Abstract
The organization of individual respiratory Complexes I, III, and IV (mammalian cells) or III and IV (yeast) of the mitochondria into higher order supercomplexes (SCs) is generally accepted. However, the factors that regulate SC formation and the functional significance of SCs are not well understood. The mitochondrial signature phospholipid cardiolipin (CL) plays a central role in formation and stability of respiratory SCs from yeast to man. Studies in yeast mutants in which the CL level can be regulated displayed a direct correlation between CL levels and SC formation. Disease states in which CL levels are reduced also show defects in SC formation. Three-dimensional density maps of yeast and bovine SCs by electron cryo-microscopy show gaps between the transmembrane-localized interfaces of individual complexes consistent with the large excess of CL in SCs over that integrated into the structure of individual respiratory complexes. Finally, the yeast SC composed of Complex III and two Complexes IV was reconstituted in liposomes from purified individual complexes containing integrated CLs. Reconstitution was wholly dependent on inclusion of additional CL in the liposomes. Therefore, non-integral CL molecules play an important role in SC formation and may be involved in regulation of SC stability under metabolic conditions where CL levels fluctuate.
Keywords: Cardiolipin, respiratory supercomplex, mitochondria, structural analysis, in vitro reconstitution
1.0 Introduction
The anionic phospholipid cardiolipin (CL1), also called diphosphatidylglycerol, (1,3-bis (sn-3′-phosphatidyl)-sn-glycerol), is uniquely localized to energy-transducing membranes, which couple generation of an electrochemical potential with ATP synthesis and substrate transport. In eukaryotes CL is a signature phospholipid of mitochondria. The unique structure of CL is composed of two phosphates, four fatty acids, three chiral centers and a free central hydroxyl (Fig. 1), which has been suggested to serve as a proton sink. In animals and higher plants the majority of acyl chains of CL contain polyunsaturated fatty acids with 18 carbons, while in Saccharomyces cerevisiae (hereafter referred to as yeast) the fatty acids are 16 and 18 carbon monounsaturated chains (Schlame and Ren, 2009). CL interacts with many membrane proteins affecting their activity, stability, level of aggregation, and compartmentalization. In this review we will focus on the specific role CL plays in organization and function of the mitochondrial respiratory chain.
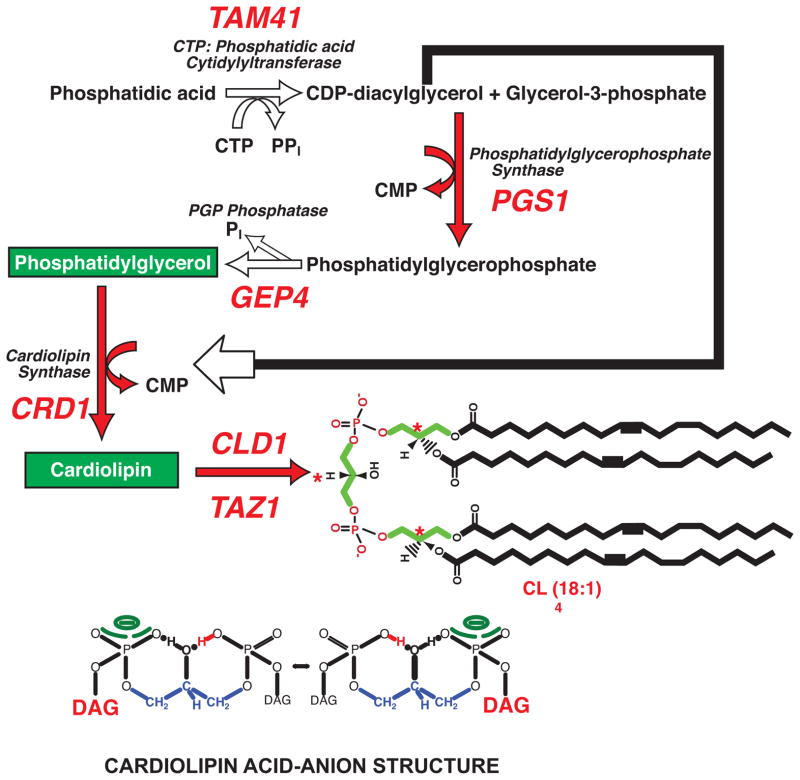
Figure 1.
Biosynthesis and structure of CL. The pathway, genes (red) and gene products responsible for CL synthesis in yeast mitochondria are shown (Henry et al., 2012; Tamura et al., 2013). The pathway in higher eukaryotes is essentially the same and is carried out by homologous genes and gene products. The CLD1 gene product (CL specific deacylase) initiates the remodeling of CL fatty acid chain composition in yeast by forming monolyso-CL (Beranek et al., 2009); higher eukaryotes utilize multiple deacylases (Baile et al., 2013). The TAZ gene product in both yeast and mammalian cells is responsible for completing the remodeling of nascent CL by transferring fatty acids to monolyso-CL from other phospholipids (Schlame et al., 2012). The result is highly unsaturated forms of CL as represented by one of the structures found in yeast and mammalian cells. The (*) in red indicates the three chiral centers of naturally occurring CLs; carbon of the central glycerol is only a chiral center if the two adjacent phosphatidyl moieties have different fatty acid compositions. The glycerol backbone (green) is indicated. The lower figure depicts the hydrogen-bonding between the central free hydroxyl of CL and the phosphate residues creating a proton sink in the lipid bilayer and raising the pKa of one phosphate near neutrality (Haines, 2009). DAG denotes the diacylglycerol lipid domain.
2.0 Synthesis of CL
The function and synthesis of mitochondrial lipids in mammalian cells and yeast are highly homologous. Yeast cells have a distinct research advantage over higher eukaryotes in ease of growth and genetic manipulation coupled with viability in the face of dramatic alterations in mitochondrial phospholipid composition. In yeast, CL and its precursor phosphatidylglycerol (PG) are synthesized from the common precursor phosphatidic acid (Fig. 1) by mitochondrial-localized enzymes that are encoded by nuclear genes, synthesized on cytoplasmic ribosomes and imported into the inner mitochondrial membrane. The fatty acid composition of newly synthesized CL is remodeled to its unique composition by transacylation reactions in which the CLD1 and TAZ1 gene products are involved. All genes of yeast involved in biosynthesis of CL have been identified and cloned with mutants available in each step of synthesis.
3.0 Mitochondrial Respiratory Chain Organization and Function
In mammalian mitochondria the respiratory chain is composed of four multi-subunit electron transfer protein complexes: Complex I (CI, NADH:ubiquinone oxidoreductase), Complex II (succinate:ubiquinone oxidoreductase), Complex III1 or cytochrome bc1 complex (CIII, ubiqunol:cytochrome c oxidoreductase), Complex IV (CIV, cytochrome c oxidase) and two small electron carriers, which transfer electrons from CI or Complex II to CIII (lipid-soluble ubiquinone (CoQ)), or from CIII to CIV (water-soluble cytochrome c). Yeast lacks CI and utilizes peripheral membrane NADH dehydrogenases lacking proton-pumping ability. Electron transfer is coupled with proton pumping by CI, CIII and CIV from the mitochondrial matrix to the mitochondrial intermembrane space (IMS) resulting in an electrochemical proton gradient across the inner mitochondrial membrane, which F1F0-ATPase (Complex V) uses for ATP synthesis (for most recent review and references see (Sun et al., 2013)
Organization of the respiratory chain as one structural and functional unit, now termed a respirasome (Schagger and Pfeiffer, 2000), was originally formulated by Chance (Chance and Williams, 1955). Once active individual respiratory complexes were purified and reconstituted into liposomes, this model was substituted by the random collision model (Hackenbrock et al., 1986), in which the respiratory chain is composed of individual complexes independently imbedded in the lipid bilayer and connected by randomly diffusing CoQ and cytochrome c. Mild solubilization of mitochondrial membranes with digitonin and development of blue-native and colorless-native polyacrylamide gel electrophoresis (BN-PAGE and CN-PAGE), demonstrated active stoichiometric assemblies of individual complexes in mitochondria from yeast to mammals. Association of CI with CIII (I1III2) or CI with CIII and one to four copies of CIV (I1III2IVn=1–4, termed “respirasome”) resulting in multiple supercomplexes (SCs) was demonstrated in mammalian mitochondria. In yeast two SCs, one composed of III2IV1, and another one of III2IV2, were found (Schagger, 2001; Schagger and Pfeiffer, 2000; Stuart, 2008).
Flux control analysis of electron transfer through the respiratory chain showed substrate channeling of CoQ in bovine heart mitochondria and of both, CoQ and cytochrome c, in mitochondria of potato tuber, thus demonstrating the existence of functional respiratory SCs (for review and refs see (Genova and Lenaz, 2013; Lenaz and Genova, 2007)). Kinetic measurements with yeast mitochondria were also consistent with substrate channeling of CoQ and cytochrome c unless mitochondria were treated with a chaotropic agent, which resulted in dissociation of SCs (Boumans et al., 1998). SCs from mouse cells isolated by BN-PAGE contained the entire respirasome along with associated CoQ and cytochrome c, which were capable of oxygen consumption after addition of NADH (Acin-Perez et al., 2008). The purified yeast SC III2IV2 also contained bound cytochrome c and catalyzed electron transfer from reduced CoQ (QH2) to O2 (Mileykovskaya et al., 2012).
In mammalian cells respirasomes composed of CI/CIII/CIV, SCs composed of CI/III and CIII/CIV and free individual respiratory complexes coexist in a dynamic equilibrium. Alteration of the ratios of SCs and individual complexes by genetic manipulation demonstrated that electron transfer occurs by a mixture of direct substrate channeling of CoQ/cytochrome c within SCs and through random collision of CoQ/cytochrome c and individual respiratory complexes dependent on the metabolic state of the cell and substrate availability (Lapuente-Brun et al., 2013).
Nevertheless, studies in whole yeast cells employing time-resolved oxidation of cytochrome c by CIV found that electron transfer between CIII and CIV is a completely random process not involving any compartmentalization of cytochrome c (Trouillard et al., 2011). The technique relied on reversible competitive inhibition of reduced CIV by carbon monoxide (CO), in which photo-dissociation of the CO-a3 heme complex allows fast O2 binding. However the approach, previously used in isolated systems, did not take into account the O2 -sensing ability of CIV, signaling systems in the cells sensitive to CO or anoxia (Kwast et al., 1999), and the ability of the electron transfer chain to dynamically reorganize with rapid dissociation of cytochrome c from respiratory SCs (Morin et al., 2003). Therefore, additional controls are required for final interpretation of results obtained by this method in the whole cells.
Several factors have been identified, which are necessary for individual respiratory complex assembly with some evidence of involvement in either formation or stability of SCs in yeast and higher eukaryotes. Respiratory SC factor (Rcf) 1 or 2 is required for late-stage assembly of the yeast Cox12p and Cox13p subunits and for cytochrome c oxidase activity in yeast; human orthologs of Rcf1p complement yeast mutants (Chen et al., 2012; Strogolova et al., 2012; Vukotic et al., 2012). However, purified yeast SC III2IV2 contained much less than stoichiometric amounts of Rcf1p (Bázan, S., Mileykovskaya, E. and Dowhan, W., unpublished results), suggesting a role in proper assembly of individual complexes for SC formation rather than stability of SCs. Mammalian mitochondria contain additional factors (Cox7a-related proteins in mice) that are required for regulation of CIV activity and SC formation (Ikeda et al., 2013; Lapuente-Brun et al., 2013).
Therefore, the preponderance of evidence indicates higher order organization of mitochondrial respiratory complexes into SCs, which may involve additional protein factors. However, even in the presence of these additional protein factors, the phospholipid CL is still required (Bázan et al., 2013; Chen et al., 2012). As will be outlined below, CL is directly involved in SC formation and stability.
4.0 Direct Involvement of CL in SC Formation and Pathological Consequences
The role of CL was first demonstrated in the formation of the yeast tetrameric SC (III2IV2) using BN- and CN-PAGE of digitonin extracts of mitochondria and kinetic analysis of substrate oxidation by the respiratory chain in intact mitochondria. In yeast Δcrd1 null mutants lacking CL, but containing elevated amounts of its precursor PG, there was no formation of the stable III2IV2 SC or substrate channeling of cytochrome c in contrast to studies with the wild type strain containing CL (Mileykovskaya et al., 2005; Pfeiffer et al., 2003; Zhang et al., 2002, 2005). Importantly, a strain in which CL content could be exogenously regulated in vivo showed a direct correlation between CL levels, the ratio of SC to individual complexes and growth-rate on non-fermentable carbon sources demonstrating dependence of energetic efficiency on CL levels.
In Barth syndrome (an X-linked mutation in the TAZ gene characterized by cardiomyopathy, skeletal myopathy, neutropenia, and growth retardation) patient mitochondria also display CL-dependent respirasome organization. Patient mitochondria also display lower CL content and a polydispersity in acyl chain composition of CL (Schlame and Ren, 2006). BN-PAGE revealed an increase in free CIV monomer and a decrease in the I1III2IV1 SC in patient lymphocytes (McKenzie et al., 2006). Dramatic changes in SC organization were observed in a pluripotent stem cell model system of this disorder (Dudek et al., 2013). An increase in mitochondrial content, which compensates for the decrease in the level of respiratory complexes and SCs, was found in immortalized patient lymphoblasts (Gonzalvez et al., 2013). Mutants in the yeast TAZ1 gene (Ma et al., 2004) also have reduced CIV and SC (Brandner et al., 2005; Li et al., 2007). Reduced formation of individual complexes and SCs was correlated with lowered CL levels due to oxidative stress and CL peroxidation in aging (Gomez and Hagen, 2012), neurodegenerative diseases (Paradies et al., 2011), cancer (Gasparre et al., 2013). For reviews see (Bogdanov et al., 2008; Joshi et al., 2009; Lenaz, 2012; Mileykovskaya and Dowhan, 2009; Schlame and Ren, 2006). Therefore, evidence from yeast to humans supports a role for CL in higher order organization of respiratory complexes.
5.0 Insights from Structural Analysis of SCs
Analysis of the structural organization of respiratory SCs can identify domains of individual complexes oriented towards each other, estimate distances between these domains, examine sites of protein contact, identify positions of tightly bound CL, and predict positions of loosely bound CLs. Three-dimensional (3D) density maps of the bovine heart respirasome (I1III2IV1) were obtained by cryo-electron tomography (Dudkina et al., 2011) of digitonin-solubilized SC and by cryo-electron microscopy (EM) and single particle analysis of the amphipol-solubilized SC (Althoff et al., 2011). Docking of available crystal structures of the individual complexes into both 3D density maps using the UCSF Chimera program (Pettersen et al., 2004) generated very similar pseudo-atomic models; see (Dudkina et al., 2010), (Althoff et al., 2011) and Fig. 2 with PDB 2YBB. Interestingly, mutual orientation of CIV and CIII in the bovine SC differs significantly from their orientation in the structure of the yeast SC (III2IV2) suggested from 2D projection averages obtained by negative stain single particle EM (Heinemeyer et al., 2007). A subsequent pseudo-atomic model derived by single particle cryo-EM and docking of the crystal structures of the yeast CIII and bovine CIV into the reconstituted yeast 3D density map demonstrated the same significant differences in the mutual orientation of CIII and CIVs in the yeast versus the bovine SC (Fig. 2 and Fig. 3; for detailed explanation see (Mileykovskaya et al., 2012)).
Figure 2.
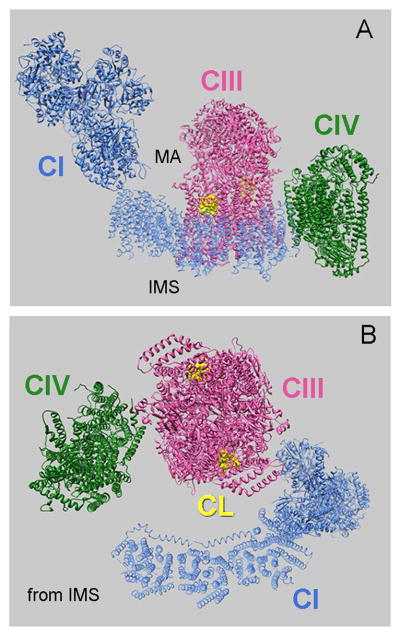
Pseudo-atomic model shows the arrangement of CI, CIII and CIV in the bovine respirasome (PDB 2YBB, (Althoff et al., 2011)). (A) Side view and (B) view from IMS. Two CL molecules (in yellow) in the external cavity formed by cytochrome c1, cytochrome b and closed by chain G in each monomer of CIII (PDB 1PP9) are shown. MA denotes the mitochondrial matrix.
Figure 3.
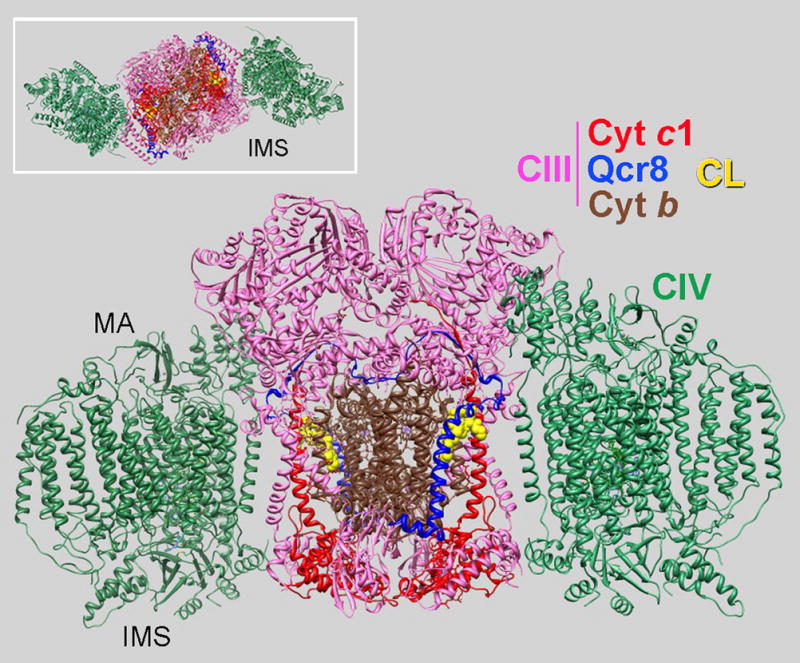
Pseudo-atomic model for the yeast tetrameric SC III2IV2. Side view (large structure) and view from the IMS (insert) showing the arrangement of CIII (PDB 3CX2) and two CIVs (PDB 1OCC) in the SC. One CL molecule (in the external cavity of CIII), cytochrome c1, cytochrome b and Qcr8 subunit (chain H, homolog of chain G in bovine) are colored in each CIII monomer as indicated. Derived from the structure reported in (Mileykovskaya et al., 2012).
The difference in arrangement of CIII and CIV in the bovine respirasome and yeast SC most likely stems from the necessity for bovine CIII to also interact with CI as shown in Fig. 2. In the yeast structure, dimeric CIII is flanked by two monomers of CIV. CIV faces the side of CIII with an external cavity formed by transmembrane domain helices of cytochrome c1 and cytochrome b (Fig. 3). This cavity contains tightly bound CL (denoted in crystal structures as L5 in (Lange et al., 2001) or CLD in PDB 1KB9; or CN3 in PDB 3CX5). The close proximity to the ubiquinone reduction site (Qi) suggests involvement of this CL in proton uptake pathway in CIII (Hunte et al., 2003).
In yeast the role of the above CL in SC stability was suggested (Pfeiffer et al., 2003) and investigated by constructing single, double and triple replacement mutants (K288L, K289L and K296L) at the site in cytochrome c1 important for CL binding (Wenz et al., 2009). The double (K288L, K289L) and triple mutants still formed CIII/CIV SCs even when expressed in the Δcrd1 background lacking CL suggesting the role of CL is in neutralization of the positive charges at this site in CIII important for interaction of CIII with CIV and stabilization of SC. However, as authors noted, the presence of high levels of PG in the Δcrd1 mutant complicated the interpretation of their results. Indeed, the results do not establish that this is the only site important for SC stabilization and direct interaction with CIV. For example, binding of PG in place of CL to other sites of CIII in the combined Δcrd1 K-mutants could be involved.
In the bovine respirasome the interface of CIII with CIV is not at the site homologous with the CL-binding site in the yeast CIII external cavity (Fig. 2 A and B). In bovine CIII this cavity contains two CLs, which appear to be stabilized by a helical belt formed by chain G (SU7 or ubiquinone-binding protein (PDB 1PP9) corresponding to Qcr8p or chain H in yeast, PDB 3CX2, Fig. 3). This belt contributes additional positive charges and closes the cavity thus more firmly stabilizing CL association. However, in yeast the belt contains fewer positive charges resulting in only one bound CL, which is probably less tightly associated (Dibrova et al., 2013).
The hydrophobic cleft close to the CIII homodimer interface also contains a conserved tightly bound CL (L7 (Palsdottir and Hunte, 2004), or CN5 in PDB 3CX5). Two CLs were resolved in the crystal structure of bovine CIV ((Shinzawa-Itoh et al., 2007); (PDB 2DYR). One of them stabilizes the CIV dimer in the crystal structure. A third CL was found by labeling with photo-reactive CL analogues near the entrance to the putative proton-pumping channel and might facilitate proton entry (Sedlak et al., 2006).
Structural studies revealed spaces between transmembrane domains at the interface between individual complexes within the bovine respirasome (Althoff et al., 2011; Dudkina et al., 2011) (Fig. 2 B) and yeast SC (Mileykovskaya et al., 2012) (Fig. 3), which may be filled with lipids. Protein contact points are outside the lipid bilayer. Consistent with these observations, about 50 CL molecules were determined in the III2IV2 SC from yeast (Mileykovskaya et al., 2012) and about 200 CL molecules were estimated to be present in the purified bovine respirasome (I1III2IV1) (Althoff et al., 2011), which is much larger due to CI presence and larger space between the three individual complexes (Fig. 2 B). In yeast the CL associated with the purified SC showed the same proportion between the five major CL species with acyl chains (16:1)4, (16:1)3(18:1)1, (16:1)2 (18:1)2 and (18:1)4 as CL found in the inner mitochondrial membrane as revealed by quantitative electrospray ionization mass spectrometry (ESI-MS) (Mileykovskaya et al., 2012).
Lipids provide a less stable and more flexible interface between the individual respiratory complexes than direct protein-protein interactions. The presence of loosely bound CLs within the spaces between individual complexes provides a means for dynamic formation and dissociation of SCs in response to CL levels either under normal or abnormal physiological states. Therefore, are the tightly bound integral CLs sufficient for SC formation or are additional CLs required?
6.0 In Vitro and In Silico CL-Dependent Reconstitution of SCs
To further study the role of CL, which may fill the spaces between transmembrane domains of CIII and CIV in the yeast IV1III2IV1 SC, a minimal system for in vitro reconstitution of the SC dependent on added lipids was developed (Bázan et al., 2013). CIII and CIV were purified using dodecyl maltoside, which dissociates the SC and removes all but tightly bound and structurally integrated lipids. The purified individual complexes contained mostly integrated CL, as was determined by quantitative ESI-MS: 8 CLs in the dimeric CIII (III2) and 2 CLs in the monomeric CIV. For the first time the reconstitution of the trimeric (III2IV1) and tetrameric (III2IV2) SCs from individual CIII and CIV in proteoliposomes was achieved (Bázan et al., 2013). Formation of the trimer was dependent on liposomes containing only phosphatidylethanolamine (PE) and phosphatidylycholine (PC) and not dependent on added CL. Phospholipase treatment of CIV abolished trimer formation, but addition of CL to PE/PC liposomes restored trimer formation. Thus CL tightly bound to CIV is important for association of CIV and CIII into a trimer. Formation of the tetramer was completely dependent on the presence of CL in liposomes, which could not be substituted by other anionic phospholipids. Negative stain EM and single particle analysis of the purified reconstituted tetramer confirmed the native structural arrangement of CIII and CIV in the SC. The individual complexes retained full function and addition of low levels of cytochrome c under conditions reported to involve its channeling within the SC (Schagger and Pfeiffer, 2000) supported electrons transfer from QH2 to O2 (our unpublished results). These experiments clearly demonstrated that the tightly bound CL found in crystal structures of the individual respiratory complexes are not sufficient for organization of CIII and CIV into the IV1III2IV1 SC. Consistent with in situ kinetic data (Zhang et al., 2005), CL is essential for the structural support of whole chain electron transport with substrate channeling within SCs.
Coarse-Grained Molecular Dynamic (CGMD) simulations of CL binding to CIII and CIV also confirmed additional CL derived from the bilayer for SC formation (Arnarez et al., 2013a; Arnarez et al., 2013b). Simulations began with crystal structures containing CL for bovine and yeast CIII as well as for bovine CIV embedded in a mixed PC/CL bilayer. Importantly, the simulations reproduced the known CL binding sites, and in addition, revealed the existence of well-defined CL binding sites enriched in positive amino acids on the membrane-exposed surfaces of CIII (Fig. 4) and CIV. Interestingly, in yeast in contrast to bovine only one CL initially filled the site close to Qi, but a second CL appeared during an extended simulation time scale. Additional simulations demonstrate how CL bound in the several specific sites on the membrane-exposed protein surfaces of bovine CIII and CIV might stabilize interactions between CIII and CIV in the bovine respirasome.
Figure 4.
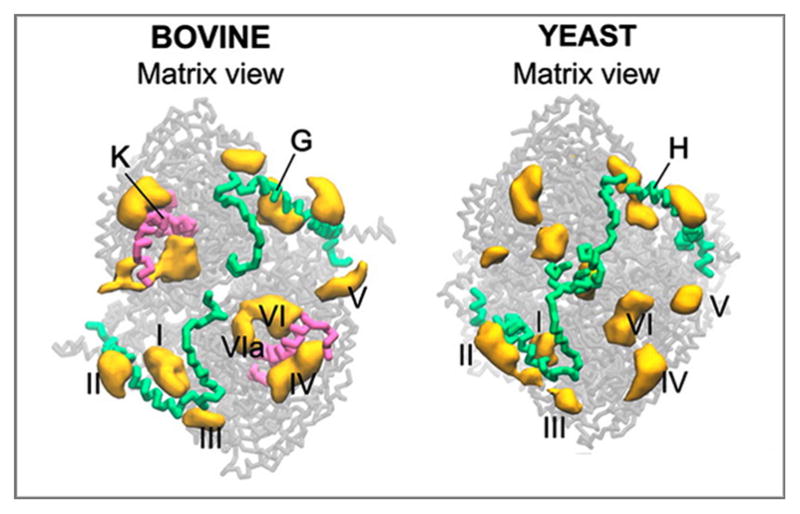
CL binding sites of bovine and yeast CIII extracted from CGMD simulation of the complexes embedded in a CL/PC bilayer. CLs bound in the sites I and VI/VIa correspond to tightly bound CLs found in the crystal structures of CIII from yeast and bovine. Site I corresponds to the CL binding site in the external cavity closed by chain H in yeast and its homolog chain G in bovine. Sites VI/VIa corresponds to the conserved site for tightly bound CL in the inner cavity located close to the CIII homodimer interface (CN5, PDB 3CX5 for yeast and CL3 in PDB 1SQP for bovine). Chain K is present in the bovine CIII close to sites VI and VIa. However, there is no homolog for this chain in yeast CIII. Sites II, III, IV and V are the sites on the membrane-exposed surfaces of CIII. The figure was adapted with permission from (Arnarez et al., 2013b). Copyright (2013) American Chemical Society.
7.0 Summary and Future Directions
The organization of individual respiratory complexes into higher order structures or SCs to form a functional respirasome has become generally accepted. These SCs contain additional CLs over those previously observed to be integrated into the structure of the individual complexes. 3D density maps of bovine and yeast SCs obtained by cryo-EM reveal spaces between the transmembrane domains of neighboring individual complexes that most likely contain many additional lipid molecules. Kinetic and biochemical evidence from mutants lacking CL coupled with successful reconstitution of SCs in yeast dependent on addition of CL over and above that integral to individual complexes confirms a direct role for CL in the higher order organization of the respiratory chain. Interestingly, yeast mutants in which PE has been depleted display higher order organization of respiratory SCs into “megacomplexes” (Bottinger et al., 2012), but the molecular basis for such organization is not known. The correlation between reduced CL levels in many disease states and disruption of respiratory SC formation strongly indicates a similar role for CL in mammalian mitochondria. Therefore, changes in CL levels might be a metabolic regulatory signal acting through the low affinity CL binding sites that support SC formation. More precise information on the location and function of CL, as well as other phospholipids within respiratory SCs, will come from a combination of structural studies, biochemical determination of lipid binding sites, predictions of lipid binding sites through molecular simulations, genetic perturbation of lipid binding sites and use of liposome reconstitution systems.
Highlights.
Organization of supercomplexes (SCs) with integral cardiolipin (CL) is analyzed.
CL content in different purified SCs correlated with spaces inside SCs is discussed.
New in vitro SC reconstitution system using CL-containing liposomes is described.
Importance of exchangeable CL bound on the membrane-exposed SC surfaces is discussed.
Change in CL level as a potential metabolic signal for SC dynamics is postulaed.
Acknowledgments
This work was supported in part by National Institutes of Health Grant GM R01 GM56389 and the John Dunn Research Foundation to W. D.
Abbreviations
- CL
cardiolipin
- IMS
intermembrane space
- CI
Complex I
- CIII
Complex III
- CIV
Complex IV
- BN-PAGE
blue-native polyacrylamide gel electrophoresis
- SC
supercomplex
- CN-PAGE
colorless-native polyacrylamide gel electrophoresis
- PE
phosphatidylethanolamine
- PC
phosphatidylcholine
- IMS
intermembrane space
- MA
matrix
- CGMD
Coarse-Grained Molecular Dynamic
Footnotes
Complex III (CIII) is a structural and functional homodimer of two cytochrome bc1 monomeric complexes and when in a supercomplex (SC) is referred to as III2.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Acin-Perez R, Fernandez-Silva P, Peleato ML, Perez-Martos A, Enriquez JA. Respiratory active mitochondrial supercomplexes. Mol Cell. 2008;32:529–539. doi: 10.1016/j.molcel.2008.10.021. [DOI] [PubMed] [Google Scholar]
- Althoff T, Mills DJ, Popot JL, Kuhlbrandt W. Arrangement of electron transport chain components in bovine mitochondrial supercomplex I1III2IV1. EMBO J. 2011;30:4652–4664. doi: 10.1038/emboj.2011.324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnarez C, Marrink SJ, Periole X. Identification of cardiolipin binding sites on cytochrome c oxidase at the entrance of proton channels. Sci Rep. 2013a;3:1263. doi: 10.1038/srep01263. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnarez C, Mazat JP, Elezgaray J, Marrink SJ, Periole X. Evidence for cardiolipin binding sites on the membrane-exposed surface of the cytochrome bc1. J Am Chem Soc. 2013b;135:3112–3120. doi: 10.1021/ja310577u. [DOI] [PubMed] [Google Scholar]
- Baile MG, Whited K, Claypool SM. Deacylation on the matrix side of the mitochondrial inner membrane regulates cardiolipin remodeling. Mol Biol Cell. 2013;24:2008–2020. doi: 10.1091/mbc.E13-03-0121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bázan S, Mileykovskaya E, Mallampalli VK, Heacock P, Sparagna GC, Dowhan W. Cardiolipin-dependent reconstitution of respiratory supercomplexes from purified Saccharomyces cerevisiae complexes III and IV. J Biol Chem. 2013;288:401–411. doi: 10.1074/jbc.M112.425876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Beranek A, Rechberger G, Knauer H, Wolinski H, Kohlwein SD, Leber R. Identification of a cardiolipin-specific phospholipase encoded by the gene CLD1 (YGR110W) in yeast. J Biol Chem. 2009;284:11572–11578. doi: 10.1074/jbc.M805511200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bogdanov M, Mileykovskaya E, Dowhan W. Lipids in the assembly of membrane proteins and organization of protein supercomplexes: implications for lipid-linked disorders. Subcell Biochem. 2008;49:197–239. doi: 10.1007/978-1-4020-8831-5_8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bottinger L, Horvath SE, Kleinschroth T, Hunte C, Daum G, Pfanner N, Becker T. Phosphatidylethanolamine and cardiolipin differentially affect the stability of mitochondrial respiratory chain supercomplexes. J Mol Biol. 2012;423:677–686. doi: 10.1016/j.jmb.2012.09.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boumans H, Grivell LA, Berden JA. The respiratory chain in yeast behaves as a single functional unit. J Biol Chem. 1998;273:4872–4877. doi: 10.1074/jbc.273.9.4872. [DOI] [PubMed] [Google Scholar]
- Brandner K, Mick DU, Frazier AE, Taylor RD, Meisinger C, Rehling P. Taz1, an outer mitochondrial membrane protein, affects stability and assembly of inner membrane protein complexes: implications for Barth Syndrome. Mol Biol Cell. 2005;16:5202–5214. doi: 10.1091/mbc.E05-03-0256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chance B, Williams GR. Respiratory enzymes in oxidative phosphorylation. IV. The respiratory chain. J Biol Chem. 1955;217:429–438. [PubMed] [Google Scholar]
- Chen YC, Taylor EB, Dephoure N, Heo JM, Tonhato A, Papandreou I, Nath N, Denko NC, Gygi SP, Rutter J. Identification of a protein mediating respiratory supercomplex stability. Cell Metab. 2012;15:348–360. doi: 10.1016/j.cmet.2012.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dibrova DV, Cherepanov DA, Galperin MY, Skulachev VP, Mulkidjanian AY. Evolution of cytochrome bc complexes: From membrane-anchored dehydrogenases of ancient bacteria to triggers of apoptosis in vertebrates. Biochim Biophys Acta. 2013 doi: 10.1016/j.bbabio.2013.07.006. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dudek J, Cheng IF, Balleininger M, Vaz FM, Streckfuss-Bomeke K, Hubscher D, Vukotic M, Wanders RJ, Rehling P, Guan K. Cardiolipin deficiency affects respiratory chain function and organization in an induced pluripotent stem cell model of Barth syndrome. Stem Cell Res. 2013;11:806–819. doi: 10.1016/j.scr.2013.05.005. [DOI] [PubMed] [Google Scholar]
- Dudkina NV, Kouril R, Peters K, Braun HP, Boekema EJ. Structure and function of mitochondrial supercomplexes. Biochim Biophys Acta. 2010;1797:664–670. doi: 10.1016/j.bbabio.2009.12.013. [DOI] [PubMed] [Google Scholar]
- Dudkina NV, Kudryashev M, Stahlberg H, Boekema EJ. Interaction of complexes I, III, and IV within the bovine respirasome by single particle cryoelectron tomography. Proc Natl Acad Sci U S A. 2011;108:15196–15200. doi: 10.1073/pnas.1107819108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gasparre G, Porcelli AM, Lenaz G, Romeo G. Relevance of mitochondrial genetics and metabolism in cancer development. Cold Spring Harb Perspect Biol. 2013;5 doi: 10.1101/cshperspect.a011411. in press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genova ML, Lenaz G. A critical appraisal of the role of respiratory supercomplexes in mitochondria. Biol Chem. 2013;394:631–639. doi: 10.1515/hsz-2012-0317. [DOI] [PubMed] [Google Scholar]
- Gomez LA, Hagen TM. Age-related decline in mitochondrial bioenergetics: does supercomplex destabilization determine lower oxidative capacity and higher superoxide production? Semin Cell Dev Biol. 2012;23:758–767. doi: 10.1016/j.semcdb.2012.04.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalvez F, D’Aurelio M, Boutant M, Moustapha A, Puech JP, Landes T, Arnaune-Pelloquin L, Vial G, Taleux N, Slomianny C, Wanders RJ, Houtkooper RH, Bellenguer P, Moller IM, Gottlieb E, Vaz FM, Manfredi G, Petit PX. Barth syndrome: cellular compensation of mitochondrial dysfunction and apoptosis inhibition due to changes in cardiolipin remodeling linked to tafazzin (TAZ) gene mutation. Biochim Biophys Acta. 2013;1832:1194–1206. doi: 10.1016/j.bbadis.2013.03.005. [DOI] [PubMed] [Google Scholar]
- Hackenbrock CR, Chazotte B, Gupte SS. The random collision model and a critical assessment of diffusion and collision in mitochondrial electron transport. J Bioenerg Biomembr. 1986;18:331–368. doi: 10.1007/BF00743010. [DOI] [PubMed] [Google Scholar]
- Haines TH. A new look at Cardiolipin. Biochim Biophys Acta. 2009;1788:1997–2002. doi: 10.1016/j.bbamem.2009.09.008. [DOI] [PubMed] [Google Scholar]
- Heinemeyer J, Braun HP, Boekema EJ, Kouril R. A structural model of the cytochrome C reductase/oxidase supercomplex from yeast mitochondria. J Biol Chem. 2007;282:12240–12248. doi: 10.1074/jbc.M610545200. [DOI] [PubMed] [Google Scholar]
- Henry SA, Kohlwein SD, Carman GM. Metabolism and regulation of glycerolipids in the yeast Saccharomyces cerevisiae. Genetics. 2012;190:317–349. doi: 10.1534/genetics.111.130286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hunte C, Palsdottir H, Trumpower BL. Protonmotive pathways and mechanisms in the cytochrome bc1 complex. FEBS Lett. 2003;545:39–46. doi: 10.1016/s0014-5793(03)00391-0. [DOI] [PubMed] [Google Scholar]
- Ikeda K, Shiba S, Horie-Inoue K, Shimokata K, Inoue S. A stabilizing factor for mitochondrial respiratory supercomplex assembly regulates energy metabolism in muscle. Nat Commun. 2013;4:2147. doi: 10.1038/ncomms3147. [DOI] [PubMed] [Google Scholar]
- Joshi AS, Zhou J, Gohil VM, Chen S, Greenberg ML. Cellular functions of cardiolipin in yeast. Biochim Biophys Acta. 2009;1793:212–218. doi: 10.1016/j.bbamcr.2008.07.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwast KE, Burke PV, Staahl BT, Poyton RO. Oxygen sensing in yeast: evidence for the involvement of the respiratory chain in regulating the transcription of a subset of hypoxic genes. Proc Natl Acad Sci U S A. 1999;96:5446–5451. doi: 10.1073/pnas.96.10.5446. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lange C, Nett JH, Trumpower BL, Hunte C. Specific roles of protein-phospholipid interactions in the yeast cytochrome bc1 complex structure. Embo J. 2001;20:6591–6600. doi: 10.1093/emboj/20.23.6591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lapuente-Brun E, Moreno-Loshuertos R, Acin-Perez R, Latorre-Pellicer A, Colas C, Balsa E, Perales-Clemente E, Quiros PM, Calvo E, Rodriguez-Hernandez MA, Navas P, Cruz R, Carracedo A, Lopez-Otin C, Perez-Martos A, Fernandez-Silva P, Fernandez-Vizarra E, Enriquez JA. Supercomplex assembly determines electron flux in the mitochondrial electron transport chain. Science. 2013;340:1567–1570. doi: 10.1126/science.1230381. [DOI] [PubMed] [Google Scholar]
- Lenaz G. Mitochondria and reactive oxygen species. Which role in physiology and pathology? Adv Exp Med Biol. 2012;942:93–136. doi: 10.1007/978-94-007-2869-1_5. [DOI] [PubMed] [Google Scholar]
- Lenaz G, Genova ML. Kinetics of integrated electron transfer in the mitochondrial respiratory chain: random collisions vs. solid state electron channeling. Am J Physiol Cell Physiol. 2007;292:C1221–1239. doi: 10.1152/ajpcell.00263.2006. [DOI] [PubMed] [Google Scholar]
- Li G, Chen S, Thompson MN, Greenberg ML. New insights into the regulation of cardiolipin biosynthesis in yeast: implications for Barth syndrome. Biochim Biophys Acta. 2007;1771:432–441. doi: 10.1016/j.bbalip.2006.06.007. [DOI] [PubMed] [Google Scholar]
- Ma L, Vaz FM, Gu Z, Wanders RJ, Greenberg ML. The human TAZ gene complements mitochondrial dysfunction in the yeast taz1 Δ mutant. Implications for Barth syndrome. J Biol Chem. 2004;279:44394–44399. doi: 10.1074/jbc.M405479200. [DOI] [PubMed] [Google Scholar]
- McKenzie M, Lazarou M, Thorburn DR, Ryan MT. Mitochondrial respiratory chain supercomplexes are destabilized in Barth Syndrome patients. J Mol Biol. 2006;361:462–469. doi: 10.1016/j.jmb.2006.06.057. [DOI] [PubMed] [Google Scholar]
- Mileykovskaya E, Dowhan W. Cardiolipin membrane domains in prokaryotes and eukaryotes. Biochim Biophys Acta. 2009;1788:2084–2091. doi: 10.1016/j.bbamem.2009.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mileykovskaya E, Penczek PA, Fang J, Mallampalli VK, Sparagna GC, Dowhan W. Arrangement of the respiratory chain complexes in Saccharomyces cerevisiae supercomplex III2IV2 revealed by single particle cryo-electron microscopy. J Biol Chem. 2012;287:23095–23103. doi: 10.1074/jbc.M112.367888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mileykovskaya E, Zhang M, Dowhan W. Cardiolipin in energy transducing membranes. Biochemistry (Mosc) 2005;70:154–158. doi: 10.1007/s10541-005-0095-2. [DOI] [PubMed] [Google Scholar]
- Morin C, Zini R, Tillement JP. Anoxia-reoxygenation-induced cytochrome c and cardiolipin release from rat brain mitochondria. Biochem Biophys Res Commun. 2003;307:477–482. doi: 10.1016/s0006-291x(03)01203-8. [DOI] [PubMed] [Google Scholar]
- Palsdottir H, Hunte C. Lipids in membrane protein structures. Biochim Biophys Acta. 2004;1666:2–18. doi: 10.1016/j.bbamem.2004.06.012. [DOI] [PubMed] [Google Scholar]
- Paradies G, Petrosillo G, Paradies V, Ruggiero FM. Mitochondrial dysfunction in brain aging: role of oxidative stress and cardiolipin. Neurochem Int. 2011;58:447–457. doi: 10.1016/j.neuint.2010.12.016. [DOI] [PubMed] [Google Scholar]
- Pettersen EF, Goddard TD, Huang CC, Couch GS, Greenblatt DM, Meng EC, Ferrin TE. UCSF Chimera--a visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- Pfeiffer K, Gohil V, Stuart RA, Hunte C, Brandt U, Greenberg ML, Schagger H. Cardiolipin stabilizes respiratory chain supercomplexes. J Biol Chem. 2003;278:52873–52880. doi: 10.1074/jbc.M308366200. [DOI] [PubMed] [Google Scholar]
- Schagger H. Respiratory chain supercomplexes. IUBMB Life. 2001;52:119–128. doi: 10.1080/15216540152845911. [DOI] [PubMed] [Google Scholar]
- Schagger H, Pfeiffer K. Supercomplexes in the respiratory chains of yeast and mammalian mitochondria. Embo J. 2000;19:1777–1783. doi: 10.1093/emboj/19.8.1777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlame M, Acehan D, Berno B, Xu Y, Valvo S, Ren M, Stokes DL, Epand RM. The physical state of lipid substrates provides transacylation specificity for tafazzin. Nat Chem Biol. 2012;8:862–869. doi: 10.1038/nchembio.1064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schlame M, Ren M. Barth syndrome, a human disorder of cardiolipin metabolism. FEBS Lett. 2006;580:5450–5455. doi: 10.1016/j.febslet.2006.07.022. [DOI] [PubMed] [Google Scholar]
- Schlame M, Ren M. The role of cardiolipin in the structural organization of mitochondrial membranes. Biochim Biophys Acta. 2009;1788:2080–2083. doi: 10.1016/j.bbamem.2009.04.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedlak E, Panda M, Dale MP, Weintraub ST, Robinson NC. Photolabeling of cardiolipin binding subunits within bovine heart cytochrome c oxidase. Biochemistry. 2006;45:746–754. doi: 10.1021/bi050870z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinzawa-Itoh K, Aoyama H, Muramoto K, Terada H, Kurauchi T, Tadehara Y, Yamasaki A, Sugimura T, Kurono S, Tsujimoto K, Mizushima T, Yamashita E, Tsukihara T, Yoshikawa S. Structures and physiological roles of 13 integral lipids of bovine heart cytochrome c oxidase. Embo J. 2007;26:1713–1725. doi: 10.1038/sj.emboj.7601618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strogolova V, Furness A, Robb-McGrath M, Garlich J, Stuart RA. Rcf1 and Rcf2, members of the hypoxia-induced gene 1 protein family, are critical components of the mitochondrial cytochrome bc1-cytochrome c oxidase supercomplex. Mol Cell Biol. 2012;32:1363–1373. doi: 10.1128/MCB.06369-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stuart RA. Supercomplex organization of the oxidative phosphorylation enzymes in yeast mitochondria. J Bioenerg Biomembr. 2008;40:411–417. doi: 10.1007/s10863-008-9168-4. [DOI] [PubMed] [Google Scholar]
- Sun F, Zhou Q, Pang X, Xu Y, Rao Z. Revealing various coupling of electron transfer and proton pumping in mitochondrial respiratory chain. Curr Opin Struct Biol. 2013;23:526–538. doi: 10.1016/j.sbi.2013.06.013. [DOI] [PubMed] [Google Scholar]
- Tamura Y, Harada Y, Nishikawa S, Yamano K, Kamiya M, Shiota T, Kuroda T, Kuge O, Sesaki H, Imai K, Tomii K, Endo T. Tam41 is a CDP-diacylglycerol synthase required for cardiolipin biosynthesis in mitochondria. Cell Metab. 2013;17:709–718. doi: 10.1016/j.cmet.2013.03.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trouillard M, Meunier B, Rappaport F. Questioning the functional relevance of mitochondrial supercomplexes by time-resolved analysis of the respiratory chain. Proc Natl Acad Sci U S A. 2011;108:E1027–1034. doi: 10.1073/pnas.1109510108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vukotic M, Oeljeklaus S, Wiese S, Vogtle FN, Meisinger C, Meyer HE, Zieseniss A, Katschinski DM, Jans DC, Jakobs S, Warscheid B, Rehling P, Deckers M. Rcf1 mediates cytochrome oxidase assembly and respirasome formation, revealing heterogeneity of the enzyme complex. Cell Metab. 2012;15:336–347. doi: 10.1016/j.cmet.2012.01.016. [DOI] [PubMed] [Google Scholar]
- Wenz T, Hielscher R, Hellwig P, Schagger H, Richers S, Hunte C. Role of phospholipids in respiratory cytochrome bc1 complex catalysis and supercomplex formation. Biochim Biophys Acta. 2009;1787:609–616. doi: 10.1016/j.bbabio.2009.02.012. [DOI] [PubMed] [Google Scholar]
- Zhang M, Mileykovskaya E, Dowhan W. Gluing the respiratory chain together. Cardiolipin is required for supercomplex formation in the inner mitochondrial membrane. J Biol Chem. 2002;277:43553–43556. doi: 10.1074/jbc.C200551200. [DOI] [PubMed] [Google Scholar]
- Zhang M, Mileykovskaya E, Dowhan W. Cardiolipin is essential for organization of complexes III and IV into a supercomplex in intact yeast mitochondria. J Biol Chem. 2005;280:29403–29408. doi: 10.1074/jbc.M504955200. [DOI] [PMC free article] [PubMed] [Google Scholar]