Abstract
Mitochondrial respiratory chain (RC) disease diagnosis is complicated both by an absence of biomarkers that sufficiently divulge all cases and limited capacity to quantify adverse effects across intermediary metabolism. We applied high performance liquid chromatography (HPLC) and mass spectrometry (MS) studies of stable-isotope based precursor–product relationships in the nematode, C. elegans, to interrogate in vivo differences in metabolic flux among distinct genetic models of primary RC defects and closely related metabolic disorders.
Methods
C. elegans strains studied harbor single nuclear gene defects in complex I, II, or III RC subunits (gas-1, mev-1, isp-1); enzymes involved in coenzyme Q biosynthesis (clk-1), the tricarboxylic acid cycle (TCA, idh-1), or pyruvate metabolism (pdha-1); and central nodes of the nutrient-sensing signaling network that involve insulin response (daf-2) or the sirtuin homologue (sir-2.1). Synchronous populations of 2000 early larval stage worms were fed standard Escherichia coli on nematode growth media plates containing 1,6-13C2-glucose throughout their developmental period, with samples extracted on the first day of adult life in 4% perchloric acid with an internal standard. Quantitation of whole animal free amino acid concentrations and isotopic incorporation into amino and organic acids throughout development was performed in all strains by HPLC and isotope ratio MS, respectively. GC/MS analysis was also performed to quantify absolute isotopic incorporation in all molecular species of key TCA cycle intermediates in gas-1 and N2 adult worms.
Results
Genetic mutations within different metabolic pathways displayed distinct metabolic profiles. RC complex I (gas-1) and III (isp-1) subunit mutants, together with the coenzyme Q biosynthetic mutant (clk-1), shared a similar amino acid profile of elevated alanine and decreased glutamate. The metabolic signature of the complex II mutant (mev-1) was distinct from that of the other RC mutants but resembled that of the TCA cycle mutant (idh-1) and both signaling mutants (daf-2 and sir-2.1). All branched chain amino acid levels were significantly increased in the complex I and III mutants but decreased in the PDH mutant (pdha-1). The RC complex I, coenzyme Q, TCA cycle, and PDH mutants shared significantly increased relative enrichment of lactate+1 and absolute concentration of alanine+1, while glutamate+1 enrichment was significantly decreased uniquely in the RC mutants. Relative intermediary flux analyses were suggestive of proximal TCA cycle disruption in idh-1, completely reduced TCA cycle flux in sir-2.1, and apparent distal TCA cycle alteration in daf-2. GC/MS analysis with universally-labeled 13C-glucose in adult worms further showed significantly increased isotopic enrichment in lactate, citrate, and malate species in the complex I (gas-1) mutant.
Conclusions
Stable isotopic/mass spectrometric analysis can sensitively discriminate primary RC dysfunction from genetic deficiencies affecting either the TCA cycle or pyruvate metabolism. These data are further suggestive that metabolic flux analysis using stable isotopes may offer a robust means to discriminate and quantify the secondary effects of primary RC dysfunction across intermediary metabolism.
Keywords: Respiratory chain mutants, Pyruvate dehydrogenase deficiency, TCA cycle mutant, Signaling pathway mutant, High performance liquid chromatography, Mass spectrometry
1. Introduction
Diagnosis of human mitochondrial respiratory chain (RC) diseases is complicated by the absence of biomarkers that divulge all cases with sufficient sensitivity or specificity [1]. For this reason, the road to diagnosis is often long and complicated, where nearly all patients in whom mitochondrial disease is suspected typically endure a complex battery of blood, urine, and radiology evaluations, frequently involving invasive biopsies to secure tissue for enzymatic and molecular analyses [2]. Unfortunately, such arduous efforts commonly fail to disclose an exact genetic or biochemical anomaly, leaving families unable to anticipate their child's prognosis or recurrence risk, and clinicians unable to develop rational therapies [3]. The emergence of massively parallel genomic sequencing is improving the ability to non-invasively establish the genetic diagnosis of mitochondrial disease [4], but does not provide a tractable means by which to quantify either the degree of primary mitochondrial dysfunction or secondary impairment in flux across interrelated metabolic pathways.
Our previous analyses in C. elegans genetic models of primary mitochondrial dysfunction identified a range of secondary metabolic alterations that occur at the level of genes and proteins, which represent stable end products of cellular adaptation [32]. These findings suggested that substantial insight into mitochondrial disease pathogenesis might be gained through investigations of intermediary metabolic flux among key biochemical pathways in C. elegans mitochondrial mutants. Stable isotopes are detected by contemporary mass spectrometers with remarkable sensitivity, have been used for decades as probes of human metabolism, and are safe in that they are non-radioactive [5]. Indeed, stable isotopic labeling strategies have been used in C. elegans to study lipogenesis [6]. We aimed to apply stable isotopic labeling in living C. elegans animals to quantify metabolic flux alterations among representative nuclear gene-based mitochondrial mutations affecting closely linked intermediary metabolic pathways.
Here, we specifically applied metabolic flux analyses utilizing stable isotopic/mass spectrometric studies of 13carbon-based precursor–product relationships [7] to sensitively differentiate metabolic flux profiles in nematode models of primary dysfunction in the mitochondrial RC and closely linked metabolic pathways. Synchronous early larval nematode populations of 2000 worms were exposed to [1,6-13C2]-glucose on standard nematode growth media (NGM) agar plates spread with live Escherichia coli throughout their developmental period (Fig. 1A). Adult worms were then washed clear of bacteria, extracted in perchloric acid with internal standard, and batched for analysis by HPLC to quantify free amino acid concentrations as well as by isotope ratio MS to quantify relative isotopic enrichment in both amino and organic acids. Although this technique permits analysis of relative isotopic enrichment in several organic acids relevant to TCA cycle flux, this method did not permit reliable interrogation of succinate nor did it permit quantitation of absolute molecular species in the TCA cycle intermediates. Therefore, we employed a novel gas chromatography (GC) MS approach to interrogate absolute isotopic incorporation in all molecular species of TCA cycle intermediates in complex I mutant relative to wild-type adult worms fed on NGM agar plates with live bacteria and universally-labeled 13C-glucose (Fig. 1B).
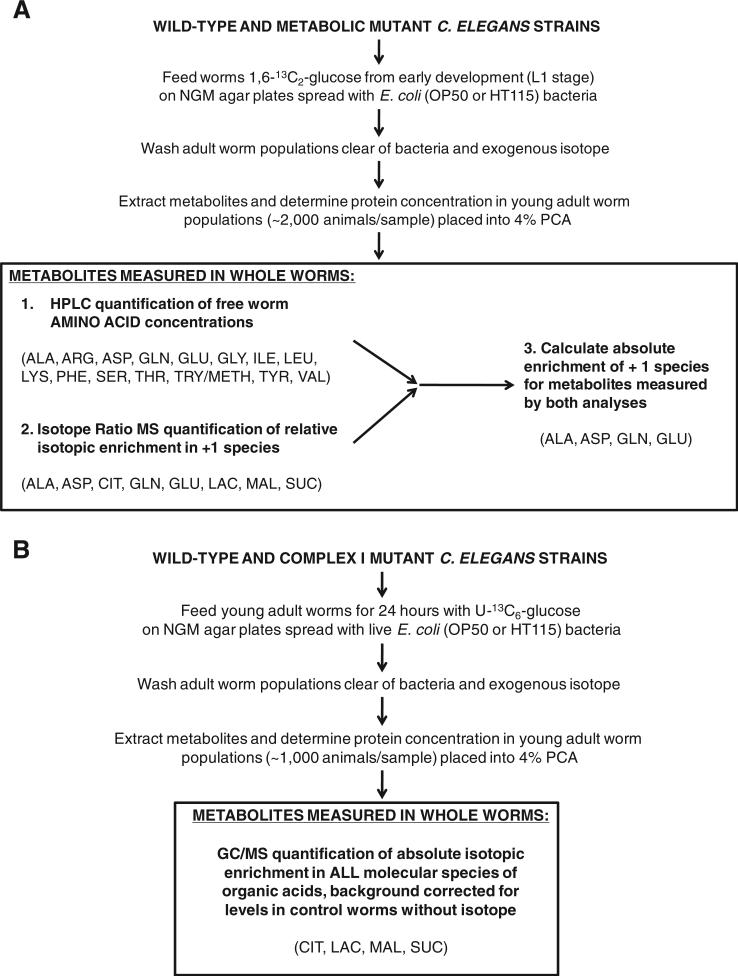
Fig. 1.
Experimental study general overview. (A) 7 metabolic mutant C. elegans strains were studied by high performance liquid chromatography (HPLC) and isotope ratio mass spectrometry (IR/MS) while fed with 1,6-13C2-glucose on standard bacterial NGM plates from early development to young adult stage. Individual metabolic mutant strains studied included mutants in RC complex I (gas-1), RC complex II (mev-1), RC complex III (isp-1), coenzyme Q biosynthesis (clk-1), TCA cycle enzyme isocitrate dehydrogenase (idh-1), sirtuin signaling (sir2.1), and the insulin receptor (daf-2) — all of which were compared to the wild-type (N2 Bristol) worms grown on OP50 E. coli. In addition, feeding RNAi-mutants were studied for the pyruvate dehydrogenase complex E1 alpha homologue (pdha-1) as compared to wild-type (N2 Bristol) worms grown on HT115 E. coli. IR/MS enrichment data was corrected for levels of a known standard run with each sample. PCA, perchloric acid. ALA, alanine; ARG, arginine; ASP, aspartate; CIT, citrate; GLN, glutamine; GLU, glutamate; GLY, glycine; ILE, isoleucine; LAC, lactate; LEU, leucine; LYS, lysine; MAL, malate; PHE, phenylalanine; SER, serine; SUC, succinate; THR, threonine; TRY/METH, tryptophan/methionine; TYR, tyrosine; VAL, valine. (B). The complex I mutant missense strain (gas-1(fc21)) was studied relative to wild-type worms following a similar strategy as detailed in Fig. 1A, with the exception that universally-labeled glucose (U-13C6-glucose) was fed to young adult worms and absolute enrichment was quantified in organic species using gas chromatography/mass spectrometry (GC/MS). GC/MS enrichment data was corrected for background 13C-enrichment in concurrently studied worms fed on standard OP50 E. coli without any isotope, which was consistently below 1% APE, corrected, for all molecular species studied.
Indeed, this in vivo approach sensitively discriminated altered flux through glycolysis, pyruvate dehydrogenase (PDH), and the tricarboxylic (TCA) cycle among different genetic mutant strains in the mitochondrial RC and related metabolic pathways (Fig. 2). These data are suggestive that a stable isotopic/mass spectrometry approach may offer a minimally-invasive means to both discriminate specific genetic-based intermediary metabolic blocks and to quantitatively characterize the secondary metabolic effects of primary mitochondrial dysfunction in humans.
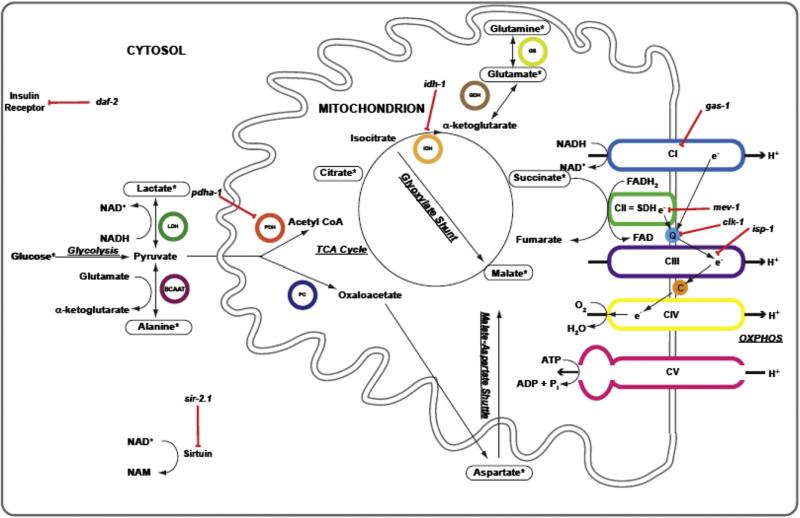
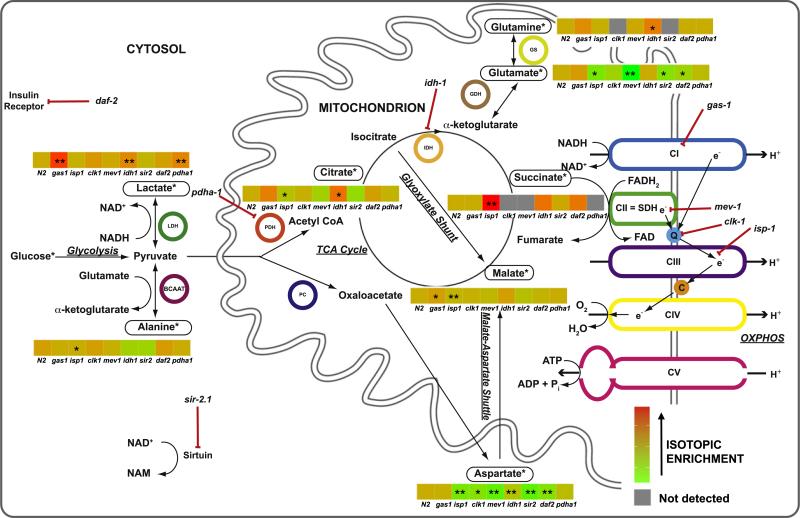
Fig. 2.
Schematic overview of relationship between biochemical analytes interrogated by 13carbon-glucose stable isotopic labeling in C. elegans. *, indicates metabolites in which isotopic enrichment can be detected following feeding worms 13C-glucose. Red lines with bars indicate the specific metabolic sites inhibited by individual mutant strains studied (italicized). Black arrows indicate direction of metabolic reactions. Colored circles highlight relevant metabolic enzymes. ADP, adenosine diphosphate; ATP, adenosine triphosphate; LDH, lactate dehydrogenase; BCAAT, branched chain amino acid transferase; C, cytochrome C; e–, electron; CI/II/III/IV/V, individual mitochondrial respiratory chain complexes, H+, proton; PDH, pyruvate dehydrogenase; PC, pyruvate carboxylase; Pi, inorganic phosphate; IDH, isocitrate dehydrogenase; GDH, glutamate dehydrogenase; GS, glutamine synthase; NAD+, nicotinamide adenine dinucleotide, oxidized; NADH, nicotinamide adenine dinucleotide, reduced; NAM, nicotinamide; Q, coenzyme Q; SDH, succinate dehydrogenase; TCA, tricarboxylic acid. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
2. Methods
2.1. C. elegans strain selection and handling
C. elegans strains were selected that harbor missense mutations in nuclear-encoded RC subunits (complex I, gas-1(fc21) [8,9]; complex II, mev-1(kn1) [10]; complex III, isp-1(qm150)) [11]; enzymes involved in coenzyme Q biosynthesis (clk-1(qm30)) [12] and the TCA cycle (idh-1(ok2832)) that was generated by the C. elegans Gene Knockout Consortium (http://celeganskoconsortium.omrf.org/variation.aspx?allele=ok2832); and central nutrient-sensing signaling network [13] components including a knockout allele of SIRT1 sirtuin homologue (sir-2.1(ok434)) [14] and a missense mutation in the insulin signaling pathway (daf-2(e1368)) [11]. N2-Bristol was the wild-type control strain studied for all analyses involving these seven classical genetic mutants, all of which were obtained as stable mutants from the Caenorhabditis Genetics Center (CGC, www.wormbase.org). In addition, as no classical allele was available, the pdha-1 mutant was generated by standard feeding RNA interference (RNAi) technique [15] with 5 mM IPTG for three generations prior to experimental analyses [16], and compared relative to wild-type (N2 Bristol) worms fed HT115 E. coli that are deficient in the RNAseIII enzyme that normally degrades most double-stranded RNAs in a bacterial cell [17].
2.1.1. Whole worm [1,6]13C2-glucose stable isotope exposure during development
Stable isotopic profiling in C. elegans young adult mutants was performed at 20 °C throughout their developmental period, as previously described [7]. In particular, worms were grown for comparative mutant strain experiments on NGM agar plates in the presence of live bacteria, since their metabolism is significantly altered when grown while shaking in liquid culture flasks due to relative starvation compared to feeding on plates, and further exacerbated by prolonged growth either with dead bacteria or without any bacteria present. Since we sought to compare the metabolic baseline of different metabolic mutants, rather than the response of one metabolic mutant to a given intervention, we sought to optimize the normal physiologic state of the mutant worms. In brief, populations of 2000 early larval stage (L1) worms per strain were grown on 10 cm NGM agar plates with 10 mMol [1,6]13C2-glucose and OP50 E. coli bacteria until adulthood. On the first day of adult life (assessed by the presence of laid eggs on the plate), worms were washed clear of bacteria in three 50 mL volumes of S. basal. Worm number was estimated by counting [18]. All sample preparations were performed in triplicate and metabolic reactions were stopped by the addition of 20 nmol internal standard (e-aminocaproic acid, 16.7 μmol/L) and 4% perchloric acid (PCA) final concentration. Samples were ground using a plastic homogenizer and motorized drill until visual inspection confirmed worm disruption. Precipitated protein was removed, re-dissolved in 1 normal NaOH, and protein concentration was determined by DC Protein Assay (Bio-Rad). The supernatant was returned to neutral pH (7) using 10 M KOH for free amino acid quantitation by HPLC analysis.
2.1.2. Whole worm free amino acid quantitation by HPLC
Whole worm free amino acid quantitation was assessed by high performance liquid chromatography (HPLC) in the Metabolomics Core Facility at the Children's Hospital of Philadelphia, as previously described [2,7]. Briefly, whole worm population free amino acid quantitation was performed by direct injection of 50 μL of neutralized sample into HPLC (Varian) using precolumn derivatization with o-phthalaldehyde and fluorescent detection [19]. Raw HPLC data was normalized to worm protein concentration and reported as nmol/mg protein. Three biologic replicate analyses were performed per strain.
2.1.3. Whole worm isotopic enrichment in free amino and organic acids by isotope ratio MS
Remaining neutralized samples after HPLC analysis were extracted using ion exchange resin (Bio-Rad) in AG50 and AG1 columns, respectively, to measure relative enrichment in amino acids and organic acids, by isotope ratio mass spectrometry, as previously described [7]. Isotope ratio MS analyses were performed in the Metabolomics Core Facility at the Children's Hospital of Philadelphia. Stable isotopic enrichment was calculated in Excel (Microsoft) for each species according to the following formula: Atom Percent Excess, corrected (APE) = (Rsa – Rst) * 100/[(Rsa – Rst) + 100], where Rsa — ratio of the sample and Rst — ratio of the standard.
2.1.4. Whole worm universally-labeled 13C-glucose isotopic exposure in gas-1 and N2 young adults
Synchronous populations of first day egg-laying, young adult stage gas-1 and N2 worms were exposed at 20 °C for 24 h on NGM plates spread with standard OP50 E. coli and universally-labeled 13C-glucose. Worms were then washed clear of bacteria in three 50 mL volumes of S. basal and worm number was estimated by counting [18]. Metabolic reactions were stopped in 4% PCA. Samples were then extracted in an AG1 column using ion exchange resin (Bio-Rad), as detailed in Section 2.1.3. Extracted samples were dried-down and stored at room temperature.
2.1.5. Relative organic acid isotopic enrichment by GC/MS
Trimethylsilyl-derivatives of each dried-down PCA worm preparation were made in the CHOP Clinical Biochemical Laboratory by addition of 150 μL of 99% bis(trimethylsilyl)trifluoroacetamide: 1% trimethylchlorosilane (Pearce Chemical Co.) and 150 μL ethylacetate. Following 30-minute incubation at 70 °C, 1 μL was injected into an Agilent 5975C gas chromatograph fitted with an Agilent 7890A mass selective detector. The running conditions included a baseline temperature of 70 °C for 2 min followed by a ramp of 5 °C per minute to 250 °C and 10 °C per minute to 300 °C. Full scan mass spectrometric data was acquired over the range mz 50–600 collected throughout the run. Positive peak identification was based on comparison with authentic standards and enrichment data derived from the total ion chromatograms. Natural abundance of stable isotopes was corrected for by subtracting the abundances of each isotope in each compound in an untreated worm sample from the respective strain.
2.2. Statistical analyses
The significance of the difference in the mean analyte level between strains was assessed by mixed-effect ANOVA, which takes into account potential batch effect due to samples being experimentally prepared (as relates to worm feeding through PCA extraction and subsequent sample separation into amino acid and organic acid fractions) and analyzed by HPLC and IR/MS machines on different days by including a batch random effect in the model. This model assumes the measures completed on the same day are more closely correlated compared to those completed on different days. For GC/MS data, two-sample t test was used to compare the mean absolute percent atoms per excess (% APE, corrected) enrichment between gas-1 and wild-type (N2) strains for lactate, malate and succinate, while one-sample t test was used for citrate since no isotopic enrichment was observed in N2. Significance level of .0025 was used to account for multiple comparisons by Bonferroni correction (20 tests performed per analyte), conveyed in Fig. 6 as “p < 0.05, corrected”. All statistical analyses were performed in SAS 9.3.
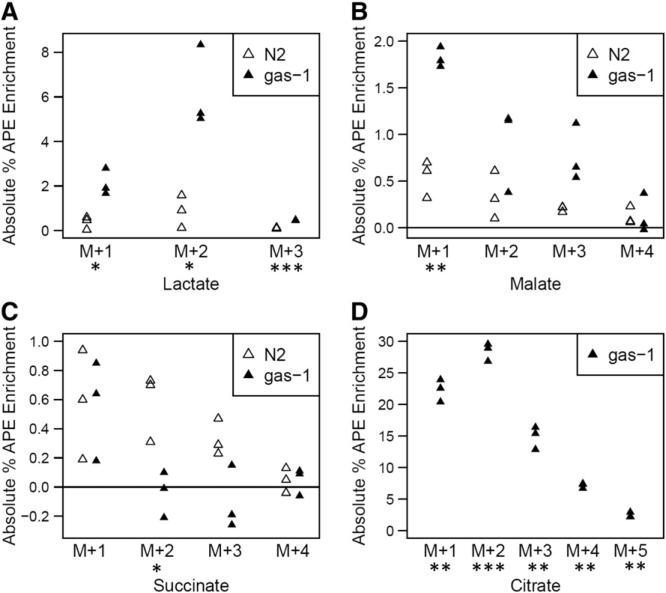
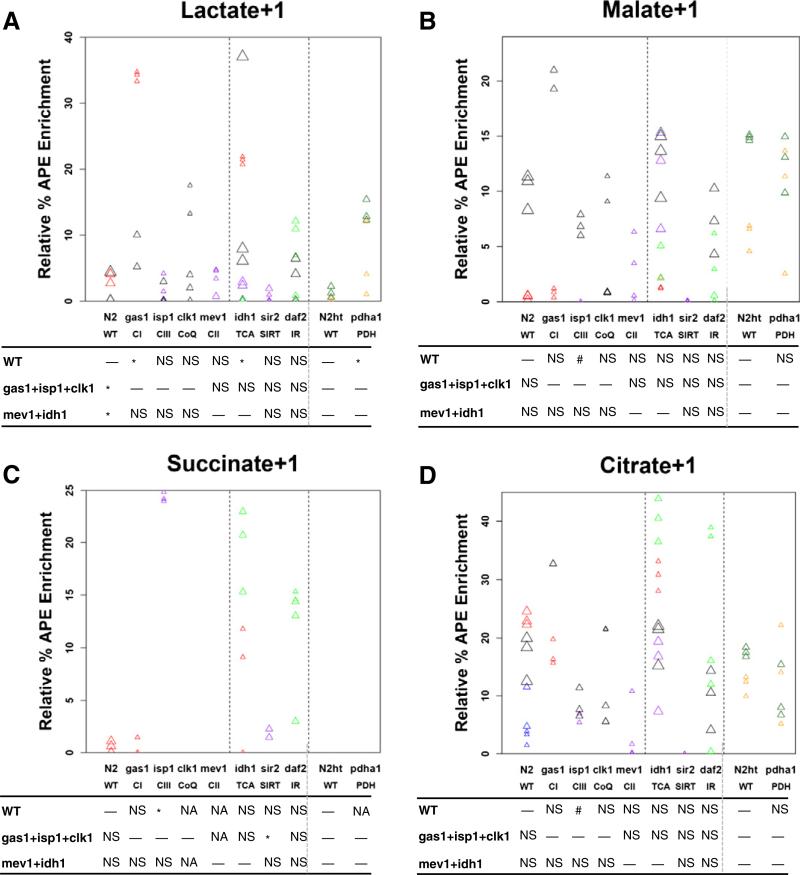
Fig. 6.
Absolute isotopic enrichment in all molecular species of lactate and key TCA cycle intermediates of RC complex I (gas-1) mutant and wild-type (N2) young adult worms following universally-labeled glucose feeding on NGM plates with live bacteria for 24 h. (A) Lactate, (B) Malate, (C) Succinate, and (D) Citrate. Absolute enrichment (percent atoms per excess, APE) in all molecular species of these four organic acids was measured by gas chromatography–mass spectrometry (GC/MS). All biological replicate data are shown, with statistical analysis performed for gas-1 mutant animals relative to N2 Bristol wild-type worms fed isotope and standard OP50 E. coli. Reported data was background-subtracted for isotopic enrichment quantified in the same strains fed live bacteria without any stable isotopic exposure. No citrate enrichment was present in wild-type worms. 3 biological replicate worm populations were analyzed per condition. *, p < 0.05; **, p < 0.01; and ***, p < 0.001.
3. Results and discussion
3.1. Free worm amino acid profiling in C. elegans metabolic mutants
3.1.1. NADH-dependent RC dysfunction (gas-1, clk-1, isp-1) yields a distinct pattern of free amino acid alterations relative to RC complex II dysfunction (mev-1)
In our earlier study of free amino acid concentrations in gas-1, mev-1, and isp-1 mutant worms relative to wild-type (N2 Bristol), we observed that RC dysfunction commonly results in elevated branched chain amino acid (BCAA) and alanine concentrations with concomitant decreases in both glutamate concentration and the ratios of individual BCAAs or alanine relative to glutamate, findings that we attributed to an increased NADH/NAD+ redox ratio [32]. We have also recently reported similar amino acid alterations in plasma of a retrospective cohort of heterogeneous human patients with definite RC disease [20]. Some of these same general trends were confirmed here upon collective analysis of free worm amino acid profiles in these same three RC mutant strains together with the coenzyme Q (CoQ) biosynthetic mutant (clk-1) (Fig. 3, Table S1, and Fig. S1). Specifically, we observed significant elevation of all three BCAAs (valine (p < 0.05), leucine (p < 0.05), and isoleucine (p < 0.01)), as well as significant decrease in glutamate (p < 0.001) across these four mutants when analyzed as a common group relative to wild-type (N2 Bristol) worms (data not shown). Collective analysis of these four RC mutants also showed significant alterations in several other free worm amino acid concentrations, with the most significant decreases evident in aspartate (p < 0.001, Fig. 3B) and glycine (p < 0.001, Fig. S1). Thus, there seem to be overall differences in RC mutant amino acid profiles regardless of the specific complex blocked within the RC. Comparison of individual free worm amino profiles of each RC mutant strain, however, revealed that the greatest similarities occurred between the complex I (gas-1), complex III (isp-1), and CoQ biosynthetic mutant worm strains, including significant reductions in glutamate, aspartate, and glycine in all three strains relative to wild-type controls (each p < 0.05, corrected for multiple testing), while BCAA levels showed a non-significant trend toward increase relative to wild-type in all three of these RC mutants and alanine was significantly increased only in the complex I (gas-1) mutant (Fig. 3 and Table S1).
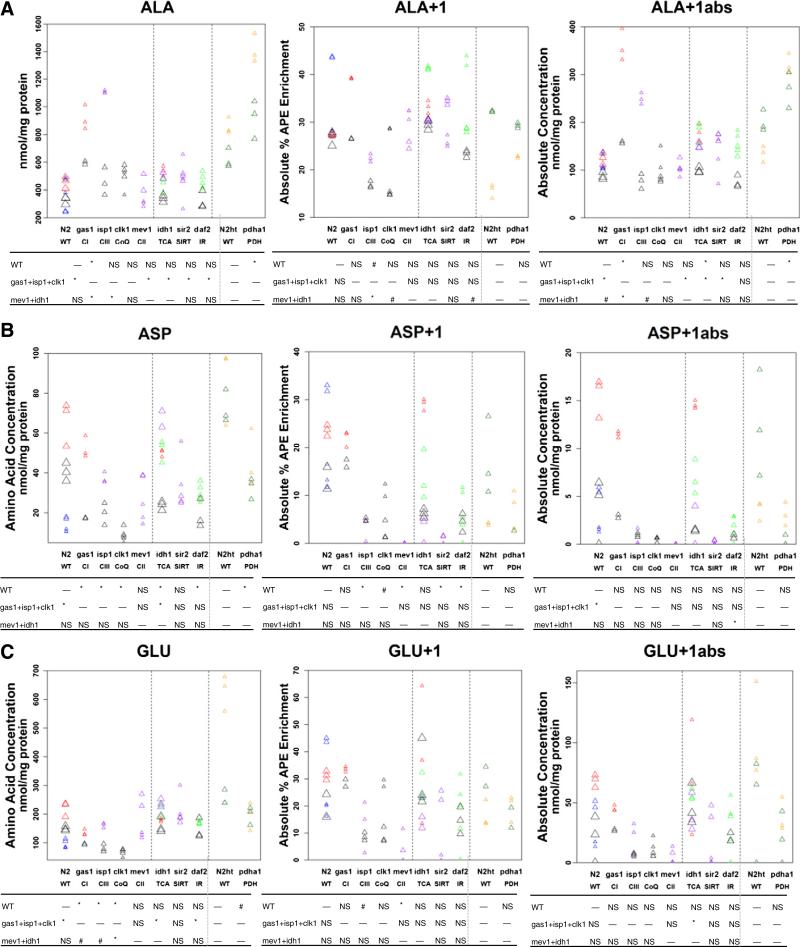
Fig. 3.
Significant alterations were identified in amino acid concentrations and isotopic enrichment of alanine, aspartate, and glutamate among the RC mutants. (A) alanine, (B) aspartate, and (C) glutamate whole worm free amino acid concentrations, as well as relative and absolute enrichment in +1 molecular species, of each metabolite was quantified in young adult worms following 1,6-13C2-glucose feeding throughout nematode development in 7 mutant worm strains (Fig. 1A) compared to wild-type (N2 Bristol) worms grown on live OP50 E. coli, and also in the pyruvate dehydrogenase mutant feeding RNAi-generated strain relative to wild-type (N2 Bristol) worms grown on live HT115 E. coli. Absolute enrichment in +1 species (right panels) was calculated in the same biological samples by both HPLC (left panels) and isotope ratio mass spectrometry (center panels). Symbol size conveys experimental set in which worm isotope exposure and sample preparation was performed. Symbol color denotes experimental day on which HPLC or MS analyses were performed, where multiple strains were typically analyzed in each experiment. Statistical significance is conveyed below each graph, where #, p < 0.05 (uncorrected for multiple testing) and *, p < 0.05 (corrected for multiple testing). All mutants were compared relative to their respective wild-type strain (N2) for all mutants except pdha1, which was compared relative to N2 grown on HT115 E. coli (N2ht). Comparisons are also shown for each mutant relative to a group of primary RC mutants (gas1, isp1, clk1) or relative to a group of TCA mutants (mev1, idh1). Dashed lines divide mutant groups by category into RC mutants, other mutants, and PDH mutant. NS, not significant. Horizontal dash indicates no comparative analysis between groups was performed. Additional whole worm amino acid concentrations, along with relative and absolute enrichment in +1 molecular species of glutamine, are shown in Fig. S1.
Most notable was the relatively distinct metabolic signature that emerged for the complex II subunit mutant (mev-1) relative to the other three primary RC mutants (gas-1, clk-1, isp-1) (Figs. 3 and S1). When analyzed as a group of RC mutants that only included gas-1, isp-1, and clk-1, a marginal increase in all BCAA levels (p < 0.05, not corrected for multiple testing) was achieved relative to wild-type controls (Fig. S1). While mev-1 showed a similar trend toward increased BCAAs as the RC mutants, statistical significance was reached only for isoleucine (p < 0.05, corrected) (Fig. S1). Neither alanine nor glutamate levels were significantly changed in mev-1 relative to wild-type worms (Fig. 3), while only mev-1 had significantly increased levels of lysine and phenylalanine (p < 0.05, corrected) (Fig. S1). When analyzed as a group of RC mutants that included gas-1, isp-1, and clk-1, significant differences at the p < 0.05 level corrected for multiple testing were seen in alanine, aspartate, glutamate, glycine, and serine relative to wild-type (N2) and in alanine, arginine, lysine, and valine relative to the complex II mutant (mev-1).
Thus, whole worm free amino acid analysis of four nematode strains harboring mutations that directly affect electron transport chain flux confirmed a common free amino acid signature of elevated alanine and decreased glutamate exists in the complex I, complex III, and CoQ biosynthetic worms relative to wild-type animals. This amino acid signature was not shared by the complex II mutant. In addition, these data support that BCAA elevation does occur in the RC mutant worms relative to wild-type, as previously reported [32]. These data are suggestive that altered glutamate and alanine levels, and possibly BCAA elevation, stem from impairment of NADH-dependent RC flux, in which RC complex II is not directly involved.
Interestingly, several of the alterations in whole worm free amino acid concentrations that we observed in the primary RC complex mutants were similar to those recently reported upon GC/MS analysis of the exometabolome, or metabolic end-products excreted in the media, of wild-type nematodes exposed to prolonged anoxia [21]. In particular, anoxia was shown to result in elevation of lactate and BCAAs. Elevations of worm lysine and tyrosine, which we observed to be at least marginally upregulated in the RC mutant worms (Fig S1) were also seen in anoxic worms, as the authors attributed to catabolic requirements for NAD+ and oxygen, respectively. Collectively, these findings are supportive that impaired aerobic respiration due to either genetic mutations or to oxygen deprivation similarly alter intermediary amino acid metabolism in C. elegans.
3.1.2. PDH mutant worms have more pronounced alanine elevation relative to primary RC mutants and significantly decreased BCAA concentrations
To permit comparison of primary RC mutant amino acid profiles with primary genetic mutants in closely interacting metabolic pathways, free worm amino acid concentrations were studied in whole worm populations deficient in pyruvate dehydrogenase (PDH) (pdha-1) (Fig. 3). Relative to the RNAi control strain, N2 grown on HT115 E. coli, the PDH mutant (pdha-1) generated by feeding RNAi showed significantly elevated alanine (p < 0.001, corrected), decreased aspartate (p < 0.001, corrected), and marginally decreased glutamate (p < 0.05, uncorrected for multiple testing) (Fig. 3 and Table S1), similarly as was seen in the primary RC mutants. A notable difference was that absolute free alanine concentration was 50% greater in pdha-1 (mean 1168 nmol/mg protein) than was seen in the RC mutants (mean 789 nmol/mg protein in gas-1, mean 785 nmol/mg protein in isp-1, and mean 504 nmol/mg protein in clk-1) (Table S1), as might be expected given the proximity of pyruvate metabolism to the site of alanine generation from pyruvate through the alanine aminotransferase reaction. Similarly, we recently observed plasma alanine levels were significantly greater in human patients with PDH deficiency relative to RC deficiency [20]. However, it is also important to consider that the N2-HT115 control strain had a mean alanine that was nearly 360 nmol/mg protein greater than the N2 control strain grown on standard OP50 E. coli, which if accounted for may well suggest a similar degree of alanine elevation was achieved in the RC and PDH mutant worm strains. Indeed, the N2-HT115 RNAi control strain showed significant elevation in nearly all free amino acid concentrations relative to N2-OP50 (Table S1), for unclear reasons, although consistent with prior demonstration that the OP50 vs HT115 bacterial food source directly impacts nematode health and metabolism, with almost one-third of metabolites found to be changed by proton nuclear magnetic resonance (NMR) based solely on nematode diet [22]. Despite this consideration, BCAA concentrations were significantly decreased in pdha-1 relative to N2-HT115 controls (p < 0.01) (Fig. S1), similarly as we also reported in plasma of human patients with PDH deficiency [20]. Thus, these findings further support that there is a clear distinction in the amino acid alterations that occur upon primary metabolic dysfunction in pyruvate metabolism versus in the RC.
3.1.3. The amino acid profile of the RC complex II mutant worm (mev-1) resembles that of a TCA cycle mutant strain (idh-1)
Given that RC complex II directly functions in the TCA cycle, we sought to determine whether the unique amino acid profile observed in the complex II mutant strain (mev-1) relative to the other RC complex mutants involved in NADH-derived electron flux might instead more closely resemble another TCA complex mutant strain. Comparative analysis of the isocitrate dehydrogenase mutant strain (idh-1), whose primary TCA cycle enzymatic defect lies in the conversion of isocitrate to alpha-ketoglutarate, showed it had significantly elevated mean glutamate concentration (p < 0.05, corrected) and a trend toward reduced mean alanine concentration (p < 0.05, uncorrected) relative to the collective group of four primary RC mutants (Fig. 3 and Table S1). idh-1 also had significantly increased levels of aspartate and glycine relative to collective analysis of the four primary RC mutants (p < 0.05, corrected). In addition, idh-1 had modest increases in concentrations of aspartate (p < 0.05, corrected), glycine (p < 0.05, corrected), serine (p < 0.05, uncorrected), glutamine (p < 0.05, uncorrected), and tyrosine (p < 0.05, uncorrected) relative to N2 wild-type controls.
Strain-level analyses showed a similar absolute level of glutamate was present in idh-1 (mean 199 nmol/mg protein) as had been evident as the primary RC mutant strain ‘outlier’ mev-1 (mean 176 nmol/mg protein), both of which were greater than in the N2 wild-type worms (mean 144 nmol/mg protein), although neither increase reached statistical significance (Fig. 3C and Table S1) Nor did either idh-1 or mev-1 have significantly changed levels of alanine relative to wild-type (Fig. 3A). idh-1 had increased concentration of one BCAA isoleucine (p < 0.05, uncorrected) relative to N2, but both isoleucine (p < 0.05, uncorrected) and valine (p < 0.05, uncorrected) were significantly decreased in idh-1 relative to the collective group of four primary RC mutants (Fig. S1). Similarly, isoleucine was significantly increased in mev-1 relative to N2 (p < 0.05, corrected) with only a non-significant trend toward increase seen in the other two BCAAs (Fig. S1). Both mev-1 and idh-1 also showed significant increases relative to wild-type in lysine with greater absolute concentration in mev-1 (Table S1), although the increased arginine only achieved statistical significance in mev-1 (p < 0.05, corrected) (Fig. S1). Overall, idh-1 had a significantly different free amino acid profile relative to either the collective group of four RC mutants or to the PDH mutant strain. However, the amino acid profile of idh-1 was notably similar to mev-1, with suggestions of increased glutamate, lysine, and isoleucine concentrations but no significant changes in alanine, leucine, or valine. Thus, analyses at the whole worm free amino acid level were suggestive that mev-1 is more appropriately classified based on its intermediary metabolic profile as a TCA cycle mutant than a RC mutant.
3.1.4. Amino acid profiles in mutants for central nutrient-sensing signaling pathways that impact mitochondrial function (daf-2 and sir-2) more closely resemble TCA cycle mutants (idh-1, mev-1) than primary RC mutants (gas-1, isp-1, clk-1)
Free amino acid concentrations were assessed in two mutants for cell signaling pathways that mediate mitochondrial function. The insulin receptor mutant has increased FOXO1 signaling (daf-2), and the SIRT1 homologue (sir-2.1) mutant has impairment of this protein deacetylase. Similarly to what was seen in the TCA mutants (idh-1 and mev-1), elevated glutamate concentration was present in both daf-2 (p < 0.05, corrected) and sir-2.1 (p < 0.05, uncorrected) relative to the collective group of three primary, non-TCA cycle RC mutants (gas-1, clk-1, isp-1) (Fig. 3C). Further, alanine concentration was not elevated in any of these mutants relative to N2 wild-type controls, but was significantly reduced in both signaling mutants relative to the same RC mutant group (both p < 0.05, corrected). Overall, few other significant amino acid changes were found in either daf-2 or sir-2.1 relative to the collective group of RC mutants (Fig. S1). However, daf-2 did show a significant reduction in free aspartate (p < 0.05, corrected) (Fig. 3B) and increases in both lysine (p < 0.05, corrected) and isoleucine (p < 0.05, uncorrected) relative to N2 wild-type controls, with a trend toward increase in the other BCAAs that did not reach statistical significance (Fig. S1 and Table S1). Indeed, daf-2 has previously been shown by NMR to have increased BCAAs relative to N2 wild-type controls [23]. sir-2.1 also showed a trend toward increased BCAA concentrations that reached significance relative to N2 only for isoleucine (p < 0.05, corrected), as well as increased lysine (p < 0.05, corrected) again similar to amino acid profiles of the idh-1 and mev-1 TCA mutants (Table S1). Thus, both daf-2 and sir-2.1 displayed whole worm free amino acid profiles that resembled those of the TCA mutants (as compared to wild-type N2), with elevations of glutamate, lysine, and isoleucine, non-significant trends toward increase in all BCAAs, and unchanged concentration of alanine.
3.2. Intermediary metabolic pathway flux analyses by isotope ratio mass spectrometry (IR/MS)
3.2.1. Complex I and PDH mutant worms have increased relative flux through glycolysis with preserved TCA cycle flux, while TCA cycle flux is reduced in complex II, complex III, and CoQ mutants
Feeding worms stable isotope in the form of 1,6-13C2-glucose permits relative “flux” analysis of enriched carbon traced from the upstream labeled carbons of glucose into +1 molecular species of eight downstream metabolites that indicate collective flux through glycolysis and the PDH complex (alanine and lactate) as well as the TCA cycle (citrate, succinate, malate, aspartate, glutamate, and glutamine). Analyses of isotope ratio MS data revealed that the collective group of four RC mutants relative to wild-type (N2) had isotopic incorporation that was significantly increased in lactate+1 (p < 0.05, corrected) and a trend toward decreased glutamate+1 and aspartate+1 (Fig. 3 and 4 and Table S2). As glutamate and aspartate are analytes that equilibrate with alpha-ketoglutarate and malate, respectively, relative enrichment in these species suggests that primary RC dysfunction may result in decreased flux at the middle and distal aspects of the TCA cycle.
Fig. 4.
Relative isotopic enrichment in +1 molecular species of key TCA cycle intermediates identified in metabolic mutant C. elegans strains studied by isotope ratio mass spectrometry (IR/MS). Relative enrichment (percent atoms per excess, APE, corrected) was measured in molecular species of four organic acids by IR/MS in young adult worms fed 1,6-13C2-glucose throughout development. (A) Lactate, (B) Malate, (C) Succinate, and (D) Citrate. Symbols and p values are the same as described in the legend for Fig. 3. Enrichment in all 8 analytes studied are shown in Table S2.
Clear differences in relative isotopic incorporation into these molecular species were evident among the four individual RC mutant strains. Increased lactate+1 enrichment reached statistical significance only in the complex I mutant (gas-1, mean 23.5% APE versus N2 mean 2.6% APE, p < 0.05, corrected), with a trend toward increased lactate+1 evident in the CoQ mutant (clk-1, mean 7.4% APE, p > 0.05) (Fig. 4A and Table S2). Relative glutamate+1 enrichment was decreased in mev-1 (p < 0.05, corrected) and isp-1 (p < 0.05, uncorrected), with a trend toward decrease in clk-1 (p = 0.07, uncorrected) but no change at all in gas-1 relative to N2 wild-type control. Similarly, relative aspartate+1 enrichment was decreased in mev-1 (p < 0.05, corrected), isp-1 (p < 0.01, corrected), and clk-1 (p < 0.05, uncorrected) but unchanged in gas-1 relative to wild-type. Interestingly, relative enrichment in the proximal TCA metabolite, citrate+1, was decreased only in the complex III mutant, isp-1 (p < 0.05, uncorrected). Thus, the complex I mutant (gas-1) showed clear evidence of increased glycolysis that likely relates to an increased cellular NADH/NAD+ redox ratio supporting significantly increased carbon flux from glucose into lactate+1, but did not display significant disruption of carbon flow into TCA cycle intermediates. Similarly, the PDH mutant that is deficient in its ability to convert pyruvate to acetyl CoA had significantly increased lactate+1 incorporation (pdha-1 mean 9.6% APE and N2-HT115 mean 0.8% APE, p < 0.05, corrected), but no inherent alteration apparent in its TCA cycle flux (Fig. 4 and Table S2). In contrast, the more distal RC mutants (mev-1, isp-1, clk-1) had only marginal (in clk-1) to no (in mev-1 and isp-1) increase evident in glycolysis and cellular NADH/NAD+ redox ratio, as discerned by analysis of lactate+1. However, these distal RC mutants each displayed a clear reduction in flux through the TCA cycle. Thus, whereas GC/MS analyses of intermediary metabolites recently reported in the exometabolome of mev-1 mutants revealed enrichment of TCA cycle intermediates [24], isotopic-based analyses allow recognition that accumulation of these analytes are the consequence of reduced carbon flux through the TCA cycle.
3.2.2. Relative TCA cycle flux is altered in the TCA cycle and nutrient-sensing signaling pathway mutants
The TCA cycle mutant, idh-1, had increased enrichment in lactate+1 (mean 10.1% APE versus N2 mean 1.7% APE, p < 0.05, corrected) as well as citrate+1 (mean 26.3% APE versus 12.3% APE in N2, p < 0.05, uncorrected), which accumulates proximal to the site of its TCA cycle enzymatic block (Fig. 4 and Table S2). The sirtuin mutant (sir-2.1) had essentially no isotopic incorporation into any TCA intermediate (Table S2), suggesting it sustains a pronounced defect in TCA cycle flux. Interestingly, the insulin signaling pathway mutant (daf-2) had a trend toward enrichment of succinate+1 (p > 0.05), but significantly reduced enrichment of aspartate+1 (p < 0.05, corrected). A similar profile of reduced TCA intermediates but substantial increase in succinate was evident in the exometabolome GC/MS analysis of anoxic worms [21]. Thus, the TCA cycle and nutrient-sensing signaling pathway mutants displayed unique patterns of relative intermediary flux through the TCA cycle that were suggestive of proximal TCA cycle disruption in idh-1, complete TCA cycle disruption in sir-2.1, and potentially distal TCA cycle alteration in daf-2. A heatmap overview of the relative amino acid and IR/MS-based isotopic changes in all mutants studied by HPLC and/or IR/MS is shown in the context of intermediary metabolic pathway flux in Fig. 5.
Fig. 5.
Heatmap of metabolite alterations observed in all metabolic mutant C. elegans strains studied by HPLC and/or IR/MS following 1,6-13C2-glucose ingestion throughout development. Heatmap conveys differences in mean isotopic enrichment for each labeled +1 metabolite tested between each metabolic mutant C. elegans strain and wild-type (N2) worms. Red and green convey increased and decreased enrichment, respectively, in the mutant strain relative to wild-type. Statistical significance determined by random effect ANOVA is conveyed by asterisks in colored heatmap boxes, where * is p < 0.05 (unadjusted for multiple comparisons) and ** is p b0.05 (adjusted for multiple comparisons). The remainder of the symbols and abbreviations is the same as detailed in the legend for Fig. 2.
3.2.3. Absolute isotopic enrichment in key metabolites demonstrates unique secondary pathophysiology of different RC mutants
Considering that a given biochemical analyte can often be generated or metabolized by more than one enzymatic reaction, absolute quantitation of molecular species following isotopic enrichment can provide insight into the relative metabolic contribution of specific pathway(s) (Table S3). Following [1,6-13C2]-glucose incorporation, quantitation of the absolute enrichment of the +1 molecular species for four analytes (alanine, aspartate, glutamate, and glutamine) revealed that the collective group of RC mutants relative to wild-type worms had decreased absolute mean concentrations of both glutamate+1 (p < 0.05, uncorrected) and aspartate+1 (p < 0.05, corrected) (Fig. 3). Dissecting the major contributing factor(s) underlying changes in absolute concentrations of the +1 molecular species of these analytes can elucidate the predominating pathophysiology. For example, dissection of changes observed in glutamate metabolism in RC mutants relative to N2 wild-type worms demonstrates: (1) whole worm free glutamate concentration was significantly reduced in gas-1, clk-1, and isp-1 but unchanged in mev-1; (2) relative glutamate+1 enrichment was reduced only in mev-1 (p < 0.05, corrected) and isp-1 (p < 0.05, uncorrected); and (3) absolute glutamate+1 concentration showed a reduced trend in all three NADH-dependent RC mutants (gas-1, clk-1, isp-1) and mev-1 (Fig. 3C). Given that the RC complex I mutant, gas-1, has impaired complex I-dependent oxidative capacity that increases the cellular ratio of NADH to NAD+ to reverse oxidation of alpha ketoacids that instead undergo transamination with glutamate to form parent amino acids [32], a reduced glutamate+1 absolute concentration in gas-1 could be attributed to its reduced glutamate pool rather than to a substantial reduction in its TCA cycle flux. Conversely, reduced glutamate+1 absolute concentration in the mev-1 mutant, whose metabolic profile more resembles a TCA cycle mutant than a primary RC mutant, has normal free glutamate pools but significantly reduced TCA cycle flux. Interestingly, the mutants in CoQ (clk-1) and complex III (isp-1), involving RC components that receive electrons from either CI-dependent or CII-dependent oxidation, show trends toward reduced absolute glutamate+1 concentration that may be attributable to both their reduced free glutamate levels and their reduced TCA cycle flux. In this way, comparative analysis of four RC mutants demonstrates that while all seem to show decreased absolute glutamate+1 mean concentration following ingestion of [1,6-13C2]-glucose, their underlying metabolic pathophysiology is distinct.
The long-term storage form of pyruvate is alanine, which is an analyte that can be elevated in many different metabolic disorders. Alanine+1 absolute mean concentration showed the most pronounced increase in the PDH mutant pdha-1 (295 nmol/mg protein versus N2-HT115 168 nmol/mg protein, p < 0.05, corrected), the complex I mutant gas-1 (279 nmol/mg protein versus N2 111 nmol/mg protein, p < 0.05, corrected), and the TCA cycle mutant idh-1 (153 nmol/mg protein, p < 0.05, corrected) (Fig. 3A). Relative to the collective group of three primary non-TCA cycle RC mutants (gas-1, clk-1, isp-1), however, lower alanine+1 absolute levels were seen in the TCA mutant (idh-1, p < 0.05, corrected) and nutrient-sensing signaling network mutants (daf-2 p > 0.05, and sir-2.1 p < 0.05, corrected) (Fig. 3A). mev-1 had no change relative to N2 control in absolute alanine+1 concentration (Fig. 3A). Thus, a direct TCA cycle block (idh-1) results in less pronounced elevation of absolute alanine+1 than does either a block at the more proximal PDH complex (pdha-1) or the more distal RC (gas-1) that also involves up-regulation of glycolysis and increase in cellular ratio of NADH to NAD+.
3.2.4. The insulin receptor signaling pathway daf-2 mutant has reduced free aspartate and aspartate+1 enrichment, suggestive of increased activity of the malate–aspartate shuttle
The daf-2 mutant worm is a model of impaired insulin receptor signaling [25], with prolonged longevity attributable to chronically elevated FOXO1 (daf-16)-pathway signaling [26]. Despite this seemingly beneficial nematode longevity phenotype, the daf-2 mutant may allow insight into the possible intermediary metabolic flux alterations occurring in insulin-resistance, which in humans can lead to diabetes mellitus and interestingly in daf-2 mutants is associated with a higher relative glucose concentration [23]. Interestingly, the most striking and consistent finding identified by our metabolomic analyses of daf-2 was its significantly reduced free aspartate levels (p < 0.05, corrected), significantly reduced relative isotopic enrichment in aspartate+1 (p < 0.05, corrected), and the suggestion of reduced absolute concentration of aspartate+1 (p = 0.06, uncorrected) (Fig. 3B). Given its otherwise apparently unchanged flux through glycolysis, the PDH complex, and the TCA cycle, this significant reduction in aspartate levels and precursor–product isotopic labeling in aspartate+1 is suggestive that daf-2 mutants have preferential glucose recycling through the gluconeogenic enzyme phosphoenolpyruvatecarboxykinase (PEPCK) of oxaloacetate to phosphoenolpyruvate. Alternatively, oxaloacetate may be preferentially converted by malate dehydrogenase to malate [21], which can then undergo dismutation to form first fumarate and then succinate (Fig. 2). Indeed, malate and fumarate supplementation have recently been shown to promote FOXO1(daf-16)-dependent lifespan extension in wild-type worms [27]. Further, a similar profile of substantial increase in succinate enrichment as we identified a trend for in the daf-2 mutant worms (Fig. 4C), was evident by GC/MS analysis of the exometabolome in anoxic worms [21]. However the other TCA cycle intermediates that were significantly decreased in their study were largely unchanged in our isotope-based daf-2 analysis, as relates to findings postulated by the authors in the setting of impaired aerobic respiration to provide support for the occurrence of malate dismutation as a means to increase the efficiency of ATP generation. The nematode phenomenon of malate dismutation requires, among other processes, an intact glyoxylate shunt to form malate from glyoxylate and fatty-acid derived acetyl-CoA [28], and results in FOXO1(daf-16) localization to the nucleus as has been shown to occur in daf-2 mutant worms [26,29]. Indeed, daf-2 mutants have previously been shown to have increased transcription of enzymes involved in both gluconeogenesis and the glyoxylate shunt, along with increased relative concentrations of glucose and succinate and decreased concentrations of malate and aspartate [23].
In addition, our findings reported here of both reduced free aspartate levels and reduced aspartate+1 enrichment following [1,6-13C2]-glucose feeding throughout development in daf-2 mutants suggests their metabolic alterations may also involve increased activity of the malate–aspartate shuttle, which serves to effectively transport NADH reducing equivalents into mitochondria across the NADH/NAD+ impermeable inner mitochondrial membrane and support NAD+ dependent glycolysis in the cytoplasm [30]. In other words, increased activity of the malate–aspartate shuttle to dilute the carbon pool of malate could potentially account for why the 13C-glucose-derived carbon label seems increased in succinate+1 but decreased in aspartate+1 in daf-2 relative to wild-type. Interestingly, over-expression of malate–aspartate shuttle components can extend lifespan in yeast models of caloric restriction [30] and daf-2 is a significantly long-lived nematode strain [26]. Collectively, we interpret these data to suggest that increased flux through the malate–aspartate shuttle in the daf-2 insulin receptor mutant accounts for their reduced aspartate+1 enrichment due to dilutional effect, and may provide a substrate for increased succinate generated by malate dismutation, as well as play a role in its pronounced longevity phenotype.
3.3. Gas chromatography/mass spectrometry (GC/MS) analysis of absolute isotopic enrichment in organic acids of complex I mutant (gas-1) and wild-type (N2) worms
3.3.1. Complex I mutant gas-1 worms accumulate increased lactate and citrate in all molecular species, increased malate+1, and decreased succinate+2 relative to wild-type worms
Absolute isotopic enrichment was assessed in a wider array of whole worm free organic acids than possible through isotope ratio MS by using GC/MS in gas-1 relative to N2 wild-type controls, where synchronous populations of first day young adult worms were studied after being fed universally-labeled glucose for 24 h (Fig. 6). This analysis confirmed gas-1 has significantly increased lactate as was seen by IR/MS analysis (Fig. 4A), which by GC/MS showed a 2-fold increase in lactate+1 (p < 0.05) and a 6-fold increase in lactate+2 (p < 0.05) relative to wild-type (Fig. 6A). Identification of increased lactate+2 species in the gas-1 mutant is suggestive that at least one other metabolic pathway besides glycolysis likely contributes to the total lactate pool, although it is not clear whether this is an NADH versus NADPH dependent process, as might potentially involve recycling through PEPCK, MDH, PC, and PDH (Fig. 2). Although enrichment in the distal TCA cycle intermediate, malate, was relatively low in both strains, overall isotopic enrichment in malate+1 species reached 1.8% APE in gas-1, which represented a 4-fold enrichment relative to wild-type (p < 0.001) (Fig. 6B). While mean enrichment was increased in malate+1 by isotope ratio MS, this change did not reach statistical significance (Fig. 4B). Interestingly, gas-1 had no enrichment among any molecular species of succinate (Fig. 6C), similarly as had been seen by isotope ratio MS analysis of succinate+1 (Fig. 6C). Instead, gas-1 had a significant decrease in succinate+2 with nearly no labeling of succinate+2, +3, or +4 species in N2. Most pronounced was the dramatic accumulation of the proximal TCA metabolite, citrate, in the gas-1 mutant worm, which was undetectable over background in N2 but reached absolute citrate+1 enrichment greater than 20% APE in gas-1 (p < 0.01) (Fig. 6D). While careful dissection of the data suggests citrate+1 enrichment may also have been increased by IR/MS analysis, this change failed to reach statistical significance (Fig. 6D). Thus, while this alternative analysis validated that there is substantial increase in lactate enrichment and no substantial alteration in succinate +1 flux in gas-1 relative to N2, GC/MS analysis with a universally-labeled glucose precursor provided additional data relative to isotope ratio MS to support that significant citrate+1 and malate+1 enrichment does indeed occur in the RC complex I mutant worms.
Overall, the finer-scale data achieved by application of GC/MS analysis to trace absolute enrichment from a universally-labeled glucose substrate in adult worms suggests that the gas-1 complex I mutant worm does indeed sustain alterations in TCA flux. Specifically, gas-1 accumulated increased glucose-derived carbon isotope in proximal TCA species (citrate) and preceding analytes (lactate) but had nearly no enrichment evident in the higher molecular species of succinate, which is located in the mid-TCA cycle. Given this apparent disruption in complete TCA cycle flux, the significant accumulation of isotope-derived label in the distal TCA intermediate, malate, may be indicative that there is some degree of alternate TCA flux involving the glyoxylate shunt to divert carbon through the middle of the TCA cycle [31]. Alternatively, it is possible this increase in malate labeling represents reverse TCA flux in the setting of elevated TCA precursors such as lactate and presumably, pyruvate.
4. Conclusion
Stable isotopic/mass spectrometric analysis of precursor–product relationships in living animals can sensitively discriminate primary RC dysfunction from genetic deficiencies in either TCA cycle or PDH enzymes. Interestingly, mutations impacting two central nodes of the nutrient-sensing signaling network that decrease sirtuin and increase FOXO1 signaling [13] yield intermediary metabolic flux profiles most similar to the TCA cycle mutants. Collectively, these data are suggestive that metabolic flux analysis using stable isotopes may offer a robust means to discriminate sites of specific intermediary metabolic disorders. In addition, this approach can potentially facilitate the noninvasive monitoring of individuals with metabolic disease to quantify the secondary effects of primary RC dysfunction across central intermediary metabolic pathways.
Supplementary data to this article can be found online at http://dx. doi.org/10.1016/j.ymgme.2013.12.011.
Supplementary Material
Acknowledgments
We are grateful to Evgueni Daikhin, MD, PhD, and Ilana Nissim for their assistance with sample analysis in the Children's Hospital of Philadelphia Metabolomics Core Facility. This work was funded in part by the National Institutes of Health (K08-DK073545, R01-HD065858, and P30-HD026979 Intellectual and Developmental Disabilities Research Center New Investigator Award to M.J.F.; and 5-K12-DK094723 to S.M.), The Philadelphia Foundation (M.J.F.), The University of Pennsylvania McCabe Award (M.J.F.), and the Tristan Mullen Fund (M.J.F. and M.Y.). The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Abbreviations
- C. elegans
Caenorhabditis elegans
- RC
respiratory chain
- PDH
pyruvate dehydrogenase
- TCA
tricarboxylic acid
- GC/MS
gas chromatography/mass spectrometry
- HPLC
high performance liquid chromatography
- RNAi
RNA interference
- BCAA
branched chain amino acids
- CoQ
coenzyme Q
- NGM
nematode growth media
- NMR
nuclear magnetic resonance
Footnotes
Conflict of interest
The authors have no competing interests to declare.
References
- 1.Haas RH, Parikh S, Falk MJ, Saneto RP, Wolf NI, Darin N, Cohen BH. Mitochondrial disease: a practical approach for primary care physicians. Pediatrics. 2007;120:1326–1333. doi: 10.1542/peds.2007-0391. [DOI] [PubMed] [Google Scholar]
- 2.Haas RH, Parikh S, Falk MJ, Saneto RP, Wolf NI, Darin N, Wong LJ, Cohen BH, Naviaux RK. The in-depth evaluation of suspected mitochondrial disease. Mol. Genet. Metab. 2008;94:16–37. doi: 10.1016/j.ymgme.2007.11.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Falk MJ. In: MITO 101 — genetic counseling for mitochondrial disease. Parikh S, DiMauro S, editors. United Mitochondrial Disease Foundation; 2008. [Google Scholar]
- 4.McCormick E, Place E, Falk MJ. Molecular genetic testing for mitochondrial disease: from one generation to the next. Neurotherapeutics. 2013;10:251–261. doi: 10.1007/s13311-012-0174-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Yudkoff M, Ah Mew N, Daikhin Y, Horyn O, Nissim I, Payan I, Tuchman M. Measuring in vivo ureagenesis with stable isotopes. Mol. Genet. Metab. 2010;100(Suppl. 1):S37–S41. doi: 10.1016/j.ymgme.2010.02.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perez CL, Van Gilst MR. A 13C isotope labeling strategy reveals the influence of insulin signaling on lipogenesis in C. elegans. Cell Metab. 2008;8:266–274. doi: 10.1016/j.cmet.2008.08.007. [DOI] [PubMed] [Google Scholar]
- 7.Falk MJ, Rao M, Ostrovsky J, Daikhin E, Nissim I, Yudkoff M. Stable isotopic pro-filing of intermediary metabolic flux in developing and adult stage Caenorhabditis elegans. J. Vis. Exp. 2011;(48):2288. doi: 10.3791/2288. http://dx.doi.org/10.3791/2288. [DOI] [PMC free article] [PubMed]
- 8.Kayser EB, Morgan PG, Hoppel CL, Sedensky MM. Mitochondrial expression and function of GAS-1 in Caenorhabditis elegans. J. Biol. Chem. 2001;276:20551–20558. doi: 10.1074/jbc.M011066200. [DOI] [PubMed] [Google Scholar]
- 9.Kayser EB, Morgan PG, Sedensky MM. GAS-1: a mitochondrial protein controls sensitivity to volatile anesthetics in the nematode Caenorhabditis elegans. Anesthesiology. 1999;90:545–554. doi: 10.1097/00000542-199902000-00031. [DOI] [PubMed] [Google Scholar]
- 10.Hosokawa H, Ishii N, Ishida H, Ichimori K, Nakazawa H, Suzuki K. Rapid accumulation of fluorescent material with aging in an oxygen-sensitive mutant mev-1 of Caenorhabditis elegans. Mech. Ageing Dev. 1994;74:161–170. doi: 10.1016/0047-6374(94)90087-6. [DOI] [PubMed] [Google Scholar]
- 11.Feng J, Bussiere F, Hekimi S. Mitochondrial electron transport is a key determinant of life span in Caenorhabditis elegans. Dev. Cell. 2001;1:633–644. doi: 10.1016/s1534-5807(01)00071-5. [DOI] [PubMed] [Google Scholar]
- 12.Jonassen T, Marbois BN, Faull KF, Clarke CF, Larsen PL. Development and fertility in Caenorhabditis elegans clk-1 mutants depend upon transport of dietary coenzyme Q8 to mitochondria. J. Biol. Chem. 2002;277:45020–45027. doi: 10.1074/jbc.M204758200. [DOI] [PubMed] [Google Scholar]
- 13.Zhang Z, Tsukikawa M, Peng M, Polyak E, Nakamaru-Ogiso E, Ostrovsky J, McCormack S, Place E, Clarke C, Reiner G, McCormick E, Rappaport E, Haas R, Baur JA, Falk MJ. Primary respiratory chain disease causes tissue-specific dysregulation of the global transcriptome and nutrient-sensing signaling network. PLoS One. 2013;8:e69282. doi: 10.1371/journal.pone.0069282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Wang Y, Tissenbaum HA. Overlapping and distinct functions for a Caenorhabditis elegans SIR2 and DAF-16/FOXO. Mech. Ageing Dev. 2006;127:48–56. doi: 10.1016/j.mad.2005.09.005. [DOI] [PubMed] [Google Scholar]
- 15.Kamath RS, Ahringer J. Genome-wide RNAi screening in Caenorhabditis elegans. Methods. 2003;30:313–321. doi: 10.1016/s1046-2023(03)00050-1. [DOI] [PubMed] [Google Scholar]
- 16.Falk MJ, Rosenjack JR, Polyak E, Suthammarak W, Chen Z, Morgan PG, Sedensky MM. Subcomplex Ilambda specifically controls integrated mitochondrial functions in Caenorhabditis elegans. PLoS One. 2009;4:e6607. doi: 10.1371/journal.pone.0006607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Timmons L, Court DL, Fire A. Ingestion of bacterially expressed dsRNAs can produce specific and potent genetic interference in Caenorhabditis elegans. Gene. 2001;263:103–112. doi: 10.1016/s0378-1119(00)00579-5. [DOI] [PubMed] [Google Scholar]
- 18.Dingley S, Polyak E, Lightfoot R, Ostrovsky J, Rao M, Greco T, Ischiropoulos H, Falk MJ. Mitochondrial respiratory chain dysfunction variably increases oxidant stress in Caenorhabditis elegans. Mitochondrion. 2010;10:125–136. doi: 10.1016/j.mito.2009.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jones BN, Gilligan JP. o-Phthaldialdehyde precolumn derivatization and reversed-phase high-performance liquid chromatography of polypeptide hydroly-sates and physiological fluids. J. Chromatogr. 1983;266:471–482. doi: 10.1016/s0021-9673(01)90918-5. [DOI] [PubMed] [Google Scholar]
- 20.Clarke C, Xiao R, Place E, Zhang Z, Sondheimer N, Bennett M, Yudkoff M, Falk MJ. Mitochondrial respiratory chain disease discrimination by retrospective cohort analysis of blood metabolites. Mol. Genet. Metab. 2013;110:145–152. doi: 10.1016/j.ymgme.2013.07.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Butler JA, Mishur RJ, Bokov AF, Hakala KW, Weintraub ST, Rea SL. Profiling the anaerobic response of C. elegans using GC–MS. PLoS One. 2012;7:e46140. doi: 10.1371/journal.pone.0046140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Reinke SN, Hu X, Sykes BD, Lemire BD. Caenorhabditis elegans diet significantly affects metabolic profile, mitochondrial DNA levels, lifespan and brood size. Mol. Genet. Metab. 2010;100:274–282. doi: 10.1016/j.ymgme.2010.03.013. [DOI] [PubMed] [Google Scholar]
- 23.Fuchs S, Bundy JG, Davies SK, Viney JM, Swire JS, Leroi AM. A metabolic signature of long life in Caenorhabditis elegans. BMC Biol. 2010;8:14. doi: 10.1186/1741-7007-8-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Butler JA, Mishur RJ, Bhaskaran S, Rea SL. A metabolic signature for long life in the Caenorhabditis elegans Mit mutants. Aging Cell. 2013;12:130–138. doi: 10.1111/acel.12029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kimura KD, Tissenbaum HA, Liu Y, Ruvkun G. daf-2, an insulin receptor-like gene that regulates longevity and diapause in Caenorhabditis elegans. Science. 1997;277:942–946. doi: 10.1126/science.277.5328.942. [DOI] [PubMed] [Google Scholar]
- 26.Lin K, Hsin H, Libina N, Kenyon C. Regulation of the Caenorhabditis elegans longevity protein DAF-16 by insulin/IGF-1 and germline signaling. Nat. Genet. 2001;28:139–145. doi: 10.1038/88850. [DOI] [PubMed] [Google Scholar]
- 27.Edwards CB, Copes N, Brito AG, Canfield J, Bradshaw PC. Malate and fumarate extend lifespan in Caenorhabditis elegans. PLoS One. 2013;8:e58345. doi: 10.1371/journal.pone.0058345. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wadsworth WG, Riddle DL. Developmental regulation of energy metabolism in Caenorhabditis elegans. Dev. Biol. 1989;132:167–173. doi: 10.1016/0012-1606(89)90214-5. [DOI] [PubMed] [Google Scholar]
- 29.Lee RY, Hench J, Ruvkun G. Regulation of C. elegans DAF-16 and its human ortholog FKHRL1 by the daf-2 insulin-like signaling pathway. Curr. Biol. 2001;11:1950–1957. doi: 10.1016/s0960-9822(01)00595-4. [DOI] [PubMed] [Google Scholar]
- 30.Easlon E, Tsang F, Skinner C, Wang C, Lin SJ. The malate–aspartate NADH shuttle components are novel metabolic longevity regulators required for calorie restriction-mediated life span extension in yeast. Genes Dev. 2008;22:931–944. doi: 10.1101/gad.1648308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Butler JA, Ventura N, Johnson TE, Rea SL. Long-lived mitochondrial (Mit) mutants of Caenorhabditis elegans utilize a novel metabolism. FASEB J. 2010;24:4977–4988. doi: 10.1096/fj.10-162941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Falk MJ, Zhang Z, Rosenjack JR, Nissim I, Daikhin E, Nissim I, Sedensky MM, Yudkoff M, Morgan PG. Metabolic pathway profiling of mitochondrial respiratory chain mutants in C. elegans. Mol Genet Metab. 2008;93(4):388–397. doi: 10.1016/j.ymgme.2007.11.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.