Abstract
The signature mitochondrial phospholipid cardiolipin plays an important role in mitochondrial function, and alterations in cardiolipin metabolism are associated with human disease. Topologically, cardiolipin biosynthesis and remodeling is complex. Precursor phospholipids must be transported from the ER, across the mitochondrial outer membrane to the matrix-facing leaflet of the inner membrane, where cardiolipin biosynthesis commences. Post-synthesis, cardiolipin undergoes acyl chain remodeling, requiring additional trafficking steps, before it achieves its final distribution within both mitochondrial membranes. This process is regulated at several points via multiple independent mechanisms. Here, we review the regulation and topology of cardiolipin biosynthesis and remodeling in the yeast Saccharomyces cerevisiae. Although cardiolipin metabolism is more complicated in mammals, yeast have been an invaluable model for dissecting the steps required for this process.
Keywords: cardiolipin, remodeling, mitochondria, yeast, regulation, lipid trafficking
1. Introduction
The unique phospholipid cardiolipin (CL) is required for the efficiency of a number of mitochondrial processes (Claypool and Koehler, 2012). CL is unique for a number of reasons: 1) unlike most other phospholipids which are synthesized in one or a few cellular locations, then disseminated throughout a cell's membranes, CL by and large remains in the mitochondrion, its site of synthesis; 2) CL is essentially a lipid dimer; it consists of two phosphate headgroups, which are attached by a glycerol moiety, and four acyl chains; and 3) after its synthesis, CL undergoes acyl chain remodeling, where acyl chains are removed by a lipase and replaced by a transacylase or acyltransferase, resulting in the establishment of only a few molecular forms of CL in a cell or tissue. Surprisingly, the acyl chain specificity of the lipase has never been demonstrated (Beranek et al., 2009), and the transacylase tafazzin has no acyl chain specificity (Schlame, 2012), although tafazzin from Drosophila has been shown to preferentially catalyze transacylation reactions on curved membranes leading to the establishment of CL with unsaturated acyl chains, which were proposed to decrease lipid disorder in areas of high curvature (Schlame et al., 2012a). Curiously, the final molecular form of CL varies between organisms and even between cell types within the same organism.
CL serves the cell in multiple capacities: it associates with all the major proteins of the mitochondrial respiratory chain and thereby increases the efficiency of electron flow and ADP/ATP exchange (Acehan et al., 2011; Bazan et al., 2013; Claypool et al., 2008b; Fry and Green, 1981; Jiang et al., 2000; Schagger et al., 1990; Schwall et al., 2012; Yu and Yu, 1980), modulates the catalytic activities and stability of interacting proteins (Claypool et al., 2008b; Gomez and Robinson, 1999; Jiang et al., 2000; Pfeiffer et al., 2003; Wenz et al., 2009), is critical for the biogenesis of mitochondrial proteins (Gebert et al., 2009; Jiang et al., 2000; Joshi et al., 2009), facilitates mitochondrial fission/fusion (Ban et al., 2010; DeVay et al., 2009; Joshi et al., 2012), and is involved in the maintenance and plenitude of cristae morphology (Acehan et al., 2009; Acehan et al., 2007; Mileykovskaya and Dowhan, 2009).
In addition to the importance of CL in promoting and maintaining normal mitochondrial function, alterations in CL metabolism have been associated with ischemia and reperfusion, heart failure, diabetic cardiomyopathy, and Barth syndrome (Chicco and Sparagna, 2007; Claypool et al., 2006; Gu et al., 2004; Paradies et al., 1997; Schlame and Ren, 2006). Barth syndrome is caused by mutations in tafazzin (TAZ1), and patients present with cardio- and skeletal myopathy, neutropenia, 3-methylglutaconic aciduria, and abnormal mitochondria (Barth et al., 1983; Schlame and Ren, 2006).
Much of the knowledge of CL biosynthesis and remodeling comes from studies in yeast. In addition to the “usual” advantages of using yeast as a model system (Baile and Claypool, 2013; Botstein and Fink, 2011), yeast are viable in the absence of CL and CL precursor phospholipids (Chang et al., 1998a; Chang et al., 1998b; Jiang et al., 1997; Osman et al., 2010; Tuller et al., 1998) whereas in higher eukaryotes CL is required for life (Zhang et al., 2011). Although CL biosynthesis and remodeling are highly conserved between yeast and higher eukaryotes, there are still a few differences. There are no orthologs of Gep4p, the phosphatidylglycerolphosphate (PGP) phosphatase, or Cld1p, a CL lipase, in higher eukaryotes (Beranek et al., 2009; Osman et al., 2010). However, the phylogenetically unrelated PTPMT1 performs the same function as Gep4p (Zhang et al., 2011); and a calcium-independent phospholipase A2 has been implicated as a CL lipase (Malhotra et al., 2009; Mancuso et al., 2007; Schlame et al., 2012b), although its exact role in CL remodeling remains nebulous (Kiebish et al., 2013). Additionally, only the TAZ1-mediated CL remodeling pathway exists in yeast, while additional remodeling enzymes have been identified in mammals (reviewed in Claypool and Koehler, 2012). Thus, while yeast have been useful in dissecting this process, the complexity of and multitude of players in mammalian CL remodeling suggest that there is still much to discover.
With the recent characterizations of Cld1p, Gep4p and Tam41p (Beranek et al., 2009; Osman et al., 2010; Tamura et al., 2013), it is likely that all of the proteins catalyzing CL synthetic or remodeling reactions have been identified in yeast; however, many questions regarding the regulation of this process, as well as the topology and trafficking of CL and its precursors, remain (Fig. 1).
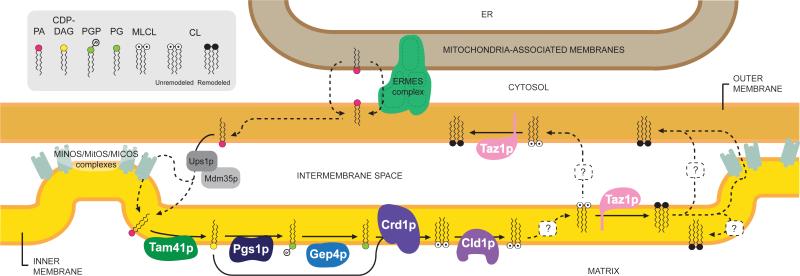
FIGURE 1. The topology of CL biosythesis and remodeling.
Phosphatidic acid (PA) is synthesized in the ER and translocates to mitochondria in a process that is influenced by the ERMES (ER-mitochondria encounter structure) complex. Ups1/Mdm35p heterodimers transport PA from the OM to the IM, potentially at contact sites (established by MINOS/MICOS/MitOS complexes). PA is converted to CDP-diacylglycerol (CDP-DAG) by Tam41p on the matrix-facing leaflet of the IM. CDP-DAG is used to generate phosphatidylglycerolphosphate (PGP) by Pgs1p. PGP is dephosphorylated to phosphatidylglycerol (PG) by Gep4p. PG and another CDP-DAG are condensed to form unremodeled CL by Crd1p. CL is deacylated by Cld1p on the matrix-facing leaflet of the IM, forming MLCL. Via an unknown mechanism, MLCL must flip to the IMS-facing leaflet of the IM or be transported to the OM to gain access to the transacylase Taz1p, which regenerates CL. Multiple rounds of deacylation/reacylation result in remodeled CL which is enriched in unsaturated acyl chains. CL achieves its final distribution on both leaflets of the IM and OM through currently ill-defined mechanisms. The depicted topology of Pgs1p has not been experimentally verified. Solid lines indicate known pathways. Dashed lines delineate potential but currently unknown phospholipid transport processes.
2. Delivering precursor phospholipids to the IM
CL biosynthesis requires CDP-diacylglycerol (CDP-DAG), which is formed from phosphatidic acid (PA) and CTP by a CDP-DAG synthase (Shen et al., 1996). Yeast contain two CDP-DAG synthases: Cds1p in the ER (Kuchler et al., 1986), and the recently characterized Tam41p in the mitochondrial inner membrane (IM; Tamura et al., 2013).
Although CDP-DAG (containing an NBD moiety) is able to be translocated from the ER to the IM in vitro, this process is inefficient (Tamura et al., 2013). The very low abundance of CL in Δtam41 yeast (Kutik et al., 2008; Tamura et al., 2012) suggests that if Cds1p-derived CDP-DAG contributes to CL biosynthesis, its role is very minor. Tam41p is peripherally associated with the matrix side of the IM (Table 1) (Gallas et al., 2006; Tamura et al., 2013). Thus, Tam41p activity requires that its substrate, PA, be transported from the ER to the matrix-facing leaflet of the IM. Phospholipid transport between the ER and mitochondrial outer membrane (OM) was suggested to be mediated by the ER-mitochondria encounter structure (ERMES) complex which physically tethers the two organelles (Kornmann et al., 2009). Indeed, loss of any ERMES complex subunit (Mdm10p, Mdm34, Mdm12p, or Mmm1p) alters the mitochondrial phospholipid profile, including reducing CL (Kornmann et al., 2009; Stroud et al., 2011; Tamura et al., 2012). However, its direct role in phospholipid transport has recently been challenged (Nguyen et al., 2012; Voss et al., 2012). Further, defects caused by the loss of a functional ERMES complex can be rescued by expressing an artificial ER-mitochondria tether, suggesting that the ERMES complex facilitates phospholipid transport by forming close contact sites between the two membranes, rather than directly transporting phospholipids (Kornmann et al., 2009; Nguyen et al., 2012; Voss et al., 2012). Notably, these studies focused on the transport of phosphatidylserine from the ER to mitochondria (and phosphatidylethanolamine to the ER after is decarboxylation in mitochondria). Thus, the mechanisms of PA and CDP-DAG transport from the ER to mitochondria, and the players involved, including a direct assessment of the role of the ERMES complex, remain to be discovered.
Table 1.
Topology of CL synthesis and remodeling enzymes
| Protein | Location/membrane association | Predicted transmembrane domainsa | Biochemical experiments | References |
|---|---|---|---|---|
| Tam41p | Matrix leaflet of IM/peripheral | 0 | Protected from protease in mitoplasts, extracted with carbonate | (Tamura et al., 2006) |
| Pgs1p | Matrix leaflet of IM/peripheral | 0, 1, 2b | N-terminal presequence imports LacZ to mitochondrial matrix | (Dzugasova et al., 1998) |
| Gep4p | Matrix leaflet of IM/peripheral | 0 | Protected from protease in mitoplasts; extracted with carbonate | (Osman et al., 2010) |
| Crd1p | Active site faces the matrix of the IM/integral | 2, 3, 4, 5 | Protected from protease in mitoplasts; blocking divalent cation entry into the matrix inhibits CL synthesisc | (Schlame and Haldar, 1993) |
| Cld1p | Matrix leaflet of IM/non-integral | 0, 1, 2 | Protected from protease in mitoplasts; partially extracted with carbonate; extracted with high salt concentrations | (Baile et al., 2013) |
| Taz1p | IMS-facing leaflet of IM and OM/non-integral | 0, 1, 2 | Degraded by protease in mitoplasts; partially extracted with carbonate; epitope tags throughout the polypeptide face the IMS | (Claypool et al., 2006) |
Transmembrane predictions were determined using the DAS-TMfilter prediction server (Cserzo et al., 2004), TMpred (Hofmann and Stoffel, 1993), HMMTOP (Tusnady and Simon, 1998), TMHMM (Krogh et al., 2001), and SPLIT (Juretic et al., 2002)
Most programs predicted Pgs1p to have 0 transmembrane domains, except TMpred which predicted either 1 or 2 transmembrane segments
Biochemical experiments have not been performed on yeast Crd1p. The experiments here analyzed the rat Crd1p homolog
To reach to IM, CL precursor phospholipids must traverse the OM, but little is known about this process. Phospholipid exchange between leaflets of purified OM vesicles is rapid, suggesting that proteins mediate this process. However, treatment with proteases or with sulfhydrul reactive compounds does not inhibit transbilayer movement across the OM (Janssen et al., 1999).
PA is transported from the OM to the IM by the intermembrane space (IMS) resident protein, Ups1p (Connerth et al., 2012). Mdm35p binds Ups1p, facilitating its import into the IMS and preventing its proteolytic degradation (Potting et al., 2010; Tamura et al., 2010). Although Ups1p/Mdm35p dimers can bind negatively charged phospholipids, only PA is transported in vitro, demonstrating the specificity of its transport activity. Ups1p is unable to dissociate from membranes containing physiological levels of CL. Thus, the higher amount of CL in the IM is modeled to confer directionality of PA transport and the ability to limit CL accumulation (Connerth et al., 2012). Once delivered to the IM, PA must traverse to the matrix side of the IM. This could be accomplished by an unidentified protein or alternatively, PA may redistribute to both leaflets of the IM based on the transmembrane pH gradient (Gallet et al., 1999; Hope et al., 1989).
3. Synthesizing CL
The first committed step of CL biosynthesis is the formation of PGP from CDP-DAG and glycerol-3-phosphate by Pgs1p (Chang et al., 1998a). While the topology of Pgs1p has never been formally investigated (Table 1), the presence of an N-terminal presequence, which is able to import the lacZ gene product to the matrix (Dzugasova et al., 1998), suggests that Pgs1p is localized on the matrix side of the IM. PGP is then dephosphorylated to phosphatidylglycerol (PG) by Gep4p, a protein peripherally attached to the matrix side of the IM (Osman et al., 2010). In the final step of CL biosynthesis, PG and another CDP-DAG are condensed to form CL by Crd1p (Chang et al., 1998b; Jiang et al., 1997). Characterization of the rat Crd1p homolog from liver indicates that it is an integral membrane protein and that its active site faces the matrix (Gallet et al., 1997; Schlame and Haldar, 1993).
Exogenous inositol downregulates phosphatidylcholine and phosphatidylinositol biosynthesis through transcriptional repression via an inositol sensitive upstream activating sequence (UASINO) (Henry et al., 2012). Pgs1p activity is similarly reduced in the presence of inositol, but it contains a mutated, nonfunctional UASINO sequence (Bachhawat et al., 1995) and PGS1 mRNA levels are unchanged in the presence of inositol (Zhong and Greenberg, 2003). Further, deletion of the UASINO-binding genes INO2, INO4, or OPI1 does not affect Pgs1p activity (Greenberg et al., 1988), suggesting its inositol-mediated regulation is independent of the INO2-INO4-OPI1 circuit. Indeed, inositol increases Pgs1p phosphorylation, leading to its repressed activity, although the kinase(s) involved has yet to be identified (He and Greenberg, 2004). Independent from its inositol-mediated regulation, Pgs1p activity is increased under conditions indicative of mitochondrial biogenesis; its mRNA abundance is highest when cells enter stationary phase, and its activity is higher in the presence of non-fermentable carbon sources and when cells contain functional mtDNA (Gaynor et al., 1991; Shen and Dowhan, 1998; Zhong and Greenberg, 2003).
Crd1p activity is similarly increased during stationary growth, in the presence of mtDNA, and in the presence of non-fermentable carbon sources, leading to increased CL levels (Baile et al., 2013; Claypool et al., 2008a; Gaynor et al., 1991; Jakovcic et al., 1971; Jiang et al., 1999; Su and Dowhan, 2006) This is not surprising considering the importance of CL in a myriad of mitochondrial functions (Claypool and Koehler, 2012); as mitochondrial biogenesis increases, CL levels concurrently increase.
Crd1p activity can be additionally regulated by the matrix pH (Gohil et al., 2004). Treatment of yeast with the protonophore CCCP (which disrupts the pH and electrical gradients across the IM), but not the K+ ionophopre valinomycin (which disrupts the electrical gradient but does not affect the matrix pH), decreases Crd1p activity. A decrease in the matrix pH is indicative of less robust electron transport chain activity, coordinating the mitochondrion's energetic requirements with CL biosynthesis. This is further exemplified by the decreases in CL levels that result from defects in respiratory complexes and/or bioenergetic function (Gohil et al., 2004; Zhao et al., 1998).
Interestingly, while steady state CL levels were reduced in Δtaz1 yeast, synthesis of CL was actually increased in the mutant, concurrent with MLCL accumulation (Gu et al., 2004). These observations led the group to suggest that de novo CL biosynthesis might be regulated by downstream CL acylation/remodeling. Crd1p's activity might therefore be negatively regulated by its own product, and under conditions where CL is decreased or aberrantly acylated, the cell compensates by promoting CL biogenesis.
CL biosynthesis is thus regulated via multiple independent mechanisms: by inositol, which regulates Pgs1p; by mitochondrial biogenesis, which affects Pgs1p and Crd1p activity; by CL, which may inhibit Crd1p activity; and by the capacity for oxidative phosphorylation, which affects Crd1p.
4. Remodeling CL
CL remodeling is initiated by the lipase Cld1p in yeast (Beranek et al., 2009), which removes an acyl chain from CL, generating MLCL. Taz1p performs an acyl-CoA independent transacylation reaction, transferring an acyl chain from a phospholipid to a lysophospholipid (generated by a phospholipase), regenerating CL (Gu et al., 2004; Xu et al., 2003; Xu et al., 2006).
Cld1p is located on the matrix-facing leaflet of the IM and does not traverse the membrane (Baile et al., 2013). Surprisingly, the localization of Taz1p is not the same as enzymes upstream in the pathway. In yeast, Taz1p was originally localized to the mitochondrial OM (Brandner et al., 2005), but was later shown to be present on both the inner and outer membrane, on leaflets facing the IMS (Claypool et al., 2006; Gebert et al., 2009). Taz1p is an interfacial membrane protein; it contains residues that are embedded in, but not through, the membrane (Claypool 2006).
Cld1p and Taz1p are localized to different sides of the IM, and neither contains transmembrane domains, suggesting that an as yet unidentified protein(s) transports MLCL generated by Cld1p to the opposite side of the IM and/or to the OM. This trafficking of MLCL is expected to occur rapidly after CL deacylation as MLCL does not accumulate in yeast with a functional Taz1p (Baile et al., 2013; Gu et al., 2004). That both Cld1p and Taz1p assemble into higher-order complexes (Baile et al., 2013; Claypool et al., 2008a; Claypool et al., 2006), and that their binding partners have been, at best, partially defined, raises the exciting possibility that the protein(s) mediating MLCL translocation physically interacts with Cld1p, Taz1p, or both enzymes, although this has yet to be tested. While proteins mediating phospholipid redistribution between membrane leaflets have been identified for the plasma membrane, Golgi, and endosomes (van Meer et al., 2008), considerably less is known about this process in the mitochondrion. CL redistribution between IM leaflets has been observed (Gallet et al., 1997; Gallet et al., 1999), but the protein(s) responsible has not been identified. So far, phospholipid scramblase 3 (PLS3) is the only mitochondrial protein suggested to facilitate transbilayer lipid trafficking (Liu et al., 2003), but this has not been formally demonstrated. Further, deletion of its predicted yeast ortholog, AIM25, in yeast does not result in MLCL accumulation (Baile, M.G., Lu, Y., Claypool, S.M. unpublished data). Importantly, the translocation of phospholipids between membrane leaflets may not be facilitated by specific proteins, but instead non-specifically by the presence of numerous transmembrane proteins, as has been suggested for bacterial membranes and the ER (Kol et al., 2004; Kol et al., 2001; van Meer et al., 2008).
Similar to Crd1p, Cld1p activity is upregulated in the presence of non-fermentable carbon sources. Cld1p expression increases in lactate-containing media, and is repressed in dextrose-containing media (Baile et al., 2013), suggesting that CL remodeling is coordinately regulated with biosynthesis. Cld1p is also regulated by changes in the electrochemical gradient, but through a different mechanism and with a different functional outcome than Crd1p regulation. While reduction of the matrix pH decreases Crd1p activity (Gohil et al., 2004), Cld1p activity increases upon dissipation of the electrical potential (Baile et al., 2013). These differences may provide a mechanism by which CL biosynthesis and remodeling activity can be independently adjusted to fit the requirements of the mitochondrion.
Cld1p is the only protein in the CL remodeling pathway whose activity is known to be regulated (Taz1p expression increases when yeast are grown in the presence of non-fermentable carbon sources; Baile et al., 2013). Despite the fact that the spatial separation of Cld1p and Taz1p provides a potential point of regulation, MLCL levels remain unchanged and very low/absent unless Taz1p is non-functional (Baile et al., 2013), suggesting that the activity of the MLCL flippase and Taz1p is never limiting.
5. Establishing the final distribution of CL
CL is enriched in the IM, but is also present on the OM (Gebert et al., 2009). How it achieves its final distribution in yeast is still unclear. Intriguingly, the presence of a subpopulation of Taz1p on the OM opens up the possibility that MLCL may be the lipid species trafficked from the IM to the OM, where it is then reacylated to form CL (Claypool et al., 2006; Gebert et al., 2009).
Phospholipid transfer between the OM and IM has been suggested to occur at contact sites between the two membranes (Blok et al., 1971; Simbeni et al., 1990). Recently, the proteins comprising this complex (termed MINOS, mitochondrial inner membrane organizing system, MitOS, or MICOS) have been identified (Harner et al., 2011; Hoppins et al., 2011; von der Malsburg et al., 2011). Loss of this complex results in abnormal cristae morphology and loss of cristae junctions. However, the effects on phospholipid transport, the import of the CL precursor PA, or the final distribution of CL, have yet to be studied in contact site mutant yeast strains.
Currently, three proteins have been described that have the ability to traffic/redistribute CL in mammals: the mitochondrial creatine kinase (MtCK; Epand et al., 2007), the mitochondrial nucleoside diphosphate kinase (NDPK-D; Epand et al., 2007; Schlattner et al., 2013), and phospholipid scramblase 3 (PLS3; Liu et al., 2003). However, MtCK has no ortholog in yeast. Yeast also do not contain a mitochondrial-specific ortholog of NDPK-D, although a small portion of the yeast nucleoside diphosphate kinase, Ynk1p, localizes to mitochondria (Amutha and Pain, 2003). However, if Ynk1p can transport CL between membranes remains to be determined. PLS3 was shown to redistribute CL between the IM and OM in mammalian mitochondria (Liu et al., 2003), but phospholipid transport between membranes is inconsistent with its role as a scramblase. Thus, it is likely that PLS3 instead coordinates with a CL transport protein, and the altered distribution of CL between the OM and IM when PLS3 is overexpressed reflects the increased availability of CL on the IMS-facing leaflet of the IM.
Three UPS isoforms exist in yeast, although a concrete function has yet to be assigned to Ups2p or Ups3p. It is tempting to speculate that, like Ups1p and PA (Connerth et al., 2012), either of these proteins can transport CL (or MLCL) between the IM and OM. Interestingly, total CL levels in UPS2 and UPS3 mutants remain largely unaffected (Osman et al., 2009; Tamura et al., 2009), but the relative distribution of CL between the IM and OM has never been analyzed.
6. Perspectives
While most, if not all, of the enzymes involved in CL biosynthesis and remodeling have been identified (at least in yeast), many questions remain. How CL precursors are trafficked, and how CL achieves its final distribution, remains incompletely resolved. The ERMES complex is undoubtedly important for the trafficking of phospholipids between the ER and OM, but whether it directly transports phospholipids or simply mediates the apposition of the two membranes remains to be answered. OM/IM contact sites are potentially important for the movement of both CL precursors and CL itself between mitochondrial membranes, but this is currently an understudied aspect of CL metabolism. Additionally, the proteins mediating CL/MLCL movement, both between membranes and between leaflets of the same membrane, await identification. The regulation of CL precursor trafficking, except for the potential ability of CL to inhibit Ups1p/Mdm35p-mediated PA import, is unknown. Trafficking steps have the potential to regulate the flux of precursors through the CL pathway, but whether this is the case has yet to be determined. Further, compared to yeast (Fig. 2 summarizes modes of regulation identified in yeast), knowledge of these processes and their regulation in mammals is lacking.
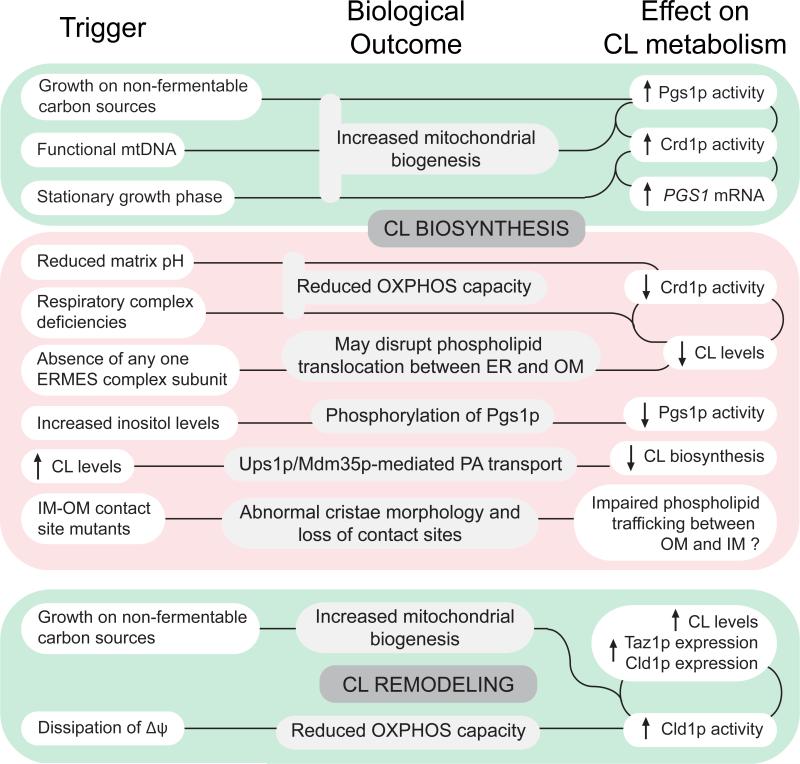
FIGURE 2. Regulatory mechanisms of CL biosynthesis and remodeling.
The CL biosynthetic pathway is upregulated under conditions favoring mitochondrial biogenesis. In contrast, deficiencies in ERMES (ER-OM), MINOS/MitOS/MICOS (OM-IM contact sites) complexes, and components of the electron transport chain, as well as increased levels of inositol and reduced matrix pH, can all lead to a down-regulation of CL biosynthesis. Additionally, CL levels can be modulated by Ups1p/Mdm35p-mediated PA transport. Similar to CL biosynthesis, growth of yeast on respiratory media also promotes CL remodeling by upregulating the activity and/or expression of enzymes in the remodeling pathway. Distinct from CL biosynthesis, dissipation of the electrical potential across the IM, indicative of reduced OXPHOS capacity, increases Cld1p activity. Green boxes indicate conditions that promote CL biosynthesis and remodeling while red boxes indicate conditions that repress CL biosynthesis.
Highlights.
Cardiolipin is required for numerous mitochondrial functions
Yeast provide an excellent model to dissect cardiolipin metabolism
The topology of cardiolipin biosynthesis and remodeling is complex
Cardiolipin metabolism is regulated by multiple independent mechanisms
ACKNOWLEDGEMENTS
Work in the authors’ laboratory is supported by National Institutes of Health grant 1R01HL108882. Y.L. is a predoctoral fellow of the American Heart Association.
ABBREVIATIONS
- CCCP
carbonyl cyanide 3-chlorophenylhydrazone
- CDP-DAG
CDP-diacylglycerol
- CL
cardiolipin
- ERMES
ER-mitochondrial encounter structure
- IM
inner membrane
- IMS
intermembrane space
- OM
outer membrane
- PA
phosphatidic acid
- PG
phosphatidylglycerol
- PGP
phosphatidylglycerolphosphate
- UASINO
inositol sensitive upstream activating sequence
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
REFERENCES
- Acehan D, Khuchua Z, Houtkooper RH, Malhotra A, Kaufman J, Vaz FM, Ren M, Rockman HA, Stokes DL, Schlame M. Distinct effects of tafazzin deletion in differentiated and undifferentiated mitochondria. Mitochondrion. 2009;9:86–95. doi: 10.1016/j.mito.2008.12.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acehan D, Malhotra A, Xu Y, Ren M, Stokes DL, Schlame M. Cardiolipin affects the supramolecular organization of ATP synthase in mitochondria. Biophysical journal. 2011;100:2184–2192. doi: 10.1016/j.bpj.2011.03.031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Acehan D, Xu Y, Stokes DL, Schlame M. Comparison of lymphoblast mitochondria from normal subjects and patients with Barth syndrome using electron microscopic tomography. Lab Invest. 2007;87:40–48. doi: 10.1038/labinvest.3700480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amutha B, Pain D. Nucleoside diphosphate kinase of Saccharomyces cerevisiae, Ynk1p: localization to the mitochondrial intermembrane space. The Biochemical journal. 2003;370:805–815. doi: 10.1042/BJ20021415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bachhawat N, Ouyang Q, Henry SA. Functional characterization of an inositol-sensitive upstream activation sequence in yeast. A cis-regulatory element responsible for inositol-choline mediated regulation of phospholipid biosynthesis. The Journal of biological chemistry. 1995;270:25087–25095. doi: 10.1074/jbc.270.42.25087. [DOI] [PubMed] [Google Scholar]
- Baile MG, Claypool SM. The power of yeast to model diseases of the powerhouse of the cell. Front Biosci (Landmark Ed) 2013;18:241–278. doi: 10.2741/4098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Baile MG, Whited K, Claypool SM. Deacylation on the matrix side of the mitochondrial inner membrane regulates cardiolipin remodeling. Molecular biology of the cell. 2013;24:2008–2020. doi: 10.1091/mbc.E13-03-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ban T, Heymann JA, Song Z, Hinshaw JE, Chan DC. OPA1 disease alleles causing dominant optic atrophy have defects in cardiolipin-stimulated GTP hydrolysis and membrane tubulation. Human molecular genetics. 2010;19:2113–2122. doi: 10.1093/hmg/ddq088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barth PG, Scholte HR, Berden JA, Van der Klei-Van Moorsel JM, Luyt-Houwen IE, Van 't Veer-Korthof ET, Van der Harten JJ, Sobotka-Plojhar MA. An X-linked mitochondrial disease affecting cardiac muscle, skeletal muscle and neutrophil leucocytes. Journal of the neurological sciences. 1983;62:327–355. doi: 10.1016/0022-510x(83)90209-5. [DOI] [PubMed] [Google Scholar]
- Bazan S, Mileykovskaya E, Mallampalli VK, Heacock P, Sparagna GC, Dowhan W. Cardiolipin-dependent reconstitution of respiratory supercomplexes from purified Saccharomyces cerevisiae complexes III and IV. The Journal of biological chemistry. 2013;288:401–411. doi: 10.1074/jbc.M112.425876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beranek A, Rechberger G, Knauer H, Wolinski H, Kohlwein SD, Leber R. Identification of a cardiolipin-specific phospholipase encoded by the gene CLD1 (YGR110W) in yeast. The Journal of biological chemistry. 2009;284:11572–11578. doi: 10.1074/jbc.M805511200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blok MC, Wirtz KW, Scherphof GL. Exchange of phospholipids between microsomes and inner and outer mitochondrial membranes of rat liver. Biochimica et biophysica acta. 1971;233:61–75. doi: 10.1016/0005-2736(71)90358-0. [DOI] [PubMed] [Google Scholar]
- Botstein D, Fink GR. Yeast: an experimental organism for 21st Century biology. Genetics. 2011;189:695–704. doi: 10.1534/genetics.111.130765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brandner K, Mick DU, Frazier AE, Taylor RD, Meisinger C, Rehling P. Taz1, an outer mitochondrial membrane protein, affects stability and assembly of inner membrane protein complexes: implications for Barth Syndrome. Molecular biology of the cell. 2005;16:5202–5214. doi: 10.1091/mbc.E05-03-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang SC, Heacock PN, Clancey CJ, Dowhan W. The PEL1 gene (renamed PGS1) encodes the phosphatidylglycero-phosphate synthase of Saccharomyces cerevisiae. The Journal of biological chemistry. 1998a;273:9829–9836. doi: 10.1074/jbc.273.16.9829. [DOI] [PubMed] [Google Scholar]
- Chang SC, Heacock PN, Mileykovskaya E, Voelker DR, Dowhan W. Isolation and characterization of the gene (CLS1) encoding cardiolipin synthase in Saccharomyces cerevisiae. The Journal of biological chemistry. 1998b;273:14933–14941. doi: 10.1074/jbc.273.24.14933. [DOI] [PubMed] [Google Scholar]
- Chicco AJ, Sparagna GC. Role of cardiolipin alterations in mitochondrial dysfunction and disease. American journal of physiology. Cell physiology. 2007;292:C33–44. doi: 10.1152/ajpcell.00243.2006. [DOI] [PubMed] [Google Scholar]
- Claypool SM, Boontheung P, McCaffery JM, Loo JA, Koehler CM. The cardiolipin transacylase, tafazzin, associates with two distinct respiratory components providing insight into Barth syndrome. Molecular biology of the cell. 2008a;19:5143–5155. doi: 10.1091/mbc.E08-09-0896. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claypool SM, Koehler CM. The complexity of cardiolipin in health and disease. Trends Biochem Sci. 2012;37:32–41. doi: 10.1016/j.tibs.2011.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claypool SM, McCaffery JM, Koehler CM. Mitochondrial mislocalization and altered assembly of a cluster of Barth syndrome mutant tafazzins. The Journal of cell biology. 2006;174:379–390. doi: 10.1083/jcb.200605043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Claypool SM, Oktay Y, Boontheung P, Loo JA, Koehler CM. Cardiolipin defines the interactome of the major ADP/ATP carrier protein of the mitochondrial inner membrane. The Journal of cell biology. 2008b;182:937–950. doi: 10.1083/jcb.200801152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Connerth M, Tatsuta T, Haag M, Klecker T, Westermann B, Langer T. Intramitochondrial transport of phosphatidic acid in yeast by a lipid transfer protein. Science. 2012;338:815–818. doi: 10.1126/science.1225625. [DOI] [PubMed] [Google Scholar]
- Cserzo M, Eisenhaber F, Eisenhaber B, Simon I. TM or not TM: transmembrane protein prediction with low false positive rate using DAS-TMfilter. Bioinformatics. 2004;20:136–137. doi: 10.1093/bioinformatics/btg394. [DOI] [PubMed] [Google Scholar]
- DeVay RM, Dominguez-Ramirez L, Lackner LL, Hoppins S, Stahlberg H, Nunnari J. Coassembly of Mgm1 isoforms requires cardiolipin and mediates mitochondrial inner membrane fusion. The Journal of cell biology. 2009;186:793–803. doi: 10.1083/jcb.200906098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dzugasova V, Obernauerova M, Horvathova K, Vachova M, Zakova M, Subik J. Phosphatidylglycerolphosphate synthase encoded by the PEL1/PGS1 gene in Saccharomyces cerevisiae is localized in mitochondria and its expression is regulated by phospholipid precursors. Curr Genet. 1998;34:297–302. doi: 10.1007/s002940050399. [DOI] [PubMed] [Google Scholar]
- Epand RF, Schlattner U, Wallimann T, Lacombe ML, Epand RM. Novel lipid transfer property of two mitochondrial proteins that bridge the inner and outer membranes. Biophysical journal. 2007;92:126–137. doi: 10.1529/biophysj.106.092353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fry M, Green DE. Cardiolipin requirement for electron transfer in complex I and III of the mitochondrial respiratory chain. The Journal of biological chemistry. 1981;256:1874–1880. [PubMed] [Google Scholar]
- Gallas MR, Dienhart MK, Stuart RA, Long RM. Characterization of Mmp37p, a Saccharomyces cerevisiae mitochondrial matrix protein with a role in mitochondrial protein import. Molecular biology of the cell. 2006;17:4051–4062. doi: 10.1091/mbc.E06-04-0366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallet PF, Petit JM, Maftah A, Zachowski A, Julien R. Asymmetrical distribution of cardiolipin in yeast inner mitochondrial membrane triggered by carbon catabolite repression. The Biochemical journal. 1997;324( Pt 2):627–634. doi: 10.1042/bj3240627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gallet PF, Zachowski A, Julien R, Fellmann P, Devaux PF, Maftah A. Transbilayer movement and distribution of spin-labelled phospholipids in the inner mitochondrial membrane. Biochimica et biophysica acta. 1999;1418:61–70. doi: 10.1016/s0005-2736(99)00022-x. [DOI] [PubMed] [Google Scholar]
- Gaynor PM, Hubbell S, Schmidt AJ, Lina RA, Minskoff SA, Greenberg ML. Regulation of phosphatidylglycerolphosphate synthase in Saccharomyces cerevisiae by factors affecting mitochondrial development. Journal of bacteriology. 1991;173:6124–6131. doi: 10.1128/jb.173.19.6124-6131.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gebert N, Joshi AS, Kutik S, Becker T, McKenzie M, Guan XL, Mooga VP, Stroud DA, Kulkarni G, Wenk MR, Rehling P, Meisinger C, Ryan MT, Wiedemann N, Greenberg ML, Pfanner N. Mitochondrial cardiolipin involved in outer-membrane protein biogenesis: implications for Barth syndrome. Curr Biol. 2009;19:2133–2139. doi: 10.1016/j.cub.2009.10.074. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gohil VM, Hayes P, Matsuyama S, Schagger H, Schlame M, Greenberg ML. Cardiolipin biosynthesis and mitochondrial respiratory chain function are interdependent. The Journal of biological chemistry. 2004;279:42612–42618. doi: 10.1074/jbc.M402545200. [DOI] [PubMed] [Google Scholar]
- Gomez B,, Jr., Robinson NC. Phospholipase digestion of bound cardiolipin reversibly inactivates bovine cytochrome bc1. Biochemistry. 1999;38:9031–9038. doi: 10.1021/bi990603r. [DOI] [PubMed] [Google Scholar]
- Greenberg ML, Hubbell S, Lam C. Inositol regulates phosphatidylglycerolphosphate synthase expression in Saccharomyces cerevisiae. Molecular and cellular biology. 1988;8:4773–4779. doi: 10.1128/mcb.8.11.4773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gu Z, Valianpour F, Chen S, Vaz FM, Hakkaart GA, Wanders RJ, Greenberg ML. Aberrant cardiolipin metabolism in the yeast taz1 mutant: a model for Barth syndrome. Molecular microbiology. 2004;51:149–158. doi: 10.1046/j.1365-2958.2003.03802.x. [DOI] [PubMed] [Google Scholar]
- Harner M, Korner C, Walther D, Mokranjac D, Kaesmacher J, Welsch U, Griffith J, Mann M, Reggiori F, Neupert W. The mitochondrial contact site complex, a determinant of mitochondrial architecture. EMBO J. 2011;30:4356–4370. doi: 10.1038/emboj.2011.379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- He Q, Greenberg ML. Post-translational regulation of phosphatidylglycerolphosphate synthase in response to inositol. Molecular microbiology. 2004;53:1243–1249. doi: 10.1111/j.1365-2958.2004.04202.x. [DOI] [PubMed] [Google Scholar]
- Henry SA, Kohlwein SD, Carman GM. Metabolism and regulation of glycerolipids in the yeast Saccharomyces cerevisiae. Genetics. 2012;190:317–349. doi: 10.1534/genetics.111.130286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hofmann K, Stoffel W. TMbase - A database of membrane spanning proteins segments. Biol. Chem. Hoppe-Seyler. 1993;374:166. [Google Scholar]
- Hope MJ, Redelmeier TE, Wong KF, Rodrigueza W, Cullis PR. Phospholipid asymmetry in large unilamellar vesicles induced by transmembrane pH gradients. Biochemistry. 1989;28:4181–4187. doi: 10.1021/bi00436a009. [DOI] [PubMed] [Google Scholar]
- Hoppins S, Collins SR, Cassidy-Stone A, Hummel E, Devay RM, Lackner LL, Westermann B, Schuldiner M, Weissman JS, Nunnari J. A mitochondrial-focused genetic interaction map reveals a scaffold-like complex required for inner membrane organization in mitochondria. The Journal of cell biology. 2011;195:323–340. doi: 10.1083/jcb.201107053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jakovcic S, Getz GS, Rabinowitz M, Jakob H, Swift H. Cardiolipin content of wild type and mutant yeasts in relation to mitochondrial function and development. The Journal of cell biology. 1971;48:490–502. doi: 10.1083/jcb.48.3.490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janssen MJ, Koorengevel MC, de Kruijff B, de Kroon AI. Transbilayer movement of phosphatidylcholine in the mitochondrial outer membrane of Saccharomyces cerevisiae is rapid and bidirectional. Biochimica et biophysica acta. 1999;1421:64–76. doi: 10.1016/s0005-2736(99)00113-3. [DOI] [PubMed] [Google Scholar]
- Jiang F, Gu Z, Granger JM, Greenberg ML. Cardiolipin synthase expression is essential for growth at elevated temperature and is regulated by factors affecting mitochondrial development. Molecular microbiology. 1999;31:373–379. doi: 10.1046/j.1365-2958.1999.01181.x. [DOI] [PubMed] [Google Scholar]
- Jiang F, Rizavi HS, Greenberg ML. Cardiolipin is not essential for the growth of Saccharomyces cerevisiae on fermentable or non-fermentable carbon sources. Molecular microbiology. 1997;26:481–491. doi: 10.1046/j.1365-2958.1997.5841950.x. [DOI] [PubMed] [Google Scholar]
- Jiang F, Ryan MT, Schlame M, Zhao M, Gu Z, Klingenberg M, Pfanner N, Greenberg ML. Absence of cardiolipin in the crd1 null mutant results in decreased mitochondrial membrane potential and reduced mitochondrial function. The Journal of biological chemistry. 2000;275:22387–22394. doi: 10.1074/jbc.M909868199. [DOI] [PubMed] [Google Scholar]
- Joshi AS, Thompson MN, Fei N, Huttemann M, Greenberg ML. Cardiolipin and mitochondrial phosphatidylethanolamine have overlapping functions in mitochondrial fusion in Saccharomyces cerevisiae. The Journal of biological chemistry. 2012;287:17589–17597. doi: 10.1074/jbc.M111.330167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Joshi AS, Zhou J, Gohil VM, Chen S, Greenberg ML. Cellular functions of cardiolipin in yeast. Biochimica et biophysica acta. 2009;1793:212–218. doi: 10.1016/j.bbamcr.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Juretic D, Zoranic L, Zucic D. Basic charge clusters and predictions of membrane protein topology. J Chem Inf Comput Sci. 2002;42:620–632. doi: 10.1021/ci010263s. [DOI] [PubMed] [Google Scholar]
- Kiebish MA, Yang K, Liu X, Mancuso DJ, Guan S, Zhao Z, Sims HF, Cerqua R, Cade WT, Han X, Gross RW. Dysfunctional cardiac mitochondrial bioenergetic, lipidomic, and signaling in a murine model of Barth syndrome. Journal of lipid research. 2013;54:1312–1325. doi: 10.1194/jlr.M034728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kol MA, de Kroon AI, Killian JA, de Kruijff B. Transbilayer movement of phospholipids in biogenic membranes. Biochemistry. 2004;43:2673–2681. doi: 10.1021/bi036200f. [DOI] [PubMed] [Google Scholar]
- Kol MA, de Kroon AI, Rijkers DT, Killian JA, de Kruijff B. Membrane-spanning peptides induce phospholipid flop: a model for phospholipid translocation across the inner membrane of E. coli. Biochemistry. 2001;40:10500–10506. doi: 10.1021/bi010627+. [DOI] [PubMed] [Google Scholar]
- Kornmann B, Currie E, Collins SR, Schuldiner M, Nunnari J, Weissman JS, Walter P. An ER-mitochondria tethering complex revealed by a synthetic biology screen. Science. 2009;325:477–481. doi: 10.1126/science.1175088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krogh A, Larsson B, von Heijne G, Sonnhammer EL. Predicting transmembrane protein topology with a hidden Markov model: application to complete genomes. J Mol Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- Kuchler K, Daum G, Paltauf F. Subcellular and submitochondrial localization of phospholipid-synthesizing enzymes in Saccharomyces cerevisiae. Journal of bacteriology. 1986;165:901–910. doi: 10.1128/jb.165.3.901-910.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kutik S, Rissler M, Guan XL, Guiard B, Shui G, Gebert N, Heacock PN, Rehling P, Dowhan W, Wenk MR, Pfanner N, Wiedemann N. The translocator maintenance protein Tam41 is required for mitochondrial cardiolipin biosynthesis. The Journal of cell biology. 2008;183:1213–1221. doi: 10.1083/jcb.200806048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu J, Dai Q, Chen J, Durrant D, Freeman A, Liu T, Grossman D, Lee RM. Phospholipid scramblase 3 controls mitochondrial structure, function, and apoptotic response. Mol Cancer Res. 2003;1:892–902. [PubMed] [Google Scholar]
- Malhotra A, Edelman-Novemsky I, Xu Y, Plesken H, Ma J, Schlame M, Ren M. Role of calcium-independent phospholipase A2 in the pathogenesis of Barth syndrome. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:2337–2341. doi: 10.1073/pnas.0811224106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mancuso DJ, Sims HF, Han X, Jenkins CM, Guan SP, Yang K, Moon SH, Pietka T, Abumrad NA, Schlesinger PH, Gross RW. Genetic ablation of calcium-independent phospholipase A2gamma leads to alterations in mitochondrial lipid metabolism and function resulting in a deficient mitochondrial bioenergetic phenotype. The Journal of biological chemistry. 2007;282:34611–34622. doi: 10.1074/jbc.M707795200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mileykovskaya E, Dowhan W. Cardiolipin membrane domains in prokaryotes and eukaryotes. Biochimica et biophysica acta. 2009;1788:2084–2091. doi: 10.1016/j.bbamem.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nguyen TT, Lewandowska A, Choi JY, Markgraf DF, Junker M, Bilgin M, Ejsing CS, Voelker DR, Rapoport TA, Shaw JM. Gem1 and ERMES do not directly affect phosphatidylserine transport from ER to mitochondria or mitochondrial inheritance. Traffic. 2012;13:880–890. doi: 10.1111/j.1600-0854.2012.01352.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman C, Haag M, Potting C, Rodenfels J, Dip PV, Wieland FT, Brugger B, Westermann B, Langer T. The genetic interactome of prohibitins: coordinated control of cardiolipin and phosphatidylethanolamine by conserved regulators in mitochondria. The Journal of cell biology. 2009;184:583–596. doi: 10.1083/jcb.200810189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Osman C, Haag M, Wieland FT, Brugger B, Langer T. A mitochondrial phosphatase required for cardiolipin biosynthesis: the PGP phosphatase Gep4. EMBO J. 2010;29:1976–1987. doi: 10.1038/emboj.2010.98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paradies G, Ruggiero FM, Petrosillo G, Quagliariello E. Age-dependent decline in the cytochrome c oxidase activity in rat heart mitochondria: role of cardiolipin. FEBS letters. 1997;406:136–138. doi: 10.1016/s0014-5793(97)00264-0. [DOI] [PubMed] [Google Scholar]
- Pfeiffer K, Gohil V, Stuart RA, Hunte C, Brandt U, Greenberg ML, Schagger H. Cardiolipin stabilizes respiratory chain supercomplexes. The Journal of biological chemistry. 2003;278:52873–52880. doi: 10.1074/jbc.M308366200. [DOI] [PubMed] [Google Scholar]
- Potting C, Wilmes C, Engmann T, Osman C, Langer T. Regulation of mitochondrial phospholipids by Ups1/PRELI-like proteins depends on proteolysis and Mdm35. EMBO J. 2010;29:2888–2898. doi: 10.1038/emboj.2010.169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schagger H, Hagen T, Roth B, Brandt U, Link TA, von Jagow G. Phospholipid specificity of bovine heart bc1 complex. European journal of biochemistry . FEBS. 1990;190:123–130. doi: 10.1111/j.1432-1033.1990.tb15554.x. [DOI] [PubMed] [Google Scholar]
- Schlame M. Cardiolipin remodeling and the function of tafazzin. Biochimica et biophysica acta. 2012;1831:582–588. doi: 10.1016/j.bbalip.2012.11.007. [DOI] [PubMed] [Google Scholar]
- Schlame M, Acehan D, Berno B, Xu Y, Valvo S, Ren M, Stokes DL, Epand RM. The physical state of lipid substrates provides transacylation specificity for tafazzin. Nature chemical biology. 2012a;8:862–869. doi: 10.1038/nchembio.1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlame M, Blais S, Edelman-Novemsky I, Xu Y, Montecillo F, Phoon CK, Ren M, Neubert TA. Comparison of cardiolipins from Drosophila strains with mutations in putative remodeling enzymes. Chem Phys Lipids. 2012b;165:512–519. doi: 10.1016/j.chemphyslip.2012.03.001. [DOI] [PubMed] [Google Scholar]
- Schlame M, Haldar D. Cardiolipin is synthesized on the matrix side of the inner membrane in rat liver mitochondria. The Journal of biological chemistry. 1993;268:74–79. [PubMed] [Google Scholar]
- Schlame M, Ren M. Barth syndrome, a human disorder of cardiolipin metabolism. FEBS letters. 2006;580:5450–5455. doi: 10.1016/j.febslet.2006.07.022. [DOI] [PubMed] [Google Scholar]
- Schlattner U, Tokarska-Schlattner M, Ramirez S, Tyurina YY, Amoscato AA, Mohammadyani D, Huang Z, Jiang J, Yanamala N, Seffouh A, Boissan M, Epand RF, Epand RM, Klein-Seetharaman J, Lacombe ML, Kagan VE. Dual function of mitochondrial Nm23-H4 protein in phosphotransfer and intermembrane lipid transfer: a cardiolipin-dependent switch. The Journal of biological chemistry. 2013;288:111–121. doi: 10.1074/jbc.M112.408633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwall CT, Greenwood VL, Alder NN. The stability and activity of respiratory Complex II is cardiolipin-dependent. Biochimica et biophysica acta. 2012;1817:1588–1596. doi: 10.1016/j.bbabio.2012.04.015. [DOI] [PubMed] [Google Scholar]
- Shen H, Dowhan W. Regulation of phosphatidylglycerophosphate synthase levels in Saccharomyces cerevisiae. The Journal of biological chemistry. 1998;273:11638–11642. doi: 10.1074/jbc.273.19.11638. [DOI] [PubMed] [Google Scholar]
- Shen H, Heacock PN, Clancey CJ, Dowhan W. The CDS1 gene encoding CDP-diacylglycerol synthase in Saccharomyces cerevisiae is essential for cell growth. The Journal of biological chemistry. 1996;271:789–795. doi: 10.1074/jbc.271.2.789. [DOI] [PubMed] [Google Scholar]
- Simbeni R, Paltauf F, Daum G. Intramitochondrial transfer of phospholipids in the yeast, Saccharomyces cerevisiae. The Journal of biological chemistry. 1990;265:281–285. [PubMed] [Google Scholar]
- Stroud DA, Oeljeklaus S, Wiese S, Bohnert M, Lewandrowski U, Sickmann A, Guiard B, van der Laan M, Warscheid B, Wiedemann N. Composition and topology of the endoplasmic reticulum-mitochondria encounter structure. J Mol Biol. 2011;413:743–750. doi: 10.1016/j.jmb.2011.09.012. [DOI] [PubMed] [Google Scholar]
- Su X, Dowhan W. Regulation of cardiolipin synthase levels in Saccharomyces cerevisiae. Yeast. 2006;23:279–291. doi: 10.1002/yea.1352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura Y, Endo T, Iijima M, Sesaki H. Ups1p and Ups2p antagonistically regulate cardiolipin metabolism in mitochondria. The Journal of cell biology. 2009;185:1029–1045. doi: 10.1083/jcb.200812018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura Y, Harada Y, Nishikawa S, Yamano K, Kamiya M, Shiota T, Kuroda T, Kuge O, Sesaki H, Imai K, Tomii K, Endo T. Tam41 is a CDP-diacylglycerol synthase required for cardiolipin biosynthesis in mitochondria. Cell Metab. 2013;17:709–718. doi: 10.1016/j.cmet.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura Y, Harada Y, Yamano K, Watanabe K, Ishikawa D, Ohshima C, Nishikawa S, Yamamoto H, Endo T. Identification of Tam41 maintaining integrity of the TIM23 protein translocator complex in mitochondria. The Journal of cell biology. 2006;174:631–637. doi: 10.1083/jcb.200603087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura Y, Iijima M, Sesaki H. Mdm35p imports Ups proteins into the mitochondrial intermembrane space by functional complex formation. EMBO J. 2010;29:2875–2887. doi: 10.1038/emboj.2010.149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tamura Y, Onguka O, Hobbs AE, Jensen RE, Iijima M, Claypool SM, Sesaki H. Role for two conserved intermembrane space proteins, Ups1p and Ups2p, [corrected] in intra-mitochondrial phospholipid trafficking. The Journal of biological chemistry. 2012;287:15205–15218. doi: 10.1074/jbc.M111.338665. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tuller G, Hrastnik C, Achleitner G, Schiefthaler U, Klein F, Daum G. YDL142c encodes cardiolipin synthase (Cls1p) and is non-essential for aerobic growth of Saccharomyces cerevisiae. FEBS letters. 1998;421:15–18. doi: 10.1016/s0014-5793(97)01525-1. [DOI] [PubMed] [Google Scholar]
- Tusnady GE, Simon I. Principles governing amino acid composition of integral membrane proteins: application to topology prediction. J Mol Biol. 1998;283:489–506. doi: 10.1006/jmbi.1998.2107. [DOI] [PubMed] [Google Scholar]
- van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nature reviews. Molecular cell biology. 2008;9:112–124. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- von der Malsburg K, Muller JM, Bohnert M, Oeljeklaus S, Kwiatkowska P, Becker T, Loniewska-Lwowska A, Wiese S, Rao S, Milenkovic D, Hutu DP, Zerbes RM, Schulze-Specking A, Meyer HE, Martinou JC, Rospert S, Rehling P, Meisinger C, Veenhuis M, Warscheid B, van der Klei IJ, Pfanner N, Chacinska A, van der Laan M. Dual role of mitofilin in mitochondrial membrane organization and protein biogenesis. Dev Cell. 2011;21:694–707. doi: 10.1016/j.devcel.2011.08.026. [DOI] [PubMed] [Google Scholar]
- Voss C, Lahiri S, Young BP, Loewen CJ, Prinz WA. ER-shaping proteins facilitate lipid exchange between the ER and mitochondria in S. cerevisiae. J Cell Sci. 2012;125:4791–4799. doi: 10.1242/jcs.105635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenz T, Hielscher R, Hellwig P, Schagger H, Richers S, Hunte C. Role of phospholipids in respiratory cytochrome bc(1) complex catalysis and supercomplex formation. Biochimica et biophysica acta. 2009;1787:609–616. doi: 10.1016/j.bbabio.2009.02.012. [DOI] [PubMed] [Google Scholar]
- Xu Y, Kelley RI, Blanck TJ, Schlame M. Remodeling of cardiolipin by phospholipid transacylation. The Journal of biological chemistry. 2003;278:51380–51385. doi: 10.1074/jbc.M307382200. [DOI] [PubMed] [Google Scholar]
- Xu Y, Malhotra A, Ren M, Schlame M. The enzymatic function of tafazzin. The Journal of biological chemistry. 2006;281:39217–39224. doi: 10.1074/jbc.M606100200. [DOI] [PubMed] [Google Scholar]
- Yu CA, Yu L. Structural role of phospholipids in ubiquinol-cytochrome c reductase. Biochemistry. 1980;19:5715–5720. doi: 10.1021/bi00566a008. [DOI] [PubMed] [Google Scholar]
- Zhang J, Guan Z, Murphy AN, Wiley SE, Perkins GA, Worby CA, Engel JL, Heacock P, Nguyen OK, Wang JH, Raetz CR, Dowhan W, Dixon JE. Mitochondrial phosphatase PTPMT1 is essential for cardiolipin biosynthesis. Cell Metab. 2011;13:690–700. doi: 10.1016/j.cmet.2011.04.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao M, Schlame M, Rua D, Greenberg ML. Cardiolipin synthase is associated with a large complex in yeast mitochondria. The Journal of biological chemistry. 1998;273:2402–2408. doi: 10.1074/jbc.273.4.2402. [DOI] [PubMed] [Google Scholar]
- Zhong Q, Greenberg ML. Regulation of phosphatidylglycerophosphate synthase by inositol in Saccharomyces cerevisiae is not at the level of PGS1 mRNA abundance. The Journal of biological chemistry. 2003;278:33978–33984. doi: 10.1074/jbc.M305242200. [DOI] [PubMed] [Google Scholar]