD3ε mRNA, 18S-normalized-perforin mRNA and 18S rRNA was 0.84 (95% confidence interval 0.76–0.91; P<0.0001), and this combination discriminated ACR with a sensitivity of 77% and a specificity of 74%. Additional analyses of the discriminating ability of single mRNA measures showed that the AUC for ACR vs. No Rejection Biopsy group was 0.70 for 18S rRNA, 0.71 for granzyme B, 0.74 for perforin, 0.75 for CD3ε, and 0.75 for IP-10. A value of 0.5 is no better than chance (the null hypothesis) and a value of 1.0 indicates a perfect discriminator. Bootstrap validation identified the optimal model to include 18S-normalized CD3ε mRNA, 18S-normalized IP-10 mRNA and 18S rRNA as predictors. Optimism-corrected estimates of the AUC, Cox’s intercept and slope statistics were: 0.830, −0.06, and 0.92, respectively. The Loess-smoothed estimates of the calibration curves of the optimism-adjusted and the unadjusted calibration curves are overlaid on a diagonal reference line representing perfect model calibration (Panel D). None of the single-marker models calibrated as well as the 3-gene model. Based on these results, the 3-gene model of 18S-normalized CD3ε mRNA, 18S-normalized IP-10 mRNA and 18S rRNA, all logged, is superior to any of the single-gene models considered and the three-gene diagnostic signature can be expected to perform well in an independent population of patients (from Suthanthiran et al. N Engl J Med 2013, with permission).
Bootstrapped model selection and internal validation
In an initial set of 500 bootstrap samples, 18S-normalized CD3ε, 18S-normalized IP-10, and 18S (all log10 transformed) remained in the final backwards elimination models more often than any of the other 6 mRNA measures. Bootstrap validation of this 3-gene model yielded an optimism-corrected estimate of the AUC of 0.83, an estimate of the expected value of the AUC in independent samples (not used to derive the diagnostic signature). The Cox’s intercept and slope statistics were −0.06 and 0.92, respectively, and the close correspondence of the unadjusted and optimism-adjusted calibration curves to the reference line demonstrates lack of overfitting and generalizability of the prediction model (Fig. 4D).
Rationale for not including perforin or granzyme B in the diagnostic signature
The further inclusion of granzyme B and/or perforin was unable to significantly improve the diagnostic signature based on IP-10, CD3ε, and 18s, due largely to their high correlations with CD3ε. This should not detract from the fact that when considered alone, 18S-normalized levels of granzyme B and perforin are strongly associated with ACR in CTOT-04, confirming the results reported in earlier publications (3, 18). Also, and as provided in the legend to Figure 4, the AUC for the combination of 18S-normalized perforin mRNA, 18S-normalized IP-10 mRNA and 18S rRNA was 0.84 (95% CI: 0.78 to 0.90; P<0.0001), only slightly lower than that of our best diagnostic signature, and this combination discriminated ACR biopsies with a sensitivity of 77% and a specificity of 76%. Importantly, this signature correlated very highly (r=0.93) with the optimal 3-gene diagnostic signature (18S-normalized CD3ε mRNA, 18S-normalized IP-10 mRNA and 18S rRNA, all logged).
Our findings that the signature is specific for ACR and distinguishes those with ACR from patients with other types of rejection such as borderline rejection and acute AMR are of considerable significance for the signature being clinically directive (3). The further observation that the diagnostic signature score is marginally higher in Banff ACR grade I compared to grades II or III is consistent with Banff grade I ACR being primarily interstitial cell infiltration and tubulitis wherein relatively more cells would be expected to have access to urine compared to grade II/III ACRs which are characterized by mild to moderate intimal arteritis (grade II) or by transmural arteritis and/or arterial fibrinoid change and necrosis of medial smooth cells with accompanying lymphocytic vascular inflammation (grade III) wherein the brunt of the injury is borne by the arterial vessel wall.
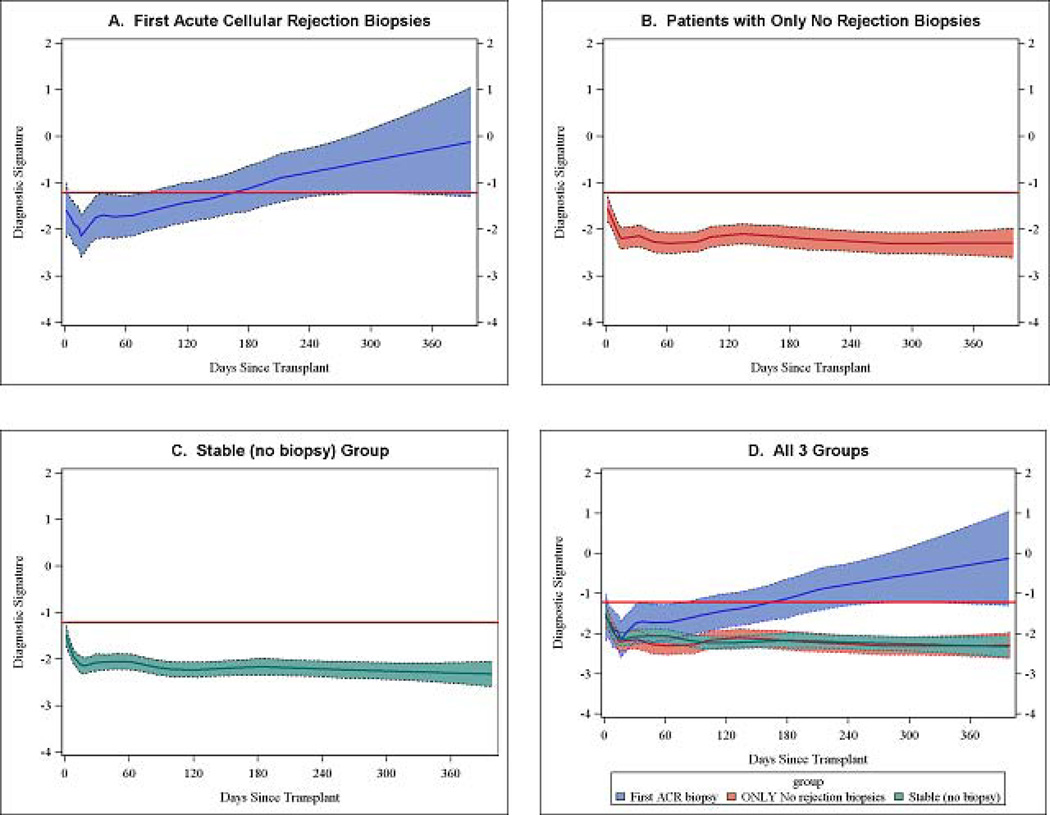
Prospective trajectory of the diagnostic signature as a function of time since transplant
Fig. 5 illustrates the LOESS-smoothed average within-person trajectories and 95% confidence bands of the diagnostic signature for the ACR group (Fig. 5A), No Rejection Biopsy group (Fig. 5B), and the Stable (no biopsy) group (Fig. 5C). It is evident that a progressive increase in the diagnostic signature is seen in the urine samples of those who went on to develop ACR whereas the trajectories in the No Rejection Biopsy group and in the Stable (no biopsy) group were flat and remained below the −1.213 threshold diagnostic for ACR throughout the first 400 days post-transplant. Thus, even after excluding the urine samples that were matched to the ACR biopsies and used to develop the diagnostic signature, we found a clear signal by about 80 days post-transplant that the future ACR patients have elevated values, and after about 160 days of transplantation the average value for the future ACR patients is greater than or equal to the threshold diagnostic for ACR (Fig. 5D).
Fig. 5. Prospective longitudinal trajectory of diagnostic signature as a function of time since kidney transplantation.
Sequential urine specimens collected from study participants as per the CTOT-04 study protocol on post-transplant days 3, 7, 15 and 30 and months 2, 3, 4, 5, 6, 9 and 12 were analyzed. The average within-person trajectories and 95% confidence bands of the diagnostic signature for the ACR group (n=195 QC-passed urine samples obtained prior to 3 days before the first ACR biopsy from 38 patients who had no abnormal biopsy prior to their first ACR biopsy) (Panel A), No Rejection Biopsy group (n=960 QC-passed urine samples from 132 patients for whom all biopsies were rejection free) (Panel B), Stable (no biopsy) group (n=1491 QC-passed urine samples from 201 patients) (Panel C). In all panels, the black lines of the LOESS curves indicate the trajectory, the colored bands the 95% confidence interval, and the red lines the diagnostic threshold. On average, the diagnostic signature in the No Rejection Biopsy group and in the Stable (no biopsy) group remained well below the −1.213 threshold diagnostic for ACR throughout the first 400 days post-transplant whereas a progressive increase in the signature was seen in the urine samples of those who went on to develop ACR. By about 80 days post-transplant there was a clear signal that the future ACR patients have elevated values (the lower bound of the 95% confidence band for this group exceeds the upper bound of the 95% confidence bands for the two other groups), and beyond about 160 days the average value for the future ACR patients is greater than or equal to the threshold diagnostic for ACR (Panel D). The y-axis values are diagnostic-signature scores without intrinsic units of measurement; they were calculated from the logistic-regression equation (−6.1487 + 0.8534 log10[CD3ε/18S] + 0.6376 log10[IP-10/18S] + 1.6464 log10[18S]). Absolute levels of CD3ε mRNA, IP-10 mRNA, and 18S rRNA in the cells from each urine sample were measured with the use of polymerase-chain reaction assays, with the units of measurement being copies per microgram of total RNA for each mRNA measure and 18S rRNA copies (×10-6) per microgram of total RNA. The mRNA copy numbers were 18S-normalized by dividing the mRNA copy number by the 18S rRNA copy number in the same sample, and the ratio was log10-transformed (from Suthanthiran et al. N Engl J Med 2013, with permission).
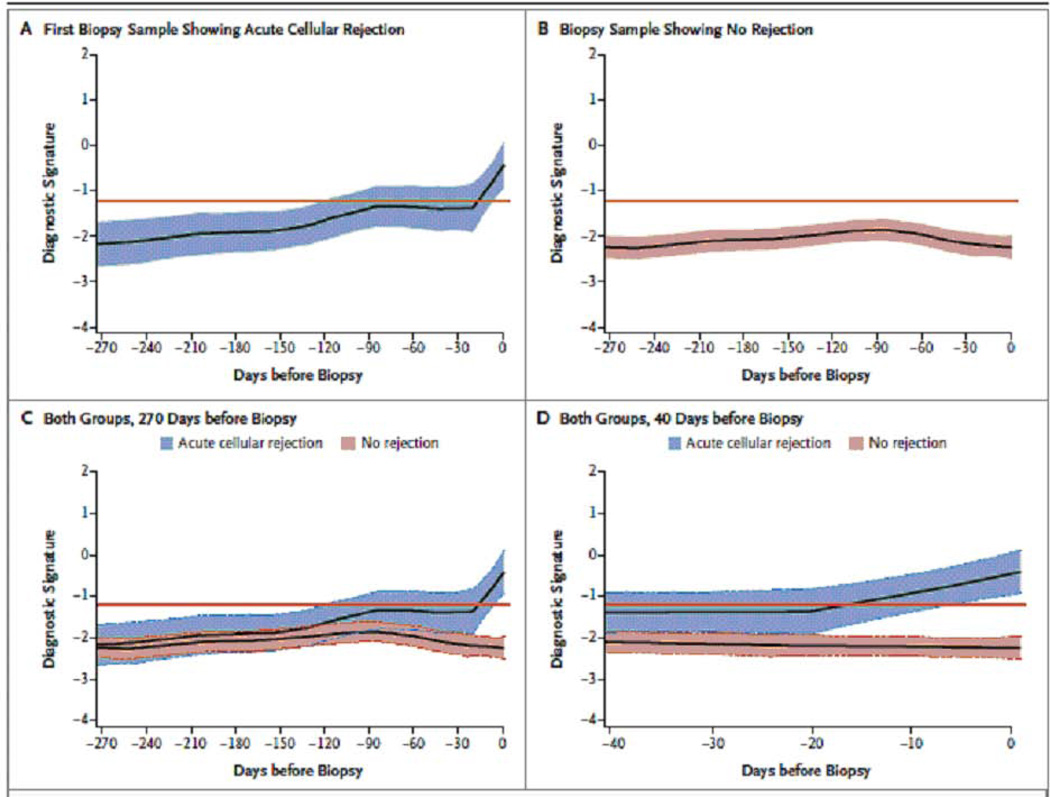
Retrospective trajectory of the diagnostic signature as a function of time prior to biopsy
Fig. 6 displays the LOESS-smoothed average within-person trajectories and 95% confidence bands of the diagnostic signature, looking backwards from the time of biopsy (time=0), for the ACR biopsy group (Fig. 6A) and the No Rejection Biopsy group (Fig. 6B). A comparison of the trajectories showed a significant difference in the trajectories between the two groups, with a marked increase in the score of the diagnostic signature during the 20-day period leading up to the first ACR biopsy whereas the signature remained relatively flat and well below the diagnostic threshold during the 270 days preceding a biopsy in the No Rejection Biopsy group (Fig. 6C and Fig. 6D) (P<0.0001).
Fig. 6. Retrospective trajectory of diagnostic signature as a function of time prior to biopsy.
The average within-person retrospective trajectory of the diagnostic signature (i.e., the trajectory as a function of the time before biopsy) in urine samples obtained at or before biopsy that passed quality control are shown for the group of 38 patients with first biopsy specimens showing acute cellular rejection (201 urine samples) (Panel A) and the group of 113 patients with specimens showing no rejection (833 urine samples) (Panel B). Only specimens obtained during the first 400 days after transplantation were included. The diagnostic signature remained relatively flat and well below the −1.213 threshold that was diagnostic of acute cellular rejection during the 270 days before biopsy in the group of patients with findings showing no rejection (Panel C). There was a significant difference in the trajectories between the two groups, with a marked increase in the diagnostic signature during the 20-day period before the first specimen showing acute cellular rejection (P<0.001) (Panel D). The y-axis values are diagnostic-signature scores without intrinsic units of measurement; they were calculated from the logistic-regression equation (−6.1487 + 0.8534 log10[CD3ε/18S] + 0.6376 log10[IP-10/18S] + 1.6464 log10[18S]). Absolute levels of CD3ε mRNA, IP-10 mRNA, and 18S rRNA in the cells from each urine sample were measured by polymerase-chain-reaction assay, with the units of measurement being copies per microgram of total RNA for each mRNA measure and copies (×10-6) per microgram of total RNA for 18S rRNA. The mRNA copy numbers were 18S-normalized by dividing the mRNA copy number by the 18S rRNA copy number in the same sample, and the ratio was log10-transformed. In all the panels, the black lines indicate the trajectory, the colored bands the 95% confidence interval, and the red lines the diagnostic threshold (from Suthanthiran et al. N Engl J Med 2013, with permission).
Specificity of the diagnostic signature
The ‘No Rejection biopsy group’ included biopsies that displayed histological features such as acute tubular necrosis (n=67), tubular atrophy (n=61), interstitial fibrosis (n=52), glomerulosclerosis (n=23), vascular narrowing (n=17), calcineurin toxicity (n=12), and/or recurrent disease (n=2) and not infrequently more than one type of allograft pathology was observed in a single biopsy. Also, 107 of the 163 biopsies were performed for suspected rejection/graft dysfunction (for-cause biopsies) and the remaining 56 biopsies were surveillance (protocol) biopsies. We investigated the performance of the 3-gene diagnostic signature after inclusion in the non-ACR group of ALL matched and QC-passed urine samples from the kidney graft recipients with biopsies showing borderline changes (19 samples from 17 patients), acute antibody mediated rejection (10 samples from 9 patients), chronic allograft nephropathy (CAN)(2 samples from 2 patients), or BKV nephropathy (BKVN)(6 samples from 5 patients).
A comparison of the levels of mRNA in the 43 matched urine specimens from 34 kidney graft recipients with biopsy confirmed Banff ACR grade 1A or higher with all available non-ACR biopsy matched, QC-passed urine samples from 143 patients (n= 200 samples) showed that the signature is diagnostic of ACR with 77% specificity (95% CI: 71 to 83%) and 79% sensitivity (95% CI: 67 to 91%) (AUC=0.83, 95% CI: 0.77 to 0.89, P<0.0001) and thus that the performance of the signature is very similar to the original analysis that was restricted to ACR versus No Rejection biopsy group (that is without inclusion of the 37 urine samples from those with borderline changes, AMR, CAN or BKVN). We further analyzed the performance of the 3-gene diagnostic signature comparing levels of mRNA in the 43 matched urine specimens from 34 kidney graft recipients with biopsy confirmed Banff ACR grade 1A or higher with levels of mRNA in just the 31 matched and QC-passed urine samples from the kidney graft recipients with biopsies showing Borderline changes (19 samples from 17 patients), AMR (10 samples from 9 patients), or CAN (2 samples from 2 patients). Despite the sample size for the comparison group being very much smaller than in the previous analyses, this analysis showed that the signature is diagnostic of ACR with 71% specificity (95% CI: 55 to 87%) and 79% sensitivity (95% CI: 67 to 91%) (AUC=0.78, 95% CI: 0.68 to 0.89, P<0.0001) and that its performance was not significantly different from that observed when comparing the ACR biopsy group with the No Rejection biopsy group.
The diagnostic signature in for-cause versus surveillance biopsies
The 3-gene signature was diagnostic of ACR with 80% specificity (95% CI: 73 to 88%) and 79% sensitivity (95% CI: 66 to 92%) (AUC=0.85, 95% CI: 0.79 to 0.92, P<0.0001) when the data were analyzed using only the 38 for-cause ACR biopsies and the 107 for-cause No Rejection biopsies. The performance characteristics of the diagnostic signature were nearly identical to the original analysis using both for-cause biopsies and surveillance biopsies [43 ACR biopsies vs. 163 No Rejection biopsies, 78% specificity (95% CI: 71 to 84%) and 79% sensitivity (95% CI: 67 to 91%) (AUC=0.85, 95% CI: 0.78 to 0.91, P<0.0001)]. The 3-gene signature was also strongly associated with the biopsy diagnosis when the analysis was restricted to surveillance No Rejection biopsies; analysis of 43 ACRs vs. 56 surveillance No Rejection biopsies showed that ACR was predicted with 73% specificity (95% CI: 62 to 85%) and 79% sensitivity (95% CI: 67 to 91%) and the AUC was 0.83 (95% CI: 0.75 to 0.91, P<0.0001).
The diagnostic signature in clinical versus subclinical ACR
Among the 43 ACR biopsies from the CTOT-04 patients, 38 biopsies were for-cause biopsies, and the remaining 5 were surveillance biopsies. Among the 163 No Rejection biopsies, 107 were for-cause biopsies, and 56 were surveillance biopsies. The mean levels of the 3-gene diagnostic signature did not differ significantly between the 38 for-cause ACR biopsies and the 5 surveillance ACR biopsies (P=0.78). The same is true for comparison of the 107 for-cause and the 56 surveillance No Rejection biopsies (P=0.66). Sensitivity of diagnostic signature was 79% in 38 for-cause biopsies and 80% in surveillance biopsies (No difference, P=1.00). Specificity of the diagnostic signature was 80% in the 107 for-cause biopsies and 73% in the surveillance biopsies (P=0.32).
Early or late ACR and the diagnostic signature
To examine whether the diagnostic signature tends to be higher (or lower) at the time of an early ACR compared to a late ACR, we arbitrarily classified the 43 ACRs with matching urine samples based on whether the ACR occurred during the first 180 days post-transplantation (early) or thereafter (late). Nineteen ACRs qualified as early ACRs, and the remaining 24 ACRs as late ACRs. The mean diagnostic signature score at the time of biopsy was −0.43 (95% CI: −1.04 to +0.18) for the early ACRs and −0.37 (95% CI: −0.88 to +0.14) for the late ACRs (P=0.87 by two tailed, unpaired t test). We also evaluated the correlation between the diagnostic signature at the time of ACR and time since transplant (treated continuously) and the analysis showed no relationship (r=0.00, P=0.97). Thus the level of the diagnostic signature at the time of an ACR did not vary by time since transplant.
Anti-rejection therapy and the diagnostic signature
We examined whether the 3-gene signature not only functions as a diagnostic signature but also as a response-predictive biomarker. Among 43 ACR biopsies with paired urine samples, 32 also had QC-passed urine samples 14±7 days after the incident ACR biopsy. A comparison of the diagnostic signature values showed a significant decrease from −0.37 at the time of ACR biopsy to −0.86 at 14±7 days post-biopsy (P=0.05). We further compared the decrease in the value for the diagnostic signature according to whether the patient was a ‘responder’, defined as the follow-up estimated glomerular filtration rate (eGFR) being no higher than it was at the time of the ACR biopsy or a non-responder to the initial anti-rejection therapy. eGFR data were available at the time of biopsy and at the follow-up time for 25 of the 32 samples and among these 25, 21 were responders and the remaining 4 were non-responders based on the above criterion. In the 21 responders, the value for the diagnostic signature decreased significantly from the baseline value of −0.43 to −1.08 at 14±7 days post biopsy (P=0.05); in the small number of non-responders, the decrease in the value from +0.12 to −0.80 was even greater than in the responders (0.92 vs. 0.65), but was not statistically significant (P=0.20). Of note, the non-responders had higher levels of the diagnostic signature at the time of biopsy and two weeks later and among the 21 responders, 7 did not show a decrease in the value of the diagnostic signature.
CTOT-04 study: clinical implications
We have developed and validated a 3-gene molecular signature that is diagnostic of ACR. The additional information regarding the exquisite specificity of the signature for diagnosing ACR and serving as a robust parameter of the immune status of the kidney allograft recipient supports the clinical utility of this new molecular signature developed and validated in the CTOT-04 study. The clinical utility of the signature is further supported by its ability to foretell the development of an episode of ACR (thereby facilitating preemptive therapy). We suggest that the results from our study will be transformative in helping to reduce the need for allograft biopsies, reducing post-transplant costs and morbidity, and improving the care of kidney graft recipients. Moreover, the renal transplant community is now better positioned to undertake an interventional trial of personalized immunosuppression based on urinary cell mRNA profiles.
Noninvasive diagnosis of interstitial fibrosis and tubular atrophy (IF/TA)
To develop urinary cell mRNA profiles diagnostic of IF/TA, we collected and profiled urine samples from kidney graft recipients whose biopsies were classified as IF/TA and from patients whose biopsies were classified as normal biopsies (28). We collected 114 urine samples at the time of an allograft biopsy and among the 114 allograft biopsies, 48 biopsies from 48 patients were classified as IF/TA, and 66 biopsies from 66 patients were classified as normal biopsies. The biopsies classified as IF/TA were clinically indicated biopsies and the normal biopsies were surveillance biopsies. The biopsy-matched urine samples were centrifuged and RNA was extracted from the urine cell pellet and reverse-transcribed to cDNA, as described above. We designed oligonucleotide primers and TaqMan probes and measured absolute levels of potentially informative mRNAs in the urine of kidney allograft recipients with the use of pre-amplification enhanced kinetic quantitative PCR assays developed in our laboratory. The sequence and location of the primers and TaqMan probes used in this study are provided in Table 3.
Table 3. Oligonucleotide Primers and TaqMan Probes used to Measure Levels of mRNA for the Noninvasive Diagnosis of IF/TA.
The sequences and the locations of the oligonucleotide primers and probes that we designed and validated for the measurement of mRNA levels and 18S ribosomal RNA (rRNA) in the urine are shown. The fluorogenic TaqMan probes were labeled with 6-carboxy-fluorscein (FAM) at the 5’ end and with 6-carboxy-tetramethylrodamine (TAMRA) or minor groove binder (MGB) at the 3’ end. FAM functioned as the reported dye and TAMRA or MGB as the quencher.
| Type of mRNA |
Sequence | Location | |
|---|---|---|---|
| GenBank | |||
| Accession Number | |||
| Vimentin | NM_003380.2 | Sense: 5’ TCAGAGAGAGGAAGCCGAAAAC 3’ | 706–727 |
| Antisense: 5’ CCAGAGACGCATTGTCAACATC 3’ | 770–749 | ||
| Probe: 5’ FAM CCCTGCAATCTTTCAGAC MGB 3’ | 729–746 | ||
| HGF | BC063485 | Sense: 5' CAAATGTCAGCCCTGGAGTTC 3' | 526–546 |
| Antisense: 5' CTGTAGGTCTTTACCCCGATAGCT 3' | 604–581 | ||
| Probe: 5’ FAM ATGATACCACACGAACACAGCTTTTTGCC TAMARA 3’ | 548–576 | ||
| α -SMA | NM_001613 | Sense: 5’ TGGGACGACATGGAAAAGATC 3’ | 288–308 |
| Antisense: 5’ CAGGGTGGGATGCTCTTCAG 3’ | 365–346 | ||
| Probe: 5’ FAM CCACTCTTTCTACAATGAGCTTCGTGTTGCC TAMRA 3’ | 314–344 | ||
| Fibronectin 1 | XM_055254 | Sense: 5' GAAAGTACACCTGTTGTCATTCAACA 3' | 2582–2607 |
| Antisense: 5' ACCTTCACGTCTGTCACTTCCA 3' | 2688–2666 | ||
| Probe: 5' FAM CCACTGGCACCCCACGCTCA TAMRA 3' | 2613–2632 | ||
| PAI1 | NM_000602.1 | Sense: 5’ AATCAGACGGCAGCACTGTCT 3’ | 716–736 |
| Antisense: 5’ GGGCGTGGTGAACTCAGTATAGT 3’ | 792–770 | ||
| Probe: 5’ FAM TGTGCCCATGATGGC MGB 3’ | 738–752 | ||
| Perforin | M28393 | Sense: 5' GGACCAGTACAGCTTCAGCACTG 3' | 492–514 |
| Antisense: 5' GCCCTCTTGAAGTCAGGGTG 3' | 587–568 | ||
| Probe: 5' FAM TGCCGCTTCTACAGTTTCCATGTGGTACAC TAMRA 3' | 526–555 | ||
| TGFI31 | NM_000660 | Sense: 5' GCGTGCTAATGGTGGAAACC 3' | 1170–1189 |
| Antisense: 5' CGGAGCTCTGATGTGTTGAAGA 3' | 1263–1242 | ||
| Probe: 5’ FAM ACAACGAAATCTATGACAAGTTCAAGCAGAGTACACA TAMRA 3’ | 1191–1227 | ||
| TIMP1 | NM003254 | Sense: 5’ GACGGCCTTCTGCAATTCC 3’ | 288–306 |
| Antisense: 5’ GTATAAGGTGGTCTGGTTGACTTCTG 3’ | 366–341 | ||
| Probe: 5’ FAM AGGGCCAAGTTCGTGG MGB 3’ | 319–334 | ||
| Granzyme B | J04071 | Sense: 5' GCGAATCTGACTTACGCCATTATT 3' | 534–557 |
| Antisense: 5' CAAGAGGGCCTCCAGAGTCC 3' | 638–619 | ||
| Probe: 5’ FAM CCCACGCACAACTCAATGGTACTGTCG TAMRA 3’ | 559–585 | ||
| FSP1 | CR450345.1 | Sense: 5’ AGGAGCTGCTGACCCGG 3’ | 104–120 |
| Antisense: 5’ GCTTCATCTGTCCTTTTCCCC 3’ | 158–138 | ||
| Probe: 5’ FAM CTGCCCAGCTTCT MGB 3’ | 124–136 | ||
| CD103 | XM_008508 | Sense: 5' CGTGCTCAGCTCCCTTCTG 3' | 211–229 |
| Antisense: 5' CCTGGTGTCCTCTTGGTTCTG 3' | 297–277 | ||
| Probe: 5’ FAM ACCAAGACCCCAGCACCAACCATACCT TAMRA 3’ | 231–257 | ||
| Collagen 1A1 | NM_000088.3 | Sense: 5' CCAGAAGAACTGGTACATCAGCAA3' | 4050–4073 |
| Antisense: 5' CGCCATACTCGAACTGGAATC3' | 4144–4124 | ||
| Probe: 5’ FAM ACAAGAGGCATGTCTGG MGB 3’ | 4085–4101 | ||
| BMP7 | NM_001719.1 | Sense: 5' GCTTCGTCAACCTCGTGGAA 3' | 526–545 |
| Antisense: 5’ CAAACCGGAACTCTCGATGGT 3’ | 597–577 | ||
| Probe: 5’ FAM ATGACAAGGAATTCTTCCACCCACGCTAC TAMRA 3’ | 547–575 | ||
| CTLA4 | BC074893 | Sense: 5’ CGCCATACTACCTGGGCATAG 3’ | 441–461 |
| Antisense: 5 GATCCAGAGGAGGAAGTCAGAATC 3’ | 529–506 | ||
| Probe: 5’ FAM CAGATTTATGTAATTGATCCAGAACCGTGCCC TAMRA 3’ | 473–504 | ||
| CTGF | NM_001901 | Sense: 5’ TGTGTGACGAGCCCAAGGA 3’ | 639–657 |
| Antisense: 5’ TAGTTGGGTCTGGGCCAAAC 3’ | 725–706 | ||
| Probe: 5’ FAM CCTGCCCTCGCGGCTTACCG TAMRA 3’ | 674–693 | ||
| FGF2 | NM_002006.3 | Sense: 5’ CCGACGGCCGAGTTGAC 3’ | 601–617 |
| Antisense: 5’ TAACGGTTAGCACACACTCCTTTG 3’ | 712–689 | ||
| Probe: 5’ FAM ACCCTCACATCAAGCTACAACTTCAAGCAGAA TAMRA 3’ | 637–668 | ||
| CD25 | NM_000417 | Sense: 5’ GACTGCTCACGTTCATCATGGT 3’ | 185–206 |
| Antisense: 5’ AATGTGGCGTGTGGGATCTC 3’ | 266–247 | ||
| Probe: 5' FAM AGAGCTCTGTGACGATGACCCGCC TAMRA 3' | 222–245 | ||
| FoxP3 | NM_014009 | Sense: 5’ GAGAAGCTGAGTGCCATGCA 3’ | 939–958 |
| Antisense: 5 GGAGCCCTTGTCGGATGAT 3’ | 1025–1007 | ||
| Probe: 5’ FAM TGCCATTTTCCCAGCCAGGTGG TAMRA 3’ | 962–983 | ||
| USAG1 | NM_015464 | Sense: 5' TGGAGGCAGGCATTTCAGTAA 3' | 364–366 |
| Antisense: 5' TTCCCGGCAACCCACTT 3' | 412–396 | ||
| Probe: 5' FAM CCCGAGTGTTCCGATCCAGTCCAGT TAMRA 3' | 392–368 | ||
| NKCC2 | BC040138.2 | Sense: 5’ TCACGAGCAACTCGCAAAGA 3’ | 588–607 |
| Antisense: 5’ TCCCATCACCGTTAGCAACTC 3’ | 658–638 | ||
| Probe: 5’ FAM TGTGGCAGTCACCCCAAGTTCAGC TAMRA 3’ | 609–632 | ||
| ITGB6 | NM_000888.3 | Sense: 5’ GGATTGAACTGCTTTGCCTGTT 3’ | 21–42 |
| Antisense: 5’ GGCACAGCCACCTTGTACGT 3’ | 69–88 | ||
| Probe: 5’ FAM TTTCTATTTCTAGGAAGGAATG MGB 3’ | 44–65 | ||
| E-cadherin | XM_007840 | Sense: 5' TGAGTGTCCCCCGGTATCTTC 3' | 2469–2489 |
| Antisense: 5' CAGCCGCTTTCAGATTTTCAT 3' | 2549–2529 | ||
| Probe: 5' FAM CCTGCCAATCCCGATGAAATTGGAAAT TAMRA 3' | 2495–2521 | ||
| 18S rRNA | K03432 | Sense: 5' GCCCGAAGCGTTTACTTTGA 3' | 929–948 |
| Antisense: 5' TCCATTATTCCTAGCTGCGGTATC 3' | 1009–986 | ||
| Probe: 5’ FAM AAAGCAGGCCCGAGCCGCC TAMRA 3’ | 965–983 |
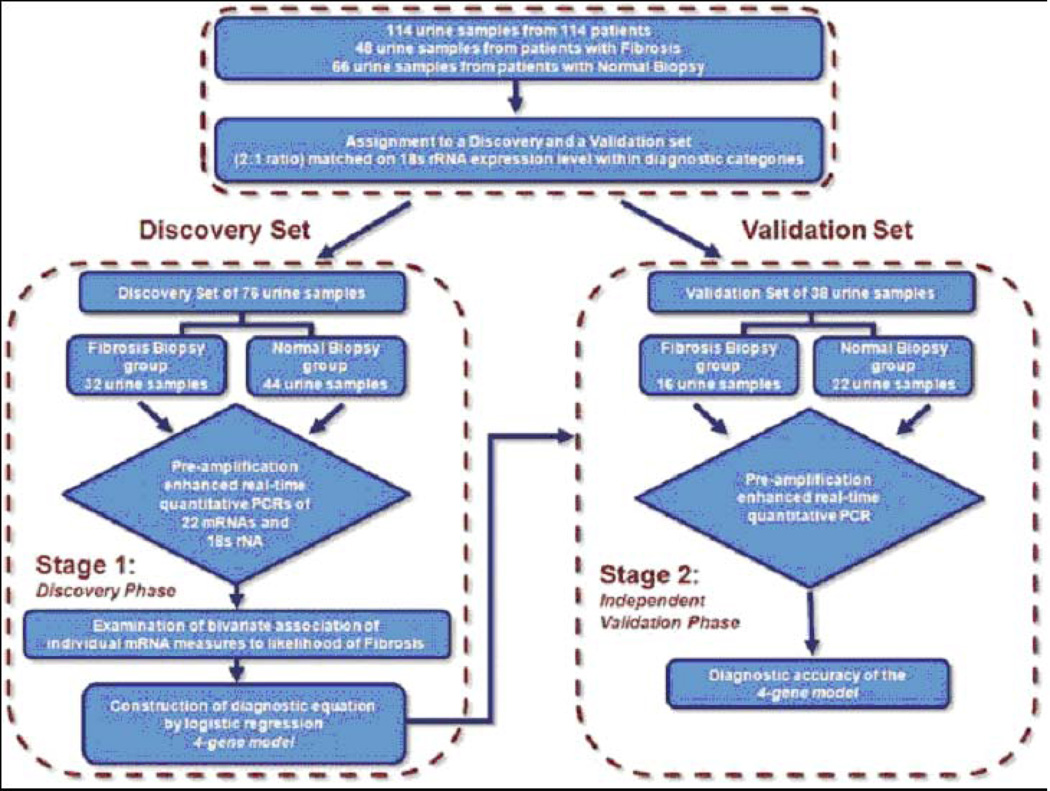
We used a two-step approach: in the first step, we developed/discovered a urinary cell signature diagnostic of IF/TA; in the second step, we validated the discovered signature (Fig. 7). To accomplish discovery and validation of the signature, the 18S rRNA copy number of the 114 patients were ranked and partitioned into consecutive triplets. Within each triplet, the first and third patients were assigned to the discovery set, and the second patient was assigned to the validation set, resulting in the two sets being exactly matched on IF/TA status and very closely matched on 18S. Twice as many patients were assigned to the discovery set to enhance statistical power for the exploratory analyses that included a procedure to protect against the risk of a Type I error.
Fig. 7. Flow chart for the discovery and validation of urinary cell mRNA profiles.
The 114 kidney allograft recipients (48 with biopsies showing fibrosis and 66 with normal biopsy results) were rank ordered within group (Fibrosis group or Normal Biopsy group) by the copy number of 18S rRNA and partitioned into triplets. Within each triplet, the first and third patients were assigned to the Discovery set and the second patient was assigned to the Validation set, resulting in the two sets being exactly matched on fibrosis status and very closely matched on 18S rRNA copy number. Twice as many patients were assigned to the Discovery set in order to enhance statistical power for the exploratory analyses, which included a procedure to protect against the risk of a Type I error (from Anglicheau et al. Transplantation 2012, with permission).
Urinary cell mRNA levels are diagnostic of IF/TA
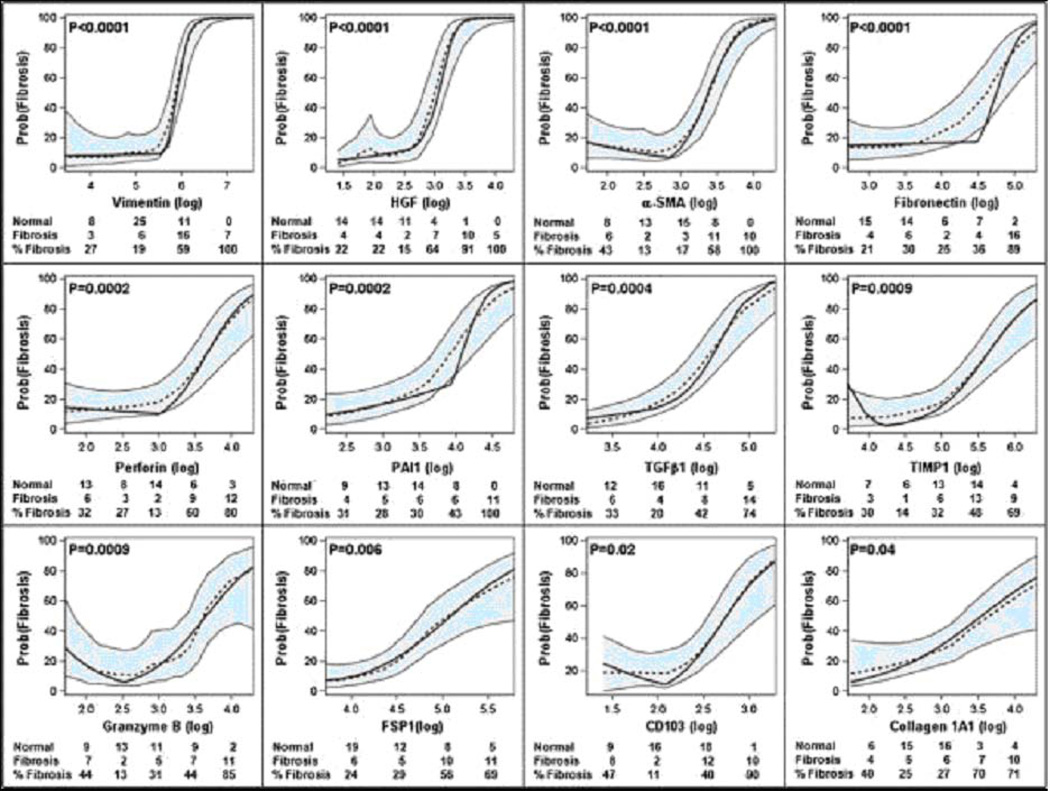
Fig. 7 is a flow chart for the discovery and validation of urinary cell mRNA profiles. Fig. 8 illustrates the predicted probability of IF/TA as a function of urinary cell mRNA copy number in the discovery set comprised of 32 kidney graft recipients with biopsy-confirmed IF/TA and 44 recipients with normal allograft biopsy results. According to piece-wise linear logistic models, the levels of 12 of the 22 mRNAs were significantly associated with the diagnosis of IF/TA after using Holm’s modified Bonferroni procedure (29) to control the risk of a Type I error: vimentin (P<0.0001), HGF (P<0.0001), α-SMA (P<0.0001), fibronectin 1 (P<0.0001), perforin (P=0.0002), PAI1 (P=0.0002), TGF-β1 (P=0.0004), TIMP1 (P=0.0009), granzyme B (P=0.0009), FSP1 (P=0.006), CD103 (P=0.02), and collagen 1A1 (P=0.04). Analysis of ROC curves showed that IF/TA is predicted quite accurately with the use of urinary cell levels of mRNA for vimentin (AUC and 95% CI = 0.90, 0.82 to 0.97), HGF (0.91, 0.84 to 0.98), α-SMA (0.88, 0.80 to 0.95), fibronectin 1 (0.83, 0.73 to 0.93), perforin (0.83, 0.74 to 0.93), TGF-β1 (0.82, 0.72 to 0.92), TIMP1 (0.81, 0.71 to 0.90), granzyme B (0.82, 0.71to 0.92), FSP1 (0.81, 0.71 to 0.91), PAI1 (0.79, 0.68 to 0.90), collagen 1A1 (0.77, 0.66 to 0.88) or CD103 (0.76, 0.65 to 0.87).
Fig. 8. Predicted probability of fibrosis as a function of urinary cell mRNA copy number in the Discovery set, for LOESS model and piece-wise linear logistic regression model, after controlling for 18S rRNA copy number.
Urine samples were collected from 32 kidney transplant recipients with graft dysfunction and biopsy-confirmed fibrosis and 44 recipients with stable allograft function and normal allograft biopsy, and levels of mRNA in urinary cells were measured with the use of pre-amplification enhanced kinetic quantitative PCR assays. The figure shows the predicted probability of fibrosis (Y-axis), controlling for 18S rRNA, as a function of individual log10-transformed mRNA copy numbers for these genes (X-axis). Each plot shows the LOESS model’s predicted probabilities (dotted line), their 95% confidence interval (shaded area) and the logistic regression model’s predicted probabilities (solid line). According to the logistic models, the levels of twelve of the twenty-two mRNAs (vimentin, HGF, α-SMA, fibronectin 1, perforin, PAI1, TGF-β1, TIMP1, granzyme B, FSP1, CD103, and collagen 1A1) were significantly (P-values <0.05 with modified Bonferroni correction) associated with the diagnosis of fibrosis. Adjusted P-value for each parametric model is shown. The number of stable patients, number of fibrosis patients, and percentage of fibrosis patients within categories of the mRNA measure appear in each plot (from Anglicheau et al. Transplantation 2012, with permission).
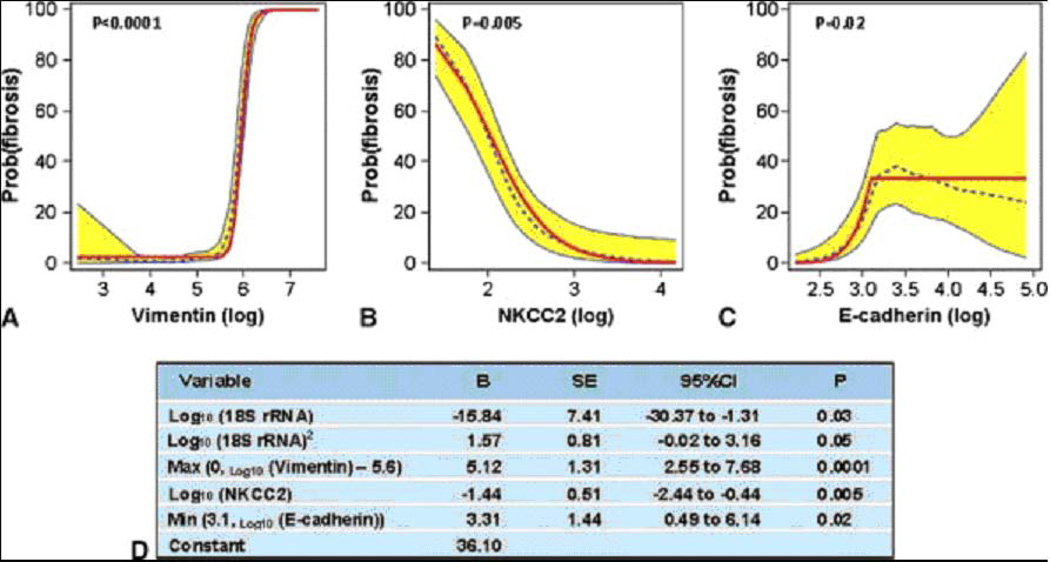
A 4-gene model diagnostic of IF/TA in the discovery set
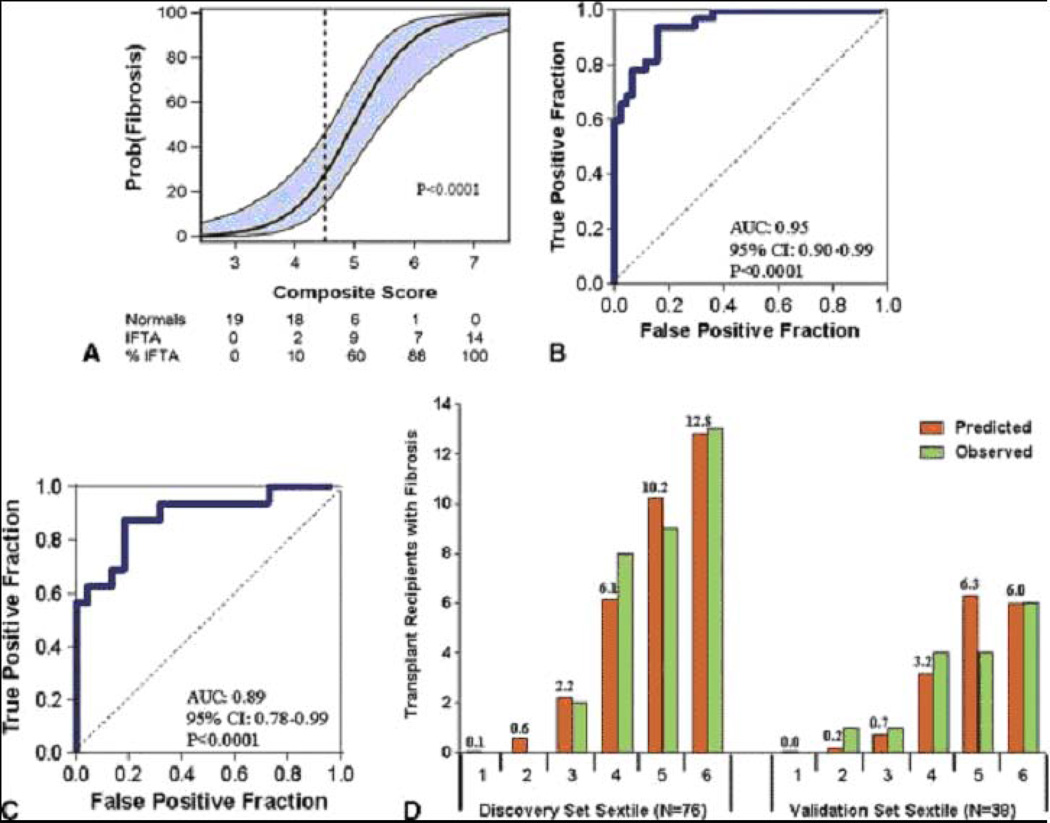
Vimentin and HGF had the highest AUCs, and we built a multigene prediction model of fibrosis around vimentin in view of the potential contribution of vimentin to the pathogenesis of fibrosis. Very interestingly, after controlling for vimentin mRNA level and the quadratic relationship of 18S rRNA level, we found that none of the previously significant predictors (HGF, TGF-β1, fibronectin 1, PAI1, FSP1, collagen 1A1, α-SMA, CD103, granzyme B, or perforin) remained significantly predictive, and two of the previously non-significant predictors, mRNA for NKCC2 and E-cadherin, became significant (Fig. 9B,C). The composite score based on the 4-gene model (Fig. 9D) was highly associated with the diagnosis of IF/TA (Fig. 10A). The ROC curve (Fig. 10B) shows, for various levels of this composite score, the fraction of true positive results (sensitivity) and false positive results (1-specificity) for distinguishing patients with IF/TA from patients with normal biopsy results. The AUC was 0.95 (95%CI: 0.90 to 0.99, P<0.0001) and with the use of the optimal cutpoint of 4.5, the composite score predicted IF/TA with a specificity of 84.1% (95%CI: 73.3 to 94.9%) and a sensitivity of 93.8% (95%CI: 85.4 to 99.9%) (Fig. 10B).
Fig. 9. Final model derived from the discovery set for the diagnosis of fibrosis.
The predicted probability of fibrosis (y-axis) as a function of individual log10-transformed messenger RNA (mRNA) copy numbers (x-axis) for vimentin (Panel A), NKCC2 (Panel B), and E-cadherin (Panel C) after controlling for the copy numbers for the other two mRNAs and 18S rRNA is shown. Each plot shows the locally weighted scatterplot smoothing (LOESS) model’s predicted probabilities (dotted line), their 95% confidence interval (CI) (shaded area), and the logistic regression model’s predicted probabilities (solid line). The parameter estimates for the four-gene model including terms accounting for the relationships, including nonlinear relationships, between the mRNAs and diagnosis are provided in Panel D (from Anglicheau et al. Transplantation 2012, with permission).
Fig. 10. Relationship of composite score to fibrosis in the Discovery set, ROC curve analysis of the composite score in the Discovery set and the Validation set and the predicted and observed number of transplant recipients with fibrosis for each sextile of the composite score within the Discovery and Validation sets.
To predict fibrosis in the Discovery set, a composite score was calculated based on a logistic model, from vimentin mRNA, NKCC2 mRNA and E cadherin mRNA as well as the 18S rRNA in urine samples obtained from the 32 subjects with biopsy-confirmed fibrosis and 44 subjects with stable graft function and normal allograft biopsy. The composite score predicted fibrosis with high accuracy. Panel A shows the predicted probability of fibrosis (Y-axis) as a logistic function of the composite score (X-axis). The blue band represents the 95% confidence interval of the model. Panel B shows the receiver-operating-characteristic curve for the diagnosis of fibrosis using the composite score. The model had an area under the curve of 0.95 (95%CI: 0.90 to 0.99, P<0.0001). At a cutpoint or 4.5, fibrosis was diagnosed with a specificity of 84.1% (95%CI: 73.3 to 94.9%) and a sensitivity of 93.8% (95%CI: 85.4 to 99.9%). The final prediction equation derived from the Discovery set was used to calculate the predicted probability of fibrosis in the Validation set of 38 kidney transplant recipients; 16 with biopsy-confirmed fibrosis and 22 with stable graft function and normal allograft biopsy. Panel C shows the receiver-operating characteristic curve of the composite score (applying the equation from Figure 9D to the urinary cell mRNA levels of vimentin, NKCC2 and E-cadherin and 18S rRNA level of those in the Validation set) for the diagnosis of fibrosis. The area under the curve for the diagnosis of fibrosis in the Validation set was 0.89 (95%CI: 0.78 to 0.99, P<0.0001). At the composite score cutpoint of 4.5 derived from the Discovery set, fibrosis was diagnosed in the Validation set with a specificity of 77.3% (95%CI: 59.8 to 94.8%) and a sensitivity of 87.5% (95%CI: 71.3 to 99.9%). Panel D shows the predicted and observed number of transplant recipients with fibrosis for each sextile of the composite score within the Discovery and Validation sets (from Anglicheau et al. Transplantation 2012, with permission).
Independent validation
The final diagnostic equation predicting IF/TA in the discovery set was then validated in an independent Validation set of 38 kidney graft recipients, 16 with biopsy-proven fibrosis and 22 with normal allograft biopsy results. Fig. 10C shows the ROC curve of this equation for the diagnosis of IF/TA. This 4-gene signature was accurately diagnostic of IF/TA; the AUC for the diagnosis of fibrosis in the independent Validation set was 0.89 (95%CI: 0.78 to 0.99, P<0.0001) (Fig. 10C). At the composite score cutpoint of 4.5 (the same cutpoint used in the discovery set), IF/TA was diagnosed in the validation set with a specificity of 77.3% (95%CI: 59.8 to 94.8%) and a sensitivity of 87.5% (95%CI: 71.3 to 99.9%).
We examined the fit of the prediction model by dividing the discovery and validation sets into sextiles of the composite score. Fig. 10D shows the predicted and observed number of kidney graft recipients with IF/TA, separately for each set, for each sextile. Based on the Hosmer-Lemeshow test, the fit between the observed and the predicted number of subjects with IF/TA in each of the sextiles was excellent (P=0.69) in the discovery set (left half of Fig. 10D). For the Validation set (right half of Fig. 10D), the P-value was 0.04, suggesting a good fit, given that this set was not involved in the estimation of the model.
IF/TA grades and urinary cell mRNA levels
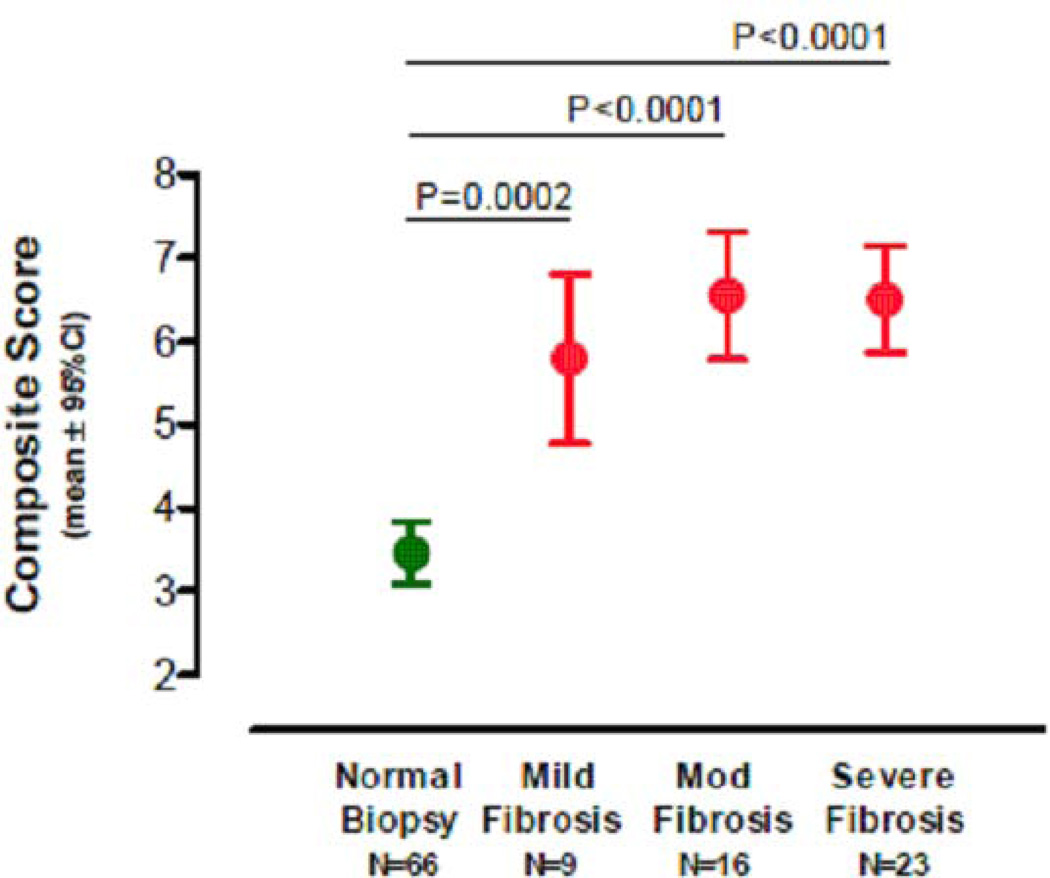
Based on the extent of kidney cortical area affected by fibrosis, IF/TA is graded as grade I (<25% of cortical area), grade II (26–50%), or grade III (>50%) in the Banff schema. We investigated whether the 4-gene composite score discriminates patients with differing grades of IF/TA from patients without IF/TA. This analysis showed that the mean composite score of the 4-gene signature was significantly different among these four groups (P<0.0001, one-way ANOVA) (Fig. 11). Pairwise comparisons revealed that the mean composite score of normal biopsies were significantly different from that of grade I fibrosis (P=0.0002), grade II fibrosis (P<0.0001), and grade III fibrosis (P<0.0001). The mean composite score, however, did not differ significantly among the three grades of fibrosis (P=0.58). A key finding from our study is that the differential expression of urinary cell mRNA levels was evident even in patients with grade I IF/TA. This finding suggests that urinary cell mRNA monitoring may be of value in detecting IF/TA before the advent of irreversible renal injury and graft dysfunction.
Fig. 11. Mean level and 95%confidence intervals of the 4-gene composite score by Banff diagnostic category.
Mean level and 95%CI of the 4-gene composite score by fibrosis grade. Kidney allograft biopsies were classified as normal, mild fibrosis (grade I, <25% of cortical area), moderate (grade II, 26–50% of cortical area), or severe (grade III, >50% of cortical area). The mean (and 95%CI) composite scores derived from urinary cell vimentin, NKCC2 and E-cadherin mRNA levels and 18S rRNA level were significantly different across the four groups (P<0.0001, one-way ANOVA). Pairwise comparisons revealed that the mean composite score of normal biopsies was significantly different from those of mild fibrosis (P=0.0002, Tukey’s honestly significant differences criterion), moderate fibrosis (P<0.0001) and severe fibrosis (P<0.0001). Within the fibrosis group, however, the mean composite scores were not significantly different (mild vs. moderate [P=0.64], mild vs. severe [P=0.65] and moderate vs. severe [P=0.99]). Values under each biopsy diagnosis show the number of kidney graft recipients from whom urine samples were collected for the measurement of urinary cell mRNA (from Anglicheau et al. Transplantation 2012, with permission).
Features contributory to successful noninvasive diagnosis of IF/TA
A number of features of our research design may have facilitated our development of a simple yet robust noninvasive classifier of fibrosis using the quantification of only 4 genes in urine samples from kidney transplant recipients. First, we measured absolute levels of mRNA copy number, avoiding some of the ambiguities inherent to the delta-delta Ct method of quantification of mRNA copy numbers. Second, we used a discovery set to develop a prediction equation and identify the composite score cutpoint, and then used the same equation and cutpoint to validate the diagnostic accuracy of the urinary cell mRNA signature in an independent cohort of kidney allograft recipients. Third, we gave consideration to the potential for non-linear relationships of gene expression measures to renal allograft diagnosis, and our approach for the discovery phase of the analysis used LOESS methods to examine the relationship of the mRNA measures to diagnosis (IF/TA vs. Normal). Another major contributor to our successful development and validation of a diagnostic biomarker of IF/TA is our pristine control group – urine samples from patients with normal biopsy findings in their surveillance biopsies. Had we used urine samples from kidney allograft recipients with acute rejection, calcineurin toxicity, or BK virus nephropathy (BKVN) as the control group, the development of a robust biomarker that distinguishes biopsies with fibrosis from biopsies without fibrosis may have been compromised since each of these conditions may be associated with some degree of fibrosis.
Our urinary cell mRNA panel was also designed with due consideration that multiple cell types and an inflammatory milieu contribute to the development of fibrosis and that fibrosis is characterized by the accumulation of extracellular matrix proteins.
Noninvasive characterization of clinical tolerance
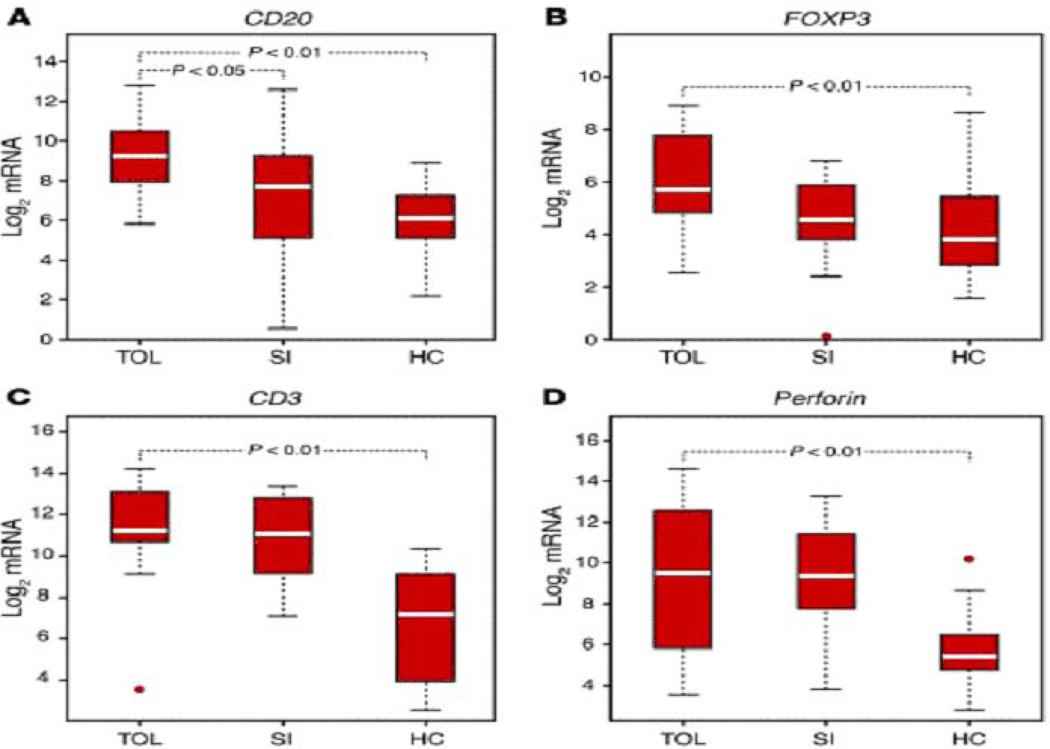
An important and yet unmet objective is to identify patients in whom immunosuppressive therapy can be minimized or completely withdrawn. In a landmark study of tolerance induction in humans, we found that intragraft levels of FoxP3 were significantly higher in the tolerant recipients compared to recipients on standard immunosuppressive therapy (30). We also found, in the largest study to date of kidney graft recipients who were not on any maintenance immunosuppressive therapy for a number of reasons, that urinary cell levels of CD20 mRNA distinguished the tolerant group from healthy volunteers and from stable patients on maintenance immunosuppressive therapy; urinary cell levels of FoxP3 mRNA were also higher in the tolerant group compared to healthy volunteers (31) (Fig. 12).
Fig. 12. Levels of mRNA in urinary cells of Tolerant, Stable, and Healthy Controls.
Total RNA was isolated from urinary cells and pre-amplification enhanced real time quantitative PCR assays were used to measure levels of mRNAs in urinary cells. Transcript levels were normalized using 18s rRNA and log transformed. Boxes depict IQR; whiskers denote 1.5 × IQR; values beyond this range are considered outliers and shown as circles. P values are shown for statistically significant differences (P<0.05) (From Newell et al Journal of Clinical Inestigation 2010, with permission).
Concluding remarks
Our urinary cell mRNA profiling studies have shown considerable robustness in diagnosing AR and IF/TA in the kidney allograft and provide mechanistic insights regarding allograft rejection and the balance between graft-destructive vs. protective forces (Fig. 13). To be clinically useful, a diagnostic test must also be reliably reproducible from center to center and the multi-center CTOT-04 trial not only validated the sensitivity and specificity of urinary cell mRNA profiling for the detection of ACR but also provided the support that this technique was feasible and collection could be performed in several transplant centers. Of note, the CTOT-04 trial utilized a single core laboratory for the processing of all urine samples for urinary cell mRNA profiling (3). A recent multicenter study by Keslar et al. (32) evaluated the reproducibility of a urinary cell mRNA protocol from center to center. In this study, six centers were sent samples, reagents, and a standardized protocol for urine gene monitor profiling. Overall, gene expression measurements had a correlation greater than 0.938 for the samples. Measurement of a known control sample among the 6 centers was able to be quantified within a factor of 1.5 (32).
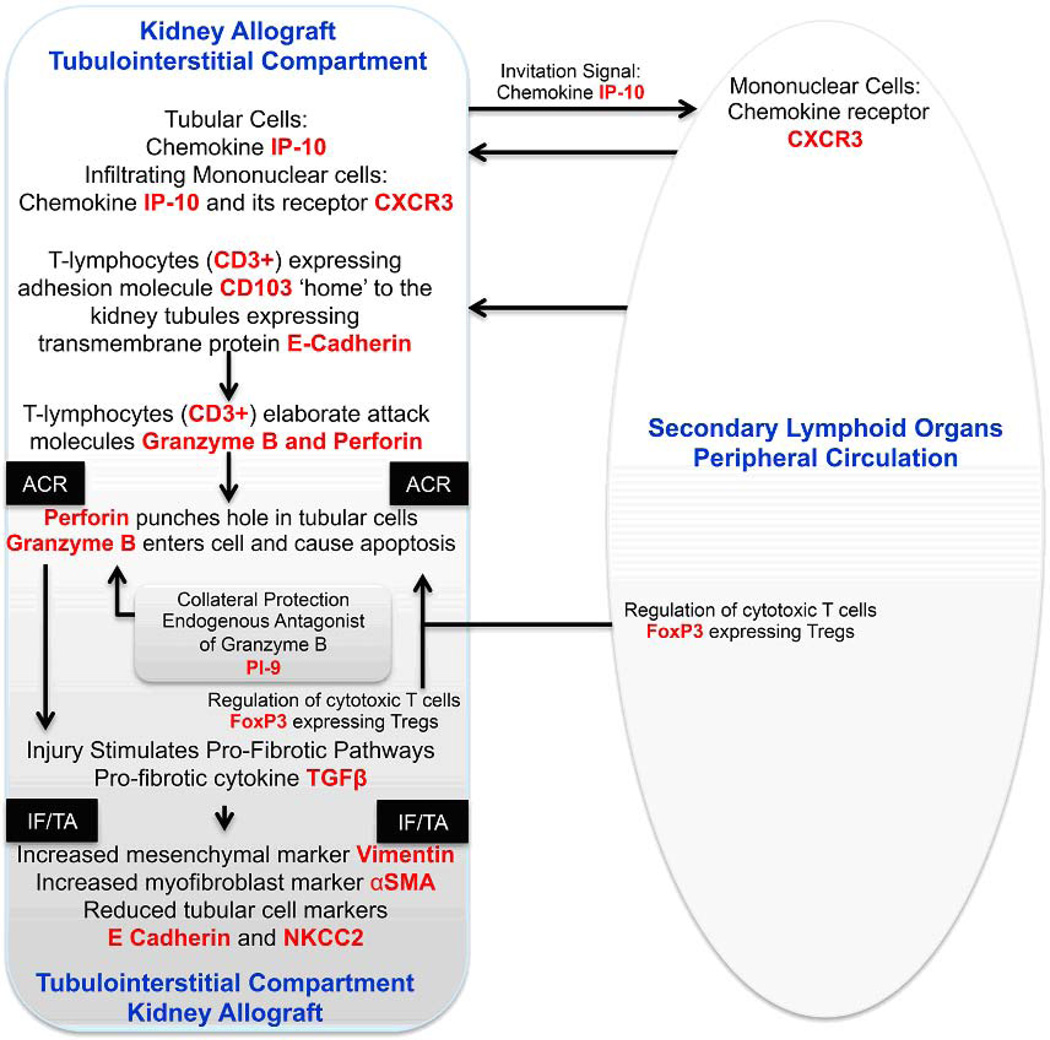
Fig. 13. Schema of acute cellular rejection in the kidney allograft.
The immune repertory contributing to acute cellular rejection involves multiple cell types, cytokines and chemokines and their ligands. T cells are the prime effector cells and dendritic cells are the primary antigen presenting cells. In this minimalist model, IP-10 and its receptor CXCR3 expressed on T cells function to facilitate the trafficking of alloreactive T cells to the kidney graft. Intraepithelial homing of the T cells is then effected by the physical interaction between CD103 expressed on the T cells and E-cadherin expressed on the tubular epithelial cells. The activated T cells employ perforin and granzyme to mediate parenchymal cell damage, which is antagonized in part by the endogenous antagonist PI-9. Counter-regulatory mechanisms such as FoxP3 expressing Tregs help dampen the anti-allograft response and the ultimate outcome of an episode of acute rejection is determined in part by the balance between the graft destructive cells and graft protective cells. Although the cells with different functional attributes appear to have different pedigrees, plasticity may exist based on environmental cues. In this T cell centric formulation, the antigen-experienced T cells provide help to B cells and facilitate antibody-mediated rejection. Acute rejection is a precursor of chronic rejection manifested histologically by IF/TA. Acute rejection associated tissue injury, de-differentiation and repair (? epithelial/endothelial-mesenchymal transition) contribute to the pathogenesis of IF/TA and progressive loss of allograft function. The levels of mRNA encoding vimentin, αSMA, E-cadherin and NKCC2 in urinary cells reflect these cellular events quantitatively. The mRNAs, colored red in the schematic illustration, have all been detected and quantified using urinary cell mRNA profiling and found to be associated with human kidney allograft status.
The Keslar et al. (32) study validated the reliability of a standardized protocol for urinary cell mRNA profiling. However, it is also worth noting that urinary cell mRNA profiling strategies have also been evaluated in several single center studies besides Weill Cornell Medical College. Galante et al. (33) evaluated urine specimens from 60 kidney transplant recipients and found that levels of granzyme B, perforins, and Fas ligand were significantly higher in the acute rejecting transplant recipients than in non-rejecting transplant recipients and reported a sensitivity that ranged from 85% to 88% and a specificity that ranged from 55% to 100% for the 3 mRNAs measured. A study by van Ham et al. investigated 70 kidney transplant recipients and reported that granzyme B and perforin were significantly higher in the acute rejection than in the acute tubular necrosis or stable control patients and reported the utility of granzyme A in the diagnosis of subclinical acute rejection (34). Both of these studies utilized a different method of RNA extraction from the CTOT-04 study: a guanidinium thiocyanate-phenol-chloroform based extraction in the Galante et al. (33) study and a GenElute Mammalian Total RNA kit based extraction in the van Ham et al. (34) study.
The benefits of urinary cell mRNA profiling include its ability to generate prognostic information. A study by Ponciano et al. identified CD20 staining in renal biopsies with acute rejection as being correlated with higher serum creatinine at 2 years (35). Urinary CD20 mRNA had a high sensitivity of 83% for intragraft CD20 infiltration on renal biopsy and thus may serve as a surrogate marker for intragraft CD20 cells (35). Our single center has reported on the utility of VP1 mRNA to detect BK virus nephropathy (36). In a follow-up study, we reported that plasminogen activator inhibitor-1 (PAI-1) was an independent predictor of graft loss at the time of BK virus nephropathy diagnosis (37). Such prognostic urine biomarkers may serve as a useful tool to individualize and personalize treatment.
Future directions
While the field of urinary cell mRNA profiling has made significant strides, there are still many potential avenues to be explored. Can urinary cell mRNA profiling replace the use of renal biopsy in many instances where a for-cause biopsy is currently performed? Can we utilize a standardized urinary gene mRNA profiling protocol as a monitoring tool to prevent the development of overt acute rejection? Can we develop a urinary mRNA gene signature that will predict the response to anti-rejection treatment? These are just some of the future steps necessary for personalizing care with urinary cell mRNA profiling.
Diagnosis and prognostication of native kidney diseases represents another potential use for urinary gene expression profiling. Several studies, primarily outside the United States, have utilized this approach to evaluate lupus nephritis. Chan et al. (38) investigated 106 lupus patients and reported that the urinary cell levels of TGF-β1 mRNA and mRNA encoding monocyte chemoattractant protein 1 (MCP-1) significantly correlate with overall lupus disease activity, as measured by the SLE Disease Activity Index, and also correlate with the histological activity index score in the lupus patients that had a kidney biopsy. A follow up study revealed that urinary MCP-1 mRNA and FoxP3 mRNA increased and IL-17 mRNA and GATA-3 mRNA decreased preceding development of a lupus flare (39).
Based on the data generated to date, we believe that urinary cell mRNA profiles have the potential to accurately reflect in-vivo immune status of the kidney graft recipient and facilitate the move from a ‘one-size fits all’ immunosuppressive regimen to transplantation precision medicine. Recent technical advances such as deep sequencing should help identify additional candidates informative of diagnosis and prognosis and our laboratory is beginning to characterize the mRNA transcriptome as well as the microRNA transcriptome at an unprecedented level of precision and incorporate the new knowledge in our urinary cell profiling protocols.
Acknowledgments
Supported in part by awards (UO1A163589, PI. A. Shaked, R37-AI051652, PI. M. Suthanthiran) from the National Institutes of Health and an award (NPRP 08-503-3-11) from the Qatar National Research Foundation to M. Suthanthiran; by an award (K08-DK087824) to T. Muthukumar from the National Institutes of Health; by an award (KL2-TR000458) to J. Lee from the National Center for Advancing Translational Sciences at the National Institutes of Health through the Weill Cornell Clinical and Translational Science Center, and by a Clinical and Translational Science Center award (ULI RR 024996) to Weill Cornell Medical College from the National Institutes of Health.
Footnotes
The authors have no conflicts of interest to declare.
References
- 1.Matas AJ, et al. OPTN/SRTR 2011 Annual Data Report: kidney. Am J Transplant. 2013;13:11–46. doi: 10.1111/ajt.12019. [DOI] [PubMed] [Google Scholar]
- 2.El Ters M, et al. Kidney allograft survival after acute rejection, the value of follow-up biopsies. Am J Transplant. 2013;13:2334–2341. doi: 10.1111/ajt.12370. [DOI] [PubMed] [Google Scholar]
- 3.Suthanthiran M, et al. Urinary-cell mRNA profile and acute cellular rejection in kidney allografts. N Engl J Med. 2013;369:20–31. doi: 10.1056/NEJMoa1215555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nankivell BJ, Alexander SI. Rejection of the kidney allograft. N Engl J Med. 2010;363:1451–1462. doi: 10.1056/NEJMra0902927. [DOI] [PubMed] [Google Scholar]
- 5.Hricik DE, et al. Multicenter validation of urinary CXCL9 as a risk-stratifying biomarker for kidney transplant injury. Am J Transplant. 2013;13:2634–2644. doi: 10.1111/ajt.12426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hariharan S, Johnson CP, Bresnahan BA, Taranto SE, McIntosh MJ, Stablein D. Improved graft survival after renal transplantation in the United States, 1988 to 1996. N Engl J Med. 2000;342:605–612. doi: 10.1056/NEJM200003023420901. [DOI] [PubMed] [Google Scholar]
- 7.Lentine KL, Gheorghian A, Axelrod D, Kalsekar A, L'Italien G, Schnitzler MA. The implications of acute rejection for allograft survival in contemporary U.S kidney transplantation. Transplantation. 2012;94:369–376. doi: 10.1097/TP.0b013e318259407f. [DOI] [PubMed] [Google Scholar]
- 8.Park WD, Griffin MD, Cornell LD, Cosio FG, Stegall MD. Fibrosis with inflammation at one year predicts transplant functional decline. J Am Soc Nephrol. 2010;21:1987–1997. doi: 10.1681/ASN.2010010049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Israni AK, et al. Inflammation in the setting of chronic allograft dysfunction post-kidney transplant: phenotype and genotype. Clin Transplant. 2013;27:348–358. doi: 10.1111/ctr.12074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Kurtkoti J, et al. The utility of 1- and 3-month protocol biopsies on renal allograft function: a randomized controlled study. Am J Transplant. 2008;8:317–323. doi: 10.1111/j.1600-6143.2007.02049.x. [DOI] [PubMed] [Google Scholar]
- 11.Rush D, et al. Beneficial effects of treatment of early subclinical rejection: a randomized study. J Am Soc Nephrol. 1998;9:2129–2134. doi: 10.1681/ASN.V9112129. [DOI] [PubMed] [Google Scholar]
- 12.Wilczek HE. Percutaneous needle biopsy of the renal allograft. A clinical safety evaluation of 1129 biopsies. Transplantation. 1990;50:790–797. doi: 10.1097/00007890-199011000-00010. [DOI] [PubMed] [Google Scholar]
- 13.Furness PN, et al. Protocol biopsy of the stable renal transplant: a multicenter study of methods and complication rates. Transplantation. 2003;76:969–973. doi: 10.1097/01.TP.0000082542.99416.11. [DOI] [PubMed] [Google Scholar]
- 14.Schwarz A, Gwinner W, Hiss M, Radermacher J, Mengel M, Haller H. Safety and adequacy of renal transplant protocol biopsies. Am J Transplant. 2005;5:1992–1996. doi: 10.1111/j.1600-6143.2005.00988.x. [DOI] [PubMed] [Google Scholar]
- 15.Gloor JM, et al. Subclinical rejection in tacrolimus-treated renal transplant recipients. Transplantation. 2002;73:1965–1968. doi: 10.1097/00007890-200206270-00023. [DOI] [PubMed] [Google Scholar]
- 16.U.S. Renal Data System. USDS 2007 Annual Data Report: Atlas of End-Stage Renal Disease in the United States. Bethesda: National Institutes of Health, National Institutes of Diabetes and Digestive and Kidney Diseases; 2007. [Google Scholar]
- 17.U.S. Renal Data System. USDS 2013 Annual Data Report: Atlas of Chronic Kidney Disease and End-Stage Renal Disease in the United States. Bethesda: National Institutes of Health, National Institutes of Diabetes and Digestive and Kidney Diseases; 2013. [Google Scholar]
- 18.Muthukumar T, et al. Messenger RNA for FOXP3 in the urine of renal-allograft recipients. N Engl J Med. 2005;353:2342–2351. doi: 10.1056/NEJMoa051907. [DOI] [PubMed] [Google Scholar]
- 19.Ding R, et al. Noninvasive diagnosis of BK virus nephritis by measurement of messenger RNA for BK virus VP1 in urine. Transplantation. 2002;74:987–994. doi: 10.1097/00007890-200210150-00016. [DOI] [PubMed] [Google Scholar]
- 20.Li B, et al. Noninvasive diagnosis of renal-allograft rejection by measurement of messenger RNA for perforin and granzyme B in urine. N Engl J Med. 2001;344:947–954. doi: 10.1056/NEJM200103293441301. [DOI] [PubMed] [Google Scholar]
- 21.Muthukumar T, et al. Serine proteinase inhibitor-9, an endogenous blocker of granzyme B/perforin lytic pathway, is hyperexpressed during acute rejection of renal allografts. Transplantation. 2003;75:1565–1570. doi: 10.1097/01.TP.0000058230.91518.2F. [DOI] [PubMed] [Google Scholar]
- 22.Ding R, et al. CD103 mRNA levels in urinary cells predict acute rejection of renal allografts. Transplantation. 2003;75:1307–1312. doi: 10.1097/01.TP.0000064210.92444.B5. [DOI] [PubMed] [Google Scholar]
- 23.Hancock WW, Gao W, Csizmadia V, Faia KL, Shemmeri N, Luster AD. Donor-derived IP-10 initiates development of acute allograft rejection. J Exp Med. 2001;193:975–980. doi: 10.1084/jem.193.8.975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Hancock WW, et al. Requirement of the chemokine receptor CXCR3 for acute allograft rejection. J Exp Med. 2000;192:1515–1520. doi: 10.1084/jem.192.10.1515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Tatapudi RR, et al. Noninvasive detection of renal allograft inflammation by measurements of mRNA for IP-10 and CXCR3 in urine. Kidney Int. 2004;65:2390–2397. doi: 10.1111/j.1523-1755.2004.00663.x. [DOI] [PubMed] [Google Scholar]
- 26.Le CT. A solution for the most basic optimization problem associated with an ROC curve. Stat Methods Med Res. 2006;15:571–584. doi: 10.1177/0962280206070637. [DOI] [PubMed] [Google Scholar]
- 27.Tripepi G, Jager KJ, Dekker FW, Zoccali C. Statistical methods for the assessment of prognostic biomarkers(part II): calibration and re-classification. Nephrol Dial Transplant. 2010;25:1402–1405. doi: 10.1093/ndt/gfq046. [DOI] [PubMed] [Google Scholar]
- 28.Anglicheau D, et al. Discovery and validation of a molecular signature for the noninvasive diagnosis of human renal allograft fibrosis. Transplantation. 2012;93:1136–1146. doi: 10.1097/TP.0b013e31824ef181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holm S. A Simple Sequentially Rejective Multiple Test Procedure. Scand J Statist. 1979;6:65–70. [Google Scholar]
- 30.Kawai T, et al. HLA-mismatched renal transplantation without maintenance immunosuppression. N Engl J Med. 2008;358:353–361. doi: 10.1056/NEJMoa071074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Newell KA, et al. Identification of a B cell signature associated with renal transplant tolerance in humans. J Clin Invest. 2010;120:1836–1847. doi: 10.1172/JCI39933. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Keslar KS, et al. Multicenter evaluation of a standardized protocol for noninvasive gene expression profiling. Am J Transplant. 2013;13:1891–1897. doi: 10.1111/ajt.12284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Galante NZ, Camara NO, Kallas EG, Salomao R, Pacheco-Silva A, Medina-Pestana JO. Noninvasive immune monitoring assessed by flow cytometry and real time RT-PCR in urine of renal transplantation recipients. Transpl Immunol. 2006;16:73–80. doi: 10.1016/j.trim.2006.03.014. [DOI] [PubMed] [Google Scholar]
- 34.van Ham SM, et al. Urinary granzyme A mRNA is a biomarker to diagnose subclinical and acute cellular rejection in kidney transplant recipients. Kidney Int. 2010;78:1033–1040. doi: 10.1038/ki.2010.274. [DOI] [PubMed] [Google Scholar]
- 35.Ponciano VC, et al. Urinary CD20 mRNA as a surrogate of CD20-positive cells infiltration during allograft dysfunction in renal transplant patients. Int Immunopharmacol. 2009;9:663–667. doi: 10.1016/j.intimp.2008.11.017. [DOI] [PubMed] [Google Scholar]
- 36.Dadhania D, et al. Validation of noninvasive diagnosis of BK virus nephropathy and identification of prognostic biomarkers. Transplantation. 2010;90:189–197. doi: 10.1097/TP.0b013e3181e2a932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Dadhania D, et al. Noninvasive prognostication of polyomavirus BK virus-associated nephropathy. Transplantation. 2013;96:131–138. doi: 10.1097/TP.0b013e31829acb38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chan RW, et al. Expression of chemokine and fibrosing factor messenger RNA in the urinary sediment of patients with lupus nephritis. Arthritis Rheum. 2004;50:2882–2890. doi: 10.1002/art.20471. [DOI] [PubMed] [Google Scholar]
- 39.Szeto CC, et al. Monitoring of urinary messenger RNA levels for the prediction of flare in systemic lupus erythematosus. Clin Chim Acta. 2012;413:448–455. doi: 10.1016/j.cca.2011.10.030. [DOI] [PubMed] [Google Scholar]