Abstract
The number of discovery proteomic studies of drug abuse has begun to increase in recent years, facilitated by the adoption of new techniques such as 2D-DIGE and iTRAQ. For these new tools to provide the greatest insight into the neurobiology of addiction, however, it is important that the addiction field has a clear understanding of the strengths, limitations, and drug abuse-specific research factors of neuroproteomic studies. This review outlines approaches for improving animal models, protein sample quality and stability, proteome fractionation, data analysis, and data sharing to maximize the insights gained from neuroproteomic studies of drug abuse. For both the behavioral researcher interested in what proteomic study results mean, and for biochemists joining the drug abuse research field, a careful consideration of these factors is needed. Similar to genomic, transcriptomic, and epigenetic methods, appropriate use of new proteomic technologies offers the potential to provide a novel and global view of the neurobiological changes underlying drug addiction. Proteomic tools may be an enabling technology to identify key proteins involved in drug abuse behaviors, with the ultimate goal of understanding the etiology of drug abuse and identifying targets for the development of therapeutic agents.
Keywords: Proteomics, Neuroproteomics, Drug abuse, Proteome fractionation, Bioinformatics, Sample quality
1. Introduction: capabilities of proteomics in addiction science
Neuroproteomic studies offer great promise for increasing understanding of the biochemical basis of addiction. While proteomics is still an evolving field, proteomic approaches have proven useful for elucidating the molecular effects of amphetamine (Freeman et al., 2005), morphine (Prokai et al., 2005; Li et al., 2006; Moron et al., 2007), cocaine (Tannu et al., 2007; Lull et al., 2009) and alcohol (Freeman et al., 2006; Matsuda-Matsumoto et al., 2007; Kashem et al., 2007, 2008; Al-Housseini et al., 2008). With a number of ongoing research programs in addiction proteomics and a growing number of investigators taking advantage of these tools, the addiction research field will benefit from a consideration of the capabilities and limitations of proteomic studies. This review is targeted toward addiction researchers new to the field of proteomics, and those wishing to gain understanding of proteomic datasets and their interpretation. Additionally, biochemists and informaticians new to addiction research may benefit from a discussion of animal models of drug use and abuse. Addiction research and neurobiology-specific challenges to proteomic studies are also examined. The specific challenges and potential effects of these technical limitations on experiment results and interpretations, and approaches to surmounting these challenges will be discussed.
When applying proteomic technologies to addiction research, an understanding of the power of proteomic analysis is essential. After genetic information is transcribed into mRNA, a template is provided to the cell from which proteins will be synthesized. Functional genomics methods, studying the steady-state levels of these mRNA species, such as quantitative RT-PCR (qRT-PCR), whole-genome microarray analysis, and next generation sequencing methods provide sensitive and high-throughput approaches to quantitatively examining mRNA (and miRNA) species present within the cells of the nervous system (for a review, see Freeman and Vrana, 2006). Functional genomic studies can help to illuminate genes involved in the development of behaviors related to drug abuse and relapse liability. However, it is not until mRNA has been translated into protein that the functional end result has been reached. It is these macromolecular species that ultimately affect cellular function. It is for this reason that analyses of mRNA and protein provide different information, and have unique values. While mRNA abundance is often correlated with protein expression, levels of regulation, including protein stability and amount of translation, exist that result in protein levels that do not always correlate to the amount of mRNA (Anderson and Seilhamer, 1997). Also, functional genomic studies cannot provide insight into post-translational modifications (e.g., phosphorylation and glycosylation of proteins after translation has occurred) or subcellular localization of the protein product. Therefore, using proteomic techniques presents the opportunity to assess the totality of gene expression, translation, modification, and localization. Unfortunately, the sensitivity of proteomic tools lag behind those of functional genomics. Moreover, examining the mRNA provides a restricted view of primarily the cell body. Indeed, from a systems biology standpoint, analysis of both mRNA and protein levels (as well as miRNA and epigenetic changes) will ultimately provide a more integrated view of the molecular underpinnings of addiction.
Previously, we and others have described the technical aspects of proteomic technologies applicable to addiction studies (Williams et al., 2004; Freeman and Vrana, 2006). While the details of existing technologies will not be reviewed here, many of the quantitative and qualitative methods applied to neurobiology and/or addiction studies have been reviewed previously. These techniques include electrophoresis-based techniques (2D-DIGE [2-dimensional differential in-gel electrophoresis]; Freeman and Hemby, 2004; Lull et al., 2009), mass spectrometry-based methods (iTRAQ [isobaric tag for relative and absolute quantitation] and ICAT [isotope coded affinity tag]; Chen et al., 2007; Martin et al., 2008), accurate mass tagging; (Chin et al., 2007), and spectral counting; (Mueller et al., 2008; Sleat et al., 2009). Finally, traditional and second generation antibody-based methods remain workhorse technologies (immunoblotting and Luminex; Vignali, 2000; Nelson et al., 2006). While reviews of each of these techniques provide exposure to the different technologies available for proteomic studies, an important area that has heretofore received limited attention is the experimental design and interpretation specific to neuroproteomic studies of drug abuse. These challenges include choice of animal model, ensuring sample quality, the complexity of brain tissue, confirming discovery findings, data analysis strategies, and integration of large data sets with the existing literature (Fig. 1). Due to the time, expense, and complexity of neuroproteomic studies, careful consideration of these issues is needed both for the investigator and eventually for the reviewers and readership of neuroproteomic reports.
Fig. 1.

Neuroproteomic workflow. Many steps exist from the planning of a neuroproteomic study to the gathering and interpretation of the resulting data. At each of these steps, a number of different variables must be considered in order to optimize the choice of a behavioral model, collection of samples, sample preparation, data collection, and data interpretation, so as to acquire the most meaningful results from a neuroproteomic experiment.
2. Animal models
2.1. Challenges of choosing the appropriate animal model
While a full discussion is beyond the scope of this review, the selection of optimal animal models of human drug abuse and addiction is an imperative. Studies of postmortem human brain tissue from drug abusing subjects provides a valuable translational element for addiction proteomic studies and, to date, a number of excellent studies have been performed (Kashem et al., 2007, 2008; Tannu et al., 2007). Naturally, these samples are difficult to obtain and a number of factors, including variations in genetic background, drug abuse history, matched controls, comorbid conditions such as psychiatric disorders or multi-drug use, the quality of samples, and limited experimental manipulability complicate their use (Hynd et al., 2003; Skold et al., 2007; Lehrmann et al., 2008). For the majority of addiction researchers, animal models provide the most tractable approach to neuroproteomic studies.
Animal models with the most utility will be those that best reflect aspects of human behaviors. However, many variables within a behavioral paradigm can influence the usefulness of data gathered in a proteomic experiment. The first of these variables is the need for a behavioral phenotype that permits correlation of proteomic changes with a specific behavior. This is, in fact, the primary consideration when choosing an animal model for a particular study.
For example, there are well documented differences between investigator-administered and self-administered drugs. Neurochemically, while equal doses of investigator-administered and self-administered cocaine and morphine can produce similar neurochemical changes, there are also profound differences (Dworkin et al., 1995; Di et al., 1996, 1998; Mark et al., 1999). Response-independent/investigator-administered drug administration can also be stressful and aversive, and these factors may contribute to differences in gene expression between self-administering and response-independent, investigator-administered animals (Kuzmin and Johansson, 1999; Mutschler et al., 2000; Thomas et al., 2003a; Jacobs et al., 2004). These response-dependent differences have also been documented in protein expression (Self et al., 1995; Graziella et al., 1998; Stefanski et al., 1999). Furthermore, differences in neuronal morphology (dendritic spine density and branching) have been reported between experimenter-administered and self-administered drugs (Robinson and Kolb, 2004). While self-administration procedures alone do not guarantee a useful animal model, a careful rationale for using investigator-administered drug models is crucial to ensure the translatability of the results.
Another challenge to consider is the influence that behavioral testing, drug challenges, and environmental conditions prior to sacrifice can have on biochemical analyses, including proteomic studies. Any behavioral interventions, such as extinction responding or breakpoint analyses, in addition to the drug administration paradigm, may themselves alter gene and protein expression in very short time periods, and can cause misleading proteomic results. It is therefore important to conduct these tests on separate groups of animals that will not be used for biochemical or molecular analysis unless the behavioral assessment is an explicit part of the experiment.
2.2. Approaches to choosing the appropriate animal models
In addition to the administration model, the behavioral outcome that proteomic changes are being associated with needs to be considered in the experimental design. Addiction research is, at its core, the investigation of behavioral pharmacology. Therefore, a behavioral outcome is required of the model system in order for the study to be addiction research and not solely pharmacology. This includes careful choice of a behavioral paradigm and time of tissue collection that will allow for proteomic changes to be more directly correlated to the observed behavior. A number of behavioral endpoints are available to researchers ranging from reward and reinforcement (conditioned place preference, progressive ratio responding, extinction responding, and reinstated responding), to cognitive function (e.g., maze or match-to-sample testing), and sensitization (psychomotor sensitization). Along these lines, researchers utilize varying doses and administration schedules to model human binging, dependence, tolerance, abstinence, and relapse behaviors (Epstein et al., 2006; Kalivas et al., 2006; Roberts et al., 2007).
Using these models, drug abuse researchers have described the behaviors discussed above with many drugs of abuse. For example, psychomotor sensitization, or an increase in sensitivity to a drug’s locomotor-activating effects, results from exposure to cocaine, amphetamine, and opioids, and has been reported in a number of studies (Miserendino and Nestler, 1995; Wachtel and DeWit, 1999; Cornish and Kalivas, 2001; Trujillo et al., 2004). Following chronic exposure, cognitive differences, including learning and memory deficits, have also been reported (Schoenbaum et al., 2004; Jovanovski et al., 2005; Ersche et al., 2006; Calu et al., 2007). In addition to focusing on drug exposure, many studies have observed behavioral changes following drug administration and periods of abstinence (Grimm et al., 2001; Morgan et al., 2002; Morgan and Roberts, 2004; Kuntz et al., 2008). Specific interest has been paid to the molecular changes that accompany the incubation of drug craving that results in an increase in relapse potential, as it is a major barrier to successful treatment in humans (Gawin and Kleber, 1986; O’Brien, 1997). The development of a model that measures one or more of the behavioral outcomes mentioned above is necessary because it becomes the variable that protein expression changes can help to explain.
3. Ensuring sample quality
3.1. Challenges to ensuring sample quality
Proteins are labile biomolecules that often exist in multiple modified forms in vivo, and begin to degrade rapidly ex vivo. Additionally, post-translational protein modifications can quickly change during sample preparation. Postmortem changes in sample pH, enzyme activity (e.g., protease, phosphatase, kinase, etc.), and structural integrity can influence the state of protein species and lead to characterization of proteomic profiles that do not fully reflect the in vivo condition. Rapid and consistent sample collection is therefore a primary concern in neuroproteomic studies to maintain sample integrity and ensure accuracy of proteomic analyses. Standard sample collection and handling protocols also facilitate comparison of large proteomic datasets between experiments and between laboratories.
Biological samples are typically preserved by freezing at the time of collection for subsequent experimentation. Postmortem human brain tissues are valuable to addiction studies, but collection conditions are often far from ideal and samples are generally maintained ex vivo at room temperature for varying durations. Studies have shown that factors such as cause of death (agonal state), postmortem interval, and time in storage can affect the structural, molecular and biochemical integrity of samples, partly through ante- and post-mortem changes in pH that can alter protein stability (Hynd et al., 2003). Together with the considerable biological, environmental, and behavioral variation inherent in the human population, sample variance introduced by these antemortem and postmortem conditions decreases intersample consistency (Hynd et al., 2003). While animal tissues are collected more rapidly and in more uniform conditions, they are also subject to similar postmortem processes. These processes can lead to decreases in sample consistency, protein yield, protein modification, and experimental sensitivity (i.e., the ability to detect non-abundant species or statistically significant differences between groups due to increased variance).
3.2. Approaches to ensure sample quality
In neuroproteomic studies of addiction, relatively small changes in protein expression and modification often possess a high degree of biological importance, but are difficult to detect. It is therefore critical that measures be taken during sample collection and preparation to minimize degradative processes that affect the sensitivity and accuracy of proteomic technologies. Snap-freezing of samples immediately following collection is standard procedure in the laboratory, but a variety of recently developed approaches offer alternatives for improving sample quality, particularly when snap-freezing is not a viable option. For example, the Stabilizor T1 (Denator AB, Gothenburg, Sweden) uses a combination of uniform conductive heat and pressure to inactivate enzymes that contribute to post-collection sample modifications. This has been demonstrated through preservation of protein phosphorylation states and minimization of protein degradation fragments (e.g., stathmin 2–20) (Svensson et al., 2009). With the Denator system, samples can be either stabilized immediately following collection, or if necessary, snap-frozen and subsequently stabilized prior to experimentation. The downside to this approach, however, is that it prevents the downstream fractionation of stabilized tissues. Alternative technologies are being developed to control ex vivo biomolecular interactions, including the Pressure Cycling Technology (for use with lipid-rich samples; Pressure Biosciences).
An alternative approach is to use focused microwave irradiation as the method of euthanasia. Previous studies have suggested that brain proteins (O’Callaghan and Sriram, 2004) and peptides (Che et al., 2005) are subject to less post mortem degradation with this method of animal sacrifice. A recent more comprehensive proteomic analysis has also compared typical snap-frozen brain tissue with samples collected from microwave-irradiated animals (Hunsucker et al., 2008). This study showed that the stability (measured as amount of protein) of a number of protein species in the brain is increased by this method. Microwave irradiation may be a very useful tool in the future, but its use is currently limited by the technical difficulties of the method.
Regardless of the manner of sample collection, assessment of proteomic sample quality and adoption of quality standards is necessary for maximally informative neuroproteomic studies. It has been proposed that levels of the stathmin 2–20 fragment be assessed as an index of sample integrity, particularly in samples isolated from human postmortem tissue (Svensson et al., 2009). Lab-on-a-chip approaches to quality assessment such as the Agilent Bioanalyzer are informative not only of general protein quality, but also of protein size, purity, and concentration. Simpler approaches, including total-protein staining of 1D gel-separated samples (Coomassie, Deep Purple, etc.) and sample quantitation (e.g., BCA and Bradford assays) aid in standardizing sample input in proteomic workflows. As with all experimental approaches, the quality of protein samples in neuroproteomic studies greatly influences the quality of the resulting data. Implementing quality control standards as a routine part of neuroproteomic study workflow improves protein resolution and quantitation, and greatly increases protein identification rates, ensuring that maximal data is obtained in these studies.
4. Neuroanatomical complexity
4.1. Challenges of neuroanatomic complexity
The brain is characterized by a high degree of anatomical, cellular, and compartment-specific specialization that contributes to proteomic complexity. Functionally related brain regions may be separated spatially, and distinct anatomical regions often contain traversing neuronal processes with no direct relation to the area of interest. Even discrete brain regions exhibit cellular heterogeneity with regard to cell types (neurons, glia, vasculature, etc.) and their biochemical characterization (e.g., primary neurotransmitter) (Doyle et al., 2008; Bateup et al., 2008). Although addiction science typically focuses on neuronal populations, these cells comprise only 20–30% of the total cellular population in brain tissue (Singh et al., 2003). This can be problematic in proteomic studies with limited sensitivity or gain in the experimental system, because rarer protein species are often masked by more abundant proteins, and significant cell type- or compartment-specific expression changes may not be detected in mixed populations. Additionally, spatial distribution of neuronal somata and their distal projections makes it difficult to ensure that the entirety of a given cell or cell population is subjected to proteomic analysis. The result of this complexity is that neuroanatomical samples can be “contaminated” by cellular material that exerts a muting effect on proteomic changes in a specific cell type or subcellular compartment or is complicated by detection of protein species attributed to sample populations irrelevant to addiction studies. This is particularly true when characterization of drug abuse-induced changes in specific subcellular structures (e.g., nuclei, synaptic terminals, etc.) is desired, or when researchers are seeking to attribute behavioral changes to a specific cell type. There are several approaches to circumventing the complexity of brain tissues to maximize precision and impact of neuroproteomic studies, ranging from anatomical dissection to subcellular fractionation to biochemical enrichment.
4.2. Approaches to resolve challenges of neuroanatomic complexity
4.2.1. Anatomical dissection
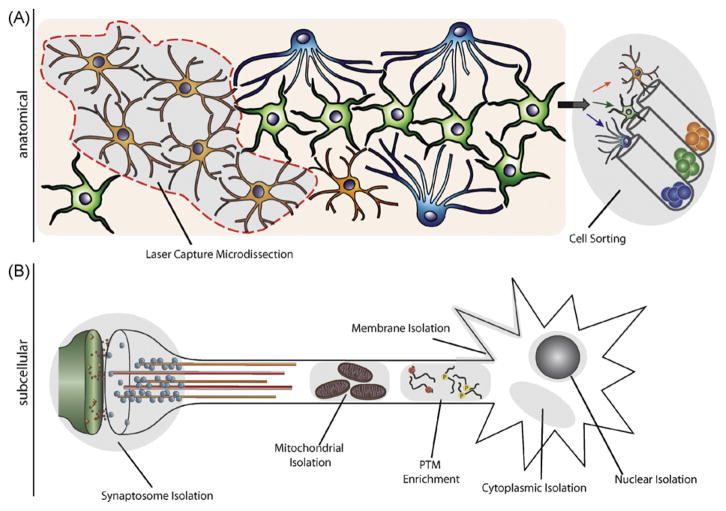
As our understanding of functional specificity of the brain advances, smaller and increasingly discrete brain subregions are identified, and accurate and consistent anatomical dissection of these distinct areas of interest is difficult. Brain atlases such as Paxinos and Watson establish standard anatomical landmarks useful for maintaining consistency between dissections (Paxinos and Watson, 2005). Small tissue punches offer a method of standardizing dissections by allowing discrete brain samples to be collected in a highly reproducible manner. When the physical size of a desired area of interest precludes routine manual dissection, or when collection of specific cell types or structures is desired, laser capture microdissection (LCM) of carefully sectioned tissue provides a valuable alternative (Fig. 2A). This approach is particularly useful in collecting samples with laminar organization (e.g., hippocampus) or distinct cytoarchitecture (e.g., nucleus accumbens). When coupled with cell labeling approaches like immunocytochemistry or transgenic expression of fluorescent proteins driven by a cell type-specific promoter, LCM enables rapid collection of individual cells from a heterogeneous population. Additionally, multiple subregions or cell types can be collected in parallel from a finite amount of tissue, enabling thorough, but specific, assessment of various aspects of a brain region of interest. Samples dissected by this method are frequently included in genomic studies, and have proven to be compatible with proteomic technologies including liquid chromatography, 2D-DIGE, and MALDI-ToF/ToF mass spectrometry (Mouledous et al., 2003a,b; Liao et al., 2004), but actual addiction research studies that use this approach have not been reported. There is a significant drawback of LCM sample collection in neuroproteomic studies, however, in that sample amounts obtained by this method are limited, and multiple rounds of LCM are often necessary to collect the amount of tissue required for proteomic analysis. This is less of an issue in genomic approaches due to availability of nucleic acid amplification and the sensitivity of the technologies, but remains to be overcome in proteomics. Currently, investigators must reconcile a trade-off between increased anatomical specificity and decreased sensitivity for rarer species when using the LCM approach for collection of samples for proteomic work.
Fig. 2.
Approaches for neuronal fractionation. To solve the problems related to tissue and cellular complexity, as well as to look at specific regions of interest, the neuroproteome may be fractionated according to anatomy (A) or subcellular compartment (B). (A) Anatomical fractionation allows for the isolation of small brain regions or cell types of interest. (B) Subcellular fractionation allows for the isolation of different subcellular compartments (e.g., synaptosomes, mitochondria, nuclei), or subcellular populations of proteins (e.g., phosphorylated or glycosylated proteins). (PTM–post-translational modifications).
Specialized cell sorting flow cytometry guided by specific parameters such as cell size, morphology, membrane potential, or fluorescence provides another approach for enrichment of specific cellular populations from complex samples (Fig. 2A). Fluorescence-activated cell sorting (FACS) uses both fluorescence and light scattering properties of labeled cells, typically by immunological or transgenic approaches, to quantify and separate distinct cell populations. FACS achieves a higher degree of sample purity than other high-throughput methods like immunopanning and magnetic bead-based sorting, and can be used with multiplexed cell staining approaches. The specificity of the cell sorting using this method closely parallels the specificity of the antigen or gene promoter selected for labeling, and can discriminate not only between classes of cells (neurons, astrocytes, microglia, etc.), but also between specific cellular subtypes characterized by biochemical and functional properties. For example, neurons can be sorted by primary neurotransmitter (e.g., dopamine vs. norepinephrine) or membrane proteins (Thy-1), while microglia can be segregated by activation state and cytokine expression. FACS has previously proven useful for isolating a wide range of brain cell populations including neural stem cells and dopaminergic neurons, that have been subsequently analyzed without requiring a period of culturing prior to experimentation (Gao and Chen, 2008; Han et al., 2008). Like LCM, cell sorting decreases anatomical complexity and increases population specificity, but does so at the expense of the amount of sample obtained for use in proteomic analyses.
In recent years, multiple mass spectrometry methods capable of analyzing limited proteomic samples have been developed to maximize data obtained from small sample amounts. Multidimensional capillary electrophoresis couples on-line reverse-phase liquid chromatography with capillary electrophoresis to separate proteins. This is then followed by mass spectrometry approaches for protein identification. Improvements in sensitivity of mass spectrometers has resulted in powerful MS-based identification methods capable of detecting proteins at low femtomole to attomole levels (Caprioli et al., 1997; Rubakhin and Sweedler, 2008), making them ideal for identifying species in small sample amounts. The Fourier-transform ion cyclotron resonance mass spectrometer, which utilizes strong magnets and nonstandard detection technology, is extremely sensitive and allows investigators to examine expression profiles of very small amounts of proteins and peptides such as those resulting from LCM/FACS collection (Rubakhin and Sweedler, 2007, 2008).
4.2.2. Subcellular fractionation
While physical dissection of neural tissue is useful in proteomic examinations of discrete brain subregions or particular cell types, subcellular fractionation also allows for specific analysis of portions of the cellular proteome (Fig. 2B). Proteomic profiling of subcellular compartments and organelles (such as the plasma membrane, nuclei, mitochondria, or synaptic terminals) has the added power of identifying localized effects of drug abuse in isolated functional milieu that may not be reflected by the whole-cell or whole-tissue proteome. Fractionation of membrane-enclosed subcellular structures is a traditional method that is often applied in preparing samples for proteomic study. A number of techniques exist for isolating subcellular compartments from homogenized tissue, including nitrogen decompression, density separation across sucrose gradients, immuno-purification, and differential centrifugation. Nuclear fractionation is generally performed by low-g centrifugation, which yields enriched fractions suitable for examination of protein regulators of chromatin, DNA modification and gene transcription. Investigation of the nuclear compartment in isolation has led to increased understanding of changes in transcription factors (Dobretsova et al., 2008), genotoxic stress (Qi et al., 2007), and indicators of cell differentiation (Salim et al., 2007; Jaishankar et al., 2008; Barthelery et al., 2009), and has the potential to reveal a great deal about long-lasting biochemical changes that correlate to addiction and relapse behaviors. Isolation of mitochondrial fractions can be achieved with higher g-force centrifugation, which enriches for intact, functional mitochondria. Proteomic analyses of this fraction have been successfully conducted in neurobiological studies of drug abuse (Cunha-Oliveira et al., 2008), as well as healthy and pathological aging (Guevara et al., 2008; Chin et al., 2008).
Plasma membrane-associated proteins (receptors, scaffolding proteins, etc.) influence neuronal function and excitability, and are therefore targets of interest in neuroproteomic studies of drug abuse. These proteins are often obscured by more abundant protein species in proteomic studies, but can be enriched for analysis through fractionation of the plasma membrane. A variety of techniques are used to isolate neuronal membranes, including differential and density gradient centrifugation (Nielsen et al., 2005; Chen et al., 2006; Olsen et al., 2007) and affinity partitioning (Schindler et al., 2006). Similar approaches (high-g ultracentrifugation) can be applied to enrich for cytosolic and microsomal fractions (Krapfenbauer et al., 2003; Stevens et al., 2008). Individual protein species identified in these compartments have been implicated in drug abuse (Hemby, 2006; Lull et al., 2009), although comprehensive examination of their proteomic profiles remains to be conducted.
As the facilitators of neurotransmission, and therefore mediators of the neuronal processes underlying behavior, synapses may represent the most relevant subcellular component in addiction research. Isolated nerve terminals, called synaptosomes, are comprised of intact presynaptic and postsynaptic membranes that enclose mitochondria, cytosolic and structural proteins, neurotransmitter-containing vesicles and the full complement of presynaptic proteins required for synaptic signaling, as well as the postsynaptic density and associated receptors and downstream signaling molecules. Modifications to synaptosome isolation techniques can further fractionate the proteome to isolate postsynaptic densities, synaptic vesicles, or synaptic lipid rafts. Removal of unwanted structures enables study of the synaptic proteome in the absence of somatic and non-neuronal material, and has aided in characterizing the composition of both synaptic vesicles and postsynaptic densities. This compartment is of particular importance in drug abuse research, as synaptic plasticity and persistent alterations in synaptic function likely result from drug abuse and contribute to drug-seeking behaviors. Synaptic alterations in response to administration of opiates, cocaine, and methamphetamine have been identified in brain tissue, and drug-related modifications have been reported in the postsynaptic density proteome and the synaptic phosphoproteome (Qin et al., 2005; Zhong et al., 2006; Abul-Husn and Devi, 2006; Eyerman and Yamamoto, 2007).
4.2.3. Biochemical fractionation
In addition to segregating compartment-specific proteomes, the neuroproteome can also be fractionated by depletion/enrichment methods to examine potential changes in classes of protein species (e.g., phosphoproteins, acidic proteins, etc.) (Fig. 2B). Focusing on a subproteome increases the likelihood of detecting proteins of interest by avoiding masking effects of abundant or irrelevant proteins, and by enriching for specific species of interest. Post-translational modification of proteins is the true regulator of protein function, and is therefore of great importance in proteomic studies. For example, in the brain, phosphorylation modulates the activity, localization, and interaction of proteins, and regulation of both basal and evoked protein phosphorylation is altered by a number of factors including aging, disease, and drug use (Tannu et al., 2008; Kruger et al., 2008). Phosphorylation-specific protein stains (e.g., Invitrogen ProQ Diamond) can be used to detect and quantify changes in phosphoprotein abundance in gel-based approaches (usually 2-dimensional gels). Similar to standard approaches, phosphoproteins of interest are then excised and identified by mass spectrometry. Alternatively, phosphoproteins can be enriched from the broader neuroproteome by immuno-isolation or chromatography methods like immobilized metal-ion affinity trapping (D’Ambrosio et al., 2006; Becker et al., 2007; Collins et al., 2008), both of which are compatible with downstream proteomic applications. This approach is particularly beneficial when studying less abundant phosphoprotein species, since they will be enriched when unphosphorylated proteins are removed. Similar approaches can be implemented to enrich for glycosylated proteins (Invitrogen ProQ Emerald gel stain, concanavalin A/lectin affinity columns) (Morelle and Michalski, 2005; Monzo et al., 2007), oxidized proteins (biotin hydrazide/avidin affinity trapping) (Meany et al., 2007), and ubiquitinated proteins (through binding to proteosome subunit sequences fused to immobilized glutathione-S-transferase columns) (Layfield et al., 2001). As technologies for biochemical enrichment continue to improve, more specific modifications can be isolated (e.g., O-linked glycosylation [isotopic and chemoenzymatic tagging] (Khidekel et al., 2007)) for standard proteomic approaches. Due to the effects of these post-translational modifications on protein structure, function, localization, and stability, biochemical fractionation to enrich for specifically modified protein species is often implemented in neurobiology studies seeking to determine the molecular basis of a pathology or behavior (i.e., drug addiction) (Hale et al., 1998; Garzon et al., 2002; Abul-Husn and Devi, 2006).
In cases where a rare target species is not restricted to a subcellular compartment or its particular modifications are less important than its total expression in a given brain subregion, samples can be fractionated into subproteomes based on protein density, charge, or other characteristics. Density gradient separation technology (e.g., enhanced density gradient extraction) separates proteins in a density-dependent manner using differential centrifugation across increasing sucrose concentrations (Lan et al., 2007). For example, the Edge 200 Separation System (Prospect Biosystems, Newark, NJ) fractionates complex protein samples into 11 density-based subproteomic components, allowing examination of distinct fractions and increasing the likelihood of detecting less abundant proteins in these simplified samples. Alternatively, technologies like the 3100 OFFGEL Fractionator (Agilent Technologies, Santa Clara, CA) separates proteins and peptides based on their isoelectric point (Horth et al., 2006). This technology is capable of simultaneously fractionating 16 samples into 12 or 24 pI-based fractions, and is compatible with both traditional in-gel and liquid-phase approaches.
5. Confirming findings from neuroproteomic studies of addiction
5.1. Challenges of confirming findings from neuroproteomic studies
The transcriptomic field has adopted a general standard that at least a selection of discovery findings need to be confirmed by an orthogonal method for a study to be complete. Confirmation experiments serve not only to validate specific discovery findings, but also lend added credence to the broader data set. For transcriptomic studies, this experimental route is now fairly clear, with a number of excellent techniques for specific, high-throughput, and quantitative confirmation by a separate method (orthogonal confirmation) (VanGuilder et al., 2008). These approaches include qPCR and bead-based multiplex assays with the flexibility to examine a large number of samples (10s–100s) and a large number of genes (10s–100s) (Shi et al., 2006). The ability to quickly transition from large scale discovery approaches to confirmation assays has greatly improved genomic studies. The proteomic field, however, still suffers from lagging technological capabilities in this area and confirmational work is often the rate-limiting step in neuroproteomic studies. This challenge is more difficult than in nucleic acid research due to the lack of easily generated protein-specific probes. Nonetheless, the proteomic field is moving towards routine integration of confirmation studies in reports (e.g., Whiteaker et al., 2007).
Confirmation of proteomic findings addresses several important aspects of discovery experiments. Even with the application of tandem mass spectrometry (and other more advanced methods such as MS3) (Olsen and Mann, 2004), decoy databases, and false discovery rates (discussed below) there always exists the potential for misidentification of proteins. Although this problem may not be as great as previously proposed (Bell et al., 2009), misidentification of proteins, especially across labs, still exists. Therefore a conservative statistical approach to protein identification is still warranted as the standards for protein identification continue to evolve. Additionally, for quantitative studies, the statistical power of proteomic studies is limited and Type I false positives of differential expression are possible. Confirmational studies address both of these concerns by validating both the identity and quantitation of a specific protein.
5.2. Approaches to confirming findings from neuroproteomic studies
Immunoblotting remains the staple for confirming proteomic results. The major limitations to antibody-based immunoblot confirmations are the relatively low throughput nature and reliance on the availability of antibodies. Commercial availability of antibodies continues to expand and there are ongoing efforts to generate proteome-wide antibodies such as the Swedish Human Proteome Resource (HPR) program (Uhlen and Hober, 2009). Immunoblotting is useful for measuring total levels of a specific protein (or a known modification if PTM-specific antibodies are available), but this may not always be directly translatable to results from a proteomic study, such as 2D-DIGE where proteins are separated both by molecular weight and isoelectric point. A differential abundance of a specific protein modification in the discovery experiment may not reflect a change in the total abundance of the protein. In these cases, further manipulations of the immunoblotting approach including Far Westerns (immunoblots of 2D gels) or isoelectric focusing (IEF) westerns, may be required.
Additional antibody-based confirmational approaches including traditional ELISAs, the newer ELISA-like approaches of Luminex technologies, and multiplexed electrochemiluminescence, provide alternatives for quickly validating discovery findings. The sample requirements for antibody-based confirmation continue to be driven to smaller and smaller amounts with new approaches that can quantify protein levels from nanoliter volumes (Fan et al., 2009). All of these technologies continue to be limited by antibody affinity reagents that remain difficult to produce and are of varying quality. Aptamer affinity probes (Zichi et al., 2008) may prove to be a useful method to circumvent the difficulties of antibody production, but the challenges of generating affinity reagents (aptamer or antibody) continue to motivate the search for confirmation technologies that do not require affinity reagents.
Multiple-reaction monitoring mass spectrometry (MRM-MS) has been used as a quantitation tool in small molecule mass spectrometry for several decades (e.g., Finlay et al., 1986; Phillips et al., 1989). Recently, a combination of advances in mass spectrometer instrumentation and software with increased research on proteomic technologies has led to the use of MRM in proteomic studies (Anderson and Hunter, 2006). The basis of the MRM approach is that with high resolution mass spectrometers such as a Q-Trap instrument, a peptide and daughter ion (from a particular protein) that are unique in the organism’s proteome can be identified to measure the level of that particular protein. This specificity is possible through initial selection of a peptide in MS mode and then the specific examination of a resultant daughter ion in MS/MS mode. These two levels of specificity ensure that only this particular peptide (and thereby protein) is being examined. For increased accuracy, multiple peptides per protein can be examined. Quantitation is achieved through calculation of area under the curve of the daughter ion compared to an exogenous peptide standard for each peptide being measured.
One method for generation of a standard is to create a synthetic peptide corresponding to each peptide of interest, but with a heavy isotope included during synthesis. The heavy isotope causes a shift in the weight of the peptide in the mass spectra so that it can be measured separately from the sample and serve as a standard concentration to which the sample is compared. This is often achieved using AQUA peptides (Sigma–Aldrich) that contain 15N and/or 13C atoms (Gerber et al., 2003). This is a very powerful approach for the quantitation of specific protein(s) but has the significant limitation that these peptides are very expensive to generate and a certain percentage will fail during development due to technical factors (ion suppression, overlapping peaks, poor fragmentation, etc.). This is most apparent when using MRM following affinity-based or electrophoresis-based discovery techniques, and is less difficult when mass spectrometry-based discovery methods are used. MRM is a valuable approach, although the assay development is typically much longer and more involved than immunoblotting approaches.
Adoption of a global internal standard (GIS) approach and mTRAQ tagging (DeSouza et al., 2008), which avoids many of the above technical issues, allows for rapid assay development. In this alternative approach, a standard sample is made, and thereby all of the component peptides are labeled with a tag. This flexibility eliminates the need to generate a synthetic peptide, and because all of the peptides in the standard sample are labeled, it is a good approach for the development phase of an MRM assay.
Whichever confirmation approach is chosen, these orthogonal data will help spur the integration of neuroproteomic data into the broader addiction literature by providing greater certainty in the discovery data results. Confirmation approaches will continue to be an area of intensive technological development as more and more investigators find this the rate-limiting step in taking discovery findings forward in their research.
6. Data analysis and reporting
6.1. Challenges in data analysis and reporting
Ultimately, the success or failure of neuroproteomic studies of addiction will be in whether they help advance the understanding of the neurobiology of addiction and potential treatments. For this to happen, there must be confidence in the protein quantitation and identification from discovery studies as well as methods for distilling large amounts of data into biological narratives and methods for sharing data sets. This is not a challenge unique to neuroproteomic studies of addiction and a number of solutions are being brought into proteomics from other ‘omic’ fields and large scale analyses.
6.2. Approaches to resolve challenges in data analysis and reporting
6.2.1. Protein identification
With the variety of different mass spectrometers, peak identification software, and data analysis/protein search engine algorithms available for protein identification in neuroproteomic studies, a common concern has been the reproducibility of these results from lab-to-lab and therefore the overall confidence that can be placed in protein identifications (Aebersold, 2009). An in-depth comparison of the many different algorithms and databases for protein identification is beyond the scope of this review but, for addiction researchers reading and reviewing neuroproteomic reports, it is important to have an understanding of the metrics available for judging protein identification data. The primary value for judging the quality of protein identification is the false positive rate. This value can be produced in a number of ways, but provides an estimation of the likelihood that the protein identification is correct. These data should be reported as part of Minimal Information Available about a Proteomics Experiment (MIAPE) compliance (Taylor et al., 2007). For example, if a protein was identified with 99% confidence, there is only a 1% chance, for each protein identification, that this identification is incorrect. While there is no universal cut-off for what can be considered a confident identification, low confidence identifications (<95%) should be treated carefully while high confidence identification (>99.9%) have minimal likelihood of being incorrect. As noted above, concerns with confidence can be obviated by rigorous post hoc confirmations using orthogonal approaches.
6.2.2. Statistical analysis of differential abundance
Initial data analysis from proteomic experiments requires the determination of change from control levels (fold-change) and statistical values. For this step, the balance of Type I and Type II errors remains a challenge for any study of protein or mRNA expression. This is especially difficult in studies of the brain, due to the highly regulated nature of neuronal tissue, as one does not expect to see large fold-changes during proteomic analyses. Nor is it expected to see a global shift in protein expression. Within the brain, for example, small fold-changes are often reported (Pawlyk et al., 2007; Pinaud et al., 2008), and have proven to be functionally significant (Sutton et al., 2003). At the same time, the large datasets generated by proteomic studies certainly contain Type I and Type II errors, especially with low magnitude changes. Therefore, as the number of neuroproteomic studies grows, it will be important to utilize analyses that will reduce the number of false positive findings, but will recognize small fold-changes that may be functionally significant within the brain region of interest. Power calculations of proteomic studies have been conducted on non-neuronal tissue (specifically for 2D-DIGE analysis) (Karp and Lilley, 2005); however, when examining brain tissue, these criteria may prove to be different. In addition, for statistical analysis of proteomic studies, multiple test corrections are often too conservative and may eliminate functionally important changes. Therefore, it is appropriate to draw from the microarray field, and use the combination of a low magnitude fold-change cut-off (e.g., 1.4) with a p-value statistical cut-off (p < 0.05 or 0.01) (Allison et al., 2006). This approach is better than complex p-value statistical analyses because it does not eliminate so many targets, but will still reduce the number of false positive findings. It is at this point that the importance of confirmation experiments is again evident. Confirmation of a differentially regulated protein by immunoblotting eliminates any lingering concerns.
6.2.3. Relationships of samples and groups to each other
Many approaches can be taken to understand what complex protein expression patterns tell us about relationships between proteins or experimental groups. A useful approach for examining how samples or groups relate to each other based on their respective proteomic profiles is to use principal component analysis (PCA). With this dimension reduction approach, the individual animals or groups can be clustered and the main effects of experimental treatments can be visualized. While PCA is not a predictive tool, it is useful to help visualize the overarching relationships. PCA has been used previously to visualize the effects of abstinence from cocaine self-administration on protein expression (Lull et al., 2009). Similarly, there are a number of different approaches to generating dendograms that provide another method for visualizing the similarity in the expression profile of different proteins, samples, or groups. There are also a number of clustering approaches (e.g., self-organizing maps, neural networks) that can cluster sets of proteins together by the similarity of their expression profiles. These more complex mathematical approaches are useful tools, but do not completely replace standard set functions such as intersection and union that can be used when comparing individual, pair-wise significant changes in a multigroup study.
6.2.4. Ontology, pathway and network analysis
To aid in placing proteomic findings in their biological context, there are a number of tools available. To accomplish this, methods initially applied in genomic studies can be applied to proteomic datasets, with some modifications. Ontological analysis determines relationships in protein data based on molecular or biological function of the proteins identified and may help to identify functions that are overrepresented within the changed proteins in a study. Common ontological categorizations are GO (Gene Ontology); (Ashburner et al., 2000), DAVID (Database for Annotation, Visualization and Integrated Discovery); (Dennis et al., 2003), Panther (Protein ANalysis THrough Evolutionary Relationships); (Thomas et al., 2003b), and KEGG (Kyoto Encyclopedia of Genes and Genomes); (Kanehisa and Goto, 2000). This type of analysis has been used for a number of different proteomic applications (Stevens et al., 2008; Nan et al., 2008). In the drug abuse field, a number of protein classes have been identified using this approach (Covarrubias et al., 2005; Rodd et al., 2008), including a proteomic study of cocaine abuse in which changes in neuronal structural proteins were found to be overrepresented following withdrawal from cocaine abuse (Lull et al., 2009).
While these categorization schemes are useful for obtaining a general picture of the proteins/changes identified, newer tools allow for the placement of protein data sets into functional pathways and networks. Pathway and network analyses, using programs such as Ingenuity (Ingenuity Systems, Redwood City, CA), GenMAPP (Gladstone Institutes—University of California, San Francisco), KEGG (Kanehisa Laboratories, Kyoto, Japan), and SPIKE (Signaling Pathway Integrated Knowledge Engine); (Elkon et al., 2008), aid in highlighting pathways and networks of proteins within the cell in which a number of members are changed. This type of analysis has been used to identify pathways and processes involved in cocaine (Lull et al., 2008, 2009), methamphetamine (Yang et al., 2008), and alcohol abuse (Hu et al., 2008).
The combined use of pathways and ontological analysis will also prove to be useful in the interface between multiple proteomic datasets, as well as mRNA and protein datasets. With the large number of studies being conducted using both large scale proteomic and microarray technology, it is important that similar studies can be compared. The meta-analysis of changes in gene and protein expression can be made by including multiple data sets within pathway or ontological analyses (Li et al., 2008). Specifically, the interface between mRNA and protein expression levels has been looked at in a number of cases (for a review, see Nie et al., 2007).
6.2.5. Data sharing
A wealth of information can be gathered from proteomic studies and it is important to standardize the reporting of such data, as well as making these data available to the public. Recently, standards have been developed for the reporting of proteomic experimental data. The development of Minimal Information Available about a Proteomics Experiment (MIAPE) standards has called for consistent reporting of the variables and methods by which proteomics experiments are conducted (Taylor et al., 2007; Robin et al., 2008). While giving insight into the ways in which proteomic experiments are conducted, this standard will also increase the reproducibility of the experiments that are reported in the literature, as well as allow for higher-order compilation and meta-analysis of similar data sets to be conducted, as seen in the microarray field (Jupiter and VanBuren, 2008).
In addition to these standards of how experiments are conducted, it is also important to make public the data that are collected. The NCBI database—Gene Expression Omnibus (GEO) was originally developed to be a public repository for gene expression data (Barrett et al., 2007). This makes gene expression data available to the public by hosting thousands of submissions regarding the abundance of species within cells. Other public repositories such as the Tranche (www.proteomecommons.org) and PRIDE databases (PRoteomics IDEntifications) have been developed to create a repository of all the protein identifications that are reported in the literature (Jones et al., 2006, 2008). These allow for protein identification and expression levels, location, and post-translational modifications to be uploaded to the database following submission to proteomics journals. In addition, the University of California, Santa Clara (UCSC) Known Genes database has allowed for the compilation of protein data from SwissProt (Uniprot) and associated mRNA data from GenBank (Hsu et al., 2006). Previously, we have applied this concept to drug abuse, basing the organization on neuroanatomical pathways (Freeman et al., 2002). As researchers continue to make available entire proteomic datasets after publication, they will be able to compile large datasets regarding specific biological questions that may lead to new and important discoveries about the roles of individual protein species in drug abuse disease processes.
7. Proteomic studies of drug abuse
There are few, if any, drug abuse studies that use all of the categories of tools discussed above. In fact, many are so new that the field of drug abuse has not yet adopted their use. A truly comprehensive proteomic analysis is a goal for the future, but this review is intended to direct drug abuse researchers to conduct complete proteomic studies that contribute significantly to the proteomic literature. There are a few examples, however, of studies that have used multiple proteomic approaches that help put the methods discussed above into perspective. In a 2D-DIGE study by Hemby and co-workers (Tannu et al., 2008), a cocaine self-administration non-human primate model was used. In this study, a small brain region was used (the nucleus accumbens) followed by cytosolic fractionation to look specifically at the soluble protein fraction. 2D-DIGE quantitation with MALDI-ToF/ToF mass spectrometry identification allowed for the identification of several proteins with altered expression profiles. In addition to this total-protein evaluation, examination of changes in protein phosphorylation were observed using Pro-Q diamond stain in a gel format. Protein identification was then examined by cellular function, taking advantage of bioinformatic methods to add value to lists of protein expression. What this study accomplishes is an examination of a self-administration model by a number of methods that focus on specific proteomes, an approach that is becoming increasingly common in the proteomic literature.
A complementary study conducted by Devi and co-workers (Moron et al., 2007) also takes advantage of sample fractionation by isolating the postsynaptic density. In contrast to the previously discussed study, this research used experimenter-administered doses of morphine in a mouse model of escalating doses. ICAT labeling of fractionated protein was used followed by liquid chromatography mass spectrometry identification and quantitation of protein species. In addition, a specific strength of this study is that Devi and co-workers undertook a series of immunoblots to validate ICAT-observed changes in protein expression. These two studies represent examples of how proteomic studies of drug abuse have been successfully conducted in the past. With all of the new tools available for sample preparation, sample analysis, and data analysis, among others, future studies will be capable of even more in-depth analyses of the drug abuse proteome.
8. Summary
As with other biomedical research fields, drug abuse research is making use of new proteomic capabilities to examine changes in protein expression and modification on a large scale. To obtain the maximum benefit and scientific advancement from these new technologies, a clear understanding of the power and limitations of neuroproteomics is necessary. With the main limitation of neuroproteomic studies being the complexity of the proteome, approaches that focus these studies need to be employed. The salient message is that there is no one best technical approach for all studies and that the principle driver of the choice of proteomic technology and experimental design should be the advancement of the understanding and treatment of drug abuse. This review has intended to provide guidance on how to determine the best manner in which to achieve the goals of a given study. This review also presents a number of experimental design and sample approaches that can be applied to neuroproteomic studies of addiction. Coupled with new technologies for data collection, analysis, and reporting, these approaches represent the future of the proteomic field and hold the key to unlocking the complex proteomic profile of drug abuse.
Acknowledgments
Role of funding source
Funding for this study was provided by NIH Grants R01DA013770-08 and R01AA016613-03 (KEV), and NRSA Grant F31-DA02281902 (MEL). The NIH and NRSA had no further role in study design; in the collection, analysis and interpretation of data; in the writing of the report; or in the decision to submit the paper for publication.
Footnotes
Conflict of interest
All authors declare that they have no conflicts of interest.
Contributors
Authors Lull and Vrana conceived the idea and outline for the manuscript. Author Lull was responsible for literature searches and preparation of a majority of the first draft. Authors VanGuilder and Freeman contributed to portions of the first draft, including development of figure artwork. Authors Lull and Freeman were responsible for a majority of the changes to the revised submission. Authors Lull, VanGuilder, Freeman, and Vrana participated in revisions of and additions to the first draft. All authors contributed to and have approved the final manuscript.
References
- Abul-Husn NS, Devi LA. Neuroproteomics of the synapse and drug addiction. J Pharmacol Exp Ther. 2006;318:461–468. doi: 10.1124/jpet.105.091520. [DOI] [PubMed] [Google Scholar]
- Aebersold R. A stress test for mass spectrometry-based proteomics. Nat Methods. 2009;6:411–412. doi: 10.1038/nmeth.f.255. [DOI] [PubMed] [Google Scholar]
- Al-Housseini AM, Sivanandam TM, Bradbury EL, Tannenberg RK, Dodd PR, Gu Q. Upregulation of beta-catenin levels in superior frontal cortex of chronic alcoholics. Alcohol Clin Exp Res. 2008;32:1080–1090. doi: 10.1111/j.1530-0277.2008.00670.x. [DOI] [PubMed] [Google Scholar]
- Allison DB, Cui X, Page GP, Sabripour M. Microarray data analysis: from disarray to consolidation and consensus. Nat Rev Genet. 2006;7:55–65. doi: 10.1038/nrg1749. [DOI] [PubMed] [Google Scholar]
- Anderson L, Hunter CL. Quantitative mass spectrometric multiple reaction monitoring assays for major plasma proteins. Mol Cell Proteomics. 2006;5:573–588. doi: 10.1074/mcp.M500331-MCP200. [DOI] [PubMed] [Google Scholar]
- Anderson L, Seilhamer J. A comparison of selected mRNA and protein abundances in human liver. Electrophoresis. 1997;18:533–537. doi: 10.1002/elps.1150180333. [DOI] [PubMed] [Google Scholar]
- Ashburner M, Ball CA, Blake JA, Botstein D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT, Harris MA, Hill DP, Issel-Tarver L, Kasarskis A, Lewis S, Matese JC, Richardson JE, Ringwald M, Rubin GM, Sherlock G. Gene ontology: tool for the unification of biology. The gene ontology consortium. Nat Genet. 2000;25:25–29. doi: 10.1038/75556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barrett T, Troup DB, Wilhite SE, Ledoux P, Rudnev D, Evangelista C, Kim IF, Soboleva A, Tomashevsky M, Edgar R. NCBI GEO: mining tens of millions of expression profiles—database and tools update. Nucleic Acids Res. 2007;35:D760–D765. doi: 10.1093/nar/gkl887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barthelery M, Jaishankar A, Salli U, Freeman WM, Vrana KE. 2D-DIGE indentification of differentially expressed HNRNPs and trancription factors during neural differentiation of human ES cells. Proteomics Clin Appl. 2009;3:505–514. doi: 10.1002/prca.200800109. [DOI] [PubMed] [Google Scholar]
- Bateup HS, Svenningsson P, Kuroiwa M, Gong S, Nishi A, Heintz N, Green-gard P. Cell type-specific regulation of DARPP-32 phosphorylation by psychostimulant and antipsychotic drugs. Nat Neurosci. 2008;11:932–939. doi: 10.1038/nn.2153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker JS, Becker JS, Zoriy MV, Dobrowolska J, Matucsh A. Imaging mass spectrometry in biological tissues by laser ablation inductively coupled plasma mass spectrometry. Eur J Mass Spectrom (Chichester, Eng) 2007;13:1–6. doi: 10.1255/ejms.833. [DOI] [PubMed] [Google Scholar]
- Bell AW, Deutsch EW, Au CE, Kearney RE, Beavis R, Sechi S, Nilsson T, Bergeron JJ, Beardslee TA, Chappell T, Meredith G, Sheffield P, Gray P, Hajivandi M, Pope M, Predki P, Kullolli M, Hincapie M, Hancock WS, Jia W, Song L, Li L, Wei J, Yang B, Wang J, Ying W, Zhang Y, Cai Y, Qian X, He F, Meyer HE, Stephan C, Eisenacher M, Marcus K, Langenfeld E, May C, Carr SA, Ahmad R, Zhu W, Smith JW, Hanash SM, Struthers JJ, Wang H, Zhang Q, An Y, Goldman R, Carlsohn E, van der PS, Hung KE, Sarracino DA, Parker K, Krastins B, Kucherlapati R, Bourassa S, Poirier GG, Kapp E, Patsiouras H, Moritz R, Simpson R, Houle B, Laboissiere S, Metalnikov P, Nguyen V, Pawson T, Wong CC, Cociorva D, Yates I, Jr, Ellison MJ, Lopez-Campistrous A, Semchuk P, Wang Y, Ping P, Elia G, Dunn MJ, Wynne K, Walker AK, Strahler JR, Andrews PC, Hood BL, Bigbee WL, Conrads TP, Smith D, Borchers CH, Lajoie GA, Bendall SC, Speicher KD, Speicher DW, Fujimoto M, Nakamura K, Paik YK, Cho SY, Kwon MS, Lee HJ, Jeong SK, Chung AS, Miller CA, Grimm R, Williams K, Dorschel C, Falkner JA, Martens L, Vizcaino JA. A HUPO test sample study reveals common problems in mass spectrometry-based proteomics. Nat Methods. 2009;6:423–430. doi: 10.1038/nmeth.1333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calu DJ, Stalnaker TA, Franz TM, Singh T, Shaham Y, Schoenbaum G. Withdrawal from cocaine self-administration produces long-lasting deficits in orbitofrontal-dependent reversal learning in rats. Learn Mem. 2007;14:325–328. doi: 10.1101/lm.534807. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caprioli RM, Farmer TB, Gile J. Molecular imaging of biological samples: localization of peptides and proteins using MALDI-TOF MS. Anal Chem. 1997;69:4751–4760. doi: 10.1021/ac970888i. [DOI] [PubMed] [Google Scholar]
- Che FY, Lim J, Pan H, Biswas R, Fricker LD. Quantitative neuropeptidomics of microwave-irradiated mouse brain and pituitary. Mol Cell Proteomics. 2005;4:1391–1405. doi: 10.1074/mcp.T500010-MCP200. [DOI] [PubMed] [Google Scholar]
- Chen P, Li X, Sun Y, Liu Z, Cao R, He Q, Wang M, Xiong J, Xie J, Wang X, Liang S. Proteomic analysis of rat hippocampal plasma membrane: characterization of potential neuronal-specific plasma membrane proteins. J Neurochem. 2006;98:1126–1140. doi: 10.1111/j.1471-4159.2006.03934.x. [DOI] [PubMed] [Google Scholar]
- Chen X, Sun L, Yu Y, Xue Y, Yang P. Amino acid-coded tagging approaches in quantitative proteomics. Expert Rev Proteomics. 2007;4:25–37. doi: 10.1586/14789450.4.1.25. [DOI] [PubMed] [Google Scholar]
- Chin MH, Geng AB, Khan AH, Qian WJ, Petyuk VA, Boline J, Levy S, Toga AW, Smith RD, Leahy RM, Smith DJ. A genome-scale map of expression for a mouse brain section obtained using voxelation. Physiol Genomics. 2007;30:313–321. doi: 10.1152/physiolgenomics.00287.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chin MH, Qian WJ, Wang H, Petyuk VA, Bloom JS, Sforza DM, Lacan G, Liu D, Khan AH, Cantor RM, Bigelow DJ, Melega WP, Camp DG, Smith RD, Smith DJ. Mitochondrial dysfunction, oxidative stress, and apoptosis revealed by proteomic and transcriptomic analyses of the striata in two mouse models of Parkinson’s disease. J Proteome Res. 2008;7:666–677. doi: 10.1021/pr070546l. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Collins MO, Yu L, Campuzano I, Grant SG, Choudhary JS. Phosphoproteomic analysis of the mouse brain cytosol reveals a predominance of protein phosphorylation in regions of intrinsic sequence disorder. Mol Cell Proteomics. 2008;7:1331–1348. doi: 10.1074/mcp.M700564-MCP200. [DOI] [PubMed] [Google Scholar]
- Cornish JL, Kalivas PW. Repeated cocaine administration into the rat ventral tegmental area produces behavioral sensitization to a systemic cocaine challenge. Behav Brain Res. 2001;126:205–209. doi: 10.1016/s0166-4328(01)00239-x. [DOI] [PubMed] [Google Scholar]
- Covarrubias MY, Khan RL, Vadigepalli R, Hoek JB, Schwaber JS. Chronic alcohol exposure alters transcription broadly in a key integrative brain nucleus for homeostasis: the nucleus tractus solitarius. Physiol Genomics. 2005;24:45–58. doi: 10.1152/physiolgenomics.00184.2005. [DOI] [PubMed] [Google Scholar]
- Cunha-Oliveira T, Rego AC, Oliveira CR. Cellular and molecular mechanisms involved in the neurotoxicity of opioid and psychostimulant drugs. Brain Res Rev. 2008;58:192–208. doi: 10.1016/j.brainresrev.2008.03.002. [DOI] [PubMed] [Google Scholar]
- D’Ambrosio C, Arena S, Fulcoli G, Scheinfeld MH, Zhou D, D’Adamio L, Scaloni A. Hyperphosphorylation of JNK-interacting protein 1, a protein associated with Alzheimer disease. Mol Cell Proteomics. 2006;5:97–113. doi: 10.1074/mcp.M500226-MCP200. [DOI] [PubMed] [Google Scholar]
- Dennis G, Sherman BT, Hosack DA, Yang J, Gao W, Lane HC, Lempicki RA. DAVID: database for annotation, visualization, and integrated discovery. Genome Biol. 2003;4:3. [PubMed] [Google Scholar]
- DeSouza LV, Taylor AM, Li W, Minkoff MS, Romaschin AD, Colgan TJ, Siu KW. Multiple reaction monitoring of mTRAQ-labeled peptides enables absolute quantification of endogenous levels of a potential cancer marker in cancerous and normal endometrial tissues. J Proteome Res. 2008;7:3525–3534. doi: 10.1021/pr800312m. [DOI] [PubMed] [Google Scholar]
- Di CP, Blaha CD, Phillips AG. Changes in dopamine oxidation currents in the nucleus accumbens during unlimited-access self-administration of d-amphetamine by rats. Behav Pharmacol. 1996;7:714–729. doi: 10.1097/00008877-199611000-00016. [DOI] [PubMed] [Google Scholar]
- Di CP, Blaha CD, Phillips AG. The relation between dopamine oxidation currents in the nucleus accumbens and conditioned increases in motor activity in rats following repeated administration of d-amphetamine or cocaine. Eur J Neurosci. 1998;10:1113–1120. doi: 10.1046/j.1460-9568.1998.00124.x. [DOI] [PubMed] [Google Scholar]
- Dobretsova A, Johnson JW, Jones RC, Edmondson RD, Wight PA. Proteomic analysis of nuclear factors binding to an intronic enhancer in the myelin proteolipid protein gene. J Neurochem. 2008;105:1979–1995. doi: 10.1111/j.1471-4159.2008.05288.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Doyle JP, Dougherty JD, Heiman M, Schmidt EF, Stevens TR, Ma G, Bupp S, Shrestha P, Shah RD, Doughty ML, Gong S, Greengard P, Heintz N. Application of a translational profiling approach for the comparative analysis of CNS cell types. Cell. 2008;135:749–762. doi: 10.1016/j.cell.2008.10.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dworkin SI, Co C, Smith JE. Rat brain neurotransmitter turnover rates altered during withdrawal from chronic cocaine administration. Brain Res. 1995;682:116–126. doi: 10.1016/0006-8993(95)00327-m. [DOI] [PubMed] [Google Scholar]
- Elkon R, Vesterman R, Amit N, Ulitsky I, Zohar I, Weisz M, Mass G, Orlev N, Sternberg G, Blekhman R, Assa J, Shiloh Y, Shamir R. SPIKE—a database, visualization and analysis tool of cellular signaling pathways. BMC Bioinform. 2008;9:110. doi: 10.1186/1471-2105-9-110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Epstein DH, Preston KL, Stewart J, Shaham Y. Toward a model of drug relapse: an assessment of the validity of the reinstatement procedure. Pschopharmacology (Berl) 2006;189:1–16. doi: 10.1007/s00213-006-0529-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ersche KD, Fletcher PC, Roiser JP, Fryer TD, London M, Robbins TW, Sahakian BJ. Differences in orbitofrontal activation during decision-making between methadone-maintained opiate users, heroin users and healthy volunteers. Psychopharmacology (Berl) 2006;188:364–373. doi: 10.1007/s00213-006-0515-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyerman DJ, Yamamoto BK. A rapid oxidation and persistent decrease in the vesicular monoamine transporter 2 after methamphetamine. J Neurochem. 2007;103:1219–1227. doi: 10.1111/j.1471-4159.2007.04837.x. [DOI] [PubMed] [Google Scholar]
- Fan AC, Deb-Basu D, Orban MW, Gotlib JR, Natkunam Y, O’Neill R, Padua RA, Xu L, Taketa D, Shirer AE, Beer S, Yee AX, Voehringer DW, Felsher DW. Nanofluidic proteomic assay for serial analysis of oncoprotein activation in clinical specimens. Nat Med. 2009;15:566–571. doi: 10.1038/nm.1903. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Finlay EM, Games DE, Startin JR, Gilbert J. Screening, confirmation, and quantification of sulphonamide residues in pig kidney by tandem mass spectrometry of crude extracts. Biomed Environ Mass Spectrom. 1986;13:633–639. doi: 10.1002/bms.1200131109. [DOI] [PubMed] [Google Scholar]
- Freeman WM, Brebner K, Amara SG, Reed MS, Pohl J, Phillips AG. Distinct proteomic profiles of amphetamine self-administration transitional states. Pharmacogenomics J. 2005;5:203–214. doi: 10.1038/sj.tpj.6500309. [DOI] [PubMed] [Google Scholar]
- Freeman WM, Dougherty KE, Vacca SE, Vrana KE. An interactive database of cocaine-responsive gene expression. Sci World J. 2002;12:701–706. doi: 10.1100/tsw.2002.145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman WM, Gooch RS, Lull ME, Worst TJ, Walker SJ, Xu AS, Green H, Pierre PJ, Grant KA, Vrana KE. Apo-AII is an elevated biomarker of chronic non-human primate ethanol self-administration. Alcohol Alcohol. 2006;41:300–305. doi: 10.1093/alcalc/agl021. [DOI] [PubMed] [Google Scholar]
- Freeman WM, Hemby SE. Proteomics for protein expression profiling in neuroscience. Neurochem Res. 2004;29:1065–1081. doi: 10.1023/b:nere.0000023594.21352.17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Freeman WM, Vrana KE. In: Quantitative functionalgenomics and proteomics of drug abuse. Madras BK, Rutter JL, Colvis CM, Shurtleff JD, et al., editors. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 2006. pp. 433–456. [Google Scholar]
- Gao X, Chen J. Direct isolation of neural stem cells in the adult hippocampus after traumatic brain injury. J Neurotrauma. 2008;25:985–995. doi: 10.1089/neu.2008.0460. [DOI] [PubMed] [Google Scholar]
- Garzon J, Rodriguez-Diaz M, Lopez-Fando A, Garcia-Espana A, Sanchez-Blazquez P. Glycosylated phosducin-like protein long regulates opioid receptor function in mouse brain. Neuropharmacology. 2002;42:813–828. doi: 10.1016/s0028-3908(02)00027-8. [DOI] [PubMed] [Google Scholar]
- Gawin FH, Kleber HD. Abstinence symptomatology and psychiatric diagnosis in cocaine abusers. Clinical observations. Arch Gen Psychiatry. 1986;43:107–113. doi: 10.1001/archpsyc.1986.01800020013003. [DOI] [PubMed] [Google Scholar]
- Gerber SA, Rush J, Stemman O, Kirschner MW, Gygi SP. Absolute quantification of proteins and phosphoproteins from cell lysates by tandem MS. Proc Natl Acad Sci USA. 2003;100:6940–6945. doi: 10.1073/pnas.0832254100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Graziella D, Montis M, Co C, Dworkin SI, Smith JE. Modifications of dopamine D1 receptor complex in rats self-administering cocaine. Eur J Pharmacol. 1998;362:9–15. doi: 10.1016/s0014-2999(98)00731-6. [DOI] [PubMed] [Google Scholar]
- Grimm JW, Hope BT, Wise RA, Shaham Y. Neuroadaptation. Incubation of cocaine craving after withdrawal. Nature. 2001;412:141–142. doi: 10.1038/35084134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guevara R, Santandreu FM, Valle A, Gianotti M, Oliver J, Roca P. Sex-dependent differences in aged rat brain mitochondrial function and oxidative stress. Free Radic Biol Med. 2008;46:169–175. doi: 10.1016/j.freeradbiomed.2008.09.035. [DOI] [PubMed] [Google Scholar]
- Hale EA, Raza SK, Ciecierski RG, Ghosh P. Deleterious actions of chronic ethanol treatment on the glycosylation of rat brain clusterin. Brain Res. 1998;785:158–166. doi: 10.1016/s0006-8993(97)01397-8. [DOI] [PubMed] [Google Scholar]
- Han BS, Iacovitti L, Katano T, Hattori N, Seol W, Kim KS. Expression of the LRRK2 gene in the midbrain dopaminergic neurons of the substantia nigra. Neurosci Lett. 2008;442:190–194. doi: 10.1016/j.neulet.2008.06.086. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hemby SE. Assessment of genome and proteome profiles in cocaine abuse. Prog Brain Res. 2006;158:173–195. doi: 10.1016/S0079-6123(06)58009-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horth P, Miller CA, Preckel T, Wenz C. Efficient fractionation and improved protein identification by peptide OFFGEL electrophoresis. Mol Cell Proteomics. 2006;5:1968–1974. doi: 10.1074/mcp.T600037-MCP200. [DOI] [PubMed] [Google Scholar]
- Hsu F, Kent WJ, Clawson H, Kuhn RM, Diekhans M, Haussler D. The UCSC known genes. Bioinformatics. 2006;22:1036–1046. doi: 10.1093/bioinformatics/btl048. [DOI] [PubMed] [Google Scholar]
- Hu W, Saba L, Kechris K, Bhave SV, Hoffman PL, Tabakoff B. Genomic insights into acute alcohol tolerance. J Pharmacol Exp Ther. 2008;326:792–800. doi: 10.1124/jpet.108.137521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunsucker SW, Solomon B, Gawryluk J, Geiger JD, Vacano GN, Duncan MW, Patterson D. Assessment of post-mortem-induced changes to the mouse brain proteome. J Neurochem. 2008;105:725–737. doi: 10.1111/j.1471-4159.2007.05183.x. [DOI] [PubMed] [Google Scholar]
- Hynd MR, Lewohl JM, Scott HL, Dodd PR. Biochemical and molecular studies using human autopsy brain tissue. J Neurochem. 2003;85:543–562. doi: 10.1046/j.1471-4159.2003.01747.x. [DOI] [PubMed] [Google Scholar]
- Jacobs EH, de Vries TJ, Smit AB, Schoffelmeer AN. Gene transcripts selectively down-regulated in the shell of the nucleus accumbens long after heroin self-administration are up-regulated in the core independent of response contingency. FASEB J. 2004;18:200–202. doi: 10.1096/fj.03-0317fje. [DOI] [PubMed] [Google Scholar]
- Jaishankar A, Barthelery M, Freeman WM, Salli U, Ritty T, Vrana K. Human embryonic and mesenchymal stem cells express different nuclear proteomes. Stem Cells Dev. 2008;18:793–802. doi: 10.1089/scd.2008.0156. [DOI] [PubMed] [Google Scholar]
- Jones P, Cote RG, Cho SY, Klie S, Martens L, Quinn AF, Thorneycroft D, Hermjakob H. PRIDE: new developments and new datasets. Nucleic Acids Res. 2008;36:D878–D883. doi: 10.1093/nar/gkm1021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones P, Cote RG, Martens L, Quinn AF, Taylor CF, Derache W, Hermjakob H, Apweiler R. PRIDE: a public repository of protein and peptide identifications for the proteomics community. Nucleic Acids Res. 2006;34:D659–D663. doi: 10.1093/nar/gkj138. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jovanovski D, Erb S, Zakzanis KK. Neurocognitive deficits in cocaine users: a quantitative review of the evidence. J Clin Exp Neuropsychol. 2005;27:189–204. doi: 10.1080/13803390490515694. [DOI] [PubMed] [Google Scholar]
- Jupiter DC, VanBuren V. A visual data mining tool that facilitates reconstruction of transcription regulatory networks. PLoS ONE. 2008;3:e1717. doi: 10.1371/journal.pone.0001717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kalivas PW, Peters J, Knackstedt L. Animal models and brain circuits in drug addiction. Mol Interv. 2006;6:339–344. doi: 10.1124/mi.6.6.7. [DOI] [PubMed] [Google Scholar]
- Kanehisa M, Goto S. KEGG: kyoto encyclopedia of genes and genomes. Nucleic Acids Res. 2000;28:27–30. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karp NA, Lilley KS. Maximising sensitivity for detecting changes in protein expression: experimental design using minimal CyDyes. Proteomics. 2005;5:3105–3115. doi: 10.1002/pmic.200500083. [DOI] [PubMed] [Google Scholar]
- Kashem MA, Harper C, Matsumoto I. Differential protein expression in the corpus callosum (genu) of human alcoholics. Neurochem Int. 2008;53:1–11. doi: 10.1016/j.neuint.2008.04.003. [DOI] [PubMed] [Google Scholar]
- Kashem MA, James G, Harper C, Wilce P, Matsumoto I. Differential protein expression in the corpus callosum (splenium) of human alcoholics: a proteomics study. Neurochem Int. 2007;50:450–459. doi: 10.1016/j.neuint.2006.10.009. [DOI] [PubMed] [Google Scholar]
- Khidekel N, Ficarro SB, Clark PM, Bryan MC, Swaney DL, Rexach JE, Sun YE, Coon JJ, Peters EC, Hsieh-Wilson LC. Probing the dynamics of O-GlcNAc glycosylation in the brain using quantitative proteomics. Nat Chem Biol. 2007;3:339–348. doi: 10.1038/nchembio881. [DOI] [PubMed] [Google Scholar]
- Krapfenbauer K, Fountoulakis M, Lubec G. A rat brain protein expression map including cytosolic and enriched mitochondrial and microsomal fractions. Electrophoresis. 2003;24:1847–1870. doi: 10.1002/elps.200305401. [DOI] [PubMed] [Google Scholar]
- Kruger M, Kratchmarova I, Blagoev B, Tseng YH, Kahn CR, Mann M. Dissection of the insulin signaling pathway via quantitative phosphoproteomics. Proc Natl Acad Sci USA. 2008;105:2451–2456. doi: 10.1073/pnas.0711713105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuntz KL, Twining RC, Baldwin AE, Vrana KE, Grigson PS. Heroin self-administration: I. Incubation of goal-directed behavior in rats. Pharmacol Biochem Behav. 2008;90:344–348. doi: 10.1016/j.pbb.2008.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kuzmin A, Johansson B. Expression of c-fos, NGFI-A and secretogranin II mRNA in brain regions during initiation of cocaine self-administration in mice. Eur J Neurosci. 1999;11:3694–3700. doi: 10.1046/j.1460-9568.1999.00795.x. [DOI] [PubMed] [Google Scholar]
- Lan W, Guhaniyogi J, Horn MJ, Xia JQ, Graham B. A density-based proteomics sample fractionation technology: folate deficiency induced oxidative stress response in liver and brain. J Biomol Tech. 2007;18:213–225. [PMC free article] [PubMed] [Google Scholar]
- Layfield R, Tooth D, Landon M, Dawson S, Mayer J, Alban A. Purification of poly-ubiquitinated proteins by S5a-affinity chromatography. Proteomics. 2001;1:773–777. doi: 10.1002/1615-9861(200106)1:6<773::AID-PROT773>3.0.CO;2-0. [DOI] [PubMed] [Google Scholar]
- Lehrmann E, Afanador ZR, ep-Soboslay A, Gallegos G, Darwin WD, Lowe RH, Barnes AJ, Huestis MA, Cadet JL, Herman MM, Hyde TM, Kleinman JE, Freed WJ. Postmortem diagnosis and toxicological validation of illicit substance use. Addict Biol. 2008;13:105–117. doi: 10.1111/j.1369-1600.2007.00085.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li CY, Mao X, Wei L. Genes and (common) pathways underlying drug addiction. PLoS Comput Biol. 2008;4:e2. doi: 10.1371/journal.pcbi.0040002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li KW, Jimenez CR, van der Schors RC, Hornshaw MP, Schoffelmeer AN, Smit AB. Intermittent administration of morphine alters protein expression in rat nucleus accumbens. Proteomics. 2006;6:2003–2008. doi: 10.1002/pmic.200500045. [DOI] [PubMed] [Google Scholar]
- Liao L, Cheng D, Wang J, Duong DM, Losik TG, Gearing M, Rees HD, Lah JJ, Levey AI, Peng J. Proteomic characterization of postmortem amyloid plaques isolated by laser capture microdissection. J Biol Chem. 2004;279:37061–37068. doi: 10.1074/jbc.M403672200. [DOI] [PubMed] [Google Scholar]
- Lull ME, Erwin M, Morgan D, Roberts DC, Vrana KE, Freeman WM. Persistent proteomic alterations in the medial prefrontal cortex with abstinence from cocaine self-administration. Proteomics Clin Appl. 2009;3:462–472. doi: 10.1002/prca.200800055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lull ME, Freeman WM, Vrana KE, Mash DC. Correlating human and animal studies of cocaine abuse and gene expression. Ann NY Acad Sci. 2008;1141:58–75. doi: 10.1196/annals.1441.013. [DOI] [PubMed] [Google Scholar]
- Mark GP, Hajnal A, Kinney AE, Keys AS. Self-administration of cocaine increases the release of acetylcholine to a greater extent than response-independent cocaine in the nucleus accumbens of rats. Psychopharmacology (Berl) 1999;143:47–53. doi: 10.1007/s002130050918. [DOI] [PubMed] [Google Scholar]
- Martin B, Brenneman R, Becker KG, Gucek M, Cole RN, Maudsley S. iTRAQ analysis of complex proteome alterations in 3xTgAD Alzheimer’s mice: understanding the interface between physiology and disease. PLoS ONE. 2008;3:e2750. doi: 10.1371/journal.pone.0002750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsuda-Matsumoto H, Iwazaki T, Kashem MA, Harper C, Matsumoto I. Differential protein expression profiles in the hippocampus of human alcoholics. Neurochem Int. 2007;51:370–376. doi: 10.1016/j.neuint.2007.04.001. [DOI] [PubMed] [Google Scholar]
- Meany DL, Xie H, Thompson LV, Arriaga EA, Griffin TJ. Identification of carbonylated proteins from enriched rat skeletal muscle mitochondria using affinity chromatography-stable isotope labeling and tandem mass spectrometry. Proteomics. 2007;7:1150–1163. doi: 10.1002/pmic.200600450. [DOI] [PubMed] [Google Scholar]
- Miserendino MJ, Nestler EJ. Behavioral sensitization to cocaine: modulation by the cyclic AMP system in the nucleus accumbens. Brain Res. 1995;674:299–306. doi: 10.1016/0006-8993(95)00030-t. [DOI] [PubMed] [Google Scholar]
- Monzo A, Bonn GK, Guttman A. Boronic acid-lectin affinity chromatography. 1 Simultaneous glycoprotein binding with selective or combined elution. Anal Bioanal Chem. 2007;389:2097–2102. doi: 10.1007/s00216-007-1627-y. [DOI] [PubMed] [Google Scholar]
- Morelle W, Michalski JC. Glycomics and mass spectrometry. Curr Pharm Des. 2005;11:2615–2645. doi: 10.2174/1381612054546897. [DOI] [PubMed] [Google Scholar]
- Morgan D, Brebner K, Lynch WJ, Roberts DC. Increases in the reinforcing efficacy of cocaine after particular histories of reinforcement. Behav Pharmacol. 2002;13:389–396. doi: 10.1097/00008877-200209000-00012. [DOI] [PubMed] [Google Scholar]
- Morgan D, Roberts DC. Sensitization to the reinforcing effects of cocaine following binge-abstinent self-administration. Neurosci Biobehav Rev. 2004;27:803–812. doi: 10.1016/j.neubiorev.2003.11.004. [DOI] [PubMed] [Google Scholar]
- Moron JA, Abul-Husn NS, Rozenfeld R, Dolios G, Wang R, Devi LA. Morphine administration alters the profile of hippocampal postsynaptic density-associated proteins: a proteomics study focusing on endocytic proteins. Mol Cell Proteomics. 2007;6:29–42. doi: 10.1074/mcp.M600184-MCP200. [DOI] [PubMed] [Google Scholar]
- Mouledous L, Hunt S, Harcourt R, Harry J, Williams KL, Gutstein HB. Navigated laser capture microdissection as an alternative to direct histological staining for proteomic analysis of brain samples. Proteomics. 2003a;3:610–615. doi: 10.1002/pmic.200300398. [DOI] [PubMed] [Google Scholar]
- Mouledous L, Hunt S, Harcourt R, Harry JL, Williams KL, Gutstein HB. Proteomic analysis of immunostained, laser-capture microdissected brain samples. Electrophoresis. 2003b;24:296–302. doi: 10.1002/elps.200390026. [DOI] [PubMed] [Google Scholar]
- Mueller LN, Brusniak MY, Mani DR, Aebersold R. An assessment of software solutions for the analysis of mass spectrometry based quantitative proteomics data. J Proteome Res. 2008;7:51–61. doi: 10.1021/pr700758r. [DOI] [PubMed] [Google Scholar]
- Mutschler NH, Miczek KA, Hammer RP., Jr Reduction of zif268 messenger RNA expression during prolonged withdrawal following “binge” cocaine self-administration in rats. Neuroscience. 2000;100:531–538. doi: 10.1016/s0306-4522(00)00298-0. [DOI] [PubMed] [Google Scholar]
- Nan Y, Yang S, Tian Y, Zhang W, Zhou B, Bu L, Huo S. Analysis of the expression protein profiles of lung squamous carcinoma cell using shot-gun proteomics strategy. Med Oncol. 2008;26:215–221. doi: 10.1007/s12032-008-9109-4. [DOI] [PubMed] [Google Scholar]
- Nelson PG, Kuddo T, Song EY, Dambrosia JM, Kohler S, Satyanarayana G, Vandunk C, Grether JK, Nelson KB. Selected neurotrophins, neuropeptides, and cytokines: developmental trajectory and concentrations in neonatal blood of children with autism or Down syndrome. Int J Dev Neurosci. 2006;24:73–80. doi: 10.1016/j.ijdevneu.2005.10.003. [DOI] [PubMed] [Google Scholar]
- Nie L, Wu G, Culley DE, Scholten JC, Zhang W. Integrative analysis of transcriptomic and proteomic data: challenges, solutions and applications. Crit Rev Biotechnol. 2007;27:63–75. doi: 10.1080/07388550701334212. [DOI] [PubMed] [Google Scholar]
- Nielsen PA, Olsen JV, Podtelejnikov AV, Andersen JR, Mann M, Wisniewski JR. Proteomic mapping of brain plasma membrane proteins. Mol Cell Proteomics. 2005;4:402–408. doi: 10.1074/mcp.T500002-MCP200. [DOI] [PubMed] [Google Scholar]
- O’Brien CP. A range of research-based pharmacotherapies for addiction. Science. 1997;278:66–70. doi: 10.1126/science.278.5335.66. [DOI] [PubMed] [Google Scholar]
- O’Callaghan JP, Sriram K. Focused microwave irradiation of the brain preserves in vivo protein phosphorylation: comparison with other methods of sacrifice and analysis of multiple phosphoproteins. J Neurosci Methods. 2004;135:159–168. doi: 10.1016/j.jneumeth.2003.12.006. [DOI] [PubMed] [Google Scholar]
- Olsen JV, Mann M. Improved peptide identification in proteomics by two consecutive stages of mass spectrometric fragmentation. Proc Natl Acad Sci USA. 2004;101:13417–13422. doi: 10.1073/pnas.0405549101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olsen JV, Nielsen PA, Andersen JR, Mann M, Wisniewski JR. Quantitative proteomic profiling of membrane proteins from the mouse brain cortex, hippocampus, and cerebellum using the HysTag reagent: mapping of neurotransmitter receptors and ion channels. Brain Res. 2007;1134:95–106. doi: 10.1016/j.brainres.2006.11.082. [DOI] [PubMed] [Google Scholar]
- Pawlyk AC, Ferber M, Shah A, Pack AI, Naidoo N. Proteomic analysis of the effects and interactions of sleep deprivation and aging in mouse cerebral cortex. J Neurochem. 2007;103:2301–2313. doi: 10.1111/j.1471-4159.2007.04949.x. [DOI] [PubMed] [Google Scholar]
- Paxinos G, Watson C. The Rat Brain in Stereotaxic Coordinates. 5. Academic Press; Burlington, MA: 2005. [Google Scholar]
- Phillips WH, Ota K, Wade NA. Tandem mass spectrometry (MS/MS) utilizing electron impact ionization and multiple reaction monitoring for the rapid, sensitive, and specific identification and quantitation of morphine in whole blood. J Anal Toxicol. 1989;13:268–273. doi: 10.1093/jat/13.5.268. [DOI] [PubMed] [Google Scholar]
- Pinaud R, Osorio C, Alzate O, Jarvis ED. Profiling of experience-regulated proteins in the songbird auditory forebrain using quantitative proteomics. Eur J Neurosci. 2008;27:1409–1422. doi: 10.1111/j.1460-9568.2008.06102.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prokai L, Zharikova AD, Stevens SM., Jr Effect of chronic morphine exposure on the synaptic plasma-membrane subproteome of rats: a quantitative protein profiling study based on isotope-coded affinity tags and liquid chromatography/mass spectrometry. J Mass Spectrom. 2005;40:169–175. doi: 10.1002/jms.736. [DOI] [PubMed] [Google Scholar]
- Qi ML, Tagawa K, Enokido Y, Yoshimura N, Wada Y, Watase K, Ishiura S, Kanazawa I, Botas J, Saitoe M, Wanker EE, Okazawa H. Proteome analysis of soluble nuclear proteins reveals that HMGB1/2 suppress genotoxic stress in polyglutamine diseases. Nat Cell Biol. 2007;9:402–414. doi: 10.1038/ncb1553. [DOI] [PubMed] [Google Scholar]
- Qin Y, Ouyang Q, Pablo J, Mash DC. Cocaine abuse elevates alpha-synuclein and dopamine transporter levels in the human striatum. Neuroreport. 2005;16:1489–1493. doi: 10.1097/01.wnr.0000175617.39054.ba. [DOI] [PubMed] [Google Scholar]
- Roberts DC, Morgan D, Liu Y. How to make a rat addicted to cocaine. Prog Neuropsychopharmacol Biol Psychiatry. 2007;31:1614–1624. doi: 10.1016/j.pnpbp.2007.08.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Robin X, Hoogland C, Appel RD, Lisacek F. MIAPEGelDB, a web-based submission tool and public repository for MIAPE gel electrophoresis documents. J Proteomics. 2008;71:249–251. doi: 10.1016/j.jprot.2008.06.005. [DOI] [PubMed] [Google Scholar]
- Robinson TE, Kolb B. Structural plasticity associated with exposure to drugs of abuse. Neuropharmacology. 2004;47 (suppl 1):33–46. doi: 10.1016/j.neuropharm.2004.06.025. [DOI] [PubMed] [Google Scholar]
- Rodd ZA, Kimpel MW, Edenberg HJ, Bell RL, Strother WN, McClintick JN, Carr LG, Liang T, McBride WJ. Differential gene expression in the nucleus accumbens with ethanol self-administration in inbred alcohol-preferring rats. Pharmacol Biochem Behav. 2008;89:481–498. doi: 10.1016/j.pbb.2008.01.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rubakhin SS, Sweedler JV. Characterizing peptides in individual mammalian cells using mass spectrometry. Nat Protoc. 2007;2:1987–1997. doi: 10.1038/nprot.2007.277. [DOI] [PubMed] [Google Scholar]
- Rubakhin SS, Sweedler JV. Quantitative measurements of cell-cell signaling peptides with single-cell MALDI MS. Anal Chem. 2008;80:7128–7136. doi: 10.1021/ac8010389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salim K, Guest PC, Skynner HA, Bilsland JG, Bonnert TP, McAllister G, Munoz-Sanjuan I. Identification of proteomic changes during differentiation of adult mouse subventricular zone progenitor cells. Stem Cells Dev. 2007;16:143–165. doi: 10.1089/scd.2006.00100. [DOI] [PubMed] [Google Scholar]
- Schindler J, Lewandrowski U, Sickmann A, Friauf E, Nothwang HG. Proteomic analysis of brain plasma membranes isolated by affinity two-phase partitioning. Mol Cell Proteomics. 2006;5:390–400. doi: 10.1074/mcp.T500017-MCP200. [DOI] [PubMed] [Google Scholar]
- Schoenbaum G, Saddoris MP, Ramus SJ, Shaham Y, Setlow B. Cocaine-experienced rats exhibit learning deficits in a task sensitive to orbitofrontal cortex lesions. Eur J Neurosci. 2004;19:1997–2002. doi: 10.1111/j.1460-9568.2004.03274.x. [DOI] [PubMed] [Google Scholar]
- Self DW, McClenahan AW, Beitner-Johnson D, Terwilliger RZ, Nestler EJ. Biochemical adaptations in the mesolimbic dopamine system in response to heroin self-administration. Synapse. 1995;21:312–318. doi: 10.1002/syn.890210405. [DOI] [PubMed] [Google Scholar]
- Shi L, Reid LH, Jones WD, Shippy R, Warrington JA, Baker SC, Collins PJ, de LF, Kawasaki ES, Lee KY, Luo Y, Sun YA, Willey JC, Setterquist RA, Fischer GM, Tong W, Dragan YP, Dix DJ, Frueh FW, Goodsaid FM, Herman D, Jensen RV, Johnson CD, Lobenhofer EK, Puri RK, Schrf U, Thierry-Mieg J, Wang C, Wilson M, Wolber PK, Zhang L, Amur S, Bao W, Barbacioru CC, Lucas AB, Bertholet V, Boysen C, Bromley B, Brown D, Brunner A, Canales R, Cao XM, Cebula TA, Chen JJ, Cheng J, Chu TM, Chudin E, Corson J, Corton JC, Croner LJ, Davies C, Davison TS, Delenstarr G, Deng X, Dorris D, Eklund AC, Fan XH, Fang H, Fulmer-Smentek S, Fuscoe JC, Gallagher K, Ge W, Guo L, Guo X, Hager J, Haje PK, Han J, Han T, Harbottle HC, Harris SC, Hatchwell E, Hauser CA, Hester S, Hong H, Hurban P, Jackson SA, Ji H, Knight CR, Kuo WP, LeClerc JE, Levy S, Li QZ, Liu C, Liu Y, Lombardi MJ, Ma Y, Magnuson SR, Maqsodi B, McDaniel T, Mei N, Myklebost O, Ning B, Novoradovskaya N, Orr MS, Osborn TW, Papallo A, Patterson TA, Perkins RG, Peters EH, Peterson R, Philips KL, Pine PS, Pusztai L, Qian F, Ren H, Rosen M, Rosenzweig BA, Samaha RR, Schena M, Schroth GP, Shchegrova S, Smith DD, Staedtler F, Su Z, Sun H, Szallasi Z, Tezak Z, Thierry-Mieg D, Thompson KL, Tikhonova I, Turpaz Y, Vallanat B, Van C, Walker SJ, Wang SJ, Wang Y, Wolfinger R, Wong A, Wu J, Xiao C, Xie Q, Xu J, Yang W, Zhang L, Zhong S, Zong Y, Slikker W., Jr The MicroArray Quality Control (MAQC) project shows inter- and intraplatform reproducibility of gene expression measurements. Nat Biotechnol. 2006;24:1151–1161. doi: 10.1038/nbt1239. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh SK, Clarke ID, Terasaki M, Bonn VE, Hawkins C, Squire J, Dirks PB. Identification of a cancer stem cell in human brain tumors. Cancer Res. 2003;63:5821–5828. [PubMed] [Google Scholar]
- Skold K, Svensson M, Norrman M, Sjogren B, Svenningsson P, Andren PE. The significance of biochemical and molecular sample integrity in brain proteomics and peptidomics: stathmin 2–20 and peptides as sample quality indicators. Proteomics. 2007;7:4445–4456. doi: 10.1002/pmic.200700142. [DOI] [PubMed] [Google Scholar]
- Sleat DE, Ding L, Wang S, Zhao C, Wang Y, Xin W, Zheng H, Moore DF, Sims KB, Lobel P. Mass spectrometry-based protein profiling to determine the cause of lysosomal storage diseases of unknown etiology. Mol Cell Proteomics. 2009 doi: 10.1074/mcp.M900122-MCP200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefanski R, Ladenheim B, Lee SH, Cadet JL, Goldberg SR. Neuroadaptations in the dopaminergic system after active self-administration but not after passive administration of methamphetamine. Eur J Pharmacol. 1999;371:123–135. doi: 10.1016/s0014-2999(99)00094-1. [DOI] [PubMed] [Google Scholar]
- Stevens SM, Duncan RS, Koulen P, Prokai L. Proteomic analysis of mouse brain microsomes: identification and bioinformatic characterization of endoplasmic reticulum proteins in the mammalian central nervous system. J Proteome Res. 2008;7:1046–1054. doi: 10.1021/pr7006279. [DOI] [PubMed] [Google Scholar]
- Sutton MA, Schmidt EF, Choi KH, Schad CA, Whisler K, Simmons D, Karanian DA, Monteggia LM, Neve RL, Self DW. Extinction-induced upregulation in AMPA receptors reduces cocaine-seeking behaviour. Nature. 2003;421:70–75. doi: 10.1038/nature01249. [DOI] [PubMed] [Google Scholar]
- Svensson M, Boren M, Skold K, Falth M, Sjogren B, Andersson M, Svenningsson P, Andren PE. Heat stabilization of the tissue proteome: a new technology for improved proteomics. J Proteome Res. 2009;8:974–981. doi: 10.1021/pr8006446. [DOI] [PubMed] [Google Scholar]
- Tannu N, Mash DC, Hemby SE. Cytosolic proteomic alterations in the nucleus accumbens of cocaine overdose victims. Mol Psychiatry. 2007;12:55–73. doi: 10.1038/sj.mp.4001914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tannu NS, Howell LL, Hemby SE. Integrative proteomic analysis of the nucleus accumbens in rhesus monkeys following cocaine self-administration. Mol Psychiatry. 2008 doi: 10.1038/mp.2008.53. (Epub ahead of print) [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taylor CF, Paton NW, Lilley KS, Binz PA, Julian RK, Jr, Jones AR, Zhu W, Apweiler R, Aebersold R, Deutsch EW, Dunn MJ, Heck AJ, Leitner A, Macht M, Mann M, Martens L, Neubert TA, Patterson SD, Ping P, Seymour SL, Souda P, Tsugita A, Vandekerckhove J, Vondriska TM, Whitelegge JP, Wilkins MR, Xenarios I, Yates JR, III, Hermjakob H. The minimum information about a proteomics experiment (MIAPE) Nat Biotechnol. 2007;25:887–893. doi: 10.1038/nbt1329. [DOI] [PubMed] [Google Scholar]
- Thomas KL, Arroyo M, Everitt BJ. Induction of the learning and plasticity-associated gene Zif268 following exposure to a discrete cocaine-associated stimulus. Eur J Neurosci. 2003a;17:1964–1972. doi: 10.1046/j.1460-9568.2003.02617.x. [DOI] [PubMed] [Google Scholar]
- Thomas PD, Campbell MJ, Kejariwal A, Mi H, Karlak B, Daverman R, Diemer K, Muruganujan A, Narechania A. PANTHER: a library of protein families and subfamilies indexed by function. Genome Res. 2003b;13:2129–2141. doi: 10.1101/gr.772403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trujillo KA, Kubota KS, Warmoth KP. Continuous administration of opioids produces locomotor sensitization. Pharmacol Biochem Behav. 2004;79:661–669. doi: 10.1016/j.pbb.2004.09.017. [DOI] [PubMed] [Google Scholar]
- Uhlen M, Hober S. Generation and validation of affinity reagents on a proteome-wide level. J Mol Recognit. 2009;22:57–64. doi: 10.1002/jmr.891. [DOI] [PubMed] [Google Scholar]
- VanGuilder HD, Vrana KE, Freeman WM. Twenty-five years of quantitative PCR for gene expression analysis. Biotechniques. 2008;44:619–626. doi: 10.2144/000112776. [DOI] [PubMed] [Google Scholar]
- Vignali DA. Multiplexed particle-based flow cytometric assays. J Immunol Methods. 2000;243:243–255. doi: 10.1016/s0022-1759(00)00238-6. [DOI] [PubMed] [Google Scholar]
- Wachtel SR, DeWit H. Subjective and behavioral effects of repeated d-amphetamine in humans. Behav Pharmacol. 1999;10:271–281. doi: 10.1097/00008877-199905000-00004. [DOI] [PubMed] [Google Scholar]
- Whiteaker JR, Zhang H, Zhao L, Wang P, Kelly-Spratt KS, Ivey RG, Piening BD, Feng LC, Kasarda E, Gurley KE, Eng JK, Chodosh LA, Kemp CJ, McIntosh MW, Paulovich AG. Integrated pipeline for mass spectrometry-based discovery and confirmation of biomarkers demonstrated in a mouse model of breast cancer. J Proteome Res. 2007;6:3962–3975. doi: 10.1021/pr070202v. [DOI] [PubMed] [Google Scholar]
- Williams K, Wu T, Colangelo C, Nairn AC. Recent advances in neuroproteomics and potential application to studies of drug addiction. Neuropharmacology. 2004;47 (suppl 1):148–166. doi: 10.1016/j.neuropharm.2004.07.009. [DOI] [PubMed] [Google Scholar]
- Yang MH, Jung MS, Lee MJ, Yoo KH, Yook YJ, Park EY, Choi SH, Suh YJ, Kim KW, Park JH. Gene expression profiling of the rewarding effect caused by methamphetamine in the mesolimbic dopamine system. Mol Cells. 2008;26:121–130. [PubMed] [Google Scholar]
- Zhong W, Dong Z, Tian M, Cao J, Xu T, Xu L, Luo J. Opiate withdrawal induces dynamic expressions of AMPA receptors and its regulatory molecule CaMKIIalpha in hippocampal synapses. Life Sci. 2006;79:861–869. doi: 10.1016/j.lfs.2006.02.040. [DOI] [PubMed] [Google Scholar]
- Zichi D, Eaton B, Singer B, Gold L. Proteomics and diagnostics: let’s get specific, again. Curr Opin Chem Biol. 2008;12:78–85. doi: 10.1016/j.cbpa.2008.01.016. [DOI] [PubMed] [Google Scholar]