Abstract
The biological relationships among self-renewal, tumorigenicity, and lineage differentiation of human osteosarcoma-initiating cells (OSIC) remain elusive, making it difficult to identify and distinguish OSIC from osteosarcoma-forming cells (OSFC) for developing OSIC-targeted therapies. Using a new inverse lineage tracking strategy coupled with serial human-to-mouse xenotransplantation, we identified a subpopulation of osteosarcoma cells with OSIC-like properties and sought to distinguish them from their progeny, OSFC. We found that serial transplantation of cells from different osteosarcoma cell lines and primary osteosarcoma tissues progressively increased the CD49f+ subpopulation composing the bulk of the osteosarcoma mass. These CD49f+ cells displayed characteristics of OSFC: limited in vivo tumorigenicity, weak lineage differentiation, more differentiated osteogenic feature, and greater chemo-sensitivity. By contrast, their parental CD49f−CD133+ cells had an inhibited osteogenic fate, together with OSIC-like properties of self-renewal, strong tumorigenicity, and differentiation to CD49f+ progeny. Hence, the CD49f−CD133+ phenotype appears to identify OSIC-like cells that possess strong tumorigenicity correlated with an impaired osteogenic fate and the ability to initiate tumor growth through generation of CD49f+ progeny. These findings advance our understanding of OSIC-like properties and, for the first time, provide a much-needed distinction between OSIC and OSFC in this cancer.
Keywords: Osteosarcoma-initiating cells, self-renewal, tumorigenicity, lineage differentiation, osteosarcoma-forming cells, osteogenic differentiation
Introduction
Osteosarcoma, generally considered a disease of cell differentiation (1, 2), is the most common malignant bone tumor (1, 3). Recent findings suggest that non-targeted, cytotoxic chemotherapy can effectively kill the vast majority of osteosarcoma cells; but a small fraction of transformed cells, OSIC, survive and thus instigate osteosarcoma re-initiation, drug resistance, and clinical relapse (2). Hence, identifying OSIC would be crucial steps toward devising OSIC-targeted therapies, thereby avoiding treatment resistance and the severe toxicity associated with current, non-selective forms of osteosarcoma chemotherapy. Investigators focusing on the identification of OSIC have reported that some subpopulations of osteosarcoma cells express prospective cancer stem cell markers, including CD117+, Stro-1+ (4), or CD133+ (5-7). Moreover, by activating an exogenous Oct-4 promoter in primary osteosarcoma cells, Oct-4/GFP+ cells display properties of cancer-initiating cells (8); whereas an osteosarcoma subpopulation with high ALDH activity also exhibits OSIC features (9). To date, the relationships among OSIC self-renewal, tumorigenicity, and lineage differentiation remain unclear, making it difficult to identify and distinguish OSIC from their OSFC for developing OSIC-targeted therapies.
The cancer stem cell hypothesis predicts that only a small fraction of transformed cells are capable of reconstituting all of the diverse cell types within a particular tumor (10, 11), as demonstrated in hematologic malignancies (12-14), central nervous system tumors (15), breast tumors (16), and colon cancer (17). An increasing number of studies show that some tumor cell subpopulations isolated by prospective stem cell markers possess the properties of cancer stem cells; however, these results are often confounded by the ability of marker-negative counterpart subpopulations to induce tumors. This often led to common controversy (11, 18-20) that is currently observed in identifying cancer stem cells (15, 21-23). For example, although the role of CD133 as a marker of cancer stem cells (15, 21) is well documented (11, 19, 20), some studies demonstrate that both CD133+ and CD133– cells can initiate tumor formation (22, 23). Other studies suggest that a combination of CD133+ with other markers or ALDH activity is needed in identifying cancer stem cells (23-26). These controversies reflect the complexity of cancer stem cells. Here, we used an inverse lineage tracking strategy coupled with serial transplantation to identify OSIC properties. Our studies show that the gain of strong tumorigenicity seen with OSIC-like CD49f−CD133+ cells correlates to diminished osteogenic fate, which distinguish them from their CD49f+ progeny that possess limited tumorigenicity in association with more differentiated osteogenic features.
Results
Serial xenotransplantation enriches self-renewal and tumorigenicity of osteosarcoma cells
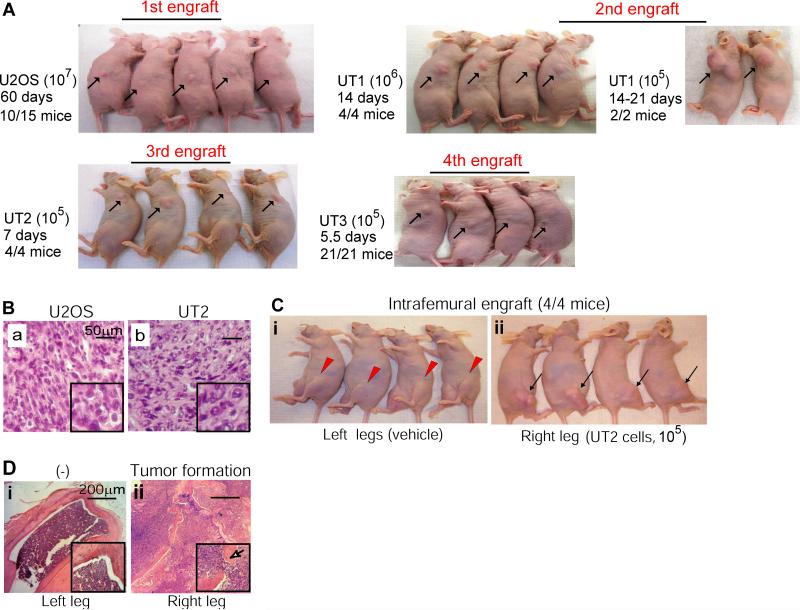
To identify osteosarcoma cells with OSIC properties, we used serial xenotransplantation as a means to enhance OSIC self-renewal and promote lineage differentiation. We first screened tumorigenicity of four human osteosarcoma cell lines and six primary osteosarcoma samples by engrafting them into nude mice with subcutaneous injection. We found that KHOS/NP cells formed tumors at 2 weeks post-injection, U2OS cells at 2 months, and TTC444 cells at 2-3 months (Supplemental Tables 1, 2). Since KHOS/NP cells are virus-transformed cells, whereas well-established U2OS cells afford the best window of time for amplifying the OSIC-like property by serial transplantation, we used these cells to derive different generations of tumor xenografts for in vivo analysis of self-renewal and tumorigenicity. The results showed that the self-renewal and tumorigenicity of U2OS cells from primary tumor xenograft to sequential progeny xenografts, designated UT1, UT2, and UT3 cells (Figure 1A), were progressively enhanced. Indeed, a reduction in cell number for transplantation (from 1 × 107 to 1 × 105 cells) was associated with a reduced time to tumor formation (from 60 days to 7 days). HE staining confirmed that the tumor mass derived from UT2 engraftment retained the same properties as the parental U2OS osteosarcoma xenograft (Figure 1B). These results show that OSIC activity is progressively enhanced by serial xenotransplantation, leading to an enhanced production of OSFC that forming the osteosarcoma mass.
Figure 1. Serial xenotransplantation enriches self-renewal and tumorigenicity of osteosarcoma cells.
(A) Self-renewal and tumorigenicity were assessed by the amount of cells injected and the time of tumor formation. Cells from the first U2OS xenograft were re-transplanted to yield the 1st, 2nd and 3rd generations of U2OS cells (designated as UT1, UT2, and UT3). (B) HE staining determined that UT2 cells retained histological features of U2OS cells, as reflected by cellular polymorphism, nuclear hyperchromasia, and mitotic activities, together with osteoid formation that recapitulated the hallmark histological phenotype of the parental osteosarcoma. (C) UT2 cells remained capable of tumorigenicity in the bone. Tumor formation occurred in the right legs of mice with injection of UT2 cells (section ii). (D) HE staining of osteosarcoma xenograft formed in the femurs of the right legs (panel C), focally breaching the cortex, and extending into soft tissue (arrow and inset).
We next examined, using orthotopic transplantation, whether the phenotype of osteosarcoma cells is maintained after serial engraftments. Because UT2 cells offered a better experimental window-time and have a similar tumorigenicity compared to UT3 cells, we intrafemorally injected 2 × 105 UT2 cells into mice. Consistent with enhanced self-renewal and tumorigenicity ascribed to UT2 cells (Figure 1A, third engraft), tumor formation in bone was taken 2 weeks (Figure 1C, panel ii). HE staining showed tumor growth in the distal femurs (Figure 1D). These results demonstrate that the enhanced OSIC activity remains capable of producing OSFC progeny able to form phenotypically recognizable osteosarcoma in bone.
Enriched OSIC activities correlate with accelerated sphere formation and expression of stem/progenitor cell-associated genes
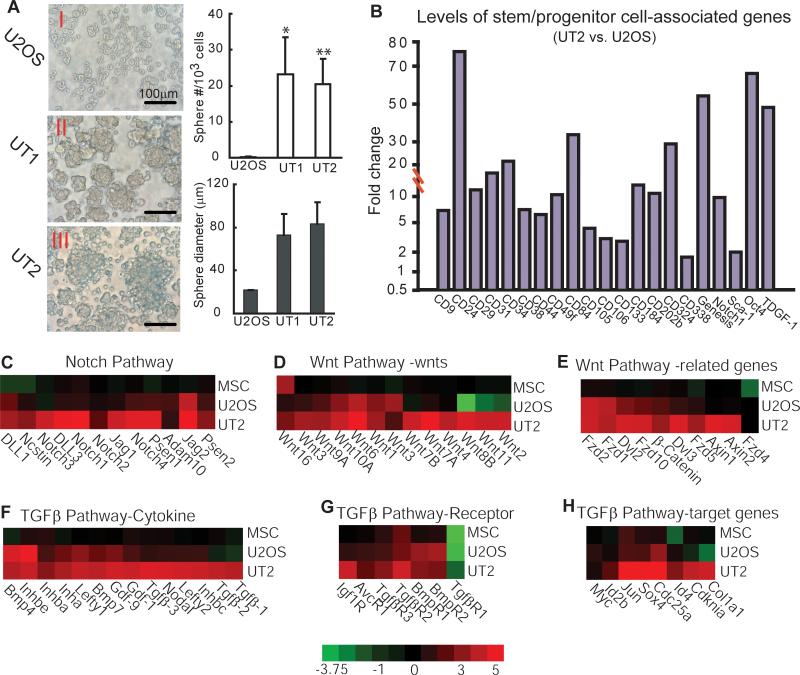
Cancer stem cells often possess the ability to produce unattached spheres in the absence of serum (27, 28). To test the enriched OSIC activities for this property, we cultured cells isolated from different xenograft tissues with basal medium containing methylcellulose. We found that after 12 days of culture, both the size and number of the floating spheres were progressively increased, from U2OS to UT1 to UT2 cells. More than 20 spheres with a 70 to 80-μm diameter were observed among each 103 of the initially seeded UT1 or UT2 cells, in contrast to spheres of <22-μm diameter formed by U2OS cells (Figure 2A). This greater stem-like sphere formation suggests that OSIC activities are enriched in UT2 cells.
Figure 2. Enriched OSIC activities correlate with accelerated sphere formation and expression of stem/progenitor cell-associated genes.
(A) Microscopy analysis of spheres cultivated in suspension for 12 days. Representative phase-contrast micrographs of spheres were shown with 400x magnification. UT1 or UT2 vs. U2OS: *P<0.05; **P<0.01. (B) Up-regulated expressions of stem cell/progenitor marker genes in UT2 cells compared to parental U2OS cells. (C-H) Serial transplantation up-regulated expressions of signaling molecules in the Notch, Wnt, and TGFβ pathways. The color bar represents the expression ratio on a log-2 scale (UT2 vs. U2OS or MSC).
Because analyses of global gene expression pattern between normal and malignant stem cell populations can provide critical insights into cancer stem cell properties (29), we determined whether enhanced self-renewal and tumorigenicity in UT2 cells correlate with increased expression of stem/progenitor cell-associated genes, using microarray analysis as described (30). Genes whose expression differed from ≥ 2 to 1,000-fold in UT2 vs. U2OS cells were identified, with more than 3,000 genes classified as up-regulated in UT2 cells (Supplemental Table 3). Among them, 21 stem or progenitor cell-associated genes were up-regulated (Figure 2B). Moreover, molecules involved in regulation of self-renewal signaling pathways in cancer stem cells (11, 18) were up-regulated in UT2 cells compared to U2OS and normal human mesenchymal stem cells (MSC), including those in the Notch, Wnt, and TGFβ pathways (Figure 2C-H). These data indicate a genetic basis for the enhanced self-renewal and tumorigenicity in UT2 cells.
Enhanced OSIC self-renewal and tumorigenicity induce a progressive expansion of CD49f+ cells forming the osteosarcoma mass
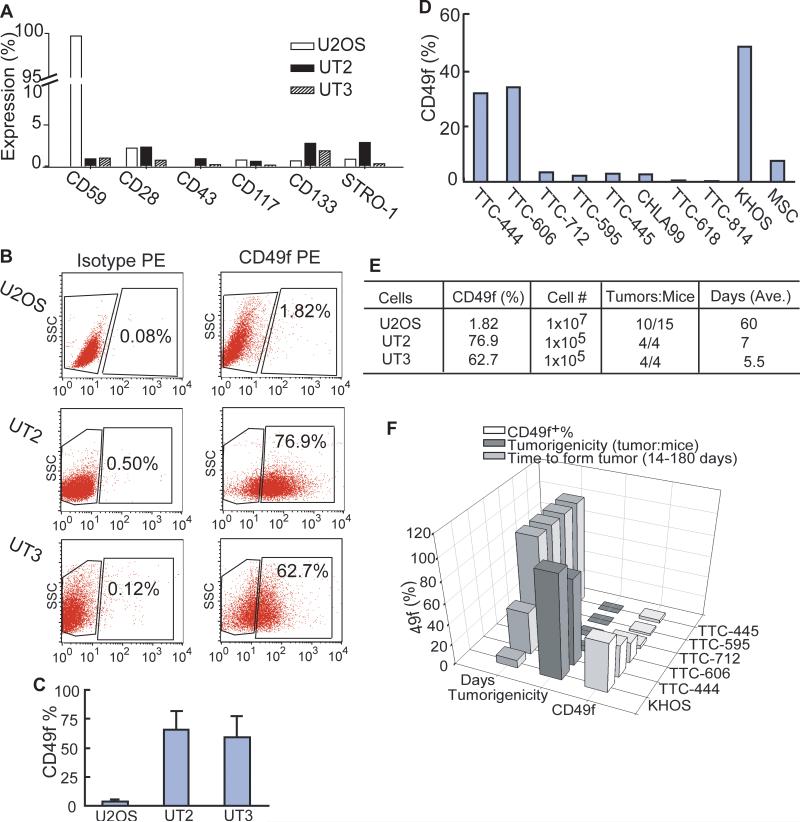
Given that cancer stem cells arise from normal stem cells (10, 11), we predict that OSIC retain some normal human MSC properties and intrinsic mesenchymal origin, expressing surface antigens commonly associated with MSC. Thus, enhanced OSIC self-renewal by serial transplantation should increase OSIC propensity to differentiate to OSFC progeny that express stem or progenitor markers and form the osteosarcoma mass. To test this idea, we analyzed the change in expression of different potential phenotypic markers of mesenchymal origin in U2OS, UT2, and UT3 cells by FACS analysis, using anti-human CD28, CD43, CD49f, CD59, CD117, CD133 and STRO-1 antibodies. We found that, while many of these markers showed no or only minor up-regulation in those cells (Figure 3A), the size of the CD49f+ subpopulation was significantly increased from U2OS to UT2 to UT3 cells (Figure 3B, 3C). Because this increase was associated with enhanced formation of the osteosarcoma mass from U2OS to UT2 to UT3 cells (Figure 1), we further investigated the expression levels of CD49f in primary osteosarcoma cells (TTC444, TTC606, TTC712, TTC595, TTC445, TTC618, TTC814, and CHLA99), MSC, and virus-transformed osteosarcoma KHOS/NP cells. Interestingly, TTC444, KHOS/NP, UT2, and UT3 cells, which show high levels of CD49f expression (Figure 3B-D; Supplemental Figure 1), formed osteosarcoma xenografts within 6-90 days (Figure 1; Supplemental Tables 1, 2). Moreover, higher levels of CD49f+ cells were inversely related to the time required to form a tumor (Figure 3E, 3F). Since CD49f is a marker of human MSC progenitors (31, 32), our studies suggest that it identifies OSFC with the capacity to proliferate actively to constitute the bulk of the osteosarcoma mass.
Figure 3. Enhanced OSIC self-renewal and tumorigenicity induce a progressive expansion of CD49f+ cells forming the osteosarcoma mass.
(A-C) Expression levels of CD59, CD28, CD43, CD117, CD133 and STRO-1 (panel A) as well as CD49f (panels B, C) were determined by FACS in U2OS, UT2 and UT3 cells. (D) FACS analysis of CD49f expression in normal (MSC), KHOS/NP, and 8 different primary osteosarcoma cells. (E & F) Inverse relationship between CD49f levels vs. time for tumor formation in serially transplanted (panel E) and the first transplanted cells (panel F).
CD49f− cells with greater tumorigenicity and drug resistance generate CD49f+ subpopulation
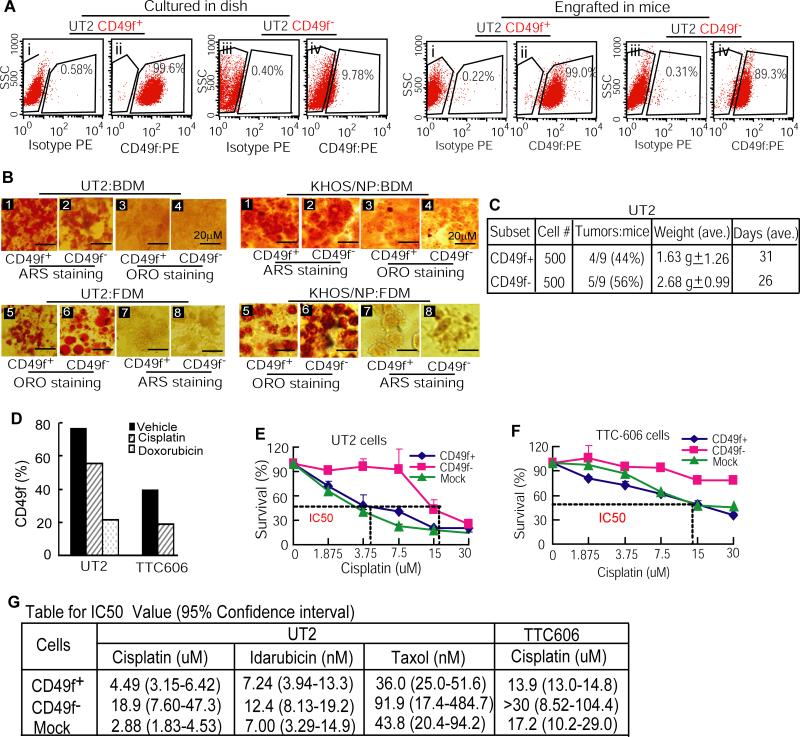
Identification of CD49f+ cells (Figure 3) forming the osteosarcoma mass raises a question: What subpopulation of U2OS cells possesses OSIC-like properties to give rise to CD49f− progeny? Because CD49f+ cells account for about 70% of the UT2 cells, we first tested in vitro whether these cells were derived from the CD49f− fraction. We cultured CD49f+ and CD49f− subpopulations of UT2 cells for 2 weeks, and then determined the changes in genetic lineage. FACS analyses showed that the CD49f− subpopulation generated about 10% of the CD49f+ cells, whereas CD49f+ cells (99.6%) remained no change (Figure 4A, left panel). We then tested the ability of CD49f− cells to generate CD49f+ subpopulation in vivo, by subcutaneously transplanting purified CD49f− and CD49f+ subpopulations into NOD/SCID mice. FACS analysis of tumor xenografts showed that the CD49f+ marker was retained on 99.0% of the xenograft cells transplanted with CD49f+ subset, whereas in xenografts generated by the CD49f− subpopulation, more than 89% of the cells became CD49f+ positive (Figure 4A, right panel). These data demonstrate that CD49f− cells can differentiate to CD49f+ progeny and that certain factors from in vivo vicinity promote tumorigenicity (33) of CD49f− cells. Together, the above results suggest that the enriched OSIC-like self-renewal and tumorigenicity in CD49f− fraction gives rise to a large number of lineage-committed CD49f+ cells (Figures 3, 4A) that ultimately form the osteosarcoma mass.
Figure 4. CD49f− cells with greater tumorigenicity and drug resistance generate CD49f+ subpopulation.
(A) CD49f− cells differentiated to CD49f+ subpopulation in culture (9.78%; left panel) and in NOD/SCID mice (>89%, right panel). (B) Both CD49f− and CD49f+ cells isolated from UT2 (left panel) or KHOS/NP cells (right panel) showed osteogenic and adipogenic potential, as judged by ARS and ORO staining. Cross staining was used as negative control (sections 1, 2 vs. 3, 4; sections 5, 6 vs. 7, 8). (C) CD49f− cells showed greater tumorigenicity than CD49f+ cells. (D) The CD49f+ subpopulation was decreased in UT2 and primary osteosarcoma TTC606 cells treated with cisplatin or doxorubicin. (E & F) CD49f− subpopulation isolated from UT2 (panel E) or TTC606 cells (panel F) resisted cisplatin treatment. Percentages of surviving cells were shown relative to the IC50 value for each of cell populations. (G) IC50 values for CD49f+ or CD49f− cells treated with different drugs for 48 hr. Data are means with 95% confidence intervals.
It has been proposed that daughter cells generated from cancer stem cells can differentiate to the various cell types seen in the bulk tumor (11, 34, 35). We therefore examined osteogenic and adipogenic potential of both parent CD49f− and daughter CD49f+ cells. Purified CD49f− or CD49f+ cells, grown to 70~80% confluence in regular medium, were further cultured with BDM or FDM for 21 days for inducing osteogenic and adipogenic differentiation. We found that by using ARS and ORO staining, both CD49f− and CD49f+ cells displayed osteoblast and adipocyte fates, whether they were isolated from UT2 or KHOS/NP cells (Figure 4B). These data suggest that CD49f+ progeny arising from the CD49f− fraction retain essentially the same differentiation potential as their parental cells.
Because CD49f− cells differentiate to CD49f+ cells (Figure 4A), we sought to test the tumorigenicity of both subpopulations in NOD/SCID mice. With subcutaneous injections of 10,000 down to 1,000 cells (3 mice per group), we observed that CD49f− cells showed higher tumorigenicity (33%) than did CD49f+ cells (17%) for up to 32 days (data not shown). Further tests with subcutaneous injection of 500 cells showed that CD49f− cells had consistently higher tumorigenic potential (Figure 4C). Since the increase in proportion of tumor formation was not striking (56% vs. 44%), there may be other subpopulations within the CD49f− fraction that possess property of OSIC-like tumorigenicity.
Cancer stem cells show greater resistance to drugs and toxins than their more differentiated progeny. We thus assessed the response of both CD49f− and CD49f+ cells to cisplatin or doxorubicin, drugs currently used to treat osteosarcoma (36). The results indicated that the CD49f+ subpopulation was drug sensitive compared to controls (Figure 4D; Supplemental Figure 2). We further determined the inhibition concentration of drug for 50% of cells (IC50) in CD49f−, CD49f+, and non-sorted (mock) cells using the XTT proliferation assay. The results showed an IC50 value of 18.9 μM for cisplatin in UT2-CD49f− cells, compared to 4.49 μM and 2.88 μM in UT2-CD49f+ and mock cells (Figure 4E). Similarly, IC50 value of >30 μM was observed in TTC606-CD49f− cells, compared to 13.9 μM and 17.2 μM in TTC606-CD49f+ and mock cells (Figure 4F). The CD49f− cells also showed greater resistance to idarubicin and taxol than did CD49f+ cells, with IC50 values that were as much as two times higher than those in CD49f+ cells (Figure 4G). Because the CD49f− subpopulation gives rise to CD49f+ cells, possesses greater tumorigenicity, and has lower sensitivity to drug treatment than its CD49f+ daughter cells (Figure 4A, 4C-G), we conclude that CD49f− subpopulation exhibits OSIC-like properties: self-renewal capacity, strong tumorigenicity, differentiation to tumor-forming CD49f+ progeny, and increased drug resistance.
CD49f− cells generate the CD133+ subpopulation
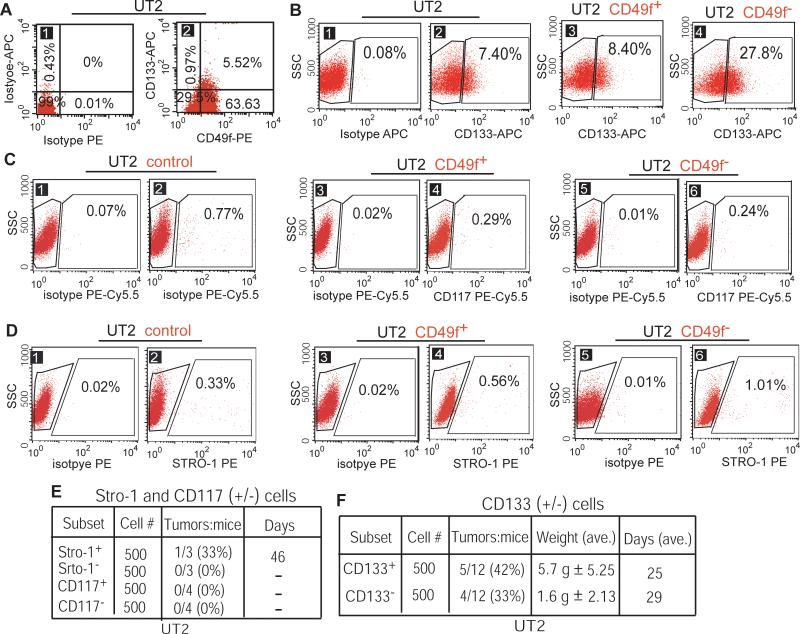
Because the intact CD49f− population does not show prominent tumorigenicity, the OSIC-like properties likely reside in one or more fractions of these cells. To address this issue, we analyzed a group of markers that have been implicated in cancer stem-like cell behavior. CD133 (alias prominin-1) is a transmembrane protein, and has been classified as a potential marker of osteosarcoma cells with stem-like cell properties (5-7) and of neural and colon cancer cells (15, 17, 37). Moreover, CD117+ and Stro-1+ subpopulations of osteosarcoma cells possess the OSIC-like properties of tumorigenicity and drug resistance (4). We thus tested whether CD133, CD117, and/or Stro-1 are potential markers of OSIC-like cells within the CD49f− fraction. FACS analysis confirmed that among UT2 cells, about two thirds (63.63% + 5.52%) are CD49f+ and 30% CD49f−; whereas 0.97% CD133+ cells were in the CD49f− subpopulation compared to 5.52% CD133+ cells in the CD49f+ subpopulation (Figure 5A). These results indicate that the majority of cells in the CD49f− subpopulation lack the CD133+ marker, and that the CD49f−CD133+ subpopulation accounts for only about 1% of UT2 cells. Interestingly, after culturing of CD49f− and CD49f+ subpopulations for 2 weeks, the proportions of CD133+ cells increased to 27.8% among CD49f− cells and to 8.4% among CD49f+ cells (Figure 5B, sections 2 vs. 3 and 4). Because CD49f− cells differentiate to both CD49f+ and CD133+ cells, we suggest that the increased smaller proportion of CD133+ cells in the CD49f+ subpopulation (Figure 5B, sections 2 vs. 3) originates from CD49f− cells. By contrast, the CD117+ subpopulation remained essentially unchanged in both the CD49f+ and CD49f− fractions of UT2 cells after 14 days of culture (Figure 5C), whereas Stro-1+ cells showed a slight increase in the CD49f− fraction compared with the CD49f+ fraction (Figure 5D). This implies that CD133+ cells within the CD49f− fraction are more promising candidate population with OSIC-like properties than CD117+ or Stro-1+ cells.
Figure 5. CD49f− cells generate the CD133+ subpopulation.
(A) FACS analyses of frequencies of the CD49f−CD133+, CD49f−CD133−, CD49f+CD133+, and CD49f+CD133− subpopulations in UT2 cells. (B) CD49f− cells differentiated to CD133+ subpopulation (27.8%) after 14 days of culture from basal level of 7.4% in UT2 cells, compared to CD49f+ cells with a slight increase (8.4%). (C & D) CD117+ (panel C) and Stro-1+ (panel D) subpopulations remained essentially unchanged after culture of CD49f+ or CD49f− cells for 14 days. (E) CD117+ cells failed to initiate tumor formation and Stro-1+ cells showed low tumorigenicity. (F) CD133+ cells showed higher tumorigenicity than CD133− cells.
Further to test whether candidate partner of CD49f− cells posses OSIC properties, we examined the tumorigenicity of CD133+, CD133−, CD117+, and Stro-1+ cells by xenotransplantation of these cells into NOD/SCID mice. The results showed that CD117+ cells did not initiate tumor formation, whereas Stro-1+ cells showed low tumorigenicity (Figure 5E). However, while both CD133+ and CD133− populations initiated tumor formation, CD133+ cells showed stronger tumorigenicity than CD133− cells (Figure 5F). Given that CD49f− cells can differentiate to both CD49f+ and CD133+ cells (Figures 4A; 5B), and that both the CD49f− and CD133+ subpopulations show greater tumorigenicity than either CD49f+ or CD133− cells (Figures 4C; 5F), we suggest that the CD49f−CD133+ phenotype serves as a marker for cells with OSIC-like properties.
Strong tumorigenicity of CD49f−CD133+ cells correlates with inhibited osteogenic capacity
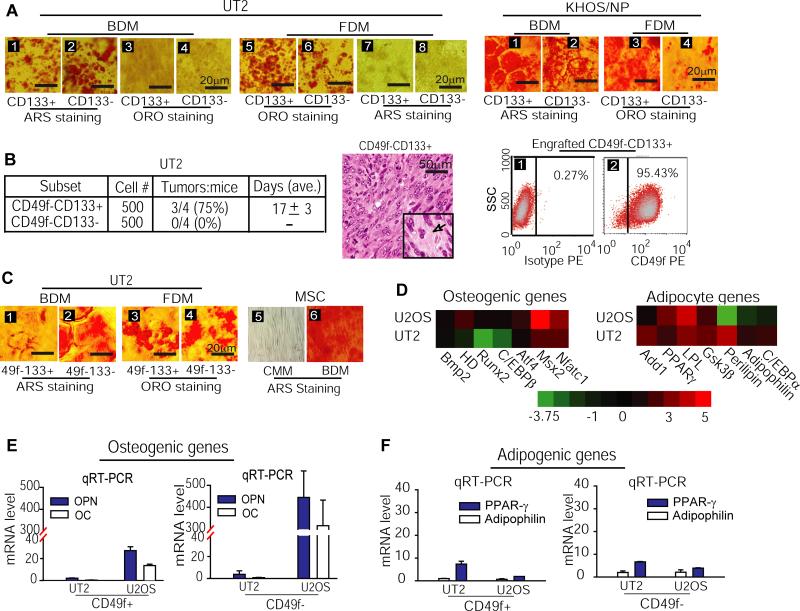
To explore OSIC-like CD49f−CD133+ properties, we first asked whether the CD133− and CD133+ cells share similar traits of multipotency. After culturing CD133+ or CD133− cells with BDM and FDM for 21 days, we performed ARS and ORO staining for analyzing osteogenic and adipogenic differentiation. We found that similar to CD49f− or CD49f+ cells, both osteogenic and adipogenic differentiation were observed in CD133+ and CD133− cells isolated from either UT2 or KHOS/NP cells (Figure 6A). Because CD49f− cells generate both CD49f+ and CD133+ subpopulations whereas both CD49f− and CD133+ cells are more tumorigenic than CD49f+ and CD133− cells, these results suggest that the ability of multipotent differentiation is preserved in both OSIC-like CD49f−CD133+ cells and their CD49f+ and CD133− progeny.
Figure 6. Strong tumorigenicity of CD49f−CD133+ cells correlates with inhibited osteogenic capacity.
(A) Both CD133+ and CD133− subpopulations of UT2 (left panel) or KHOS/PN cells (right panel) displayed osteogenic and adipogenic differentiation, as judged by ARS and ORO staining. (B) CD49f−CD133+ cells but not CD49f−CD133− cells formed tumors in NOD/SCID mice (left panel); HE staining of CD49f−CD133+ xenograft cells showed the histological features of U2OS cells with osteoid (extra-cellular matrix) formation, as reflected by spindle cells with a vesicular nucleus often containing prominent nucleoli, frequent mitotic activities, and focal areas of osteoid formation (middle panel); engrafted CD49f−CD133+ cells differentiated to CD49f+ cells in mice (right panel). (C) While both CD49f−CD133+ and CD49f−CD133− cells showed a similar capacity for fat differentiation (sections 3 and 4), CD49f−CD133+ cells showed a reduced potential for bone differentiation, compared to CD49f−CD133− cells and MSC (sections 1 vs. 2 and 6). (D) Microarray analysis revealed a decreased expression of osteogenic marker genes in UT2 cells. (E & F) qRT-PCR analysis of osteogenic (panel E) and adipogenic marker gene expressions (panel F) in CD49f− and CD49f+ subpopulations.
To test the OSIC-like properties of CD49f−CD133+ cells, we compared the tumorigenicity of CD49f−CD133+ versus CD49f−CD133− subpopulations of UT2 cells in NOD/SCID mice. The results showed that with transplantation of 500 cells, tumors were induced at day 17 in 3 of 4 mice given the CD49f−CD133+ cells, whereas tumors were not induced (0/4) in any of the mice given the CD49f−CD133− cells for up to 6 months (Figure 6B, left panel). HE staining confirmed that the tumor mass derived from CD49f−CD133+ xenografts (Figure 6B, middle panel) retained the same properties of the parental U2OS osteosarcoma (Figure 1B). Importantly, analysis of phenotypic changes in CD49f−CD133+ xenografts showed that more than 95% of the engrafted cells gained the CD49f+ marker (Figure 6B, right panel), the hallmark of cells constituting the bulk of osteosarcoma (Figures 1,3,4). Hence, CD49f−CD133+ cells appear to identify the OSIC-like properties: self-renewal, osteosarcoma initiation, and differentiation to CD49f+ progeny that form the osteosarcoma mass. Because the tumorigenicity of transplanted CD49f−CD133+ cells remained less than 100% (Figure 6B, left panel), we conclude that the OSIC-like CD49f−CD133+ subset of osteosarcoma cells likely coexists with an additional tumorigenic subpopulation(s) within the CD49f− fraction.
Since multipotency is an accepted classical feature of cancer stem cells (11), we investigated the potential for bone and fat differentiation by CD49f−CD133+ and CD49f−CD133− cells, using ARS and ORO staining analyses. Interestingly, while the two fractions had similar adipogenic capacities (Figure 6C, section 3 vs. 4), the CD49f−CD133+ cells showed reduced osteogenic differentiation compared to both CD49f−CD133− cells and MSC in the presence of BDM (Figure 6C, sections 1 vs. 2 and 6). Because this reduction of osteogenic fate in CD49f−CD133+ cells correlated with an increased rate of tumor production (Figures 6B, left panel), we investigated osteogenic and adipogenic gene expressions during the transition from U2OS through the UT2 cells, using microarray analysis. We found that in contrast to non-inhibition of adipogenic gene expression, osteogenic gene expression was essentially blocked in UT2 cells compared to U2OS cells (Figure 6D). We further analyzed changes in the expression of osteogenic and adipogenic target genes in both CD49f− and CD49f+ subpopulations during the transition from U2OS to UT2 cells. qRT-PCR analyses of osteoblastic differentiation genes, osteopontin (OPN) and osteocalcin (OC), showed that in contrast to the adipogenic genes PPAR-γ and adipophilin that had only a modest change in CD49f− and CD49f+ subpopulations, the expression of OPN and OC decreased strikingly over the same transition (Figure 6E vs. 6F). Hence, these findings suggest that the self-renewal and tumorigenicity of OSIC properties exhibited by OSIC-like CD49f−CD133+ subpopulation are associated with an inhibited osteogenic potential, in contrast to their CD49f+ progeny that display limited tumorigenicity with more differentiated osteogenic feature.
The CD49f−CD133+ phenotype identifies a small fraction of transformed cells in patient samples of either primary or metastatic osteosarcoma
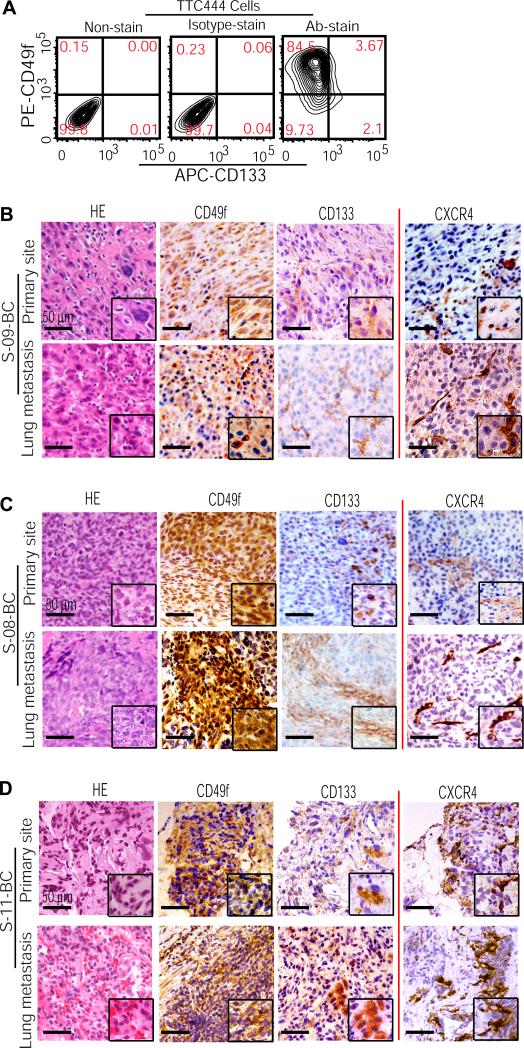
Previous studies have suggested that cancer stem cells constitute a small fraction of transformed cells. They are capable of extensive self-renewal and can repeatedly generate copious numbers of progeny that ultimately form the bulk of the tumor mass, while the initial proportion of cancer stem cells remains essentially the same (11). We find that during the transition from the low tumorigenicity of U2OS cells to the high tumorigenicity of UT2 and UT3 cells, OSIC-like CD49f−CD133+ cells consistently represent a much smaller proportion of osteosarcoma cells than do their CD49f+ progeny (Figures 4-6). To investigate whether this phenomenon exists in human osteosarcoma patient samples, we first examined CD49f− versus CD49f+ versus CD49f−CD133+ populations in TTC444 cells, which were derived from the primary bone tumor and their earlier passage was proven to have the tumorigenicity in vivo (Figure 3F; Supplemental Table 2). FACS analysis showed that in contrast to the 84.5% of cells expressing CD49f, 9.7% and 2.1% expressed CD49f− and CD49f−CD133+, respectively (Figure 7A). Further to determine such populations in osteosarcoma tissues, the paired samples from both primary and concordant metastatic pulmonary sites in 3 osteosarcoma patients were examined with HE staining and immunohistochemical analysis. The results showed that in contrast to scant expression of CD133 in both the primary and metastatic sites, a massive CD49f expression was detected in these tissues (Figure 7B-D, left panels). Because osteosarcoma most commonly metastasizes to the lungs (3) whereas cancer stem cells are thought to be responsible for establishing metastasis (38, 39), we examined in these patient tissues whether CD133+ subpopulation generated by CD49f− cells (Figure 5) co-existed with a subpopulation expressing CXCR4, a marker identified with metastatic property (38, 40, 41). Immunohistochemical analysis showed that similar to CD133, CXCR4 was expressed by a small number of localized cells in either the primary or metastatic site (Figure 7B-D, right panels). Together, these results demonstrate that OSIC-like CD49f−CD133+ cells consistently represent small proportion of transformed cells in both the primary and metastatic lesion, whereas CXCR4+ cells could be a potential tumorigenic subpopulation together with CD49f−CD133+ cells to initiate osteosarcoma growth and induce metastasis. Moreover, OSFC-like CD49f+ progeny, making up the tumor bulk whether formed in the primary or metastatic site, could be the progeny derived from the localized OSIC-like parental population.
Figure 7. The OSIC-like CD49f−CD133+ phenotype identifies a small fraction of transformed cells in patient samples of either primary or metastatic osteosarcoma.
(A) FACS analysis of CD49f−CD133+ versus CD49f+ cells in TTC444 cells. Ab, antibody. (B-D) HE staining of the paired tissue samples from both primary and concordant metastatic pulmonary sites in 3 osteosarcoma patients. Parallel immunohistochemical assay detected the expression of CD49f, CD133, and CXCR4 in these samples.
Discussion
The identity of the cells giving rise to OSFC has been elusive. By defining the lineage relationship between OSFC and their parental cells using an inverse lineage-tracking strategy, we determined that the CD49f+ progeny, which constitute the osteosarcoma mass, are derived from CD49f− or CD49f−CD133+ cells (Figures 4A; 6B, right panel). CD49f (also called integrin α6) has been used as a CD34-like protein to identify early MSC progenitors with increased clonogenicity in mice (31). Moreover, studies from Rich's group found high levels of CD49f expression in surgical biopsy specimens of glioblastoma multiforme (32). We found that although the expanded CD49f+ progeny shorten the time of tumor formation (Figures 1, 3), their tumorigenicity and drug resistance are reduced compared to the same features of their parental CD49f− cells (Figure 4C-G). Thus, expansion of the CD49f+ population is driven by the enhanced self-renewal of their parental CD49f− cells, while the limited tumorigenicity of these progeny may be related to expression of the CD49f marker. CD49f−CD133+ cells appear important because they have a greater capacity than CD49f− cells to generate CD49f+ OSFC (Figures 6B vs. 4A) and induce tumors in vivo more readily (75%) than the CD49f− (56%), CD133+ (42%), CD49f+ (44%), CD133− (33%), or CD49f−CD133− (0%) subpopulation (Figures 4C vs. 5F vs. 6B). Those CD49f−CD133+ cells are consistently found as a small fraction within UT2 xenografts, primary tumors, or metastatic lesions and produce an abundance of more differentiated CD49f+ progeny (Figures 5-7). Moreover, CD49f−CD133+ cells co-exist with a small subpopulation expressing CXCR4, a marker possessing metastatic property of cancer stem cells (38-41), in both the primary and metastatic sites of osteosarcoma patient samples (Figure 7). Altogether, these findings suggest that CD49f−CD133+ cells possess the OSIC-like properties of self-renewal and tumorigenicity, and are capable of initiating and sustaining osteosarcoma growth (Supplemental Figure 3).
The phenotype of cancer stem cells has become increasingly complex, despite the publication of well-designed and properly executed studies (10, 11, 19). The role of CD133 as a robust marker of cancer stem cells is a case in point (18, 19). In some studies, only CD133+ cells are able to initiate tumors in vivo, whereas in others CD133– cells also possess tumor-initiating potential (11, 19, 22, 23). Our analysis demonstrates that whereas CD49f−CD133+, CD49f−, CD49f+, CD133+, and CD133− subpopulations can all form osteosarcoma in vivo, CD49f−CD133+ cells possess the strongest tumorigenicity and have a substantial capacity to differentiate to CD49f+ cells (Figures 4-6). Interestingly, while CD49f−CD133+ cells retain the capacity for adipogenic differentiation, their strong tumorigenicity is associated with decreased osteogenic potential (Figure 6B-F). This inverse relationship is observed during the transition from the low tumorigenicity of U2OS cells to the high tumorigenicity of UT2 cells induced by serial transplantation (Figure 1), in which osteogenic but not adipogenic differentiation marker genes are decreased in UT2 or CD49f− cells (Figure 6D-F). Thus, our studies reveal that the gain of tumorigenicity correlates with a diminished osteogenic potential in OSIC-like CD49f−CD133+ cells, while the decreased tumorigenicity of OSFC progeny is associated with more differentiated osteogenic features. What, then, is the mechanism of the inhibited osteogenic fate of CD49f−CD133+ cells? One explanation is that CD133 expression promotes the attachment of stem cells to their niche (18, 19), where they could preserve self-renewal potential by sustaining an aberrant differentiation program that represses osteogenic potential, as we hypothesize (Supplemental Figure 3). This hypothesis, although attractive, will require rigorous testing with CD49f−CD133+ versus CD49f−CD133− cells and more differentiated CD49f+ progeny.
Currently, the mechanisms of CD49f−CD133+ contributing to OSIC properties remain unknown. Several groundbreaking studies with mouse transgenic models have shown the importance of the Wnt, Hedgehog, Notch, BMI1, and TGF-β pathways in activating and maintaining cancer stem cells (19, 37, 42, 43). We found that in UT2 cells generated by serial transplantation (Figure 1), the molecules crucial to mediating Wnt, Notch, and TGF-β signaling pathways are up-regulated, compared to findings in MSC and/or U2OS cells (Figure 2B-H). This suggests that in UT2 cells, CD49f−CD133+ subpopulation maintains these pathways that are known to be associated with cancer stem cells. Hence, determining such molecular basis for self-renewal of the CD49f−CD133+ cells will be an important step toward devising targeted therapeutics that inhibit the tumorigenicity of CD49f−CD133+ cells.
Recent studies from Iwakuma's group show that by using human osteosarcoma KHOS/NP (transformed with Kirsten murine sarcoma virus) and MNNG/HOS (transformed with carcinogenic nitrosamine) cells, both CD117+ and Stro-1+ subpopulations exhibit OSIC properties, whereas Stro-1 likely contributes more than CD117 (4). We found that neither CD117+ nor Stro-1+ cells show appreciable tumorigenicity compared to CD133+, CD133−, CD49f−, CD49f+, or CD49f−CD133+ cells (Figures 5E vs. 4C and 6B). Moreover, whereas unsorted KHOS/NP cells induced tumor formation (Supplemental Table 1), CD117+CD49+ cells isolated from KHOS/NP cells still lack tumorigenicity (0/4 mice; 500 cells/mouse), in contrast to the CD117−CD49f− population, which induced tumor formation in 2/4 mice tested. We suggest that the discrepancies between Iwakuma's work and ours may result from the differences in cellular background. However, we do not rule out the contribution of the Stro-1+ marker to the OSIC properties of CD49f−CD133+ cells, as our initial studies indicate that the Stro-1+ subpopulation isolated from UT2 cells does possess some tumorigenicity. Further dissection of the CD49f− subpopulation among primary osteosarcoma cells, using a serial single cell clonogenic assay and human-to-mouse xenotransplantation, should determine the still-missing counterpart(s) of the OSIC-like CD49f−CD133+ cells, leading to identifying of OSIC. Such studies could, in turn, point the way to treatments that eradicate osteosarcoma by expunging its root cause.
Our studies show that the gain of strong tumorigenicity, seen with OSIC-like CD49f−CD133+ cells, correlates with a diminished osteogenic fate. This new phenotype distinguishes CD49f−CD133+ subpopulation from their progeny, CD49f+ cells, which compose the bulk of the tumor mass and possess more differentiated osteogenic features with limited tumorigenicity. Moreover, CD49f+ progeny remain capable of inducing osteosarcoma in vivo. This finding provides new insight that may help to resolve the current controversy over whether some daughter cells or marker-negative counterpart subpopulations of cancer stem cells possess limited tumorigenicity in vivo. Hence, these studies advance our understanding of cancer stem cell properties and suggest new approaches to targeted therapy in patients with osteosarcoma.
Materials and Methods
Patient samples, cells, and cell culture
Three osteosarcoma patient samples taken before chemotherapy were provided by CHLA Pathology Tissue Bank in accord with the protocol approved by the CHLA Committee on Clinical Investigations. Human osteosarcoma cell lines U2OS, MG63, SAOS-2 were from ATCC (Manassas, VA, USA) and cultured as described (30). UT1, UT2, and UT3 cells were derived from the second, third, and fourth human-to-mouse xenotransplantations of parental U2OS xenograft tissues as described (30). Other primary cells and cell culture are detailed in Supplemental Information.
Flow cytometric analysis and cell sorting
Flow cytometric analysis was performed as described (44), with details in Supplemental Information.
Xenotransplantation
Cells (5 × 102 to 1 × 107) were injected subcutaneously into the shoulder of 4-6 week old BALB/c nude mice or 7-week old NOD/SCID mice (Jackson Laboratory, Bar Harbor, Maine). All mouse work was performed according to guidelines under protocols approved by the Children's Hospital Los Angeles Institutional Animal Care and Use Committee. More details of animal work are described in Supplemental Information.
Intrafemoral implantation
Seven-week old BALB/c nude mice were anesthetized with a mixture of xylazine and ketamine. The knee of mice was bent and placed facing the experimenter. A 26-gauge needle was then used to perforate the femur head and, through the punctured hole once the needle was removed, a second needle coupled to a 1 ml syringe containing cells (1 × 105 in 20 μl of PBS) was carefully introduced into the medullary cavity of the femur. Control groups were injected with PBS. The surgical procedure was completed with a stitch of the knee skin.
Drugs and drug resistance test
Anti-osteosarcoma drugs (36), including cisplatin (Pharmachemie B.V., Haarlem, The Netherlands) and doxorubicin (Ben Venue Laboratories, Benford, OH), as well as other anti-cancer toxins, including taxol (Bristol-Myers Squibb, Princeton, NJ) and idarubicin (Hisun Pharmaceutical, Hangzhou, China), were diluted to final concentrations with corresponding culture medium. Drug resistance assay is detailed in Supplemental Information.
Tumor processing and histology
Tumor processing, hematoxylin-eosin (HE) staining, and immunohistochemical assay were performed by CHLA Pathology Core, as described (30).
Sphere culture
Cells (3 × 105 /ml) were plated into 6-well plate coated with 1.5% Noble Agar (Difco, Detroit, MI) as described (45), with details in Supplemental Information.
Microarray and Cluster analyses
Microarray and cluster analyses were performed as described (30), with more details in Supplemental Information.
In vivo and in vitro tracking of genetic lineage
Cells were injected subcutaneously into NOD/SCID mice or cultured in dishes. The xenograft tissues or cells were harvested for assessing changes in surface immunophenotypes with FACS using CD49f, CD133, CD117, or Stro-1 antibodies.
Osteogenic and adipogenic differentiation
As described (46, 47), Alizarin Red S (ARS) and Oil Red O (ORO) (Sigma) were used for differentiation evaluation (see Supplemental Information).
Quantitative real-time polymerase chain reaction (qRT-PCR)
qRT-PCR, using QuantiTectTM SYBR Green PCR kit (Qiagen, Inc., Valencia, CA), is detailed in Supplemental Information.
Supplementary Material
Acknowledgments
This work was supported by grants from the National Institutes of Health (R01 CA120512 and ARRA-R01CA120512 to Lingtao Wu).
Footnotes
Conflict of interest
The authors declare no conflict of interest.
Data deposition. The microarray data from the manuscript have been submitted to the GEO database (http://www.ncbi.nlm.nih.gov/geo). Series: GSE30807; 3 samples: GSM764199, GSM764200, and GSM764201. Title: Gene expression of normal human mesenchymal stem cells, osteosarcoma U2OS cells, and U2OS cell-derived UT2 cells by serial transplantation. File format, GEOarchive; user name, Lingtao Wu. Confidential access for peer-reviewers: user ID, USC007; password, usc007.
Supplemental Information: Supporting information includes: a) Supplemental Materials and Methods; b) Supplemental Tables (4), and c) Supplemental Figures (3).
References
- 1.Thomas D, Kansara M. Epigenetic modifications in osteogenic differentiation and transformation. J Cell Biochem. Jul 1. 2006;98(4):757–69. doi: 10.1002/jcb.20850. [DOI] [PubMed] [Google Scholar]
- 2.Tang N, Song WX, Luo J, Haydon RC, He TC. Osteosarcoma development and stem cell differentiation. Clin Orthop Relat Res. 2008 Sep;466(9):2114–30. doi: 10.1007/s11999-008-0335-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bakhshi S, Radhakrishnan V. Prognostic markers in osteosarcoma. Expert Rev Anticancer Ther. 2010 Feb;10(2):271–87. doi: 10.1586/era.09.186. [DOI] [PubMed] [Google Scholar]
- 4.Adhikari AS, Agarwal N, Wood BM, Porretta C, Ruiz B, Pochampally RR, et al. CD117 and Stro-1 identify osteosarcoma tumor-initiating cells associated with metastasis and drug resistance. Cancer Res. 2010 Jun 1;70(11):4602–12. doi: 10.1158/0008-5472.CAN-09-3463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Di Fiore R, Santulli A, Ferrante RD, Giuliano M, De Blasio A, Messina C, et al. Identification and expansion of human osteosarcoma-cancer-stem cells by long-term 3-aminobenzamide treatment. J Cell Physiol. 2009 May;219(2):301–13. doi: 10.1002/jcp.21667. [DOI] [PubMed] [Google Scholar]
- 6.Tirino V, Desiderio V, d'Aquino R, De Francesco F, Pirozzi G, Graziano A, et al. Detection and characterization of CD133+ cancer stem cells in human solid tumours. PLoS ONE. 2008;3(10):e3469. doi: 10.1371/journal.pone.0003469. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Tirino V, Desiderio V, Paino F, De Rosa A, Papaccio F, Fazioli F, et al. Human primary bone sarcomas contain CD133+ cancer stem cells displaying high tumorigenicity in vivo. FASEB J. 2011 Jun;25(6):2022–30. doi: 10.1096/fj.10-179036. [DOI] [PubMed] [Google Scholar]
- 8.Levings PP, McGarry SV, Currie TP, Nickerson DM, McClellan S, Ghivizzani SC, et al. Expression of an exogenous human Oct-4 promoter identifies tumor-initiating cells in osteosarcoma. Cancer Res. 2009 Jul 15;69(14):5648–55. doi: 10.1158/0008-5472.CAN-08-3580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Wang L, Park P, Zhang H, La Marca F, Lin CY. Prospective identification of tumorigenic osteosarcoma cancer stem cells in OS99-1 cells based on high aldehyde dehydrogenase activity. Int J Cancer. 2011;128(2):294–303. doi: 10.1002/ijc.25331. [DOI] [PubMed] [Google Scholar]
- 10.Dalerba P, Cho RW, Clarke MF. Cancer stem cells: models and concepts. Annu Rev Med. 2007;58:267–84. doi: 10.1146/annurev.med.58.062105.204854. [DOI] [PubMed] [Google Scholar]
- 11.O'Brien CA, Kreso A, Jamieson CH. Cancer stem cells and self-renewal. Clin Cancer Res. 2010 Jun 15;16(12):3113–20. doi: 10.1158/1078-0432.CCR-09-2824. [DOI] [PubMed] [Google Scholar]
- 12.Hong D, Gupta R, Ancliff P, Atzberger A, Brown J, Soneji S, et al. Initiating and cancer-propagating cells in TEL-AML1-associated childhood leukemia. Science. 2008 Jan 18;319(5861):336–9. doi: 10.1126/science.1150648. [DOI] [PubMed] [Google Scholar]
- 13.Cox CV, Evely RS, Oakhill A, Pamphilon DH, Goulden NJ, Blair A. Characterization of acute lymphoblastic leukemia progenitor cells. Blood. 2004 Nov 1;104(9):2919–25. doi: 10.1182/blood-2004-03-0901. [DOI] [PubMed] [Google Scholar]
- 14.Cox CV, Martin HM, Kearns PR, Virgo P, Evely RS, Blair A. Characterization of a progenitor cell population in childhood T-cell acute lymphoblastic leukemia. Blood. 2007 Jan 15;109(2):674–82. doi: 10.1182/blood-2006-06-030445. [DOI] [PubMed] [Google Scholar]
- 15.Singh SK, Hawkins C, Clarke ID, Squire JA, Bayani J, Hide T, et al. Identification of human brain tumour initiating cells. Nature. 2004 Nov 18;432(7015):396–401. doi: 10.1038/nature03128. [DOI] [PubMed] [Google Scholar]
- 16.Al-Hajj M, Wicha MS, Benito-Hernandez A, Morrison SJ, Clarke MF. Prospective identification of tumorigenic breast cancer cells. Proc Natl Acad Sci U S A. 2003 Apr 1;100(7):3983–8. doi: 10.1073/pnas.0530291100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ricci-Vitiani L, Lombardi DG, Pilozzi E, Biffoni M, Todaro M, Peschle C, et al. Identification and expansion of human colon-cancer-initiating cells. Nature. 2007 Jan 4;445(7123):111–5. doi: 10.1038/nature05384. [DOI] [PubMed] [Google Scholar]
- 18.Bomken S, Fiser K, Heidenreich O, Vormoor J. Understanding the cancer stem cell. Br J Cancer. 2010 Aug 10;103(4):439–45. doi: 10.1038/sj.bjc.6605821. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wu Y, Wu PY. CD133 as a marker for cancer stem cells: progresses and concerns. Stem Cells Dev. 2009 Oct;18(8):1127–34. doi: 10.1089/scd.2008.0338. [DOI] [PubMed] [Google Scholar]
- 20.Mizrak D, Brittan M, Alison MR. CD133: molecule of the moment. J Pathol. 2008 Jan;214(1):3–9. doi: 10.1002/path.2283. [DOI] [PubMed] [Google Scholar]
- 21.Rountree CB, Ding W, He L, Stiles B. Expansion of CD133-expressing liver cancer stem cells in liver-specific phosphatase and tensin homolog deleted on chromosome 10-deleted mice. Stem Cells. 2009 Feb;27(2):290–9. doi: 10.1634/stemcells.2008-0332. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shmelkov SV, Butler JM, Hooper AT, Hormigo A, Kushner J, Milde T, et al. CD133 expression is not restricted to stem cells, and both CD133+ and CD133- metastatic colon cancer cells initiate tumors. J Clin Invest. 2008 Jun;118(6):2111–20. doi: 10.1172/JCI34401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Joo KM, Kim SY, Jin X, Song SY, Kong DS, Lee JI, et al. Clinical and biological implications of CD133-positive and CD133-negative cells in glioblastomas. Lab Invest. 2008 Aug;88(8):808–15. doi: 10.1038/labinvest.2008.57. [DOI] [PubMed] [Google Scholar]
- 24.Dylla SJ, Beviglia L, Park IK, Chartier C, Raval J, Ngan L, et al. Colorectal cancer stem cells are enriched in xenogeneic tumors following chemotherapy. PLoS One. 2008;3(6):e2428. doi: 10.1371/journal.pone.0002428. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang EH, Hynes MJ, Zhang T, Ginestier C, Dontu G, Appelman H, et al. Aldehyde dehydrogenase 1 is a marker for normal and malignant human colonic stem cells (SC) and tracks SC overpopulation during colon tumorigenesis. Cancer Res. 2009 Apr 15;69(8):3382–9. doi: 10.1158/0008-5472.CAN-08-4418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Joo KM, Nam DH. Prospective identification of cancer stem cells with the surface antigen CD133. Methods Mol Biol. 2009;568:57–71. doi: 10.1007/978-1-59745-280-9_5. [DOI] [PubMed] [Google Scholar]
- 27.Lee J, Kotliarova S, Kotliarov Y, Li A, Su Q, Donin NM, et al. Tumor stem cells derived from glioblastomas cultured in bFGF and EGF more closely mirror the phenotype and genotype of primary tumors than do serum-cultured cell lines. Cancer Cell. 2006 May;9(5):391–403. doi: 10.1016/j.ccr.2006.03.030. [DOI] [PubMed] [Google Scholar]
- 28.Dontu G, Abdallah WM, Foley JM, Jackson KW, Clarke MF, Kawamura MJ, et al. In vitro propagation and transcriptional profiling of human mammary stem/progenitor cells. Genes Dev. 2003 May 15;17(10):1253–70. doi: 10.1101/gad.1061803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Majeti R, Becker MW, Tian Q, Lee TL, Yan X, Liu R, et al. Dysregulated gene expression networks in human acute myelogenous leukemia stem cells. Proc Natl Acad Sci U S A. 2009 Mar 3;106(9):3396–401. doi: 10.1073/pnas.0900089106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Luo P, Yang X, Ying M, Chaudhry P, Wang A, Shimada H, et al. Retinoid-suppressed phosphorylation of RARalpha mediates the differentiation pathway of osteosarcoma cells. Oncogene. 2010 May 13;29(19):2772–83. doi: 10.1038/onc.2010.50. [DOI] [PubMed] [Google Scholar]
- 31.Lee RH, Seo MJ, Pulin AA, Gregory CA, Ylostalo J, Prockop DJ. The CD34-like protein PODXL and alpha6-integrin (CD49f) identify early progenitor MSCs with increased clonogenicity and migration to infarcted heart in mice. Blood. 2009 Jan 22;113(4):816–26. doi: 10.1182/blood-2007-12-128702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lathia JD, Gallagher J, Heddleston JM, Wang J, Eyler CE, Macswords J, et al. Integrin alpha 6 regulates glioblastoma stem cells. Cell Stem Cell. 2010 May 7;6(5):421–32. doi: 10.1016/j.stem.2010.02.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Noel AC, Lefebvre O, Maquoi E, VanHoorde L, Chenard MP, Mareel M, et al. Stromelysin-3 expression promotes tumor take in nude mice. J Clin Invest. 1996 Apr 15;97(8):1924–30. doi: 10.1172/JCI118624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ferrandina G, Petrillo M, Bonanno G, Scambia G. Targeting CD133 antigen in cancer. Expert Opin Ther Targets. 2009 Jul;13(7):823–37. doi: 10.1517/14728220903005616. [DOI] [PubMed] [Google Scholar]
- 35.Woodward WA, Sulman EP. Cancer stem cells: markers or biomarkers? Cancer Metastasis Rev. 2008 Sep;27(3):459–70. doi: 10.1007/s10555-008-9130-2. [DOI] [PubMed] [Google Scholar]
- 36.Chou AJ, Geller DS, Gorlick R. Therapy for osteosarcoma: where do we go from here? Paediatr Drugs. 2008;10(5):315–27. doi: 10.2165/00148581-200810050-00005. [DOI] [PubMed] [Google Scholar]
- 37.O'Brien CA, Pollett A, Gallinger S, Dick JE. A human colon cancer cell capable of initiating tumour growth in immunodeficient mice. Nature. 2007 Jan 4;445(7123):106–10. doi: 10.1038/nature05372. [DOI] [PubMed] [Google Scholar]
- 38.Dewan MZ, Ahmed S, Iwasaki Y, Ohba K, Toi M, Yamamoto N. Stromal cell-derived factor-1 and CXCR4 receptor interaction in tumor growth and metastasis of breast cancer. Biomed Pharmacother. 2006 Jul;60(6):273–6. doi: 10.1016/j.biopha.2006.06.004. [DOI] [PubMed] [Google Scholar]
- 39.Charafe-Jauffret E, Ginestier C, Iovino F, Tarpin C, Diebel M, Esterni B, et al. Aldehyde dehydrogenase 1-positive cancer stem cells mediate metastasis and poor clinical outcome in inflammatory breast cancer. Clin Cancer Res. 2010 Jan 1;16(1):45–55. doi: 10.1158/1078-0432.CCR-09-1630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hermann PC, Huber SL, Herrler T, Aicher A, Ellwart JW, Guba M, et al. Distinct populations of cancer stem cells determine tumor growth and metastatic activity in human pancreatic cancer. Cell Stem Cell. 2007 Sep 13;1(3):313–23. doi: 10.1016/j.stem.2007.06.002. [DOI] [PubMed] [Google Scholar]
- 41.Oda Y, Yamamoto H, Tamiya S, Matsuda S, Tanaka K, Yokoyama R, et al. CXCR4 and VEGF expression in the primary site and the metastatic site of human osteosarcoma: analysis within a group of patients, all of whom developed lung metastasis. Mod Pathol. 2006 May;19(5):738–45. doi: 10.1038/modpathol.3800587. [DOI] [PubMed] [Google Scholar]
- 42.Ikushima H, Todo T, Ino Y, Takahashi M, Miyazawa K, Miyazono K. Autocrine TGF-beta signaling maintains tumorigenicity of glioma-initiating cells through Sry-related HMG-box factors. Cell Stem Cell. 2009 Nov 6;5(5):504–14. doi: 10.1016/j.stem.2009.08.018. [DOI] [PubMed] [Google Scholar]
- 43.Wu Y, Cain-Hom C, Choy L, Hagenbeek TJ, de Leon GP, Chen Y, et al. Therapeutic antibody targeting of individual Notch receptors. Nature. 2010 Apr 15;464(7291):1052–7. doi: 10.1038/nature08878. [DOI] [PubMed] [Google Scholar]
- 44.Luo P, Wang A, Payne KJ, Peng H, Wang JG, Parrish YK, et al. Intrinsic retinoic acid receptor alpha-cyclin-dependent kinase-activating kinase signaling involves coordination of the restricted proliferation and granulocytic differentiation of human hematopoietic stem cells. Stem Cells. 2007 Oct;25(10):2628–37. doi: 10.1634/stemcells.2007-0264. [DOI] [PubMed] [Google Scholar]
- 45.Kang HG, Jenabi JM, Zhang J, Keshelava N, Shimada H, May WA, et al. E-cadherin cell-cell adhesion in ewing tumor cells mediates suppression of anoikis through activation of the ErbB4 tyrosine kinase. Cancer Res. 2007 Apr 1;67(7):3094–105. doi: 10.1158/0008-5472.CAN-06-3259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zhang W, Deng ZL, Chen L, Zuo GW, Luo Q, Shi Q, et al. Retinoic acids potentiate BMP9- induced osteogenic differentiation of mesenchymal progenitor cells. PLoS One. 2010;5(7):e11917. doi: 10.1371/journal.pone.0011917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Maxson S, Burg KJ. Conditioned media cause increases in select osteogenic and adipogenic differentiation markers in mesenchymal stem cell cultures. J Tissue Eng Regen Med. 2008 Mar-Apr;2(2-3):147–54. doi: 10.1002/term.76. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.