Abstract
Light-driven oxidation of water to dioxygen in plants, algae, and cyanobacteria is catalyzed within photosystem II (PS II) by a Mn4Ca cluster. Although the cluster has been studied by many different methods, its structure and mechanism have remained elusive. X-ray absorption and emission spectroscopy and extended X-ray absorption fine structure studies have been particularly useful in probing the electronic and geometric structures and the mechanism of the water oxidation reaction. Recent progress, reviewed here, includes polarized X-ray absorption spectroscopy measurements of PS II single crystals. Analysis of those results has constrained the Mn4Ca cluster geometry to a set of three similar high-resolution structures. The structure of the cluster from the present study is unlike either the 3.0- or 3.5-Å-resolution X-ray structures or other previously proposed models. The differences between the models derived from X-ray spectroscopy and crystallography are predominantly because of damage to the Mn4Ca cluster by X-rays under conditions used for the structure determination by X-ray crystallography. X-ray spectroscopy studies are also used for studying the changes in the structure of the Mn4Ca catalytic center as it cycles through the five intermediate states known as the Si states (i = 0–4). The electronic structure of the Mn4Ca cluster has been studied more recently using resonant inelastic X-ray scattering spectroscopy (RIXS), in addition to the earlier X-ray absorption and emission spectroscopy methods. These studies are revealing that the assignment of formal oxidation states is overly simplistic. A more accurate description should consider the charge density on the Mn atoms, which includes the covalency of the bonds and delocalization of the charge over the cluster. The geometric and electronic structures of the Mn4Ca cluster in the S states derived from X-ray spectroscopy are leading to a detailed understanding of the mechanism of O–O bond formation during the photosynthetic water-splitting process.
I. Introduction
Photosynthetic water oxidation is a fundamental chemical reaction that sustains the biosphere. It takes place at a catalytic Mn4Ca site within the oxygen-evolving complex (OEC) of photosystem II (PS II), which is embedded in the thylakoid membranes of green plants, cyanobacteria, and algae.1 Life-supporting oxygen, currently present at 20% in the Earth's atmosphere, is a consequence of this process by oxygenic photosynthetic organisms that are able to use sunlight as the energy source and water as the primary reductant for the fixation of carbon. Only because of the constant regeneration of oxygen through the photosynthetic water oxidation reaction is it abundant in the atmosphere. Although 3 billion years of evolution have created many differences between oxygenic photosynthetic organisms, remarkably the OEC has remained, to our current knowledge, practically unchanged.
The heart of the OEC is a cluster of four Mn ions and one Ca ion (Mn4Ca) that have been shown by X-ray absorption spectroscopy (XAS) to be linked by mono- and di-μ-oxo or hydroxo bridges.2–4 The specific protein environment and probably one Cl ion are also essential for optimal water-splitting activity, which is driven by light-induced charge separation at the P680 chlorophyll pigments of the PS II reaction center. During water oxidation, the OEC cycles through five different oxidation states (Figure 1), which are known as Si states (i = 0–4), coupling the one-electron photochemistry of the reaction center with the four-electron chemistry of water oxidation.5 The OEC in the cluster undergoes oxidation in response to electron transfers driven by photon absorption by the reaction center pigments. The intermediate oxidized species are sufficiently stable to accumulate the oxidizing equivalents needed for the four-electron transfer of the final step. Thus, nature avoids releasing harmful chemical intermediates such as superoxide or peroxide during the water oxidation. Once four oxidizing equivalents are accumulated in the OEC (S4 state), a spontaneous reaction occurs that results in the release of dioxygen and the formation of the S0 state.
Figure 1.
S-state cycle and the proposed oxidation states of the Mn cluster in the S states. The MLSs for the S0 and S2 states and the g values of the EPR signals identified with S0, S1, S2, and S3 (for spinach) are also shown.
The structure of the catalytic Mn4Ca complex has been the subject of intense study ever since Mn was identified as an essential element for oxygenic photosynthesis.1,2,6–9 The critical questions related to this process concern the structural and electronic state and the changes in the Mn4Ca complex as the OEC proceeds through the S-state cycle. The urgent need to develop renewable energy resources that are essentially carbon-neutral has highlighted the importance of learning how water is oxidized to oxygen during photosyn-thesis.10 The focus of this review is on recent results from our laboratory that address these questions, using polarized extended X-ray absorption fine structure (EXAFS) of single crystals of PS II,11 range-extended EXAFS,12,13 and resonant inelastic X-ray scattering spectroscopy (RIXS).14,15
II. Structure of the Mn4Ca Cluster in Photosynthesis
A. Single-Crystal X-ray Spectroscopy
A key to understanding the mechanism of water oxidation by the OEC in PS II is the determination of the structure of the Mn4Ca cluster, which has eluded both Mn EXAFS3,4,16,17 and X-ray diffraction (XRD) studies18–22 until now. XRD studies have provided critical information about its structure; however, even the XRD data of the highest resolution presently available at 3.0 Å are insufficient to accurately determine the positions of Mn, Ca, and the bridging and terminal ligands. This is reflected by the differences in the placement of the metal ions and putative ligands in the 3.022 and 3.5 Å21 structures. Moreover, there is a serious concern of X-ray-induced damage to the Mn4Ca cluster; hence, it is important to review the issue of the X-ray dose used in X-ray crystallography and spectroscopy studies. Therefore, before addressing the issue of Mn4Ca models derived from single-crystal X-ray spectroscopy,11 we present a brief review of the Mn XANES and EXAFS data that are directly relevant to the problem of X-ray damage to the Mn4Ca cluster.23 It is a case study for other redox-active metalloproteins.
1. Damage by X-rays: A Case Study for Metallo-protein Crystallography
At present, there are discrepancies among the models of the structure of the Mn4Ca complex in the published X-ray crystallography studies and inconsistencies with X-ray,24–26 electron paramagnetic resonance (EPR),27–29 and Fourier transform IR (FTIR)30–32 spectroscopy data (Figure 2) This disagreement is predominantly a function of X-ray-induced damage to the catalytic metal site.
Figure 2.
Structural models of the Mn cluster containing short 2.7–2.8 Å Mn–Mn distances characterized by di-μ-oxo bridges and 3.3 Å Mn–Mn distances characterized by mono-μ-oxo bridges. The numbers in parentheses denote the number of short (2.7–2.8 Å) and long Mn–Mn (~3.3 Å) distances. The topologically equivalent motifs to those proposed from X-ray crystallographic analyses at 3.0 and 3.5 Å are (A) on top left22 and (B) right.21 (C and D) Three short (2.7–2.8 Å) and one long (3.3 Å) Mn–Mn distances (bottom) are required to satisfy the solution12,13 and single-crystal EXAFS (bottom).11 Of these motifs, only one option, on the extreme right (D, bottom), has the requisite numbers of short and long Mn–Mn distances with the correct relative orientation to satisfy the dichroism observed in the single-crystal polarized EXAFS spectra.11
It is therefore imperative to monitor the X-ray-induced effects on the electronic and geometric structures of the Mn4Ca complex in situ to monitor the X-ray-induced damage or lack thereof of the Mn4Ca complex to validate the structures based on X-ray crystallography.
In order to determine a safe level of X-ray dose for our single-crystal X-ray spectroscopy studies, we conducted a comprehensive survey of the effects of the radiation dose, energy of the X-rays, and temperature of irradiation on the structure of the Mn4Ca cluster in single crystals and solutions of PS II.23 These studies have revealed that the conditions used for structure determination by XRD methods are damaging to the metal-site structure, as monitored by X-ray absorption near-edge structure (XANES) and EXAFS spectroscopy, for both cyanobacteria and spinach at room and lower temperatures.23,33 With the advent of third-generation synchrotron sources, the intensity of X-rays available for crystallography has increased considerably, and X-ray damage to biomolecules has been a continuing concern. This is a well-recognized problem, and various steps have been taken to minimize such damage.34 In most crystallography studies, it is the loss of diffractivity, as evidenced by the loss of resolution, that is used as a measure of X-ray damage.35 However, as shown in our study,23 this method is not sensitive to localized damage, as is prevalent in redox-active metal-containing proteins. We used in situ XANES and EXAFS to show that for the Mn4Ca cluster in PS II the damage to the metal site precedes the loss of diffractivity by more than an order of magnitude of the X-ray dose. Because structure–function correlations are routinely made using the structures determined by X-ray crystallography, it is important to know that the metal-site structures are indeed intact. This is particularly important for radiation-sensitive metalloproteins.
The Mn XANES data from PS II single crystals show that, following X-ray doses characteristic of the XRD measurements, Mn is reduced to Mn(II) from Mn4(III2,IV2) present in the S1 state (Figure 3). The reduction of Mn is similar in PS II crystals (Figure 3A) and in solution (Figure 3B), and the quantity of Mn(II) can be accurately quantitated as shown in Figure 3C. Figure 3D shows typical XANES spectra from Mn complexes in oxidation states II-IV. The X-ray dose used for exposure is from 0.14 to 5.0 × 1010 photons/μm2 (light-blue to black lines, Figure 3A). An average dose of ~3.5 × 1010 photons/μm2 was used for XRD studies. The exposure was at 100 K, and all XANES data were collected at 10 K at low dose (1 × 107 photons/μm2). XANES and difference spectra show that the increase in the amplitude at ~6552 eV provides definite evidence for the photoreduction to Mn(II) in PS II crystals by exposure to X-rays. At a dose of 2.3 × 1010 photons/μm2, equal to 66% of the average dose used for diffraction measurements, 80% of the Mn ions in PS II are present as Mn(II). The damage results are summarized in Figure 4. The dose on the abscissa is given in grays and in photons/unit area, units that are commonly used for crystallography and spectroscopy experiments, respectively.
Figure 3.
(A) Mn K-edge shift of PS II crystals as a function of the X-ray dose at 13.3 keV (0.933 Å) and 100 K (top) and the difference spectra (below). The spectrum at the highest inflection point energy (IPE) is from an undamaged PS II crystal. (B) Similar trend in the XANES (top) and the difference spectra (below) as seen for PS II solutions. (C) Changes in the XANES spectra from an intact PS II sample as a function of the addition of 10% increments of the XANES spectrum of aqueous Mn(II) (top) and the corresponding difference spectra (below) (light-blue to black lines). The similarity between the difference spectra in A–C is striking, showing the photoreduction of Mn(III2,IV2) in native PS II to Mn(II). (D) Comparison of Mn K-edge spectra from two tetranuclear complexes, [Mn4O3(OAc)4(dbm)3]104,105 and [(Mn2O2)2(tphpn)2](ClO4)4,106,107 in oxidation states Mn4(III3,IV), Mn4(III2,IV2), and Mn(II) in aqueous solution. The K-edge spectra from the complexes in higher oxidation states are similar to that from an intact PS II sample. The figure was adapted from Yano et al.23
Figure 4.
Increasing Mn(II) content in PS II due to radiation damage. (Solid blue line) Mn(II) content in PS II crystals as a function of X-ray irradiation at 13.3 keV (0.933 Å). The irradiation was carried out at 100 K. The conditions are similar to those during XRD data collection. (Dashed blue line) The damage profile for PS II solution samples, which is very similar to that seen for crystals although slightly higher for the same dose. EXAFS spectra of samples shown in Figure 5 are at the levels of damage denoted (on the right) by A–D (0.05, 0.1, 0.3, and 1.4 × 1010 photons/μm2, respectively, at 13.3 keV and 100 K). An average dose of ~3.5 × 1010 photons/μm2 was used for XRD studies. (Dashed green line) Generation of Mn(II), which is considerably greater when the X-ray irradiation is at 6.6 keV (1.89 Å), which is the energy at which the anomalous diffraction measurements for PS II were conducted. (Solid blue line) Mn(II) produced by damage in crystals, which is considerably decreased when the irradiation is conducted at 10 K. The figure is adapted from Yano et al.23
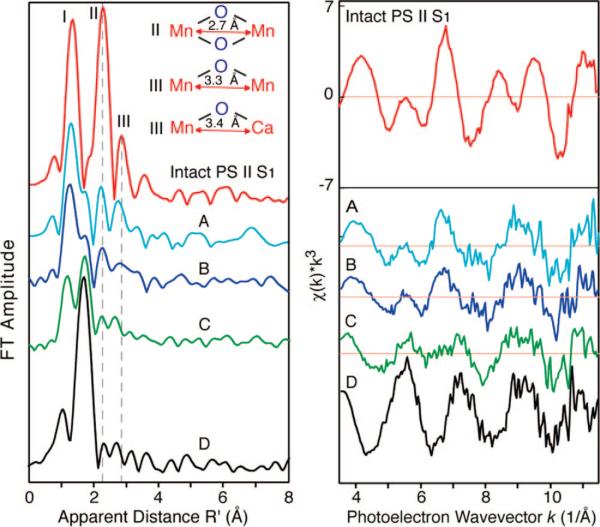
In addition to Mn reduction, the EXAFS spectrum changes significantly at the dose that was used for X-ray crystallography experiments. The spectrum changes from one that is characteristic of a multinuclear oxo-bridged Mn4Ca complex to one that is typical of mononuclear hexacoordinated Mn(II) in solution (Figure 5).23 The three Fourier peaks (Figure 5, left in red) are characteristic of a bridged Mn4Ca complex. Peak I derives from bridging Mn–oxo and Mn–terminal ligand atoms, peak II is from Mn–Mn distances at 2.7 Å characteristic of di-μ-oxo-bridged moieties, and peak III is from mono-μ-oxo-bridged Mn–Mn distances at 3.3 Å and Mn–Ca distances at 3.4 Å. The dosages corresponding to A–D are labeled in Figure 4. The Fourier peaks exhibit drastic changes as the percent of Mn(II) increases. Peaks II and III (vertical dashed gray lines) that are characteristic of Mn–Mn distances at 2.7, 3.3, and 3.4 Å decrease and disappear along with peak I, which is due to Mn–O bridging atoms. At the same time, the appearance of a longer Mn–ligand interaction at 2.2 Å is observed. The EXAFS modulations (Figure 5, right) from the intact PS II spectrum contain several sinusoidal components, as expected from a bridged multinuclear Mn4Ca complex. The spectrum of sample D is significantly different in both frequency and phase of the modulations with just one sinusoidal oscillation, as one would expect from a symmetric hexacoordinated Mn(II) species with one shell of backscattering atoms. The spectra from samples A–C are intermediate, with considerable damping in the amplitude and changes in the frequency components to the spectra. The results are similar for PS II solution and crystal samples.
Figure 5.
Spectral changes of PS II Mn EXAFS due to radiation damage. (left) The FT of the EXAFS spectrum from an intact PS II solution sample is on top (red). The FTs from PS II solution samples exposed to 0.05, 0.1, 0.3, and 1.4 × 1010 photons/μm2 at 13.3 keV and 100 K and containing 25 (blue), 35 (dark blue), 45 (green), and 90% (black) photoreduced Mn(II) are labeled A–D, respectively. An average dose of ~3.5 × 1010 photons/μm2 was used for XRD studies. (right) Corresponding k3-space EXAFS spectra of PS II In addition to Mn reduction, the EXAFSsamples. The figure is adapted from Yano et al.23
These X-ray spectroscopy results present clear evidence that there are no specific metal-bridging or metal–metal interactions in PS II samples irradiated at the average dose used for X-ray crystallography. This is shown by the disappearance of the Fourier peaks that are characteristic of Mn-bridging oxo, and Mn–Mn/Mn–Ca interactions with di-μ-oxo or mono-μ-oxo bridges, and by the appearance of Mn–O distances characteristic of Mn(II)–O ligand bond lengths.
The density seen in the XRD results is in all probability from the Mn(II) hexacoordinated ions that are distributed in the binding pocket, presumably bound to carboxylate ligands and free water. This produces a distribution of metal binding sites and the resultant unresolved electron density.
These studies reveal that the conditions used for structure determination by X-ray crystallography cause serious damage specifically to the metal-site structure, resulting not only in the reduction of Mn (Figures 3 and 4) but also in the disruption of the bridged Mn structure (Figure 5). The results show, furthermore, that the damage to the active metal site occurs at a much lower X-ray dose than that causing the loss of diffractive power of the crystals as established by X-ray crystallography. The damage is significantly higher at wavelengths used for anomalous diffraction measurements and is much lower at liquid He temperatures (10 K) compared to 100 K, where the crystallography experiments were conducted (Figure 4). For future X-ray crystallography work on the Mn4Ca complex, it will therefore be imperative to develop protocols that mitigate the X-ray-induced damage. More generally, these data show that in redox-active metalloproteins careful evaluations of the structural intactness of the active site(s) are required before structure–function correlations can be made on the basis of high-resolution X-ray crystal structures.
2. Structure of the Mn4Ca Complex from Polarized EXAFS of PS II Crystals
PS II membranes can be oriented on a substrate such that the membrane planes are roughly parallel to the substrate surface. This imparts a one-dimensional order to these samples; while the z axis for each membrane (collinear with the membrane normal) is roughly parallel to the substrate normal, the x and y axes remain disordered.36 Exploiting the plane-polarized nature of synchrotron radiation, spectra can be collected at different angles between the substrate normal and the X-ray e vector. The dichroism of the absorber–backscatterer pair present in the oriented samples is reflected in, and can be extracted from, the resulting X-ray absorption spectra.17,24,37–39 This is possible because the EXAFS amplitude is proportional to ~cos2 θ, where θ is the angle between the X-ray e vector and the absorber–backscatterer vector. The EXAFS of the oriented PS II samples exhibits distinct dichroism, from which we have deduced the relative orientations of several interatomic vector directions relative to the membrane normal and derived a topological representation of the metal sites in the OEC. However, because the samples are ordered in only one dimension, the dichroism information is available only in the form of an angle with respect to the membrane normal. For EXAFS measurements, this means that the absorber–backscatterer vectors can lie anywhere on a cone defined by the angle that the vector forms with the membrane normal.
Further refinement can be performed if samples with three-dimensional order, i.e., single crystals, are examined instead of oriented membranes.40,41 These studies have been able to significantly expand the X-ray absorption spectroscopic information available for these systems over what is gleaned from studies of isotropic samples.
We have developed the methodology for collecting single-crystal XAS data from PS II. XAS spectra were measured at 10 K using a liquid He cryostat or cryostream, and an imaging plate detector placed behind the sample was used for in situ collection of diffraction data and determination of the crystal orientation (Figure 6). EXAFS experiments with PS II require a substantially lower X-ray dose than XRD measurements, and the onset of radiation damage can be precisely determined and controlled by monitoring the Mn K-edge position, thus allowing the collection of data from the intact Mn4Ca cluster of PS II. In addition, EXAFS provides metal-to-metal and metal-to-ligand distances with high accuracy (~0.02 Å) and a resolution of ~0.1 Å. Mn and Ca EXAFS studies of frozen solutions of PS II preparations have provided accurate distances and numbers of Mn–Mn, Mn–Ca, and Mn/Ca–ligand vectors in the Mn4Ca cluster and have led to the development of several possible structural models for the Mn4Ca cluster.
Figure 6.
Experimental setup used for single-crystal X-ray absorption experiments. The PS II crystal was placed in a liquid-He cryostat at a temperature of 10 K at ambient pressure. The Mn XANES or EXAFS spectrum was measured as a fluorescence excitation spectrum using a 30-element energy-discriminating solid-state Ge detector placed at 90° to the incident beam. Downstream of the X-ray beam and the PS II crystal is the MAR 345 image-plate detector that was used for obtaining the diffraction pattern from which the orientation matrix of the PS II crystal was determined by indexing the reflections. The incident X-ray beam is defined by the slits in the left foreground, and the I0 ion chamber, which is behind the slits, is used to measure the intensity of the incident beam. A shutter that protects the sample from damage during spectrometer motion is behind the ion chamber and before the cryostat.
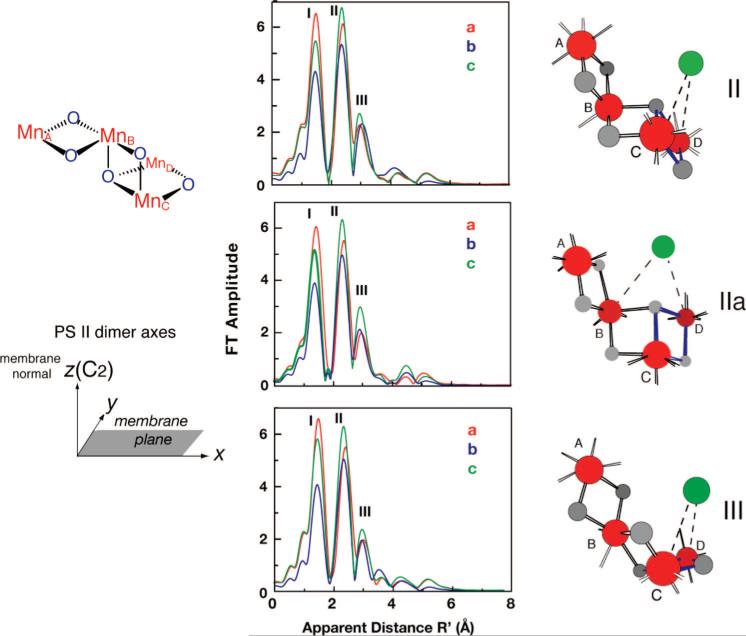
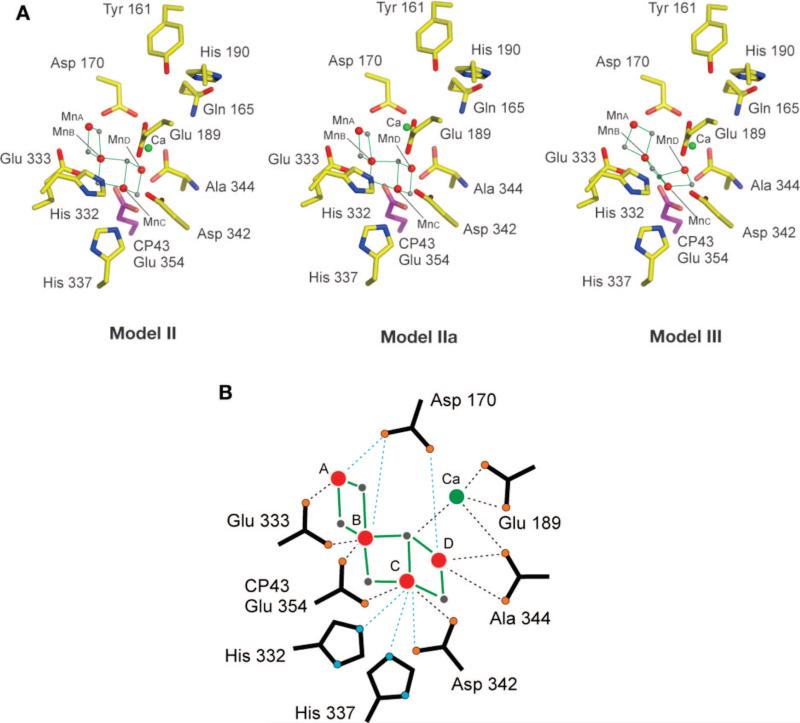
We have reported the polarized EXAFS spectra of oriented PS II single crystals, which result in a set of similar high-resolution structures for the Mn4Ca cluster (Figure 7).11 These were derived from simulations of single-crystal polarized spectra based on the experimentally observed dichroism. These structural models are dependent only on the spectroscopic data and are independent of the electron density from XRD studies. In order to discriminate between symmetry-related structural models that are possible because of the presence of a noncrystallographic C2 axis, the cluster was placed within PS II, taking into account the overall shape of the electron density of the metal site and the putative ligand information from X-ray crystallography, including IR and EPR studies. The center of mass of the structural models was translated to the center of the electron density attributed to the Mn4Ca cluster in the 3.0 Å XRD structure.22 The models were not optimized with respect to the electron density or rotated with respect to the crystal axes because that would change the dichroic characteristics. Of the four options shown by Yano et al.,11 it is possible to rule out model I on the basis of oriented-membrane EXAFS data.13,24 The other three models, II, IIa, and III, in the putative ligand environment are shown in Figure 8. However, we caution that the ligand positions are tentative because they are likely to be affected by radiation damage, as are the positions of the metal atoms from XRD.23,33
Figure 7.
High-resolution structural models for the Mn4Ca cluster in PS II from polarized EXAFS. The Mn4Ca models II, IIa, and III (from Yano et al.11) are shown on the right. Each model is compatible with the polarized Mn EXAFS spectra of single crystals of PS II (see Figure 9A, bottom). The Mn4 motif common to the three structures is shown at upper left. The models are shown in the orientation in which they should be placed in the PS II membrane according to the axis system shown at the upper right. The Mn atoms are shown in red. The distance between the MnC and MnD atoms is ~2.8 Å (indicated by blue oxo bonds), and the distance between the MnA and MnB atoms, as well as the MnB and MnC atoms, is ~2.7 Å. The distance between MnB and MnD is ~3.3 Å. The Ca atom (green sphere) is ~3.4 Å from two Mn atoms. The bridging motif to Ca is not well-defined by our experiments; therefore, dashed lines connect the Ca atom to the two Mn atoms at ~3.4 Å. In the middle are the FTs from the calculated polarized EXAFS spectra of the three models, and these compare well with the FTs from the experimental polarized EXAFS spectra shown in the bottom of Figure 9A.
Figure 8.
(A) Structural models II, IIa, and III placed in the ligand environment from the XRD data of Loll et al.22 at a resolution of 3.0 Å. The center of mass was translated to the center of the electron density attributed to the Mn4Ca cluster in the XRD structure. This was carried out without any rotation of the models. (B) Schematic diagram of model II with the view along the membrane plane, with putative ligands from the electron density. Bonds between Mn and the bridging oxo are shown as solid green lines. Bonds to putative terminal ligand atoms from amino acids and to Ca atoms are shown as dotted lines (black, <3.0 Å; blue, >3.0 Å).
The structure of the Mn4Ca cluster from the polarized EXAFS study of PS II single crystals is unlike either the 3.0-22 or 3.5-Å21-resolution X-ray structures and other previously proposed models. The comparison of the experimental data with the polarized EXAFS spectra calculated for the XRD models at 3.521 and 3.0 Å22 resolution is shown in Figure 9A. A comparison of model II from polarized single-crystal EXAFS is overlaid from models derived from the X-ray crystal structures at 3.0 and 3.5 Å in Figure 9B. The disagreement is notable; it is most likely due to the X-ray damage to the Mn4Ca cluster during XRD measurements.23 We emphasize that these differences are in the overall structure and orientation of the cluster and not just in the details of the coordinate positions of the atoms.
Figure 9.
(A) Comparison of the experimental Mn EXAFS dichroism from single crystals of PS II with the polarized spectrum calculated from the proposed structures from XRD studies. There are clear differences between the experimental spectra shown in the bottom panel and those calculated from the XRD coordinates for the Mn4Ca cluster at 3.5 Å resolution (top panel)21 and 3.0 Å resolution (middle panel).22 The intensity and the dichroism pattern of FT peaks I–III (3.5 Å structure) and of FT peaks II and III (3.0 Å structure) are significantly different from the experimental data (bottom panel). For the 3.0 Å resolution structure, the μ-oxo bridges were not resolved in the electron density. Hence, only FT peaks II and III, with contributions from Mn and/or Ca, are shown. (B) Comparison of model II for the Mn4Ca cluster derived from polarized Mn EXAFS with the models derived from the X-ray crystal structures at 3.022 and 3.5 Å21 resolution. The spheres represent Mn (red), Ca (green), and the bridging oxygen ligand atoms (gray) for model II from polarized EXAFS of single crystals and Mn (violet), Ca (cyan), and the bridging oxygen ligand atoms (yellow) for the X-ray crystal structures. (a and b) Comparison of model II with the 3.0-Å-resolution structure for the Mn4Ca cluster. a: The view is along the membrane plane. b: The view is onto the membrane plane from the cytoplasmic (stromal) side. (c and d) Comparison of model II with the 3.5-Å-resolution X-ray structure for the Mn4Ca cluster. c: The view is along the membrane plane. d: The view is onto the membrane plane from the cytoplasmic side.
The polarized EXAFS study of PS II single crystals demonstrates that the combination of XRD and polarized EXAFS can be used to construct a high-resolution model of the Mn4Ca cluster that is not compromised by radiation-induced damage to the cluster.
B. Range-Extended XAS
EXAFS spectra of metallo-protein samples are normally collected as an excitation spectrum by electronically windowing the KR emission (2p to 1s) from the metal atom.42–44 The solid-state detectors used over the past decade have a resolution of 150–200 eV, making it impossible to discriminate the fluorescence of the absorbing atom (Mn in our case for PS II) from that of the following element (Fe in PS II) in the periodic table. Unfortunately, because of the presence of Fe in the PS II samples, we have been limited to a k of about 12 Å–1, which places an inherent limit on the resolution of Mn distances to ~ 0.15 Å because spatial resolution in EXAFS is inversely related to the spectral range. We have overcome this limitation by using a high-resolution crystal monochromator45 that has a resolution of 1 eV. Using the crystal monochromator, we were able to collect EXAFS spectra well beyond the Fe K edge to k = 16.5 Å–1, improving the distance resolution to ~0.1 Å (Figure 10) We have shown the feasibility of the range-extended EXAFS method and collected data without interference from Fe in the sample.12
Figure 10.
(A) Schematic for the crystal monochromator used in a backscattering configuration. (B) Left: X-ray fluorescence of Mn and Fe. Above: Mn Kα1 and Kα2 fluorescence peaks, with a natural line width of ~5 eV, split by 11 eV. The multicrystal monochromator with 1 eV resolution is tuned to the Mn Kα1 peak (red spectrum). Below: fluorescence peaks of Mn and Fe as detected using a Ge detector. The fluorescence peaks are convoluted with the electronic window resolution of 150–200 eV of the Ge detector (black and green spectra for Mn and Fe fluorescence). This method of detection cannot resolve Mn Kα1 and Kα2 fluorescence peaks. Note different energy scales for the schemes are shown above and below. Iron is an obligatory element in functional PS II complexes. Right: Comparison of the PS II Mn K-edge EXAFS spectrum from an S1 state PS II sample obtained with a traditional 30-element energy-discriminating Ge detector with a spectrum collected using the high-resolution crystal monochromator. Use of the high-resolution detector eliminates the interference of Fe and removes the limit of the energy range for Mn EXAFS data collection.
We can now fit two short Mn–Mn distances in the S1 and S2 states, while earlier solution EXAFS studies could discern only one distance of ~2.7 Å. We have also collected range-extended polarized EXAFS data using oriented PS II membranes. These studies also show the heterogeneity clearly; the Mn–Mn distance is ~2.7 or 2.8 Å depending on the angle that the X-ray e vector makes with the membrane normal. Interestingly, for the first time, we were able to resolve the FT peak at ~3.3 Å into one Mn–Mn vector at ~3.2 Å and two Mn–Ca vectors at ~3.4 Å that are aligned at different angles to the membrane normal.13
In Figure 11A, FTs of the range-extended EXAFS for PS II in an isotropic solution and in oriented membranes in the S1 state are compared. Peaks, labeled I–III, correspond to the shells of backscatterers from the Mn absorber. Peak I contains the Mn–O bridging and terminal interactions, peak II corresponds to the di-μ-oxo-bridged Mn–Mn moieties, and peak III has information about the mono-μ-oxo-bridged Mn–Mn and Mn–Ca interactions. Increased spectral resolution results in the detection of the orientation dependence of peaks II and III.
Figure 11.
(A) FT of Mn K-edge EXAFS spectra from isotropic solution and oriented PS II membrane samples in the S1 state obtained with a high-resolution spectrometer (range-extended EXAFS). Angles indicate orientation of the membrane normal with respect to the X-ray e-field vector. (B) Polar plot of the X-ray absorption linear dichroism of PS II samples in the S1 state. The Napp values (solid circles) for oriented membranes are plotted at their respective detection angles (θ). The best fits of Napp vs θ are shown for the various Mn–Mn vectors as solid lines. (C) Orientation of the average Mn–Ca (~3.4 Å) and Mn–Mn (~3.2 Å) vectors relative to the membrane normal. The figure was adapted from Pushkar et al.13
Figure 11B shows polar plots of Napp plotted (solid circles) with respect to the detection angle (θ). By fitting the angle dependence of the amplitude (Napp), the number of back-scatterers (Niso) and the average orientation 〈φ〉 for that shell of scatterers can be determined. Analysis of the orientation dependence of the ~2.7 Å Mn–Mn vector results in two Mn–Mn interactions at an average angle of 〈φ〉 = 61 ± 5°, with respect to the membrane normal. The ~2.8 Å Mn–Mn vector has one Mn–Mn interaction at an angle of 〈φ〉 = 64 ± 10° with respect to the membrane normal. In this way, we independently derived that the Mn–Mn distances of ~2.7 and ~2.8 Å are present in a 2:1 ratio in PS II samples in the S1 state.
The range-extended EXAFS method allowed us to resolve the complex nature of peak III containing at least two peaks, IIIA and IIIB, having different distances of 3.2 and 3.4 Å. Previous Ca EXAFS studies of native PS II,46 Sr EXAFS of Sr-substituted PS II,47 and Mn EXAFS of Ca-depleted PS II48,49 have shown the contribution of both Mn–Mn (~3.2 Å) and Mn–Ca (~3.4 Å) vectors to FT peak III. Peak~IIIB can be assigned to the Mn–Ca vector and peak IIIA to the Mn–Mn vector; moreover, the dichroic behavior of peak IIIB is strikingly similar to that reported previously for the Mn–Sr vector.24 The dichroism of these peaks shows that the Mn–Mn vector at ~3.2 Å is at ~20° to the membrane plane, while the Mn–Ca vector at ~3.4 Å is aligned at ~18° to the membrane normal (Figure 11C).
We have extended these studies to the S0 and S3 intermediate states of the enzymatic cycle, and the preliminary results show that there is a significant change in the structure of the Mn complex in the S3 state. This result has important implications for choosing between the many mechanisms that have been proposed for the water oxidation reaction. In conjunction with the data from single-crystal XAS studies, these studies promise to reveal the changes that occur in the structure of the Mn4Ca catalyst as it cycles through the S states and the mechanism of water oxidation.
III. Electronic Structure of the Mn4Ca Cluster
In addition to the geometric structure, a detailed electronic structure of the Mn4Ca cluster is required before some key questions for understanding the mechanism of photosynthetic water oxidation can be addressed. They are as follows: (1) Do all four oxidizing equivalents necessary to oxidize water to dioxygen accumulate on the four Mn ions of the OEC? (2) Do ligand-centered oxidations take place before the formation and release of dioxygen? (3) What are the oxidation-state assignments for the Mn ion during S-state advancement.50
To understand the mechanism of water oxidation in detail, it is crucial to know whether the extracted electrons accompanying each S-state transition are directly derived from bound water, from the Mn atoms, or from any other parts of the OEC.51 Several spectroscopic methods, such as EPR,52–58 FTIR,30–32,59–62 XAS,2,16,63 and UV spectroscopy,64 have been used to address the oxidation state of Mn in each S state (Figure 1). EPR spectroscopy has shown that the S0 and S2 states are characterized by spin S = ½ ground states, exhibiting multiline EPR signals (MLSs).65–69 The S1 and S3 states are characterized by parallel-polarized EPR signals, indicating integral spin ground states.70–73
XANES has been used extensively to investigate the oxidation state of Mn during the S-state transition.74–76 There is a consensus that Mn-centered oxidation occurs during the S0 to S1 and S1 to S2 transitions. However, there is still controversy concerning the involvement of Mn oxidation in the S2 to S3 transition.50,63,75,76 Within the context of localized oxidation, the formal oxidation state of the native S1 state has been assigned to Mn(III2,IV2) and S2 to Mn(III,IV3). Some groups have proposed Mn(III3,IV) in the S2 state and therefore Mn(III4) for the S1 state,53,77 although most spectroscopic evidence does not support this assignment.29,78 Uncertainty remains in the case of the S0 and S3 states. In the S0 state, one of the questions is whether Mn(II) is present, in which case the oxidation states are Mn(II,III,IV2),69 or whether the oxidation states are Mn(III3,IV).26 In the S3 state, there is controversy about whether a Mn-centered oxidation occurs63,76 or whether a ligand-centered oxidation takes place before O–O bond formation and release of molecular oxygen.74,75,79,80
The disagreement concerning the S2 to S3 transition has led to two different types of proposed dioxygen evolution mechanisms, with one type incorporating the oxidation of the ligand or substrate in the S3 state and the other type invoking Mn oxidation during the S2 to S3 transition.51 Fundamental differences in the chemistry of O–O bond formation and dioxygen evolution exist between these two types of mechanisms.
Mn XANES (1s–3d,4p) and Kβ X-ray emission spectroscopy (XES) (3p–1s) of Mn, including the newly introduced RIXS (1s–3d,2p–1s), have been used to address these questions about the oxidation states and the electronic structure of the Mn4Ca cluster.50
A. Mn K-Edge Absorption and Kβ Emission Spectroscopy
The XANES and Kβ XES flash patterns from a comprehensive study75 are shown in Figure 12, which has led to important conclusions about Mn oxidation states. Both spectroscopies show a clear shift in the spectra between the samples given 0 and 1 flashes. This shift is to higher energy for XANES spectroscopy and to lower energy for Kβ XES, indicating the presence of Mn oxidation during the S1 to S2 transition. The small change between samples given 1 and 2 flashes in both spectroscopies provides strong support for the S2 to S3 transition proceeding without a Mn-centered oxidation. On the next transition, S3 → [S4] → S0, dioxygen is released, shifting the position of the spectra of samples given 3 flashes to lower energy for XANES spectroscopy and to higher energy for Kβ XES. These flash patterns are explained by the proposed oxidation states of the Mn cluster in the S0, S1, S2, and S3 states, as shown in Figure 1. These oxidation states are as follows: S0, Mn4(II,III,IV2) or Mn4(III3,IV); S1, Mn4(III2,IV2); S2, Mn4(III,IV3); S3, Mn4-(III,IV3)•, where the superscript dot represents an oxidation that is not Mn-centered. Recent 55Mn ENDOR studies show that the oxidation states in the S0 state are Mn4(III3,IV).28,29
Figure 12.
Oscillation of XANES IPEs and that of first moments (E) of the Kβ emission spectra from the 0 to 3 flash samples of spinach PS II (BBY). The figure was adapted from Messinger et al., 2001.75
However, one has to be aware that the Mn K-edge position cannot be simply an indicator of the oxidation state. Because of the extent of the metal 4p orbital, this orbital overlaps with p orbitals of the ligands, either through σ or π bonding. Consequently, XANES is sensitive not only to the oxidation state but also to the ligand environment of the metal. However, compared to XANES, Kβ XES is primarily dependent on changes in the oxidation state rather than differences in the overall ligand environment. This was illustrated by a detailed XANES and XES study of multi-nuclear Mn complexes prepared in different oxidation states and with different geometric structures.81,82
B. X-ray Resonant Raman Scattering or RIXS
The description of the Mn4Ca cluster in the various S states in terms of the formal oxidation states is very useful, and Mn K-edge XANES has been the conventional X-ray spectroscopic method for determining the oxidation states. However, it is also important to understand the electronic structure of the Mn cluster in more detail, and we are using the technique of X-ray resonant Raman scattering or RIXS that has the potential for addressing this issue. The RIXS data are collected by scanning the incident energy [1s-3d/4p absorption (v) followed by 2p–1s emission (f)] to yield two-dimensional plots that can be interpreted along the incident energy axis or the energy-transfer axis, which is the difference between the incident and emission energies (Figure 13).83 In the case of a 3d transition-metal ion like Mn, the decay with the highest probability after 1s core hole creation by X-ray absorption is a radiative 2p–1s transition (therefore, called 1s2p RIXS). This process can be viewed as an inelastic scattering of the incident photon at the Mn atom. The energy difference between the initial and intermediate states is equivalent to the K-edge preedge transition (v), and the difference between the initial and final states (v – f) is comparable to L-edge spectroscopy. While the K-edge spectrum is a measure of the charge density of the metal, the L-edge spectrum has information about the spin state of the metal also, due to the strong (2p, 3d) multiplet interaction. Hence, RIXS is sensitive not only to the metal charge density but also to the metal spin state. In general, L-edge spectroscopy is difficult for biological samples because of severe radiation damage caused by the higher X-ray absorption at lower energy, and additionally experiments need to be carried out under ultrahigh-vacuum conditions. In RIXS, L-edge-like spectra are obtained, but the excitation energy is the same as that for K-edge spectroscopy. It also allows us measurement of spectra at 10 K in the presence of an exchange gas atmosphere. Hence, RIXS spectroscopy can circumvent some difficulties in the K- and L-edge spectroscopy, and the method is suitable for biological samples.
Figure 13.
(Left) Energy-level diagram for the RIXS experiment. The excitation is from a 1s–3d level (v), and the emission detected is from a 2p–1s level (f). In RIXS, which is two-dimensional spectroscopy, both the excitation energy (v) is scanned (1s–3d and 1s–4p) using the beamline monochromator and the emission energy (f) is scanned across the Ka lines (2p–1s) with a high-resolution analyzer. Both the monochromator and analyzer have a ~1 eV bandwidth. The difference (v – f) between the incident energy (v) and the emission energy (f) gives the L-edge-like spectrum. Thus, using K-edge energies, one can collect L-edge-like (2p–3d) spectra. (Right) Three-dimensional plot showing the RIXS spectrum. Abscissa is the excitation energy (v) across the 1s–3d energy range of the spectrum (K preedge). The 1s–3d K-edge fluorescence excitation spectrum is plotted in the back of the three-dimensional spectrum for reference. Ordinate is the difference between the excitation (v) and emission energy (f) (L-edge-like).
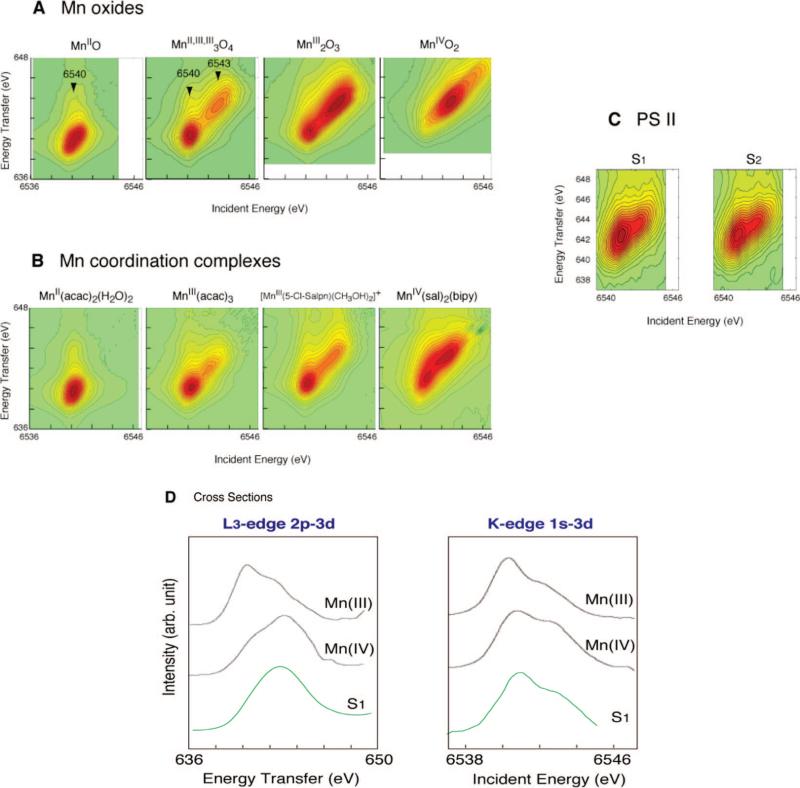
We have compared the spectral changes in the RIXS spectra between the Mn4Ca cluster in the S states,14 Mn oxides,15 and Mn coordination complexes (Figure 14) The results indicate clear differences in how the protein environment modulates the Mn atoms, leading to stronger covalency (see below) for the electronic configuration in the Mn4Ca cluster compared to that in the oxides or coordination complexes.
Figure 14.
(A) Contour plots of the 1s2p3/2 RIXS planes for four Mn oxides in oxidation states II–IV. (B) Four molecular complexes MnII(acac)2(H2O)2, MnIII(acac)3, [MnIII(5-Cl-Salpn)(CH3OH)2]+, and MnIV(sal)2(bipy). (C) S1 and S2 states. The abscissa is the excitation energy, and ordinate is the energy-transfer axis. (D) On the left are integrations of the 2D plot parallel to the ordinate, which yields L-edge-like spectra. The intense feature at 640 eV corresponds to transitions to like states (L3 edges) and the feature at 655 eV corresponds to J = ½ like states (L2 edges). On the right are integrations parallel to the energy-transfer axis that yield K-preedge spectra. The figure was adapted from Glatzel et al., 2004.14
The RIXS two-dimensional data are best shown as contour plots (Figure 14A–C). The comparison of Mn(II), Mn(III), Mn(IV), and PS II in the S1 and S2 states shows that both states contain a mixture of oxidation states Mn(III) and Mn(IV). The integrated cross sections along the Raman or energy-transfer axis are the L3-like edge (2p–3d) (Figure 14D, left). The integrated cross sections along the incident energy are the Mn K-preedge (1s–3d) transition (Figure 14D, right). It is clear from Figure 14 that Mn in the S1 state contains oxidation states III and IV, thus providing confirmation, in combination with EPR results, for the Mn(III2,IV2) assignment for the S1 state.
The first moment of the spectrum integrated along the incident energy axis (see Figure 14D) was calculated for all of the S states and compared with the first moments obtained from Mn oxides and Mn coordination compounds in formal oxidation states of II–IV.
In RIXS, the first moments along the incident energy and energy-transfer axes define “the center of gravity” energy. The first moments were calculated for each spectrum using the following equation: first moment = ∑EjIj/∑Ij, where Ej and Ij are the energy and fluorescence intensities of the jth data point.
As observed in Mn oxides, the first moment along the incident energy (Mn K preedge) increases as the Mn oxidation state increases as a result of the decrease in the charge density on the Mn (Figure 15A). The first moment shift for the series of Mn oxides follows the formal oxidation state in a linear manner. This suggests that the charge density or effective number of 3d electrons, , is directly connected to the effective number of unpaired 3d electrons (spins) in these compounds. The concept of using the effective number of electrons, , localized on a specific atom has been suggested before by numerous authors (reviewed by Glatzel and Bergmann83) to explain spectral shapes in core hole spectroscopy (see below), such as 1s2p RIXS spectroscopy at the K preedge. Covalency can also be deduced from the RIXS spectra because it can be included in the ligand-field multiplet theory, which has been used for simulating the spectra. This is done by considering ligand-to-metal charge transfer using configuration interaction, where the Mn K-preedge configurations can be described by a c1|1s3d5〉 + c2|1s3dn+1 L〉 excited-state configuration mixing (L denotes a hole in a ligand orbital), where c1 and c2 are coefficients describing the magnitude of the admixture of the respective configuration (covalency). The coefficients c1 and c2 can be derived in a fit of the experimental spectra.84–86 It can be more illustrative to describe this using an electron density picture, where it can be expanded in terms of atomic orbitals that can be used to determine the charge (and spin) with fractional orbital population numbers because valence electrons in covalent systems are shared between several atoms. We can thus assign an effective number of electrons to the metal ion 3d shell, , in the 1s excited state. A more complete description of this is included in work by Glatzel et al.14 and references cited therein.
Figure 15.
(A) First moment positions in the 1s2p3/2 RIXS plane for the Mn oxides. (B) Coordination complexes and PS II in S1 and S2 states. A linear fit for the Mn oxides is shown as a dotted line in part A. The same Mn oxide line is also indicated in part B for a comparison to the slope of Mn coordination complexes and PS II data. Along the incident energy axis, the plot gives an ordering of the systems in terms of the effective number of 3d electrons. The figure was adapted from Glatzel et al., 2004.14
The interpretation of the first moment shift along the energy-transfer direction is more complicated because it is also affected by the final-state interaction. This arises from the (2p, 3d) electron–electron interaction, which is the exchange energy for electrons with parallel spins (see Glatzel et al., 2004, for details14).
For Mn coordination compounds, the first moment position along the incident energy axis (Mn preedge) shifts more toward lower energy compared to those of the Mn oxides (Figure 15B), which indicates that Mn coordination compounds have much stronger covalency compared to the Mn oxides. The changes per oxidation state in the first moment positions are more pronounced between the Mn oxides than those between the Mn coordination compounds, consistent with a stronger covalency in the coordination complexes. It is worth noting that the two Mn(III) complexes do not have similar first moment positions. This can be explained by the presence of one counterion in MnIII(Cl-Salp). [MnIII(Cl-Salp)(CH3OH)2] is positively charged. When the complex contains a counterion, it behaves like a more “oxidized” form than its formal oxidation state; therefore, the energy difference between MnIII(Cl-Salp) and MnIV(sal)2(bipy) is much smaller. This, in fact, confirms that the first moment analysis reflects the effective number of electrons on Mn.
The first moment of the incident energy for PS II is lower than those for the Mn(IV) coordination complex and larger than or similar to those for the Mn(III) coordination complexes (Figure 15B), which supports the mixed oxidation states of Mn(III) and Mn(IV) in the S1 and S2 states assigned earlier by the EPR and XANES studies. As observed in the PS II S1 and S2 contour plots and line plots (Figure 14), the spectral changes during the S1 to S2 transition are subtle becauase of the oxidation state changes of one Mn out of four Mn. The first moment shift between S1 and S2 states is a factor of 7–8 smaller than that between MnIII(acac)3 and MnIV(Sal)2(bipy). The magnitude of the spectral change “per Mn ion” between MnIII(acac)3 and MnIV(Sal)2(bipy) (both are neutral complexes, without any counterion) is twice as large as that between the S1 and S2 states when we consider that there are four Mn ions present in PS II.
We thus find that the electron that is extracted from the OEC in PS II between S1 and S2 is strongly delocalized, consistent with strong covalency for the electronic configuration in the OEC. The data collection of the S0 and S3 RIXS spectra is currently in progress (unpublished data). The preliminary result indicates that the S0 spectrum has a weaker peak compared to the S1 state, having a more pronounced low-energy component. In the S2 to S3 transition, the peak intensity becomes stronger and the spectral feature is altered. The orbital population change per change in the oxidation state between the S2 and S3 states is half as much as that between S0 and S1 or S1 and S2 transitions, indicating that the electron is removed from a more covalent form or even more delocalized orbital during this transition.
IV. Mechanism of Photosynthetic Water Oxidation
There are many proposed mechanisms for the photosynthetic water oxidation reaction and the formation of the all-important O–O bond.51,87,88 The two favored mechanisms for water oxidation by the OEC are (i) nucleophilic attack on a MnV≡O or MnIV≡O• terminal ligand by Ca-bound water or hydroxide89 (Scheme 1B) and (ii) reaction of a bridging/terminal Mn–O unit with predominant radical character with an oxo/hydroxo/water ligand of Ca/Mn or an exogenous H2O (Scheme 1A).79
Scheme 1.
The RIXS data show that the trend for the OEC/Mn4Ca cluster as it is oxidized is toward more delocalization of charge. These data,50 along with other studies,75,90–92 run counter to the idea of one highly charged Mn(V) in the cluster as an intermediate involved in water oxidation. Most MnV≡O complexes known at present are all low-spin,93,94 contrary to the Mn cluster in PS II, which is high-spin all the way to the S3 state.65–70,72,73,95–100 It has also been noted that breaking a MnV≡O bond, a prerequisite for the release of dioxygen, would be more difficult from a low-spin configuration.
The structural changes observed by EXAFS during the S2 to S3 transition, as observed in the Mn–Mn79 and Mn–Ca(Sr)101 distances, are significant. It is hard to rationalize such structural changes in the Mn–Mn/Ca distances without significant involvement of the bridging O atoms.
We hypothesize that during the S2 to S3 transition the oxidation is predominantly from a bridging oxygen ligand triggering the structural changes in the OEC. We think that the observed structural changes in the S2 to S3 transition are compatible with primarily a ligand-centered oxidation. It is hard to rationalize the changes in the Mn–Mn and Mn–Ca distances during the S-state transitions, especially during the S2 to S3 transition, without significant involvement of the bridging O atoms. Theoretical analysis of the OEC indicates that the formation of a low-lying ligand oxygen radical precursor state may be required for formation of the O–O bond.102,103 To reach this state,a structural rearrangement is needed at the S2 to S3 transition and the changes in the Mn–Mn distances require a model that involves a bridging O atom.
Future Studies
As presented in this review, the combination of XRD and polarized EXAFS on single crystals has several advantages for unraveling structures of X-ray-damage-prone, redox-active metal sites in proteins. X-ray crystal structures at medium resolution are sufficient to determine the overall shape and placement of the metal site within the ligand sphere, and refinement by means of polarized EXAFS can provide accurate metal-to-metal and metal-to-ligand vectors. In addition, different intermediate states of the active site (including different metal oxidation states), which may be difficult to study with X-ray crystal-lography at high resolution, can be examined. The role of ligands in determining the structure and activity of the cluster is also a subject for X-ray spectroscopic investigations using site-directed mutants. The structural model from polarized EXAFS from the S1 state presented here will provide a framework for studying the structure of the higher S states. The study of the structural and electronic structure changes in the catalytic Mn4Ca cluster in the higher S states of solution and crystals by X-ray spectroscopic methods, now underway, promises to provide a reliable foundation for the investigation of the mechanism of the O–O bond formation during photosynthetic water oxidation. Understanding nature's design in the structure of the Mn4Ca cluster and the mechanism of water oxidation will help in the design of biomimetic catalysts for water splitting.
Acknowledgment
The research presented here was supported by the NIH Grant GM 55302 and by the Director, Office of Science, Office of Basic Energy Sciences (OBES), Division of Chemical Sciences, Geosciences, and Biosciences of the Department of Energy (DOE), under Contract DEAC02-05CH11231. Synchrotron facilities were provided by the Stanford Synchrotron Radiation Laboratory (SSRL), the Advanced Light Source (ALS), and the Advanced Photon Source (APS) operated by DOE OBES. The SSRL Biomedi cal Technology program is supported by NIH, the National Center for Research Resources (NCRR), and the DOE Office of Biological and Environmental Research, and the BioCAT at APS is supported by NCRR. We are grateful to Jan Kern, Athina Zouni, Johannes Messinger, Yulia Pushkar, Ken Sauer, Uwe Bergmann, Pieter Glatzel, Steve Cramer, Matthew Latimer, Jacek Biesiadka, Bernard Loll, and Wolfgang Saenger for their contributions to the work presented in this review. We thank Ken Sauer for a careful reading of the manuscript.
References
- 1.Wydrzynski T, Satoh S. Photosystem II: The Light-DriVen Water: Plastoquinone Oxidoreductase. Springer; Dordrecht, The Netherlands: 2005. [Google Scholar]
- 2.Yachandra VK, Sauer K, Klein MP. Chem. ReV. 1996;96:2927–2950. doi: 10.1021/cr950052k. [DOI] [PubMed] [Google Scholar]
- 3.Sauer K, Yano J, Yachandra VK. Coord. Chem. ReV. 2007 doi: 10.1016/j.ccr.2007.08.009. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Penner-Hahn JE. Struct. Bonding (Berlin) 1998;90:1–36. [Google Scholar]
- 5.Kok B, Forbush B, McGloin M. Photochem. Photobiol. 1970;11:457–476. doi: 10.1111/j.1751-1097.1970.tb06017.x. [DOI] [PubMed] [Google Scholar]
- 6.Sauer K. Acc. Chem. Res. 1980;13:249–256. [Google Scholar]
- 7.Debus RJ. Biochim. Biophys. Acta. 1992;1102:269–352. doi: 10.1016/0005-2728(92)90133-m. [DOI] [PubMed] [Google Scholar]
- 8.Rutherford AW, Zimmermann J-L, Boussac A. Oxygen Evolution. In: Barber J, editor. The Photosystems: Structure, Function, and Molecular Biology. Elsevier; Amsterdam, The Netherlands: 1992. pp. 179–229. [Google Scholar]
- 9.Ort DR, Yocum CF. Oxygenic Photosynthesis: The Light Reactions. Kluwer Academic Publishers; Dordrecht, The Netherlands: 1996. [Google Scholar]
- 10.Lewis NS, Nocera DG. Proc. Natl. Acad. Sci. U.S.A. 2006;103:15729–15735. doi: 10.1073/pnas.0603395103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yano J, Kern J, Sauer K, Latimer M, Pushkar Y, Biesiadka J, Loll B, Saenger W, Messinger J, Zouni A, Yachandra VK. Science. 2006;314:821–825. doi: 10.1126/science.1128186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yano J, Pushkar Y, Glatzel P, Lewis A, Sauer K, Messinger J, Bergmann U, Yachandra VK. J. Am. Chem. Soc. 2005;127:14974–14975. doi: 10.1021/ja054873a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pushkar Y, Yano J, Glatzel P, Messinger J, Lewis A, Sauer K, Bergmann U, Yachandra VK. J. Biol. Chem. 2007;282:7198–7208. doi: 10.1074/jbc.M610505200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glatzel P, Bergmann U, Yano J, Visser H, Robblee JH, Gu WW, de Groot FMF, Christou G, Pecoraro VL, Cramer SP, Yachandra VK. J. Am. Chem. Soc. 2004;126:9946–9959. doi: 10.1021/ja038579z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Glatzel P, Yano J, Bergmann U, Visser H, Robblee JH, Gu W, de Groot FMF, Cramer SP, Yachandra VK. J. Phys. Chem. Solid. 2005;66:2163–2167. doi: 10.1016/j.jpcs.2005.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Yachandra VK. The Catalytic Manganese Cluster: Organization of the Metal Ions. In: Wydrzynski T, Satoh S, editors. Photosystem II: The Light-DriVen Water: Plastoquinone Oxidoreductase. Vol. 22. Springer; Dordrecht, The Netherlands: 2005. pp. 235–260. [Google Scholar]
- 17.Dau H, Iuzzolino L, Dittmer J. Biochim. Biophys. Acta. 2001;1503:24–39. doi: 10.1016/s0005-2728(00)00230-9. [DOI] [PubMed] [Google Scholar]
- 18.Zouni A, Witt H-T, Kern J, Fromme P, Krauss N, Saenger W, Orth P. Nature. 2001;409:739–743. doi: 10.1038/35055589. [DOI] [PubMed] [Google Scholar]
- 19.Kamiya N, Shen JR. Proc. Natl. Acad. Sci. U.S.A. 2003;100:98–103. doi: 10.1073/pnas.0135651100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Biesiadka J, Loll B, Kern J, Irrgang KD, Zouni A. Phys. Chem. Chem. Phys. 2004;6:4733–4736. [Google Scholar]
- 21.Ferreira KN, Iverson TM, Maghlaoui K, Barber J, Iwata S. Science. 2004;303:1831–1838. doi: 10.1126/science.1093087. [DOI] [PubMed] [Google Scholar]
- 22.Loll B, Kern J, Saenger W, Zouni A, Biesiadka J. Nature. 2005;438:1040–1044. doi: 10.1038/nature04224. [DOI] [PubMed] [Google Scholar]
- 23.Yano J, Kern J, Irrgang K-D, Latimer MJ, Bergmann U, Glatzel P, Pushkar Y, Biesiadka J, Loll B, Sauer K, Messinger J, Zouni A, Yachandra VK. Proc. Natl. Acad. Sci. U.S.A. 2005;102:12047–12052. doi: 10.1073/pnas.0505207102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Cinco RM, Robblee JH, Messinger J, Fernandez C, Holman KLM, Sauer K, Yachandra VK. Biochemistry. 2004;43:13271–13282. doi: 10.1021/bi036308v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Dau H, Liebisch P, Haumann M. Phys. Chem. Chem. Phys. 2004;6:4781–4792. [Google Scholar]
- 26.Robblee JH, Messinger J, Cinco RM, McFarlane KL, Fernandez C, Pizarro SA, Sauer K, Yachandra VK. J. Am. Chem. Soc. 2002;124:7459–7471. doi: 10.1021/ja011621a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Peloquin JM, Campbell KA, Randall DW, Evanchik MA, Pecoraro VL, Armstrong WH, Britt RD. J. Am. Chem. Soc. 2000;122:10926–10942. [Google Scholar]
- 28.Kulik LV, Epel B, Lubitz W, Messinger J. J. Am. Chem. Soc. 2005;127:2392–2393. doi: 10.1021/ja043012j. [DOI] [PubMed] [Google Scholar]
- 29.Kulik LV, Epel B, Lubitz W, Messinger J. J. Am. Chem. Soc. 2007 in press. [Google Scholar]
- 30.Debus RJ, Strickler MA, Walker LM, Hillier W. Biochemistry. 2005;44:1367–1374. doi: 10.1021/bi047558u. [DOI] [PubMed] [Google Scholar]
- 31.Kimura Y, Mizusawa N, Ishii A, Nakazawa S, Ono T. J. Biol. Chem. 2005;280:37895–37900. doi: 10.1074/jbc.M505887200. [DOI] [PubMed] [Google Scholar]
- 32.Chu HA, Hillier W, Debus RJ. Biochemistry. 2004;43:3152–3166. doi: 10.1021/bi035915f. [DOI] [PubMed] [Google Scholar]
- 33.Grabolle M, Haumann M, Muller C, Liebisch P, Dau H. J. Biol. Chem. 2006;281:4580–4588. doi: 10.1074/jbc.M509724200. [DOI] [PubMed] [Google Scholar]
- 34.Garman E. Curr. Opin. Struct. Biol. 2003;13:545–551. doi: 10.1016/j.sbi.2003.09.013. [DOI] [PubMed] [Google Scholar]
- 35.Henderson R. Philos. Trans. R. Soc. London, Ser. B: Biol. Sci. 1990;241:6–8. doi: 10.1098/rstb.1990.0019. [DOI] [PubMed] [Google Scholar]
- 36.Rutherford AW. Biochim. Biophys. Acta. 1985;807:189–201. [Google Scholar]
- 37.Mukerji I, Andrews JC, DeRose VJ, Latimer MJ, Yachandra VK, Sauer K, Klein MP. Biochemistry. 1994;33:9712–9721. doi: 10.1021/bi00198a042. [DOI] [PubMed] [Google Scholar]
- 38.Dau H, Andrews JC, Roelofs TA, Latimer MJ, Liang W, Yachandra VK, Sauer K, Klein MP. Biochemistry. 1995;34:5274–5287. doi: 10.1021/bi00015a043. [DOI] [PubMed] [Google Scholar]
- 39.George GN, Prince RC, Cramer SP. Science. 1989;243:789–791. doi: 10.1126/science.2916124. [DOI] [PubMed] [Google Scholar]
- 40.Flank AM, Weininger M, Mortenson LE, Cramer SP. J. Am. Chem. Soc. 1986;108:1049. [Google Scholar]
- 41.Scott RA, Hahn JE, Doniach S, Freeman HC, Hodgson KO. J. Am. Chem. Soc. 1982;104:5364–5369. [Google Scholar]
- 42.Scott RA. X-ray absorption spectroscopy. In: Rousseau DL, editor. Structural and Resonance Techniques in Biological Research. Academic Press; Orlando, FL: 1984. pp. 295–362. [Google Scholar]
- 43.Cramer SP. Biochemical Applications of X-ray Absorption Spectroscopy. In: Koningsberger DC, Prins R, editors. X-ray Absorption: Principles, Applications and Techniques of EXAFS, SEXAFS, and XANES. Wiley-Interscience; New York: 1988. pp. 257–326. [Google Scholar]
- 44.Yachandra VK. Methods Enzymol. 1995;246:638–675. doi: 10.1016/0076-6879(95)46028-4. [DOI] [PubMed] [Google Scholar]
- 45.Bergmann U, Cramer SP. SPIE Conference on Crystal and Multilayer Optics. Vol. 3448. SPIE; San Diego, CA: 1998. A high-resolution large-acceptance analyzer for X-ray fluorescence and Raman spectroscopy. pp. 198–209. [Google Scholar]
- 46.Cinco RM, Holman KLM, Robblee JH, Yano J, Pizarro SA, Bellacchio E, Sauer K, Yachandra VK. Biochemistry. 2002;41:12928–12933. doi: 10.1021/bi026569p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Cinco RM, Robblee JH, Rompel A, Fernandez C, Yachandra VK, Sauer K, Klein MP. J. Phys. Chem. B. 1998;102:8248–8256. doi: 10.1021/jp981658q. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Latimer MJ, DeRose VJ, Mukerji I, Yachandra VK, Sauer K, Klein MP. Biochemistry. 1995;34:10898–10909. doi: 10.1021/bi00034a024. [DOI] [PubMed] [Google Scholar]
- 49.Latimer MJ, DeRose VJ, Yachandra VK, Sauer K, Klein MP. J. Phys. Chem. B. 1998;102:8257–8265. doi: 10.1021/jp981668r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Yano J, Yachandra VK. Photosynth. Res. 2007;92:289–303. doi: 10.1007/s11120-007-9153-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Messinger J. Phys. Chem. Chem. Phys. 2004;6:4764–4771. [Google Scholar]
- 52.Peloquin JM, Britt RD. Biochim. Biophys. Acta. 2001;1503:96–111. doi: 10.1016/s0005-2728(00)00219-x. [DOI] [PubMed] [Google Scholar]
- 53.Carrell TG, Tyryshkin AM, Dismukes GC. J. Biol. Inorg. Chem. 2002;7:2–22. doi: 10.1007/s00775-001-0305-3. [DOI] [PubMed] [Google Scholar]
- 54.Brudvig GW. AdV. Chem. Ser. 1995;246:249–263. [Google Scholar]
- 55.Styring SA, Rutherford AW. Biochemistry. 1988;27:4915–4923. [Google Scholar]
- 56.Ioannidis N, Sarrou J, Schansker G, Petrouleas V. Biochemistry. 1998;37:16445–16451. doi: 10.1021/bi981724e. [DOI] [PubMed] [Google Scholar]
- 57.Ioannidis N, Schansker G, Barynin VV, Petrouleas V. J. Biol. Inorg. Chem. 2000;5:354–363. doi: 10.1007/pl00010664. [DOI] [PubMed] [Google Scholar]
- 58.Ioannidis N, Nugent JHA, Petrouleas V. Biochemistry. 2002;41:9589–9600. doi: 10.1021/bi0159940. [DOI] [PubMed] [Google Scholar]
- 59.Chu H-A, Hillier W, Law NA, Babcock GT. Biochim. Biophys. Acta. 2001;1503:69–82. doi: 10.1016/s0005-2728(00)00216-4. [DOI] [PubMed] [Google Scholar]
- 60.Kimura Y, Mizusawa N, Ishii A, Ono T. Biochemistry. 2005;44:16072–16078. doi: 10.1021/bi051306r. [DOI] [PubMed] [Google Scholar]
- 61.Kimura Y, Mizusawa N, Yamanari T, Ishii A, Ono T. J. Biol. Chem. 2005;280:2078–2083. doi: 10.1074/jbc.M410627200. [DOI] [PubMed] [Google Scholar]
- 62.Strickler MA, Hillier W, Debus RJ. Biochemistry. 2006;45:8801–8811. doi: 10.1021/bi060583a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Iuzzolino L, Dittmer J, Dörner W, Meyer-Klaucke W, Dau H. Biochemistry. 1998;37:17112–17119. doi: 10.1021/bi9817360. [DOI] [PubMed] [Google Scholar]
- 64.Dekker JP. Optical studies on the oxygen-evolving complex of photosystem II. In: Pecoraro VL, editor. Manganese Redox Enzymes. VCH Publishers; New York: 1992. pp. 85–103. [Google Scholar]
- 65.Dismukes GC, Siderer Y. FEBS Lett. 1980;121:78–80. [Google Scholar]
- 66.Hansson O, Andréasson LE. Biochim. Biophys. Acta. 1982;679:261–268. [Google Scholar]
- 67.Messinger J, Nugent JHA, Evans MCW. Biochemistry. 1997;36:11055–11060. doi: 10.1021/bi9711285. [DOI] [PubMed] [Google Scholar]
- 68.Åhrling KA, Peterson S, Styring S. Biochemistry. 1998;37:8115–8120. doi: 10.1021/bi980117o. [DOI] [PubMed] [Google Scholar]
- 69.Messinger J, Robblee JH, Yu WO, Sauer K, Yachandra VK, Klein MP. J. Am. Chem. Soc. 1997;119:11349–11350. doi: 10.1021/ja972696a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Dexheimer SL, Klein MP. J. Am. Chem. Soc. 1992;114:2821–2826. [Google Scholar]
- 71.Yamauchi T, Mino H, Matsukawa T, Kawamori A, Ono T-A. Biochemistry. 1997;36:7520–7526. doi: 10.1021/bi962791g. [DOI] [PubMed] [Google Scholar]
- 72.Campbell KA, Peloquin JM, Pham DP, Debus RJ, Britt RD. J. Am. Chem. Soc. 1998;120:447–448. [Google Scholar]
- 73.Matsukawa T, Mino H, Yoneda D, Kawamori A. Biochemistry. 1999;38:4072–4077. doi: 10.1021/bi9818570. [DOI] [PubMed] [Google Scholar]
- 74.Roelofs TA, Liang W, Latimer MJ, Cinco RM, Rompel A, Andrews JC, Sauer K, Yachandra VK, Klein M. Proc. Natl. Acad. Sci. U.S.A. 1996;93:3335–3340. doi: 10.1073/pnas.93.8.3335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Messinger J, Robblee JH, Bergmann U, Fernandez C, Glatzel P, Visser H, Cinco RM, McFarlane KL, Bellacchio E, Pizarro SA, Cramer SP, Sauer K, Klein MP, Yachandra VK. J. Am. Chem. Soc. 2001;123:7804–7820. doi: 10.1021/ja004307+. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Haumann M, Muller C, Liebisch P, Iuzzolino L, Dittmer J, Grabolle M, Neisius T, Meyer-Klaucke W, Dau H. Biochemistry. 2005;44:1894–1908. doi: 10.1021/bi048697e. [DOI] [PubMed] [Google Scholar]
- 77.Kuzek D, Pace RJ. Biochim. Biophys. Acta. 2001;1503:123–137. doi: 10.1016/s0005-2728(00)00218-8. [DOI] [PubMed] [Google Scholar]
- 78.Yachandra VK, DeRose VJ, Latimer MJ, Mukerji I, Sauer K, Klein MP. Science. 1993;260:675–679. doi: 10.1126/science.8480177. [DOI] [PubMed] [Google Scholar]
- 79.Liang W, Roelofs TA, Cinco RM, Rompel A, Latimer MJ, Yu WO, Sauer K, Klein MP, Yachandra VK. J. Am. Chem. Soc. 2000;122:3399–3412. doi: 10.1021/ja992501u. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Siegbahn PEM. Chem.sEur. J. 2006;12:9217–9227. doi: 10.1002/chem.200600774. [DOI] [PubMed] [Google Scholar]
- 81.Visser H, Anxolabéhère-Mallart E, Bergmann U, Glatzel P, Robblee JH, Cramer SP, Girerd JJ, Sauer K, Klein MP, Yachandra VK. J. Am. Chem. Soc. 2001;123:7031–7039. doi: 10.1021/ja004306h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Pizarro SA, Glatzel P, Visser H, Robblee JH, Christou G, Bergmann U, Yachandra VK. Phys. Chem. Chem. Phys. 2004;6:4864–4870. doi: 10.1039/B407513G. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Glatzel P, Bergmann U. Coord. Chem. ReV. 2005;249:65–95. [Google Scholar]
- 84.de Groot FMF. J. Electron Spectrosc. Relat. Phenom. 1994;67:529–622. [Google Scholar]
- 85.de Groot F. Chem. ReV. 2001;101:1779–1808. doi: 10.1021/cr9900681. [DOI] [PubMed] [Google Scholar]
- 86.de Groot FMF, Glatzel P, Bergmann U, van Aken PA, Barrea RA, Klemme S, Havecker M, Knop-Gericke A, Heijboer WM, Weckhuysen BM. J. Phys. Chem. B. 2005;109:20751–20762. doi: 10.1021/jp054006s. [DOI] [PubMed] [Google Scholar]
- 87.Renger G. Biochim. Biophys. Acta. 2001;1503:210–228. doi: 10.1016/s0005-2728(00)00227-9. [DOI] [PubMed] [Google Scholar]
- 88.McEvoy JP, Brudvig GW. Chem. ReV. 2006;106:4455–4483. doi: 10.1021/cr0204294. [DOI] [PubMed] [Google Scholar]
- 89.Vrettos JS, Limburg J, Brudvig GW. Biochim. Biophys. Acta. 2001;1503:229–245. doi: 10.1016/s0005-2728(00)00214-0. [DOI] [PubMed] [Google Scholar]
- 90.Weng TC, Hsieh WY, Uffelman ES, Gordon-Wylie SW, Collins TJ, Pecoraro VL, Penner-Hahn JE. J. Am. Chem. Soc. 2004;126:8070–8071. doi: 10.1021/ja0494104. [DOI] [PubMed] [Google Scholar]
- 91.Haumann M, Liebisch P, Muller C, Barra M, Grabolle M, Dau H. Science. 2005;310:1019–1021. doi: 10.1126/science.1117551. [DOI] [PubMed] [Google Scholar]
- 92.Yano J, Robblee J, Pushkar Y, Marcus MAM, Bendix J, Workman JM, Collins TJ, Solomon EI, George SD, Yachandra VK. J. Am. Chem. Soc. 2007;129:12989–13000. doi: 10.1021/ja071286b. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Workman JM, Powell RD, Procyk AD, Collins TJ, Bocian DF. Inorg. Chem. 1992;31:1548–1550. [Google Scholar]
- 94.Bendix J, Deeth RJ, Weyhermüller T, Bill E, Wieghardt K. Inorg. Chem. 2000;39:930–938. doi: 10.1021/ic990971j. [DOI] [PubMed] [Google Scholar]
- 95.Dismukes GC, Siderer Y. Proc. Natl. Acad. Sci. U.S.A. 1981;78:274–278. doi: 10.1073/pnas.78.1.274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Casey JL, Sauer K. Biochim. Biophys. Acta. 1984;767:21–28. doi: 10.1016/0005-2728(84)90208-1. [DOI] [PubMed] [Google Scholar]
- 97.Zimmermann J-L, Rutherford AW. Biochim. Biophys. Acta. 1984;767:160–167. [Google Scholar]
- 98.Boussac A, Un S, Horner O, Rutherford AW. Biochemistry. 1998;37:4001–4007. doi: 10.1021/bi9728710. [DOI] [PubMed] [Google Scholar]
- 99.Horner O, Rivière E, Blondin G, Un S, Rutherford AW, Girerd J-J, Boussac A. J. Am. Chem. Soc. 1998;120:7924–7928. [Google Scholar]
- 100.Boussac A, Sugiura M, Inoue Y, Rutherford AW. J. Biol. Chem. 2000;39:13788–13799. doi: 10.1021/bi001159r. [DOI] [PubMed] [Google Scholar]
- 101.Pushkar Y, Yano J, Sauer K, Boussac A, Yachandra VK. unpublished data. [Google Scholar]
- 102.Siegbahn PEM, Crabtree RH. J. Am. Chem. Soc. 1999;121:117–127. [Google Scholar]
- 103.Siegbahn PEM. Inorg. Chem. 2000;39:2923–2935. doi: 10.1021/ic9911872. [DOI] [PubMed] [Google Scholar]
- 104.Wemple MW, Adams DM, Folting K, Hendrickson DN, Christou G. J. Am. Chem. Soc. 1995;117:7275–7276. [Google Scholar]
- 105.Cinco RM, Rompel A, Visser H, Aromi G, Christou G, Sauer K, Klein MP, Yachandra VK. Inorg. Chem. 1999;38:5988–5998. doi: 10.1021/ic991003j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chan MK, Armstrong WH. J. Am. Chem. Soc. 1991;113:5055–5057. [Google Scholar]
- 107.Chan MK. Ph.D. Dissertation. University of California; Berkeley, Berkeley, CA: 1986. [Google Scholar]