Abstract
Histone deacetylase (HDAC)-inhibitors (HDACis) are well characterized anti-cancer agents with promising results in clinical trials. However, mechanistically little is known regarding their selectivity in killing malignant cells while sparing normal cells. Gene expression-based chemical genomics identified HDACis as being particularly potent against Down syndrome associated myeloid leukemia (DS-AMKL) blasts. Investigating the anti-leukemic function of HDACis revealed their transcriptional and posttranslational regulation of key autophagic proteins, including ATG7. This leads to suppression of autophagy, a lysosomal degradation process that can protect cells against damaged or unnecessary organelles and protein aggregates. DS-AMKL cells exhibit low baseline autophagy due to mTOR activation. Consequently, HDAC inhibition repressed autophagy below a critical threshold, which resulted in accumulation of mitochondria, production of reactive oxygen species, DNA-damage and apoptosis. Those HDACi-mediated effects could be reverted upon autophagy activation or aggravated upon further pharmacological or genetic inhibition. Our findings were further extended to other major acute myeloid leukemia subgroups with low basal level autophagy. The constitutive suppression of autophagy due to mTOR activation represents an inherent difference between cancer and normal cells. Thus, via autophagy suppression, HDACis deprive cells of an essential pro-survival mechanism, which translates into an attractive strategy to specifically target cancer cells.
Keywords: HDAC inhibitor, autophagy, AML, Down syndrome, ATG7
Introduction
Pediatric acute myeloid leukemia (AML) is a heterogeneous disease, characterized by recurrent cytogenetic and molecular genetic aberrations that result in alterations in the transcriptional program of normal hematopoietic cells. Most frequently, numerical and structural aberrations involving chromosome 21 can be detected. Despite the recent progress in AML therapy, a high percentage of patients succumb to this fatal disease or to the consequences of high-dose polychemotherapy. Particularly, children with Down syndrome (DS; i.e. trisomy 21), which are at high risk to develop acute megakaryoblastic leukemia (AMKL) [1], suffer from chemotherapy related toxicity and mortality [2; 3]. Thus, DS-AMKL patients would especially benefit from less toxic but equally effective treatment alternatives. DS-AMKL and the antecedent transient leukemia (DS-TL) are characterized by GATA1 mutations and over-activation of mammalian target of rapamycin (mTOR), as a consequence of aberrant activation of insulin-like growth factor (IGF) signaling cascade [4]. Novel insights into this well-characterized AML subgroup would also guide the identification of novel treatment modalities in other AML subtypes harboring aberrations in chromosome 21 or constitutive mTOR activation [5; 6].
Protein acetylation is a reversible process regulated by histone acetyl transferases (HATs) and histone deacetylases (HDACs) [7]. Histone acetylation is an important epigenetic modification regulating the transcription of various genes [7]. In addition, several non-histone proteins can serve as a substrate for HATs and HDACs, whereby their activity and stability is modified [8]. This important posttranslational modification has been found to be deregulated in many tumors [9; 10]. Thus, targeting this process by FDA-approved HDAC inhibitors (HDACis), such as vorinostat and romidepsin, has been the focus of many clinical studies [11–14]. Different mechanisms of HDACi-induced apoptosis in cancer cells have been proposed, such as acetylation of p53 [15; 16]. However, despite the promising results in clinical trials, little is known regarding their selectivity in killing malignant cells while sparing normal cells. Thus, the precise mechanism of action of these inhibitors in human malignancies is still unclear.
In this study, we identified HDACis as potent anti-leukemic agents in DS-AMKL. In addition to known anti-cancer effects [14], we found a previously unrecognized effect of HDACis in blocking autophagy, a lysosomal degradation process that takes place constitutively at a basal level [17]. This is achieved via acetylation and suppression of ATG7 and other proteins of the autophagy interaction network. Those observations were unexpected as previous studies proposed HDACis as autophagy activators [18–22]. Our findings suggest that repression of autophagy by HDACis below a critical threshold in tumor cells with a low basal level of autophagy may constitute an effective treatment option.
Materials and Methods
Cell lines and patients samples
Human leukemia cell lines were obtained from the German Collection of Microorganisms and Cell Cultures (DSMZ). Culturing and maintenance were performed according to the supplier's instructions. For in vitro studies, patient samples were provided by the AML-'Berlin-Frankfurt-Münster' Study Group (AML-BFM-SG, Hannover, Germany). CD34+-HSPCs from donors were positively selected by immunomagnetic labeling with corresponding magnetic cell-sorting beads (Miltenyi Biotech). Cells were maintained or used for colony-forming assays (MethoCult GF H4434, StemCell Technologies) as described [23; 24]. All investigations had been approved by the local Ethics Committee. VPA (SIGMA Life Science), SAHA (Biomol) and TSA (Applichem) were dissolved according to the manufacturer’s instructions and used in the indicated concentration. JQ2 was kindly provided by Dr. Bradner (Boston) and dissolved in DMSO.
Microarray data collection and analysis
Microarray expression profiles were collected using Affymetrix chips and analyzed using dChip [25] and GSEA [26]. Gene expression based chemical genomics was performed using the Connectivity Map [27] and the previously published DS-AMKL gene signature [28]. All microarray data have been deposited in NCBI’s Gene Expression Omnibus (GEO; http://www.ncbi.nlm.nih.gov/geo/) with GEO Series accession number GSE30517.
Xenograft mouse model
NOD.Cg-Rag1tm1Mom Il2rgtm1Wjl/SzJ mice (Jackson Laboratory) were maintained in a pathogen free environment. All experimental procedures using these mice were performed in accordance with protocols approved by the local authorities (Niedersächsisches Landesamt für Verbraucherschutz und Lebensmittelsicherheit). For the in vivo drug trial, 1 × 107 CMY cells or 5×106 K562LC3-GFP cells were injected i.f. per animal into a cohort of recipients that were treated with 400 mg/kg/d VPA i.p. or PBS for 28 days starting 7 days post transplantation or 10 mg/kg/d Panobinostat i.p. or vehicle for 3 days. Recipients were randomly assigned. K562LC3-GFP-transplanted mice were finally analyzed on day four.
Statistics
The statistical analyses were done by unpaired Student's t-test. Comparisons of more than two groups was performed by ANOVA with Duncan’s post-hoc analysis. The level of significance was set at P<0.05. All data are presented as mean±s.d. Calculations were performed using GraphPad Prism 4 or 6.
Results
HDACis revert the DS-AMKL gene expression signature
We previously defined the DS-AMKL gene expression signature compared to non-DS-AMKL consisting of 500 up- and 500 down-regulated genes [28]. In order to identify novel treatment options against DS-AMKL, we connected the DS-AMKL gene expression signature to a reference collection of gene-expression profiles from cultured human cells treated with bioactive small molecules (Connectivity Map) [27]. Using this approach, the HDACis Valproic acid (VPA), Trichostatin A (TSA) and vorinostat (SAHA) were identified to reverse the DS-AMKL gene expression program (Figure 1a and data not shown). In fact, Gene Set Enrichment Analysis (GSEA) [26] validated the conversion of the transcriptome of DS-AMKL cell lines (CMK and CMY) upon VPA treatment. Thus, the top 500 up-regulated genes of the DS-AMKL signature were repressed and the top 500 down-regulated genes were activated (Supplementary Figure 1).
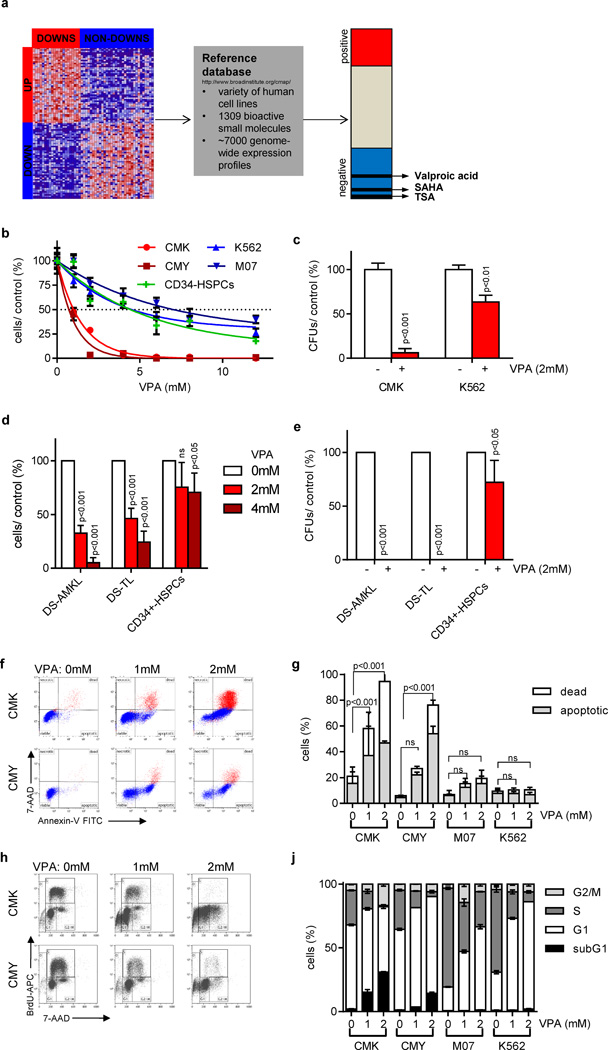
Figure 1. Gene expression-based chemical genomics identifies Down syndrome leukemia to be highly sensitive against HDACis.
(a) Scheme illustrating the setup of the gene expression-based chemical genomic screening, which identified VPA, TSA and SAHA to reverse the DS-AMKL gene signature (17). (b) Number of cells 48h after incubation with the indicated concentrations of VPA relative to the untreated control (=100%). (c) Number of CFUs per 5×102 plated CMK and K562 cells with and without VPA (2mM) relative to the untreated control (=100%). (d) Number of blasts from patients with DS-AMKL (n=3), DS-TL (n=4) and of CD34+-HSPCs (n=2) grown in liquid culture 48h after addition of VPA (2 and 4mM) in relation to the untreated control (white bar; =100%). (e) Number of CFUs of DS-AMKL (n=2) and DS-TL (n=3) blasts as well as CD34+-HSPCs (n=2) in the presence of VPA (2mM) in relation to the untreated control cells (=100%). (d–e) Data are presented as mean±s.d. (f–g) Representative dot plots of Annexin V/ 7-AAD apoptotic assay (f) and diagram showing the percentage of Annexin V+/ 7-AAD- apoptotic and 7-AAD+ dead cells (g) 48h after addition of VPA in comparison to the untreated control cells. (h–j) Representative dot plots (h) and diagram (j) of BrdU/ 7-AAD cell cycle analysis of cells 48h after addition of VPA in comparison to the untreated control cells. (b–c; f–j) All experiments are presented as mean±s.d of at least two independent experiments performed in replicates.
DS-AMKL cell lines and primary patient samples are highly sensitive to HDACi treatment
Next, we sought to understand the biologic effects of the reversion of the DS-AMKL gene signature by HDACis. Growth curves and colony-forming assays revealed an exceptional sensitivity of DS-AMKL cell lines (CMK, CMY; IC50: both 1mM) to VPA treatment compared to control cell lines K562 (IC50: 4.75mM), M07 (IC50: 6.75mM) and CD34+-HSPCs from healthy donors (IC50: 4.75mM; Figure 1b–c).
Most importantly, in vitro growth and colony-forming capacity of primary DS-AMKL and DS-TL blasts were greatly reduced upon VPA treatment, whereas normal CD34+-HSPCs were moderatly affected (Figure 1d–e). We found that VPA treatment induced apoptosis in DS-AMKL cells by performing Annexin V/ 7-AAD, caspase 3/7 activity assays and hypodiploidy staining (Figure 1f–g, Supplementary Figure 2a–b). In addition, BrdU-incorporation and CFSE assays showed that VPA treatment triggered cell cycle arrest in CMK and CMY cell lines (Figure 1h–j, Supplementary Figure 2c). Thus, VPA exerts its growth inhibitory effect on DS-AMKL cells by inducing apoptosis and cell cycle arrest. On the other hand, although K562 and M07 cells were resistant to VPA-induced apoptosis, they still have undergone cell cycle arrest (Figure 1j and Supplementary Figure 2c). The decrease in the proliferation of DS-AMKL cell lines upon VPA treatment is not due to differentiation (Supplementary Figure 2d), which is in accordance to previous studies in AML [29].
Several studies reported the presence of aberrant p53 expression in a fraction of DS-AMKL patients [5; 13; 30]. In addition to that, p53 can serve as a direct substrate of HDACs [8; 16]. However, we did not detect any differences in the expression level or the acetylation status of p53 upon VPA treatment. Moreover, shRNA-mediated knockdown of p53 did not rescue HDACi-induced apoptosis or cell cycle arrest, implicating that both are p53-independent (Supplementary Figure 2e–f).
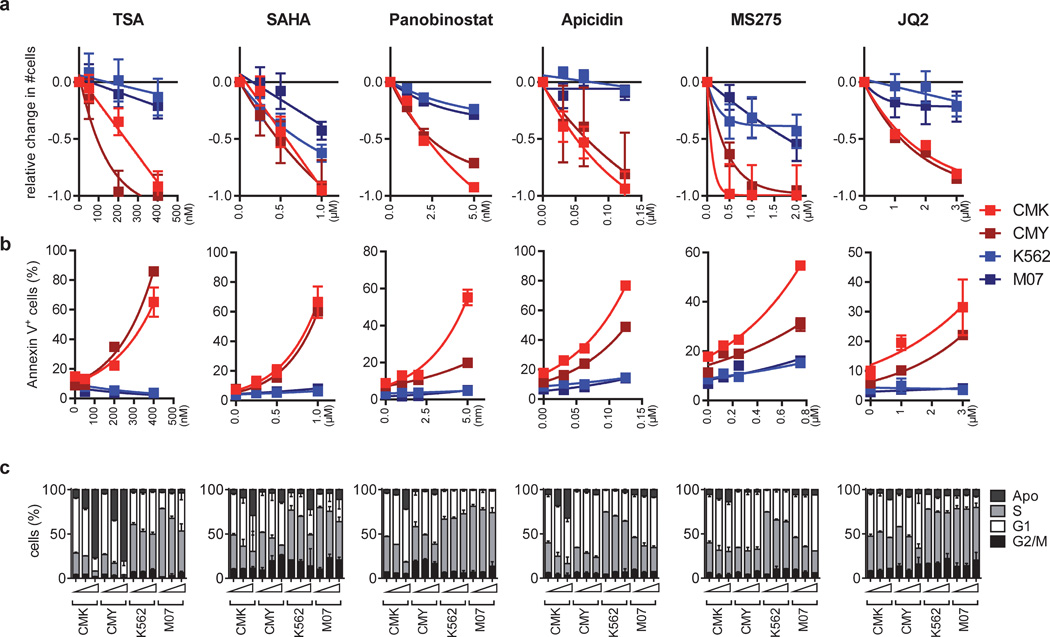
To further confirm that the observed effects reflect VPA’s function in inhibiting HDACs, we assessed acetylation of histones H3 and H4. Both were highly acetylated by dose and timing of VPA (Supplementary Figure 2e). Accordingly, we observed similar effects on growth, cell cycle and viability of DS-AMKL cells using the HDAC class I/II inhibitors TSA, SAHA and Panobinostat (Figure 2a–c and Supplementary Figure 3a–b; for overview about the selectivity of the inhibitors see Supplementary Table 1). More specifically, CMK and CMY cells were highly sensitive to HDAC class I inhibitor Apicidin, the selective HDAC1-3 inhibitor MS-275 [31] and the selective HDAC1/2 inhibitor JQ2 (Figure 2a–c), but not to HDAC8 inhibitor PCI-34051 [32] or the HDAC6 inhibitor Tubastatin A (Supplementary Figure 3c–d). Consistently, shRNA- or siRNA-mediated knockdown of either HDAC1 or HDAC2 partially recapitulated HDACis’ pro-apoptotic effects on CMY cells (Supplementary Figure 4a–c). It also synergized with low doses of VPA to induce apoptosis (Supplementary Figure 4a–b).
Figure 2. Global and specific HDACis induce cell cycle arrest and apoptosis in DS-AMKL cell lines.
(a) Fractional change of the number of viable CMK, CMY, K562, M07 cells 48h after incubation with the indicated HDACis relative to the untreated control (=0). (b) Percentage of Annexin V+ CMK, CMY, K562 and M07 apoptotic cells 48h after incubation with the indicated HDACis. (c) BrdU/ 7-AAD cell cycle analysis of CMK, CMY, K562 and M07 cells 48h after incubation with increasing concentrations of the indicated HDACis in comparison to the untreated control. All experiments are presented as mean±s.d. and are representative for at least two independent experiments performed in replicates.
Taken together, our data support the conclusion that HDACis efficiently invert the DS-AMKL gene signature by inhibition of HDAC1/2, thus resulting in cell cycle arrest and apoptosis.
HDACis transcriptionally and posttranslationally repress ATG7
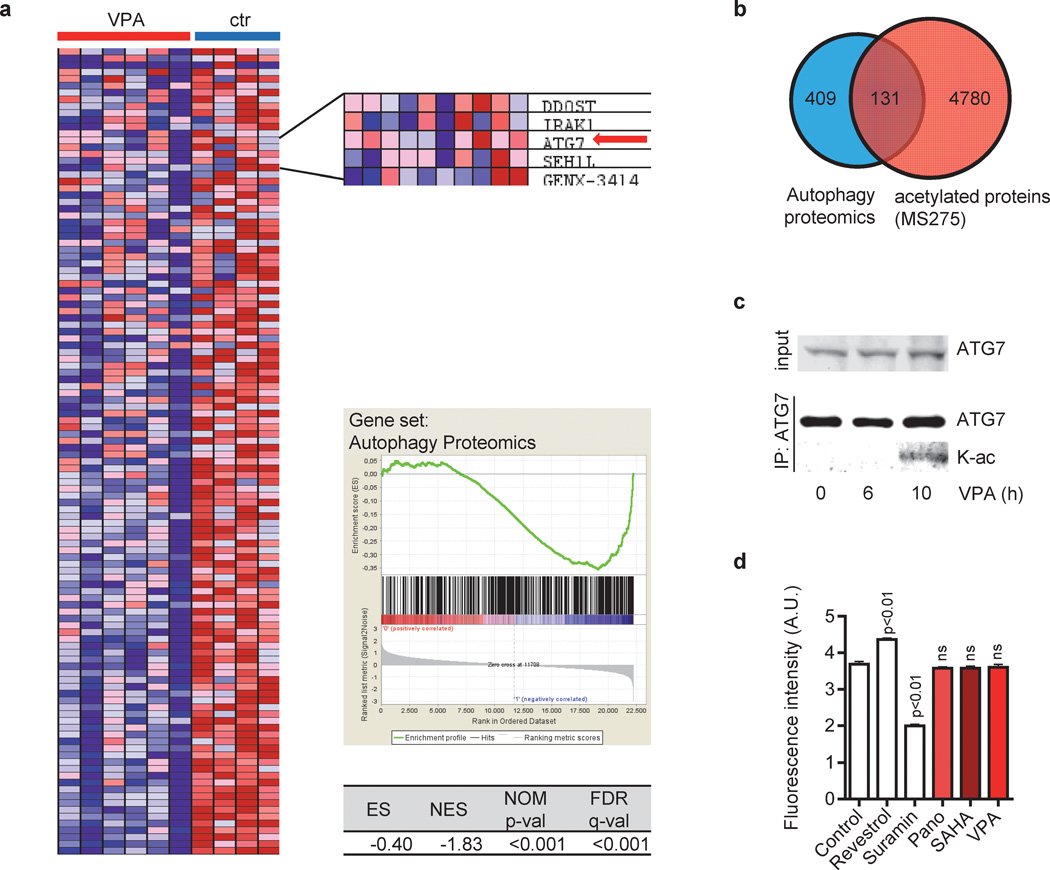
To gain more insights into the molecular mechanisms of the anti-leukemic effects of HDACis, we performed GSEA in CMK and CMYs cell lines. Upon HDAC inhibition, a set of 409 genes belonging to the autophagy interaction network was significantly downregulated (Figure 3a). This gene set was defined by a proteomic approach done by Behrends et al. [33]. Autophagy requires the evolutionarily conserved autophagy-related (ATG) proteins that are essential to envelope intracellular organelles in autophagosomes fusing to lysosomes. Among the leading edge dataset was ATG7 [34], which functions in the ATG12-ATG5-ATG16L1 complex and the LC3-conjugation complex. Both complexes play important roles in the elongation and closure of the autophagosome [35]. It was shown that ATG7 is regulated by acetylation [36; 37]. In fact, upon integration of the AML blasts’ acetylome [38] and the autophagy interaction network [33], we identified, besides ATG7, 130 proteins belonging to the autophagy interaction network to be acetylated in response to HDAC inhibition (Figure 3b). Blocking autophagy by acetylation of ATG7 has been linked to NAD-dependent deacetylase Sirt1 (HDAC Class III) [36; 37]. However, immunoprecipitation of ATG7 followed by immunoblotting with an acetyl-Lysine specific antibody confirmed a SIRT1-independent acetylation of endogenous ATG7 in VPA-treated CMK cells (10h) (Figure 3c–d). Therefore, HDACis can transcriptionally and posttranslationally modulate the autophagy interaction network including the core protein ATG7.
Figure 3. HDACis target the autophagy interaction network.
(a) GSEA of CMY and CMK cells with (VPA) and without (ctr) VPA treatment using a set of 409 genes of the autophagy interaction network, as determined by a proteomic approach (20). GSEA indicated the repression of those key autophagy genes by VPA. Cells were treated for 24h and 48h with 1mM or 2mM VPA. Left - heat map. Right – inlet showing selected genes including ATG7 (red arrow), enrichment plots and statistics. (b) Venn diagram showing the overlap of 409 proteins of the autophagy interaction network (20) and 4780 proteins which are acetylated in response to HDACi treatment of leukemic cell lines (25). (c) Western blots of immunoprecipitated ATG7 using ATG7 and acetyl-Lysin antibody of CMK cells with or without incubation with 2mM VPA for 6h and 10h. (d) SIRT1 activity was measured using the SIRT1 Fluorometric Drug Discovery Kit. Recombinant human SIRT1 was pre-incubated with Panobinostat (2µM), SAHA (2 µM) or VPA (2mM) for 2h. Resveratrol and suramin sodium were included as positive controls for SIRT1 activation and inhibition, respectively. Experiments are presented as mean±s.d. and are representative for at least three independent experiments with two replicates.
HDACis repress autophagy
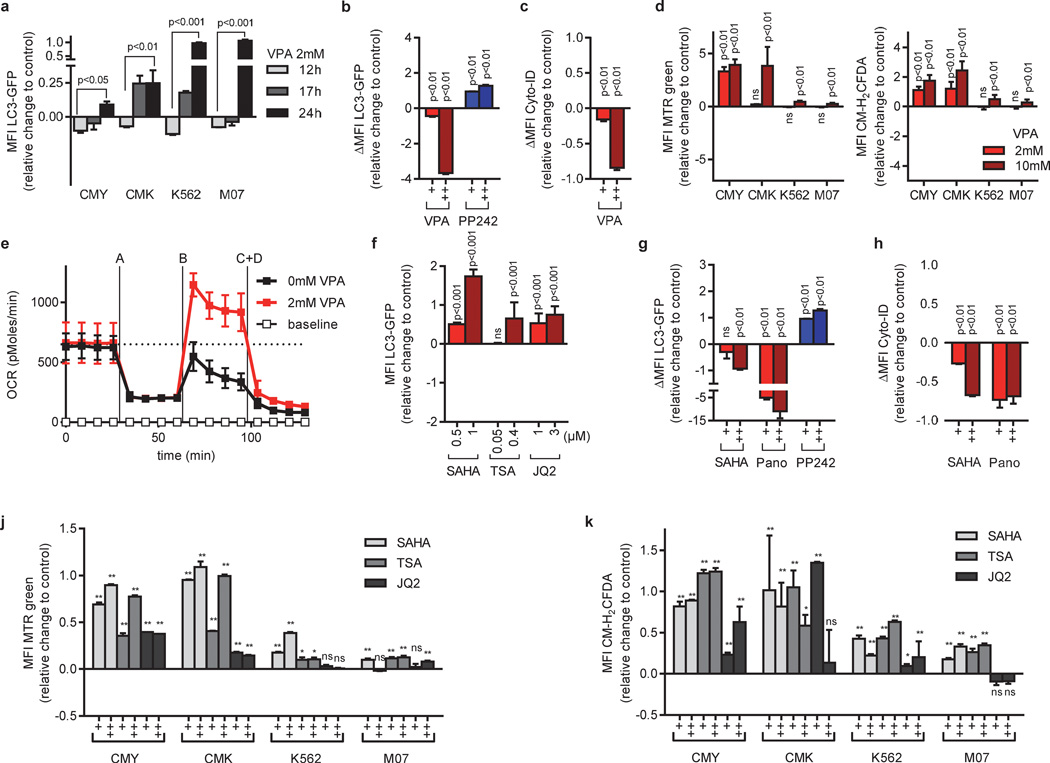
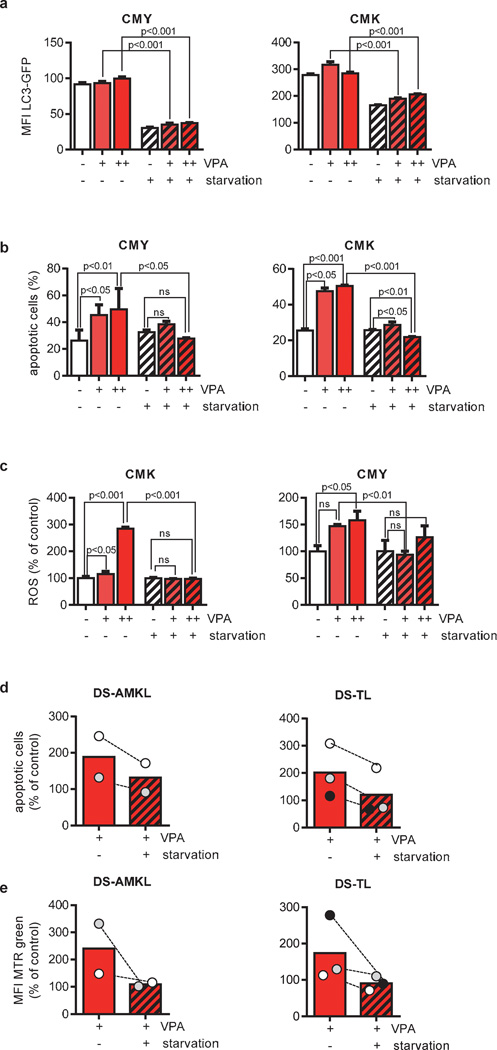
We hypothesized that HDACis exert an anti-leukemic effect through interference with autophagy and assessed this connection more directly. To quantify autophagic flux we used a GFP-tagged LC3 reporter. In cells expressing LC3-GFP, increased autophagic flux leads to a progressive delivery of LC3-GFP to autolysosomes. Upon the fusion with the lysosome, LC3 is degraded by lysosomal enzymes leading to a decrease in the level of LC3-GFP. Conversely, decreased autophagic flux results in LC3-GFP accumulation [34]. Consistent with previous reports [18–22; 39], we detected an initial dose-dependent decrease in total LC3-GFP cellular abundance 12h after VPA treatment (Figure 4a and data not shown), indicating the induction of autophagy. Additionally, we validated our results using mCherry-LC3 reporter. We could observe an increase in mCherry-LC3 punctae - i.e. formation of autophagosomes - by VPA (Supplementary Figure 5).
Figure 4. HDACis inhibit autophagy leading to the accumulation of mitochondria and ROS.
(a) Flow cytometric analysis (MFI±s.d) of autophagic flux in CMY, CMK, K562 and M07 cells expressing LC3-GFP after VPA (2mM) treatment. The bar graph represents the level of LC3 GFP cellular abundance with respect to the control. Values above 0 indicate inhibition of autophagic flux, while values below 0 indicate activation of autophagic flux. (b) Autophagic flux of LC3-GFP cells treated with VPA (0.2, 2 mM) or PP242 (2, 5µM) in the presence or absence of chloroquine (CQ). ΔMFI LC3-GFP= MFI LC3-GFP (+CQ) - MFI LC3-GFP (-CQ). Values below 0 indicate inhibition of autophagic flux, while values above 0 indicate autophagy activation. (c) ΔMFI Cyto-ID measured in CMY cells incubated with VPA (0.4 and 2mM). ΔMFI Cyto-ID= MFI Cyto-ID (+NH4Cl) - MFI Cyto-ID (-NH4Cl). Values below 0 indicate inhibition of autophagic flux, while values above 0 indicate autophagy activation. (d) Mitochondrial pool (left) or ROS production (right) analyzed by flow cytometry (MFI±s.d.) of MitoTracker Green (MTR)-stained or CM-H2DCFDA treated CMY, CMK, K562 and M07 cells (VPA: 2 and 10mM), respectively. (e) Diagram showing the oxygen consumption rate (OCR). The basal respiration rate, oligomycin-sensitive respiration rate (A), the maximal respiration rate (B) and the non-mitochondrial respiration rate (C+D) measured in CMK cells. PANOVA= 5×10−7 (AUC 2mM VPA vs AUC 0mM VPA). (f) Flow cytometric analysis of autophagic flux in CMY cells expressing LC3-GFP (MFI±s.d) after SAHA (0.5 and 1µM), TSA (0.05, 0.4µM) or JQ2 (1, 3µM) treatment. As in (A), values above 0 indicate inhibition of autophagic flux, while values below 0 indicate activation of autophagic flux. (g) Autophagic flux of LC3-GFP expressing cells treated with SAHA (0.2 and 2µM), Panobinostat (0.2 and 2µM) or PP242 (2, 5µM) in the presence and absence of CQ. (h) ΔMFI Cyto-ID signal in CMY cells treated with SAHA (0.4, 2) or Panobinostat (0.1, 0.4). (j–k) Diagrams showing the (j) mitochondrial pool (MTR; MFI±s.d.) and (k) ROS production (CM-H2DCFDA; MFI ±s.d.) analyzed by flow cytometry of CMY, CMK, K562 and M07 cells treated with SAHA (0.5, 1µM), TSA (0.05, 0.4µM) or JQ2 (1, 3µM). (a–d) and (f–k) Data are presented as change relative to the control (=0). (a–k) Data are representative for three independent experiments with two to eight replicates. *P<0.05; **P<0.01.
However, prolonged exposure to VPA up to 24h led to a block of autophagy shown by a gradual accumulation of LC3-GFP (Figure 4a). The accumulation of total LC3-GFP signal upon chloroquine-mediated blockage of autophagolysosomal degradation (ΔMFI LC3-GFP) was reduced in cells pretreated with VPA, indicating a reduction in autophagic flux by VPA (Figure 4b). The results using the LC3-GFP reporter were confirmed by directly labeling the autophagic compartment (pre-autophagosomes, autophagosomes, and autophagolysosomes) with a previously published selective dye (Figure 4c and Supplementary Figure 6) [40].
HDACis lead to autophagic substrate accumulation
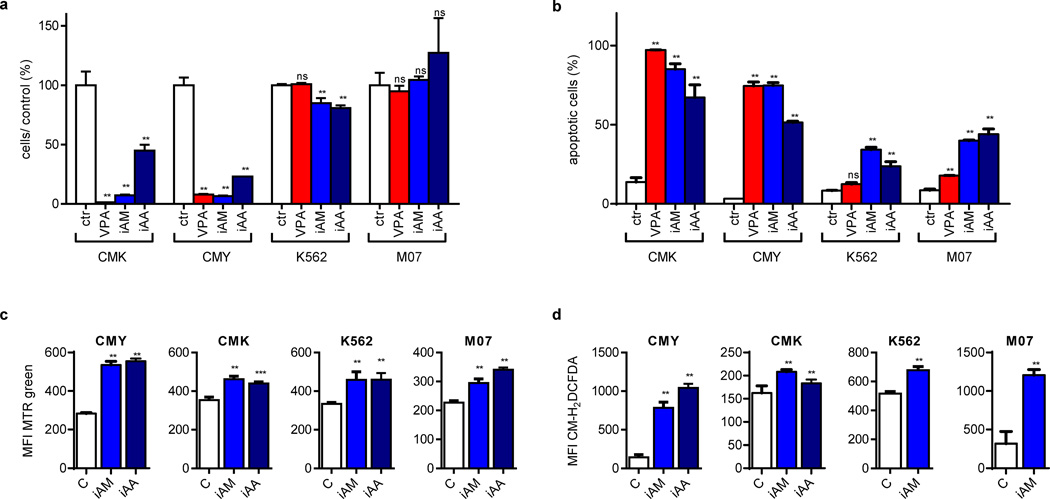
Autophagy participates in the removal of damaged mitochondria, which prevents the initiation of intrinsic apoptotic pathway or DNA damage by increased ROS generation [41–43]. To test whether the observed blockage of autophagy by VPA is functionally relevant, we tested the effects of VPA on mitochondrial mass, ROS formation, DNA damage and the cellular oxygen consumption rate. Our results demonstrated a dose-dependent accumulation of mitochondrial mass and increased ROS formation (Figure 4d). This was mostly pronounced in CMK and CMY cells, which are highly sensitive to HDACi-induced apoptosis, and moderately in the resistant K562 and M07 cells (Figure 1b–c, f–g and Figure 2). The increase in ROS formation due to VPA treatment in CMK cells led to an increase in phosphorylated H2AX (γH2AX; Supplementary Figure 2e), a marker for DNA-double strand breaks (DSBs).
Using real-time measurement of the oxygen consumption rate (OCR), we noticed an increase in the maximal mitochondrial respiratory capacity of CMK cells after VPA treatment (Figure 4e). Neither the basal OCR nor the ATP content per cell were increased, indicating that the mitochondrial mass accumulation was not a consequence of increased energy consumption (Figure 4e and Supplementary Figure 7). This is underscored by the observation that the ATP production in CMK cells is mainly dependent on aerobic glycolysis (Warburg effect); the cellular ATP level only dropped specifically after addition of glycolysis inhibitor 2-deoxy-D-glucose (2-DG) and not upon addition of mitochondrial electron chain inhibitor oligomycin (Supplementary Figure 7). Thus, mitochondrial mass accumulation is not beneficial to the leukemic cells, but causes DNA damage due to ROS production.
Similar to VPA, the global HDACis SAHA, TSA and Panobinostat as well as the specific HDAC1/2 inhibitor JQ2 blocked autophagy and led to autophagic substrate accumulation (i.e. dysfunctional mitochondria; Figure 4f–k). Again, shRNA-mediated knockdown of HDAC2 reduced autophagic flux and synergized with low doses of VPA (Supplementary Figure 4a–b).
Re-activation of autophagy reverts the effects of HDACis
If autophagy inhibition is a critical step in mediating HDACis’ anti-leukemic function, then other autophagy inhibitors should recapitulate HDACis’ effects. On the other hand, autophagy activation should mitigate it. In fact, similar to HDACis, pharmacological inhibitors of either autophagosome maturation (vinblastine and nocodazole) or acidification (ammonium chloride, chloroquine and hydroxychloroquine) suppressed leukemic growth, induced apoptosis, resulted in mitochondrial mass accumulation and increased ROS formation in DS-AMKL cell lines (Figure 5a–d). In contrast, physiological autophagy activation through starvation [34] reversed VPA-mediated suppression of autophagic flux (Figure 6a). This resulted in the rescue of ROS accumulation and a complete prevention of apoptosis in DS-AMKL cell lines (Figure 6b–c). We chose not to use pharmacological activators such as rapamycin and PP242 to rescue HDACi-mediated apoptosis due to their nonspecific toxic effects on leukemic blasts [23; 44–46].
Figure 5. Pharmacologic inhibition of autophagy recapitulates the effects of HDACis on DS-AMKL cells.
(a) Number of viable cells 48h after incubation with VPA (2mM), inhibitors of autophagosome maturation (iAM: vinblastine and nocodazole) and inhibitors of autophagosome acidification (iAA: ammonium chloride, chloroquine and hydroxychloroquine) in relation to the untreated control cells (ctr; =100%). (b) Percentage of Annexin V+ cells 48h after addition of VPA (2mM), iAM or iAA in comparison to the untreated control cells (ctr). (c–d) Diagrams showing (c) the mitochondrial pool of (MTR; MFI±s.d.) and (d) ROS production (CM-H2DCFDA; MFI±s.d.) of CMY, CMK, K562 and M07 cells after addition of iAM or iAA as analyzed by flow cytometry (MFI±s.d.). (a–d) All experiments are presented as mean±s.d of at least two independent experiments with two to four replicates. *P<0.05; **P<0.01; ***P<0.001.
Figure 6. Starvation reversed VPA-mediated suppression of autophagic flux.
(a) Flow cytometric analysis of autophagic flux in CMY and CMK cells expressing LC3-GFP (MFI±s.d.) after incubation with VPA (0.5, 2mM) for 24h in cell culture or starvation medium. (b) Percentage of apoptotic CMY and CMK cells treated with VPA (2, 10mM) for 30h in cell culture or starvation medium. The subdiploid fraction was detected by flow cytometry after PI nuclear staining. (c) ROS production as analyzed by flow cytometry (MFI±s.d.) of CM-H2DCFDA-treated CMK and CMY cells 30h after treatment with VPA (2, 10mM) in cell culture or starvation medium. (d) Bars represent the mean percentage of apoptotic primary DS-AMKL (n=2) or DS-TL cells (n=3) 16 to 24h after treatment with VPA (10mM) under normal or starvation condition in relation to the untreated control (=100%). The subdiploid fraction was detected by flow cytometry after PI nuclear staining. (e) Mitochondrial pool analyzed by flow cytometry (MFI±s.d.) of MTR-stained DS-AMKL (n=2) or DS-TL cells (n=3) 16 to 24h after treatment with VPA (10mM) in cell culture or in starvation medium in relation to the untreated control (=100%). (d–e) Data from one experiment with triplicates presented as distribution of the mean value.
Despite the fact that studying primary leukemic blasts is particularly challenging, they are crucial to validate the effects observed in cell lines. Treatment of DS-AMKL (n=2) and DS-TL (n=3) blasts with VPA resulted in the induction of apoptosis and accumulation of mitochondria (Figure 6d–e). Similar to the effects seen in cell lines, starvation abrogated the effect of VPA on both processes (Figure 6d–e).
These results highlight the existence of a threshold for the basal level of autophagy below which cells undergo apoptosis.
Knockdown of ATG7 recapitulates the effects of HDACis
Our results led us to hypothesize that inhibition of ATG7 by acetylation is, at least in part, responsible for the proapoptotic effect of HDACis. Indeed, siRNA-mediated knockdown of ATG7 resulted in increased apoptosis in CMY cells (Figure 7a). Autopahgic flux was decreased (Figure 7b and Supplementary Figure 6c), suggesting that the elevated rate of apoptosis was due to autophagy inhibition by ATG7 knockdown. Consequently, this caused an increase in mitochondrial mass accumulation and ROS formation (Figure 7c–d) in CMY cells. These data suggest that HDACis transcriptionally and posttranslationally regulate autophagy by modulating the acetylation status of ATG7.
Figure 7. ATG7 knockdown mimics the effect of HDACis on apoptosis and autophagy.
(a) Percentage of apoptotic cells after ATG7 knockdown. VPA treatment was performed for the last 48h. Right: Western blots showing the knockdown of ATG7 by siRNA. (b) Basic and PP242 activated autophagic flux (compared to si-Ctr [=0]). Values below 0 indicate inhibition of autophagic flux, while values above 0 indicate autophagy activation. (c) Mitochondrial pool (MTR; MFI±s.d.) and (d) ROS production (CM-H2DCFDA; MFI ±s.d.) as analyzed by flow cytometry of siATG7 or the empty vector control (si-Ctr) transfected cells 72h after electroporation.
Low constitutive autophagic activity determines the susceptibility of AML blasts to HDACis
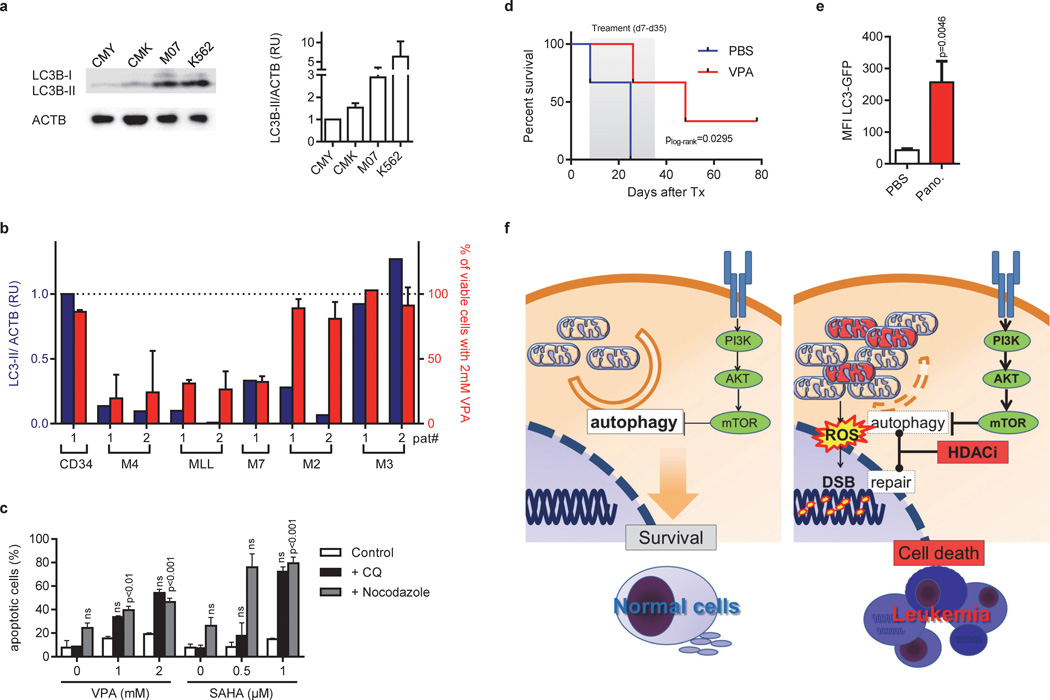
Our results indicate that maintaining a certain level of autophagy is important for the survival and growth of DS-AMKL cells. Recently, it has been shown that the activation of the IGF/IGF1R/PI3K/mTOR pathway is a distinguishing feature of DS-AMKL and DS-TL [23]. In order to confirm a lowered basal autophagic level in DS-AMKL cells due to mTOR activation, we measured endogenous LC3-I to LC3-II conversion levels, a specific posttranslational modification during autophagy [34], by Western blot (Figure 8A). We found that DS-AMKL cells exhibited significantly less LC3-II compared to control cell lines, K562 and M07 (Figure 8a). This suggests the presence of a lowered basal autophagic activity [34]. Consistently, we observed a weaker stimulation of autophagy by PP242 (mTOR inhibitor) in CMK and CMY cells as compared to M07 and K562 cells (Supplementary Figure 6b and 8a) and a weaker increase of mCherry-LC3 punctae (Supplementary Figure 5) in CMK and CMY. Collectively, these results prove the presence of a lower basal autophagy level in DS-AMKL cells associated with the activation of the IGF/IGF1R/PI3K/mTOR pathway.
Figure 8. Low constitutive autophagic activity determines the susceptibility to HDACis in vitro and in vivo.
(a) Immunoblot analysis of LC3-I and LC3-II isoforms in CMY, CMK, M07 and K562 cells using anti-LC3B antibody, normalized to ACTB. The right diagram shows quantification of LC3-II expression relative to ACTB assessed by densitometry. Data are representative for at least three independent experiments performed in replicates. *P<0.05; **P<0.01. (b) Diagram showing the level of LC3-II normalized to ACTB assessed by densitometry (blue bars and blue Y-axis) and the percent of viable cells determined by luminescent cell viability assay (red bars and red Y-axis) from patients with AML FAB M4eo (n=2), AML with MLL rearrangement (n=2), AML FAB M3 (n=2), AML FAB M2 (n=2), AML FAB M7 (or non-DS-AMKL; n=1) and in CD34+-HSPCs (n=1) samples. Viability was determined for cells grown in liquid culture for 48h after addition of 2mM VPA in relation to the untreated control (100%). (c) Percentage of Annexin V+ apoptotic K562 cells treated with indicated concentrations VPA or SAHA for 48h with or without addition of chloroquine (CQ) or nocodazol. (a–c) The data is a representative of at least two independent experiments with two replicates. (d) Kaplan-Meier analysis of NRG mice transplanted i.f. with CMY cells treated with PBS (n=3) or 400mg/kg/day VPA (n=3) i.p. for 28 days, starting 7 days after transplantation. (e) Flow cytometric analysis (MFI±s.d.) of autophagic flux in K562 cells expressing LC3-GFP isolated from NRG mice transplanted i.f. and treated with PBS (n=3) or 10 mg/kg/day Panobinostat (n=4) i.p. for 3 days, starting 28 days after transplantation. (f) Model illustrating the mechanism of VPA-induced apoptosis in DS-AMKL cells with PI3K/mTOR overactivation. In comparison to normal cells (left), PI3K/mTOR activation suppresses basal autophagy to a critical threshold that still allows equilibrium of organelle/ mitochondria and protein aggregate turnover or clearance. Further suppression by HDAC inhibition results in the accumulation of dysfunctional mitochondria, which trigger the intrinsic pathway of apoptosis and ROS production, that causes DNA-damage (right). The repair of DSBs is hampered by direct inhibition of key repair enzymes by HDAC inhibition (16;34).
Thus, constitutive autophagic activity might be an indicator of the cancer cell’s susceptibility to HDACis; i.e. low constitutive autophagic activity determines susceptibility to HDACis while high constitutive autophagic activity confers resistance. To further confirm this hypothesis, we investigated six additional AML cell lines and additional pediatric AML patient samples (AML FAB M4eo with inv(16) [n=2], AML with MLL rearrangement [n=2], AML FAB M3 with t(15;17) [n=2], AML FAB M7 [or AMKL; n=1], AML FAB M2 with t(8;21) [n=2]). In the cell lines, we observed a positive correlation between the IC50 of VPA and the LC3-II/ACTB coefficient (Supplementary Figure 3a and 8b; r2=0.735; p=3.1×10−3). All AML patient samples except two AML FAB M3 samples with t(15;17) had a lower LC3-II level than CD34+-HSPCs from a healthy donor (Figure 8b). Accordingly, the two AML FAB M3 samples did not respond to VPA treatment. In contrast, the AML FAB M4eo, AML FAB M7 and the MLL rearranged patient samples with a low baseline level of autophagy were highly sensitive to VPA (Figure 8b and Supplementary Figure 8c; r2=0.897; p=4×10−4). Cell death was accompanied by mitochondrial mass accumulation and ROS production (Supplementary Figure 9a–b). Only in two patients with AML FAB M2 and t(8;21), the response to VPA was not correctly predicted by the LC3-II level, suggesting an alternative mechanism of resistance in this subgroup.
If a low constitutive autophagic activity determines the susceptibility of cancerous cells to HDACis, then lowering it by pharmacologic and genetic interaction should render resistant cells susceptible to HDACi-induced apoptosis. Consistent with this hypothesis and previous reports [20; 21], suppression of autophagy by chloroquine or nocodazol sensitized the resistant K562 cells to the proapoptotic effects of HDACis such as VPA and SAHA (Figure 8c). The same effects were observed by siRNA-mediated knockdown of ATG7 (Figure 7A and data not shown).
These results indicate the possibility of targeted therapy by VPA against leukemic cells with low basal autophagy level. On the other hand, they indicate that lowering the basal autophagy level can resensitize resistant cells against HDACi-induced apoptosis.
HDACis inhibited the growth of DS-AMKL in xenograft mouse model
To confirm our results in vivo, we transplanted CMY cells intrafemorally into NRG-mice. VPA treatment for 28 days (starting 7 days post transplantation) prolonged the survival of these mice (Figure 8d). In addition, we could show that treatment of NRG-mice with Panobinostat for 3 days repressed autophagy of transplanted K562 cells, which is confirmed by the accumulation of the LC3-GFP reporter signal (Figure 8e).
These results led us to conclude that VPA-induced inhibition of autophagic flux promotes anti-leukemic activity of HDACis in vivo.
Discussion
In this study, we have identified HDACis as potent anti-leukemic agents in DS-AMKL. We showed that autophagy inhibition is a critical step in mediating HDACis’ anti-leukemic function, which is achieved by targeting ATG7 -the core autophagic component. Thereby, HDACis induce apoptosis specifically in AML cells with a low basal level of autophagy [47]. The constitutive suppression of autophagy by PI3K/mTOR activation represents an inherent difference between cancer and normal cells (Figure 8f). This specific vulnerability can be exploited to eliminate malignant cells while sparing normal cells [43]. We hypothesize that, in AML with PI3K/mTOR activation, basal autophagy is reduced to a critical threshold that still allows the equilibrium of organelle’s turnover or clearance. Further suppression of basal autophagy by HDACis breaks this equilibrium, resulting in accumulation of dysfunctional mitochondria. This triggers ROS production and DNA damage, which induces the intrinsic pathway of apoptosis. In turn, the leukemic cells with low basal level of autophagy will undergo apoptosis.
ROS production, DNA damage and induction of apoptosis upon HDACi treatment have previously been shown in various hematologic and non-hematologic malignancies [11; 14; 48–50]. Therefore, our results can provide a molecular explanation to the HDACis’ mechanisms of action in different malignancies. HDACis are particularly potent against several lymphoid malignancies, including cutaneous T-cell lymphoma (CTCL), with constitutive PI3K/mTOR activation [51]. Thus, the autophagic level of different cancerous cells might explain the cell context-dependent effects of HDACis and various outcomes of clinical studies testing HDACi-treatment.
Autophagy requires the evolutionarily conserved ATG proteins, which includes ATG7, to envelope intracellular organelles in autophagosomes fusing to lysosome [34]. We uncovered that HDACis transcriptionally and posttranslationally modulate the autophagy interaction network [33] including the core protein, ATG7. The knockdown of ATG7 in DS-AMKL cell lines recapitulates the effects of HDACis. This is consistent with previous reports showing that ATG7 knockdown sensitizes cells to HDACis [21]. However, to what extent transcriptional and posttranslational modulation of the autophagy interaction network accounts for HDACi-mediated autophagy repression requires further investigation. The importance of transcriptional regulation of autophagy by histone acetylation was recently underscored by Füllgrabe et al [52]. Interestingly, we have found that DS-AMKL blasts express autophagy genes despite their low basal autophagy activity. We speculate that this transcriptional program is required to ensure basal autophagy above a critical threshold despite posttranslational repression of autophagy by PI3K/mTOR activation [53]. Our findings that Atg proteins are involved in HDACis mediated inhibition of autophagy argue against the possibility that HDACis only target mitophagy, the specific autophagic elimination of mitochondria. The methods applied measure autophagy in general and not a specific subset of autophagy. Thus, further studies need to be conducted to understand which type of autophagy is mainly affected by HDACis.
Potent anticancer therapies, such a radiotherapy and certain cytotoxic drugs, trigger the pro-survival activity of autophagy, which protects cancer cells against their killing action [54]. In this context, HDACis have been shown to trigger their antineoplastic effects by inducing metabolic stress in cancerous cells with high metabolic demand [54]. Thus, when used in combination, autophagy inhibitors can enhance the anticancer activity of those agents by depriving malignant cells of this pro-survival mechanism [48]. Similarly, autophagy inhibitors such as chloroquine can sensitize HDACi-resistant cancerous cell lines (with high or normal autophagy level) to HDACis. Here we showed that this synergism further suppresses autophagy to a level lower than the critical threshold, which consequently results in cell death. This synergism is represented in studies done on colon cancer cell lines, chronic myeloid leukemia and mouse embryonic fibroblasts [20–22]. Those studies demonstrated that autophagy inhibition by chloroquine enhances SAHA-induced apoptosis [20–22].
Whether HDACis induce [18–22] or inhibit autophagy [55] has been a controversial issue. Our study demonstrated a dual effect of HDACis on the autophagy of leukemic blasts: after an initial period of activation, HDAC inhibitors reduced autophagic flux. Based on this initial phase, previous studies proposed HDACis as autophagy activators [18–22]. In contrast, we provide strong evidence that HDACi-mediated autophagy repression, at a later time point, is responsible for apoptosis in leukemic cells. First, HDACis caused dose-dependent autophagic substrate accumulation, which was replicated using pharmacological and genetic inhibition of autophagy. In contrast, pharmacological induction did not lead to a later breakdown of autophagic flux (Figure 4B and 4G). Secondly, and most importantly, physiological activation of autophagy through starvation reversed the effect of HDACis. We validated these results by using xenotransplantation models and primary samples from AML patients, which excludes confounding variables introduced by studyin cell lines. Moreover, inhibition of autophagy upon prolonged treatment with multiple HDACis or combined knockout of Hdac1 and Hdac2 was previously reported in the context of cardiomyocytes [56; 57]. Also Oehme et al. recently reported the ability of HDAC10 to induce autophagy [55]. We speculate that the initial activation of autophagy upon HDAC inhibitory treatment may represent a general unspecific cellular pro-survival response. The second stage of autophagy inhibition might represent a time point at which drug-mediated suppression overcomes the pro-survival effect. Accordingly, we observed acetylation of the key autophagic protein ATG7 in response to HDAC inhibition only after a latency period of 10h.
In conclusion, in the context of AML we identified autophagy inhibition as a novel mechanism of HDACi-induced apoptosis that selectively targets leukemic cells with low basal autophagic activity. In addition, we could show that the fine tuning of autophagy to a level lower than the critical basal level level can specifically target cancerous cells. Understanding those context-dependent networks resulted in the initiation of the VPA DS-AMKL 2011 trial (EudraCT: 2011-001838-42) and will open the avenue for new personalized cancer therapies.
Supplementary Material
Acknowledgements
We thank Dr. Bradner for sharing reagents, the Mizushima and Levine laboratories for valuable methodological advice and C. Reimer and L. Queißer for technical assistance. This work was supported by a grant to J.H.K. and D.R. from the Wilhelm Sander-Foundation (2011.057.1), the German Research Foundation (DFG; KL-2374/1-1) and to S.H.O. from the NIH. J.H.K is a fellow of the Emmy Noether-Programme from the DFG (KL-2374/2-1). S.H.O. is an Investigator of the Howard Hughes Medical Institute. G.M.N.B and M.V.S. were supported by the DFG (BE-2089/2-1 and EXC62/1), N.S. by the SFB/TR77, M.L. by the DFG (LE-953/8-1 and LE-953/6-1) and the Mildred-Scheel-Stiftung (#109891) and K.H. by the Hannover Biomedical Research School. Z.L. was a Fellow of the Leukemia & Lymphoma Society and is supported by NIH grant R01 HL107663.
Footnotes
Conflict of interest
The authors declare no competing financial interests.
Author contribution
J.H.K., M.V.S. and M.E.-K. designed and performed experiments, analyzed data and wrote the manuscript. G.M.N.B., S.H.O. and Z.L. designed experiments, interpreted data and wrote the manuscript. K.H., N.S., M.E.K. and D.P.D. designed and performed experiments, analyzed data and revised the manuscript. B.T. designed experiments and revised the manuscript. J.S. performed and analyzed experiments. J.P.B. performed gene expression-based chemical genomics screening. D.R., K.W. and M.L. provided materials/reagents and revised the manuscript.
Supplementary information are available at Leukemia's website.
References
- 1.Klusmann JH, Creutzig U, Zimmermann M, Dworzak M, Jorch N, Langebrake C, et al. Treatment and prognostic impact of transient leukemia in neonates with Down syndrome. Blood. 2008 Mar 15;111(6):2991–2998. doi: 10.1182/blood-2007-10-118810. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Creutzig U, Reinhardt D, Diekamp S, Dworzak M, Stary J, Zimmermann M. AML patients with Down syndrome have a high cure rate with AML-BFM therapy with reduced dose intensity. Leukemia. 2005 Aug;19(8):1355–1360. doi: 10.1038/sj.leu.2403814. [DOI] [PubMed] [Google Scholar]
- 3.O'Brien MM, Cao X, Pounds S, Dahl GV, Raimondi SC, Lacayo NJ, et al. Prognostic features in acute megakaryoblastic leukemia in children without Down syndrome: a report from the AML02 multicenter trial and the Children's Oncology Group Study POG 9421. Leukemia. 2013 Mar;27(3):731–734. doi: 10.1038/leu.2012.223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Klusmann JH, Godinho FJ, Heitmann K, Maroz A, Koch ML, Reinhardt D, et al. Developmental stage-specific interplay of GATA1 and IGF signaling in fetal megakaryopoiesis and leukemogenesis. Genes Dev. 2010 Aug;24(1)(15):1659–1672. doi: 10.1101/gad.1903410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Blink M, Buitenkamp TD, van den Heuvel-Eibrink MM, Danen-van Oorschot AA, de H, V, Reinhardt D, et al. Frequency and prognostic implications of JAK 1–3 aberrations in Down syndrome acute lymphoblastic and myeloid leukemia. Leukemia. 2011 Aug;25(8):1365–1368. doi: 10.1038/leu.2011.86. [DOI] [PubMed] [Google Scholar]
- 6.Klusmann JH, Reinhardt D, Hasle H, Kaspers GJ, Creutzig U, Hahlen K, et al. Janus kinase mutations in the development of acute megakaryoblastic leukemia in children with and without Down's syndrome. Leukemia. 2007 Jul;21(7):1584–1587. doi: 10.1038/sj.leu.2404694. [DOI] [PubMed] [Google Scholar]
- 7.Barbetti V, Gozzini A, Cheloni G, Marzi I, Fabiani E, Santini V, et al. Time- and residue-specific differences in histone acetylation induced by VPA and SAHA in AML1/ETO-positive leukemia cells. Epigenetics. 2013 Feb;8(2):210–219. doi: 10.4161/epi.23538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Minucci S, Pelicci PG. Histone deacetylase inhibitors and the promise of epigenetic (and more) treatments for cancer. Nat Rev Cancer. 2006 Jan;6(1):38–51. doi: 10.1038/nrc1779. [DOI] [PubMed] [Google Scholar]
- 9.Ungewickell A, Medeiros BC. Novel agents in acute myeloid leukemia. Int J Hematol. 2012 Aug;96(2):178–185. doi: 10.1007/s12185-012-1151-5. [DOI] [PubMed] [Google Scholar]
- 10.He LZ, Tolentino T, Grayson P, Zhong S, Warrell RP, Jr, Rifkind RA, et al. Histone deacetylase inhibitors induce remission in transgenic models of therapy-resistant acute promyelocytic leukemia. J Clin Invest. 2001 Nov;108(9):1321–1330. doi: 10.1172/JCI11537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Trus MR, Yang L, Suarez SF, Bordeleau L, Jurisica I, Minden MD. The histone deacetylase inhibitor valproic acid alters sensitivity towards all trans retinoic acid in acute myeloblastic leukemia cells. Leukemia. 2005 Jul;19(7):1161–1168. doi: 10.1038/sj.leu.2403773. [DOI] [PubMed] [Google Scholar]
- 12.Chuang DM, Leng Y, Marinova Z, Kim HJ, Chiu CT. Multiple roles of HDAC inhibition in neurodegenerative conditions. Trends Neurosci. 2009 Nov;32(11):591–601. doi: 10.1016/j.tins.2009.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Garcia-Manero G. Can we improve outcomes in patients with acute myelogenous leukemia? Incorporating HDAC inhibitors into front-line therapy. Best Pract Res Clin Haematol. 2012 Dec;25(4):427–435. doi: 10.1016/j.beha.2012.10.005. [DOI] [PubMed] [Google Scholar]
- 14.Quintas-Cardama A, Santos FP, Garcia-Manero G. Histone deacetylase inhibitors for the treatment of myelodysplastic syndrome and acute myeloid leukemia. Leukemia. 2011 Feb;25(2):226–235. doi: 10.1038/leu.2010.276. [DOI] [PubMed] [Google Scholar]
- 15.Roy S, Packman K, Jeffrey R, Tenniswood M. Histone deacetylase inhibitors differentially stabilize acetylated p53 and induce cell cycle arrest or apoptosis in prostate cancer cells. Cell Death Differ. 2005 May;12(5):482–491. doi: 10.1038/sj.cdd.4401581. [DOI] [PubMed] [Google Scholar]
- 16.McCormack E, Haaland I, Venas G, Forthun RB, Huseby S, Gausdal G, et al. Synergistic induction of p53 mediated apoptosis by valproic acid and nutlin-3 in acute myeloid leukemia. Leukemia. 2012 May;26(5):910–917. doi: 10.1038/leu.2011.315. [DOI] [PubMed] [Google Scholar]
- 17.Youle RJ, Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol. 2011 Jan;12(1):9–14. doi: 10.1038/nrm3028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Insinga A, Monestiroli S, Ronzoni S, Gelmetti V, Marchesi F, Viale A, et al. Inhibitors of histone deacetylases induce tumor-selective apoptosis through activation of the death receptor pathway. Nat Med. 2005 Jan;11(1):71–76. doi: 10.1038/nm1160. [DOI] [PubMed] [Google Scholar]
- 19.Robert T, Vanoli F, Chiolo I, Shubassi G, Bernstein KA, Rothstein R, et al. HDACs link the DNA damage response, processing of double-strand breaks and autophagy. Nature. 2011 Mar 3;471(7336):74–79. doi: 10.1038/nature09803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carew JS, Nawrocki ST, Kahue CN, Zhang H, Yang C, Chung L, et al. Targeting autophagy augments the anticancer activity of the histone deacetylase inhibitor SAHA to overcome Bcr-Abl-mediated drug resistance. Blood. 2007 Jul 1;110(1):313–322. doi: 10.1182/blood-2006-10-050260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Carew JS, Medina EC, Esquivel JA, Mahalingam D, Swords R, Kelly K, et al. Autophagy inhibition enhances vorinostat-induced apoptosis via ubiquitinated protein accumulation. J Cell Mol Med. 2010 Oct;14(10):2448–2459. doi: 10.1111/j.1582-4934.2009.00832.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Gammoh N, Lam D, Puente C, Ganley I, Marks PA, Jiang X. Role of autophagy in histone deacetylase inhibitor-induced apoptotic and nonapoptotic cell death. Proc Natl Acad Sci U S A. 2012 Apr 24;109(17):6561–6565. doi: 10.1073/pnas.1204429109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Klusmann JH, Godinho FJ, Heitmann K, Maroz A, Koch ML, Reinhardt D, et al. Developmental stage-specific interplay of GATA1 and IGF signaling in fetal megakaryopoiesis and leukemogenesis. Genes Dev. 2010 Aug 1;24(15):1659–1672. doi: 10.1101/gad.1903410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Klusmann JH, Li Z, Bohmer K, Maroz A, Koch ML, Emmrich S, et al. miR-125b-2 is a potential oncomiR on human chromosome 21 in megakaryoblastic leukemia. Genes Dev. 2010 Mar 1;24(5):478–490. doi: 10.1101/gad.1856210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Li C, Wong WH. Model-based analysis of oligonucleotide arrays: expression index computation and outlier detection. Proc Natl Acad Sci U S A. 2001 Jan 2;98(1):31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Subramanian A, Tamayo P, Mootha VK, Mukherjee S, Ebert BL, Gillette MA, et al. Gene set enrichment analysis: a knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci U S A. 2005 Oct 25;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lamb J, Crawford ED, Peck D, Modell JW, Blat IC, Wrobel MJ, et al. The Connectivity Map: using gene-expression signatures to connect small molecules, genes, and disease. Science. 2006 Sep 29;313(5795):1929–1935. doi: 10.1126/science.1132939. [DOI] [PubMed] [Google Scholar]
- 28.Bourquin JP, Subramanian A, Langebrake C, Reinhardt D, Bernard O, Ballerini P, et al. Identification of distinct molecular phenotypes in acute megakaryoblastic leukemia by gene expression profiling. Proc Natl Acad Sci U S A. 2006 Feb 28;103(9):3339–3344. doi: 10.1073/pnas.0511150103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tonelli R, Sartini R, Fronza R, Freccero F, Franzoni M, Dongiovanni D, et al. G1 cell-cycle arrest and apoptosis by histone deacetylase inhibition in MLL-AF9 acute myeloid leukemia cells is p21 dependent and MLL-AF9 independent. Leukemia. 2006 Jul;20(7):1307–1310. doi: 10.1038/sj.leu.2404221. [DOI] [PubMed] [Google Scholar]
- 30.Kiyoi H, Yamaji S, Kojima S, Naoe T. JAK3 mutations occur in acute megakaryoblastic leukemia both in Down syndrome children and non-Down syndrome adults. Leukemia. 2007 Mar;21(3):574–576. doi: 10.1038/sj.leu.2404527. [DOI] [PubMed] [Google Scholar]
- 31.Lucas DM, Davis ME, Parthun MR, Mone AP, Kitada S, Cunningham KD, et al. The histone deacetylase inhibitor MS-275 induces caspase-dependent apoptosis in B-cell chronic lymphocytic leukemia cells. Leukemia. 2004 Jul;18(7):1207–1214. doi: 10.1038/sj.leu.2403388. [DOI] [PubMed] [Google Scholar]
- 32.Balasubramanian S, Ramos J, Luo W, Sirisawad M, Verner E, Buggy JJ. A novel histone deacetylase 8 (HDAC8)-specific inhibitor PCI-34051 induces apoptosis in T-cell lymphomas. Leukemia. 2008 May;22(5):1026–1034. doi: 10.1038/leu.2008.9. [DOI] [PubMed] [Google Scholar]
- 33.Behrends C, Sowa ME, Gygi SP, Harper JW. Network organization of the human autophagy system. Nature. 2010 Jul 1;466(7302):68–76. doi: 10.1038/nature09204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Mizushima N, Yoshimori T, Levine B. Methods in mammalian autophagy research. Cell. 2010 Feb 5;140(3):313–326. doi: 10.1016/j.cell.2010.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Mizushima N. Autophagy: process and function. Genes Dev. 2007 Nov 15;21(22):2861–2873. doi: 10.1101/gad.1599207. [DOI] [PubMed] [Google Scholar]
- 36.Lee IH, Cao L, Mostoslavsky R, Lombard DB, Liu J, Bruns NE, et al. A role for the NAD-dependent deacetylase Sirt1 in the regulation of autophagy. Proc Natl Acad Sci U S A. 2008 Mar 4;105(9):3374–3379. doi: 10.1073/pnas.0712145105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lee IH, Finkel T. Regulation of autophagy by the p300 acetyltransferase. J Biol Chem. 2009 Mar 6;284(10):6322–6328. doi: 10.1074/jbc.M807135200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Choudhary C, Kumar C, Gnad F, Nielsen ML, Rehman M, Walther TC, et al. Lysine acetylation targets protein complexes and co-regulates major cellular functions. Science. 2009 Aug 14;325(5942):834–840. doi: 10.1126/science.1175371. [DOI] [PubMed] [Google Scholar]
- 39.Dalby KN, Tekedereli I, Lopez-Berestein G, Ozpolat B. Targeting the prodeath and prosurvival functions of autophagy as novel therapeutic strategies in cancer. Autophagy. 2010 Apr;6(3):322–329. doi: 10.4161/auto.6.3.11625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Chan LL, Shen D, Wilkinson AR, Patton W, Lai N, Chan E, et al. A novel image-based cytometry method for autophagy detection in living cells. Autophagy. 2012 Sep;8(9):1371–1382. doi: 10.4161/auto.21028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Nakahira K, Haspel JA, Rathinam VA, Lee SJ, Dolinay T, Lam HC, et al. Autophagy proteins regulate innate immune responses by inhibiting the release of mitochondrial DNA mediated by the NALP3 inflammasome. Nat Immunol. 2011 Mar;12(3):222–230. doi: 10.1038/ni.1980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang Y, Qi H, Taylor R, Xu W, Liu LF, Jin S. The role of autophagy in mitochondria maintenance: characterization of mitochondrial functions in autophagy-deficient S. cerevisiae strains. Autophagy. 2007 Jul;3(4):337–346. doi: 10.4161/auto.4127. [DOI] [PubMed] [Google Scholar]
- 43.White E. Deconvoluting the context-dependent role for autophagy in cancer. Nat Rev Cancer. 2012 Jun;12(6):401–410. doi: 10.1038/nrc3262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Janes MR, Limon JJ, So L, Chen J, Lim RJ, Chavez MA, et al. Effective and selective targeting of leukemia cells using a TORC1/2 kinase inhibitor. Nat Med. 2010 Feb;16(2):205–213. doi: 10.1038/nm.2091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Aronson LI, Davenport EL, Mirabella F, Morgan GJ, Davies FE. Understanding the interplay between the proteasome pathway and autophagy in response to dual PI3K/mTOR inhibition in myeloma cells is essential for their effective clinical application. Leukemia. 2013 May 14; doi: 10.1038/leu.2013.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kojima K, Shimanuki M, Shikami M, Samudio IJ, Ruvolo V, Corn P, et al. The dual PI3 kinase/mTOR inhibitor PI-103 prevents p53 induction by Mdm2 inhibition but enhances p53-mediated mitochondrial apoptosis in p53 wild-type AML. Leukemia. 2008 Sep;22(9):1728–1736. doi: 10.1038/leu.2008.158. [DOI] [PubMed] [Google Scholar]
- 47.Wullschleger S, Loewith R, Hall MN. TOR signaling in growth and metabolism. Cell. 2006 Feb 10;124(3):471–484. doi: 10.1016/j.cell.2006.01.016. [DOI] [PubMed] [Google Scholar]
- 48.Maiso P, Colado E, Ocio EM, Garayoa M, Martin J, Atadja P, et al. The synergy of panobinostat plus doxorubicin in acute myeloid leukemia suggests a role for HDAC inhibitors in the control of DNA repair. Leukemia. 2009 Dec;23(12):2265–2274. doi: 10.1038/leu.2009.182. [DOI] [PubMed] [Google Scholar]
- 49.Rosato RR, Almenara JA, Yu C, Grant S. Evidence of a functional role for p21WAF1/CIP1 down-regulation in synergistic antileukemic interactions between the histone deacetylase inhibitor sodium butyrate and flavopiridol. Mol Pharmacol. 2004 Mar;65(3):571–581. doi: 10.1124/mol.65.3.571. [DOI] [PubMed] [Google Scholar]
- 50.Ungerstedt JS, Sowa Y, Xu WS, Shao Y, Dokmanovic M, Perez G, et al. Role of thioredoxin in the response of normal and transformed cells to histone deacetylase inhibitors. Proc Natl Acad Sci U S A. 2005 Jan 18;102(3):673–678. doi: 10.1073/pnas.0408732102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Batty N, Malouf GG, Issa JP. Histone deacetylase inhibitors as anti-neoplastic agents. Cancer Lett. 2009 Aug 8;280(2):192–200. doi: 10.1016/j.canlet.2009.03.013. [DOI] [PubMed] [Google Scholar]
- 52.Fullgrabe J, Lynch-Day MA, Heldring N, Li W, Struijk RB, Ma Q, et al. The histone H4 lysine 16 acetyltransferase hMOF regulates the outcome of autophagy. Nature. 2013 Jul 17; doi: 10.1038/nature12313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Mizushima N. The role of the Atg1/ULK1 complex in autophagy regulation. Curr Opin Cell Biol. 2010 Apr;22(2):132–139. doi: 10.1016/j.ceb.2009.12.004. [DOI] [PubMed] [Google Scholar]
- 54.Mathew R, Karantza-Wadsworth V, White E. Role of autophagy in cancer. Nat Rev Cancer. 2007 Dec;7(12):961–967. doi: 10.1038/nrc2254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Oehme I, Linke JP, Bock BC, Milde T, Lodrini M, Hartenstein B, et al. Histone deacetylase 10 promotes autophagy-mediated cell survival. Proc Natl Acad Sci U S A. 2013 Jul 9;110(28):E2592–E2601. doi: 10.1073/pnas.1300113110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Cao DJ, Wang ZV, Battiprolu PK, Jiang N, Morales CR, Kong Y, et al. Histone deacetylase (HDAC) inhibitors attenuate cardiac hypertrophy by suppressing autophagy. Proc Natl Acad Sci U S A. 2011 Mar 8;108(10):4123–4128. doi: 10.1073/pnas.1015081108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Moresi V, Carrer M, Grueter CE, Rifki OF, Shelton JM, Richardson JA, et al. Histone deacetylases 1 and 2 regulate autophagy flux and skeletal muscle homeostasis in mice. Proc Natl Acad Sci U S A. 2012 Jan 31;109(5):1649–1654. doi: 10.1073/pnas.1121159109. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.