Abstract
Nek2 is a serine/threonine protein kinase that localizes to the centrosome and is implicated in mitotic regulation. Overexpression of Nek2 induces premature centrosome separation and nuclear defects indicative of mitotic errors, while depletion of Nek2 interferes with cell growth. As Nek2 expression is upregulated in a range of cancer cell lines and primary human tumors, inhibitors of Nek2 may have therapeutic value in cancer treatment. We used a radiometric proximity assay in a high-throughput screen to identify small molecule inhibitors of Nek2 kinase activity. The assay was based on the measurement of the radiolabelled phosphorylated product of the kinase reaction brought into contact with the surface of wells of solid scintillant-coated microtitre plates. Seventy non-aggregating hits were identified from approximately 73,000 compounds screened and included a number of toxoflavins and a series of viridin/wortmannin-like compounds. The viridin-like compounds were >70-fold selective for Nek2 over Nek6 and Nek7 and inhibited the growth of human tumor cell lines at concentrations consistent with their biochemical potencies. An automated mechanism-based microscopy assay in which centrosomes were visualised using pericentrin antibodies confirmed that two of the viridin inhibitors reduced centrosome separation in a human tumor cell line. The data presented show pharmacological inhibition of Nek2 kinase results in the expected phenotype of disruption to centrosome function associated with growth inhibition and further supports Nek2 as a target for cancer drug discovery.
Keywords: Cell cycle, mitosis, centrosome separation, Nek2 kinase inhibitors, automated immunofluorescence
INTRODUCTION
Mitotic protein kinases are of considerable interest as chemotherapeutic targets for hyperproliferative diseases, such as cancer.1 Cyclin-dependent kinase 1 (Cdk1), Aurora A, Aurora B and Polo-like kinase 1 (Plk1) are all required for normal progression through mitosis, exhibit deregulated activity in tumor cells and lead to cell cycle arrest and/or apoptosis when depleted. A less well characterized, but similarly conserved, mitotic kinase is the NIMA-related kinase, Nek2. Based on recent validation studies, Nek2 is now receiving attention as another putative anti-cancer target.
Nek2 is a serine/threonine protein kinase that is regulated in a cell cycle-dependent manner.2 It is the closest relative in the human genome of the NIMA kinase of Aspergillus nidulans, which is an essential regulator of mitotic progression. Nek2 is activated by dimerization and autophosphorylation, and inhibited through interaction with and dephosphorylation by protein phosphatase 1.3 Nek2 is localized to the centrosome where it regulates spindle pole separation at the onset of mitosis through phosphorylation and displacement of proteins, including C-Nap1 and rootletin.3 There is also evidence that it contributes to chromatin condensation and spindle checkpoint function.3
Nek2 is abnormally expressed in cancer cells.4 Initially, microarray studies revealed increased expression of Nek2 mRNA in Ewings tumor cell lines and diffuse large B-cell lymphomas. Subsequently, elevated levels of Nek2 protein have been identified in a wide variety of cancer cell lines as well as in a significant proportion of primary human cancers, including breast tumors, cholangiocarcinomas and testicular seminomas.5-8 The mechanism for upregulation of Nek2 expression remains to be determined. However, the locus that carries the Nek2 gene, 1q32, is amplified in both breast and gastric tumors.9; 10
Experimental studies suggest that abnormal Nek2 expression may contribute to the classic tumor hallmarks of aneuploidy and chromosome instability. Overexpression of active Nek2 leads to premature centrosome separation and the accumulation of cells with multiple nuclei and supernumerary centrosomes, while overexpression of kinase-inactive Nek2 or depletion by RNAi of the wild type enzyme interferes with centrosome separation and bipolar spindle formation.6; 11; 12 These data support the hypothesis that Nek2 activity is carefully regulated in normal cells to promote accurate cell division. Importantly, total Nek2 depletion in HeLa cells results in the arrest of cell proliferation, raising the possibility that Nek2 inhibitors might block cancer progression.13 Also, RNAi-based depletion of Nek2 selectively interfered with the proliferation of cholangiocarcinoma cell lines but not normal fibroblast cell lines, and led to a reduction in tumor size and peritoneal dissemination of cholangiocarcinoma tumor xenografts.7 Meanwhile RNAi knockdown of Nek2 in ER positive and negative human breast cancer cell lines reduced cell growth and migration and the size of human breast tumor xenografts.8
Although an inhibitor of the interaction of the spindle checkpoint protein, Hec1, with Nek2 has been described,14 no selective inhibitors of Nek2 kinase activity have been reported. The purpose of this work was to identify in a non-biased high-throughput screen small molecule inhibitors of Nek2 kinase activity. Here we describe the outcome of the screen and the chemical properties and cellular effects of some of the compounds identified.
MATERIALS AND METHODS
Materials and Km determination are described in Supplementary Material.
FlashPlate assay
384-well FlashPlates were coated overnight at 4°C with myelin basic protein (MBP; 25 μg/ml in phosphate-buffered saline, PBS), washed twice with PBS on a 16 pin plate washer (ELX50 Biotek Instruments Ltd., Northstar, UK) and compound (3 μl) in 2% dimethyl sulfoxide (DMSO) added to each well (final nominal concentration 32 μM in 0.3% DMSO,) followed by master mix (8 μl) consisting of buffer (50 mM Hepes pH 7.4±0.2, 5 mM MnCl2, 5 mM ß-glycerophosphate, 5 mM NaF, 10 mM MgCl2, 1 mM DTT) with or without active Nek2 enzyme (20 pg/well). Finally, a mix of 10 μM ATP and 0.2 μCi γ-33P-ATP (8 μl) was added (total volume 19 μl). These additions were made using a MiniTrak™ 5 (Perkin Elmer Life Sciences). Plates were shaken (~1 minute), incubated at room temperature (4 hours), washed twice (10 mM sodium pyrophosphate) and then counted on a Topcount-NXT™ (Perkin Elmer Life Sciences). Plates were formatted to contain total activity controls (n=32), positive controls (staurosporine at 25 μM, n=8), and no enzyme blanks (n=24) on the outside two columns either side of the plate. Signal to noise (S/N) and signal to background (S/B) ratios were calculated using the equations S/N=mean S − mean B / √ (sdS)2 + (sdB)2 and S/B = mean S / mean B, respectively where sd=standard deviation. The Z′ factor was used to assess assay performance.15 Screening data were analyzed using ActivityBase version 5.2 (IDBS, Guildford, UK). The activity of hits was confirmed in triplicate. Potency (IC50) was determined over a concentration range of 0.07 to 159 μM.
Filter assay
The kinase reaction buffer was as described above. Compounds (10 μM) were added to a 96-well polypropylene microtitre plate, followed by substrate (10 μl of PLM peptide at 20 μM) and ATP (10 μl of 20 μM) containing 0.25 μCi γ-33P-ATP and the reaction initiated by adding Nek2 (20 pg/well; 50 μl volume). The plate was incubated at room temperature (90 minutes) and the reaction stopped by the addition of 2% orthophosphoric acid (30 μl). The solution was transferred to the wells of a pre-wetted phosphocellulose filter plate, the contents of each well filtered and washed twice with orthophosphoric acid (0.5%). Microscint 20 scintillant was added to each well (25 μl) and the plate read for 30 seconds per well using the Topcount NXT.
Kinase specificity assays
Specificity assays were performed using modification of an established method.16 Recombinant kinases (1 μg/ml) were mixed on ice with compounds dissolved in DMSO, or DMSO alone (2.5% DMSO final concentration), in a microtitre plate. Kinase buffer (KB; 50 mM Hepes. KOH pH 7.4, 5 mM MgCl2 (for Cdk1, Plk1 and Aurora A) or MnCl2 (for Nek2, Nek6 and Nek7), 5 mM β-glycerophosphate, 5 mM NaF, 4 μM ATP, 1 mM DTT, 20 nCi γ32P-[ATP]/ml) containing 0.5 mg/ml β-casein (for Nek2, Nek6, Nek7 and Plk1) or histone H1 (for Cdk1 and Aurora A) as substrate was added and the components mixed on ice by gentle pipetting. The plate was sealed and transferred to a waterbath (30°C) for 30 minutes, and the reactions stopped at 4°C by the addition of 0.5 M EDTA containing 0.2% bromophenol blue. Reaction products were slot-blotted as arrays onto nitrocellulose transfer membrane (Whatman, UK), washed three times in blotting buffer (25 mM Tris, 192 mM glycine, 10% MeOH (v/v)) and air-dried, exposed to a storage phosphoscreen (60 minutes), and analyzed using a Cyclone Phosphorimager and Optiquant analysis software (Perkin Elmer, USA). IC50 values calculated based on two independent experiments performed in triplicate using Prism 4.00 software with the bottom constraint set to zero (GraphPad Software Inc, San Diego, CA).
Cell culture and growth inhibition assays
HeLa, U2OS and tetracycline Nek2-inducible U2OS:GFP-Nek2A cells were cultured and induced as previously described.11 For growth inhibition assays, cells were seeded (2.5-6 × 106 cells per well) into 96-well plates. Compounds (dissolved in DMSO) at a range of concentrations or DMSO (1-2% final concentration) alone were added after 24 hours and the cells incubated for 72 hours. Cell numbers were measured using WST-1 reagents or sulphorhodamine B (SRB). The concentration of inhibitor that reduced cell growth by 50% (GI50) of DMSO control was calculated.
Immunofluorescence microscopy
Cells were fixed and processed for indirect immunofluorescence microscopy,11 using rabbit anti-pericentrin (1:1000; Abcam), mouse anti-γ-tubulin (1:500; Sigma), Alexa Fluor 594 goat anti-mouse or Alexa Fluor 488 goat anti-rabbit (1 μg/ml; Molecular Probes) antibodies. DNA was stained with Hoechst 33258 (0.2 μg/ml; Calbiochem) or DAPI (0.1 μg/ml; Invitrogen).
Automated cell-based assay for Nek2 activity
Tetracycline-inducible U2OS:GFP-Nek2A cells were seeded (4000 cells/well) in a black clear bottomed-plate and cultured overnight. Nek2A expression was induced with 1 μg/ml doxycycline (100 μl per well). PBS was added to the control (uninduced) wells. Simultaneously, test compound in 1% DMSO (v/v) final concentration or 1 % DMSO alone was added and incubated overnight. Cells were washed once with PBS, fixed for 10 minutes on ice with cold 100% methanol (−20°C) and the methanol removed by washing (3× with PBS). An antibody to the centrosomal marker, pericentrin, was added (1.3 μg/ml in 5% bovine serum albumin (BSA)/PBS), the plate incubated with gentle agitation for 1 hour at room temperature and washed as before. Goat anti-rabbit Alexa-488 (2 μg/ml) antibody and the nuclear stain DAPI (0.1 μg/ml) were added in 5% BSA/PBS, the plate incubated for 1 hour at room temperature then washed (3 × PBS) and refrigerated until ready to image. Plates were read on an IN Cell Analyser 1000 (see Supplemental Material for details of image acquisition). An algorithm was designed to identify the DAPI-stained nuclei, and to segment the cells setting a radius of 10 μm as the cell outline or ‘collar’. The pericentrin-identified centrosomes present only within the 10 μm cell radius were counted by adding another segmentation parameter (organelles). The size of the objects was optimized to be between 0.5-10 μm, with a pixel intensity (or sensitivity) set to 70. A threshold filter was added to the algorithm in order to classify the number/percentage of cells which had 1 or >1 centrosome.
RESULTS
Determination of Km for ATP
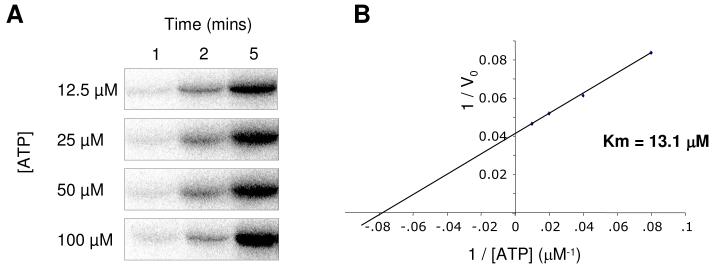
The Km of ATP for Nek2A kinase was determined. Kinase reactions were performed using baculovirus-expressed Nek2A, β-casein as substrate and buffer conditions as previously optimized.17 By varying the ATP concentration and determining the initial rates of reaction, the Km for ATP was calculated to be 13.1 μM (Figure 1A & B).
Figure 1. Determination of the Km for ATP for the Nek2 kinase.
A. Kinase assays were performed with baculovirus-expressed His6-Nek2A, β-casein as substrate and different concentrations of ATP as indicated. From these data, initial reaction rates (V0) were calculated. B. A Lineweaver-Burk plot showing I/V0 against 1/[ATP] where the x-axis intercept indicates −1/Km.
Optimization of high-throughput screen for Nek2 inhibitors
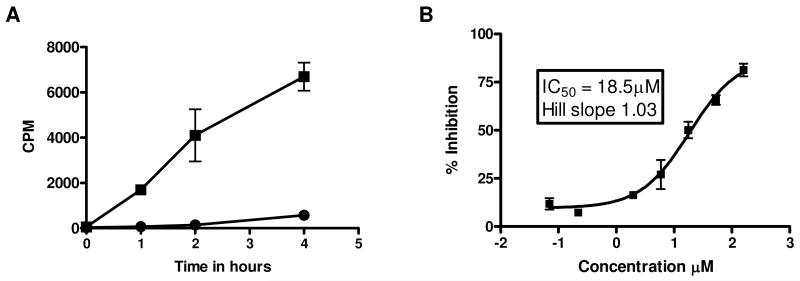
A FlashPlate assay was designed as described in the Materials and Methods. MBP (25 g/ml) provided a higher signal compared to either β-casein or phospholemman (PLM) peptide (data not shown). Also as expected, increasing the amount of unlabelled ATP (1-8 μM) competed with radioactively-labeled ATP thereby reducing the extent to which MBP was radioactively labeled and thus the signal obtained. A concentration of 2 μM ATP (plus 0.2 μCi γ-33P-ATP) was used subsequently. The assay was DMSO-tolerant up to 1% (0.3% final concentration). Enzymatic linearity was observed for up to 4 hours (S/B=40) using 20 ng/well Nek2 (Figure 2A). The IC50 for the general kinase inhibitor staurosporine (Figure 2B) was (13.6 ± 5.7 μM; mean ± sd) and was comparable to previously reported data18 and to that determined in the filter assay (13.0 ± 12.4 μM).
Figure 2. Optimization of conditions for Nek2 kinase biochemical assay.
The conditions for the FlashPlate assay were established according to the Materials and Methods. A. Enzyme reaction time-course; mean ± sd of n=3 wells (■ enzyme reaction, ● no enzyme blanks) B. Inhibition of Nek2 kinase activity by staurosporine. IC50 was calculated using GraphPad Prism.
High-throughput screen for inhibitors of Nek2 kinase activity
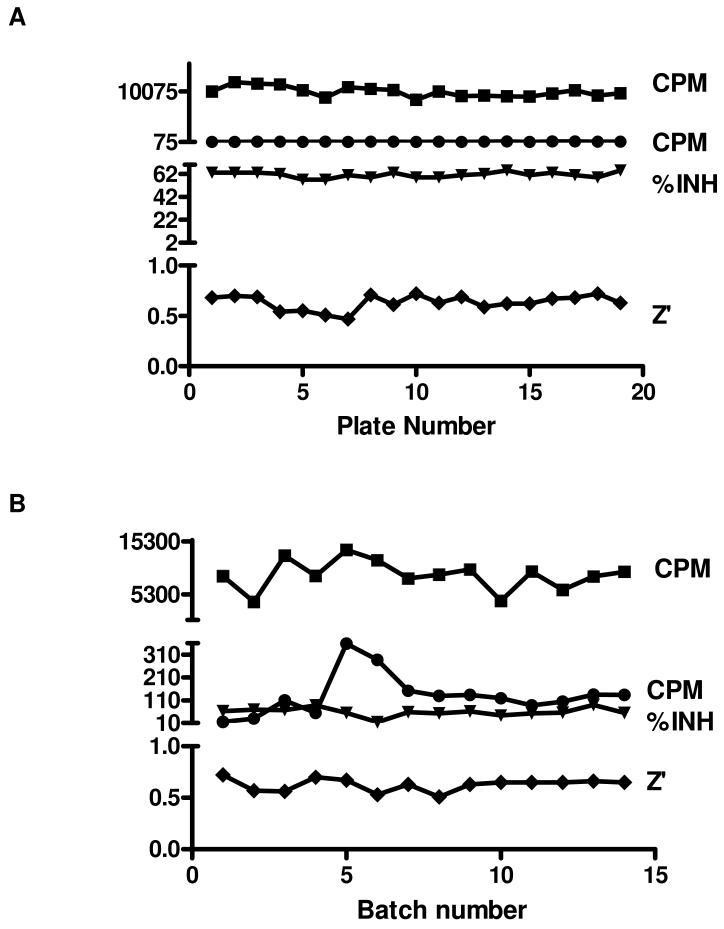
The overall S/B and S/N ratios for the library screen were 68 and 4.7 respectively. The Z′ factor was used to pass or fail each plate: plates were retested if the Z′ factor was <0.5. In a typical batch of 19 plates (Figure 3A), the Z′ factor was 0.63 ± 0.07 (coefficient of variance (CV) 11.6%) with a within-batch signal for enzyme activity of 9998 ± 975 cpm (CV = 9.8%). No enzyme blanks were 134 ± 36 cpm. Staurosporine inhibited Nek2 activity by 61.2 ± 2.4% at a concentration of 25 μM. The robust nature of the entire screen is illustrated in Figure 3B. The mean Z′ factor was 0.63 ± 0.06 and staurosporine inhibition was 59 ± 18%.
Figure 3. Assay performance achieved in the high-throughput screen.
A. Assay reproducibility for the screen of one batch of ~6080 compounds on nineteen 384-well plates. The mean values for total activity ■ (CPM), no enzyme blanks ● (CPM) and the positive control staurosporine (25 μM) expressed as % inhibition (▼) are shown, with the mean Z′ factor ♦ for each plate. B. The batch-to-batch variation of the same variables throughout the screen of ~73,000 compounds.
Compounds that inhibited enzymatic activity by >50% were selected as hits. The activity of 169 compounds was confirmed in triplicate (0.23% hit rate). Confirmed hits were screened to determine if the compounds were likely to be inhibiting Nek2 non-selectively via aggregation.19 The assay used was identical to that described for the primary screen except that 0.01% Triton X-100 was added to the buffer. Seventy of the confirmed hits retained activity in this assay and were therefore likely to be non-aggregators. It should be noted, however, that the aggregating properties of early stage compounds may depend not only on the protein target but also the type and concentration of the detergent used and the assay format20 and thus some compounds with aggregating properties may not have been identified. The potency (IC50) of these 70 compounds ranged from 0.65 to 91 μM.
Several series of compounds were identified as Nek2 inhibitors. These included a group of steroid-like viridins and another of toxoflavins. The chemical structures of the toxoflavins are shown in Table 1 and these were approximately equipotent against Nek2. Toxoflavins have activity against a number of kinases,21 and we have previously reported these compounds as hits in a screen for inhibitors of PKB22 and compounds of this type were also identified as inhibitors of Aurora A (data not shown). Because of their promiscuous activity against other kinase targets this series of compounds was not pursued further.
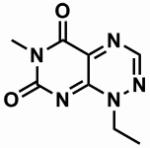
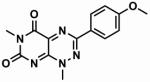
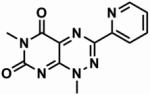
Table 1. The chemical structures of five toxoflavin compounds identified as potent inhibitors of Nek2 in the HTS.
| Compound | Structure | % Nek2 inhibition at 30 μM | PKB inhibition IC50(μM)* |
|---|---|---|---|
| CCT039836 |
|
90.6 ± 0.9 | 0.9 |
| CCT039838 |
|
85.2 ± 3.4 | n.d. |
| CCT039839 |
|
87.5 ± 0.7 | 1.8 |
| CCT039840 |
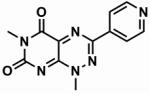
|
86.7 ± 1.2 | 2.2 |
| CCT039841 |
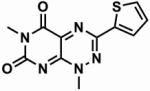
|
81.9 ± 0.9 | 8.5 |
The IC50 values were previously determined for four of these toxoflavins identified as hits in a screen for PKB inhibitors.22 n.d., not determined.
The activity of a selection of the viridin-like compound series was confirmed in both the FlashPlate (IC50 = 0.5-2.9 μM) and the filter assay (IC50 = 1.4-11.9 μM) (Table 2). Interestingly, these steroidal compounds have a similar structure to the non-specific PI3 kinase inhibitor, wortmannin,23 although the analogs found in the screen were up to 10-fold more potent inhibitors of Nek2 than wortmannin (Table 2). The structurally unrelated compound LY294002, a non-specific PI3 kinase inhibitor, 23; 24 was inactive against Nek2 in both enzymatic assays. It is interesting to note that both the toxoflavins and the viridins have the potential for inhibition through chemical reaction with proteins, particularly with exposed thiol groups, as observed for toxoflavins,21; 25 or with lysine, as observed with the PI3 kinase inhibitor, wortmannin.26
Table 2. The chemical structures, Nek2 kinase IC50 values and anti-proliferative effects (GI50) of the viridin-like compounds.
| Compound | Structure | Nek2 inhibition (IC50 μM) |
Growth inhibition (GI50 μM) |
||
|---|---|---|---|---|---|
| FlashPlate* | Filter* | U2OS Mean ± sem |
HeLa n=1 |
||
| CC004729 |
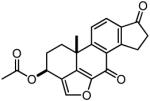
|
2.9 (3.6, 2.2) |
11.9 | n.d. | n.d. |
| CC0047311 |
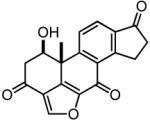
|
9 ± 2.7 | 1.4 (1.1, 1.6) |
11.0 ± 3.0 | 1.8 |
| CC004733 |
|
2.5 (3.3, 1.6) |
4.4 (1.0, 7.7) |
10.6 ± 2.9 | 10.2 |
| CC004730 |
|
8.25% inhibition at 32 μM |
n.d. | 50% inhibition at 100μM |
n.d. |
| Wortmannin |
|
14 | 15 | 23.7 | 15.0 |
mean ± sd or average of 2 determinations with individual values shown in brackets. n.d., not determined.
In addition, small molecules from a kinase-focused compound set were also identified as Nek2 inhibitors and their activities confirmed in the FlashPlate (IC50 = 11-47 μM) or the filter (IC50 = 7.3-39 μM) assays. Those hits (MW = 295-355, ClogP 1.1-4.1) did not contain potentially reactive chemical functionality and included a 2-amino pyridine, 4-(2-amino-5-(thiophen-3-yl)pyridin-3-yl)benzamide). This compound (4) belongs to a chemical class that has been successfully progressed as inhibitors of CHK2 kinase.27 The primary screen was thus able to identify more drug-like compounds than those described above.
Selectivity of viridin compounds for mitotic kinases
Although the viridin-like compounds are known to inhibit members of different kinase families, we were specifically interested to compare their activity against Nek2 with their activity against other kinases that are implicated in mitosis.3 Interestingly, the two viridin-like compounds, CC004731 and CC004733, showed a 70-1000-fold selectivity for Nek2 compared with Nek6 and Nek7 in this assay (Table 3). Similarly, neither staurosporine nor wortmannin inhibited Nek6 or Nek7, whereas they had IC50 values of 3.7 and 14.8 μM, respectively, against Nek2. These IC50 concentrations compare with those obtained using the FlashPlate and filter assays and with those previously described for Nek2 inhibition using a non-radioactive kinase assay (14 and 47 μM at 100 μM ATP for staurosporine and wortmannin respectively).18 Hence, there are significant differences in the ability of these types of compounds to inhibit different members of the Nek family. In contrast, the viridin-like compounds were more modestly selective against Nek2 (3-8-fold) compared to Aurora A, Plk1 and Cdk1.
Table 3. Selectivity of viridin compounds for mitotic kinases.
IC50 values (± Standard Error, logEC50) were determined for staurosporine, wortmannin, CC004731 and CC004733 for the mitotic kinases Nek2, Nek6, Nek7, Aurora A, Plk1 and Cdkl/Cyclin B in a biochemical kinase assay. >400 μM denotes no inhibition at 400 μM inhibitor, the highest concentration used in the assay.
Antiproliferative effects of the viridin-like compounds
Nek2 depletion using RNAi approaches results in an anti-proliferative phenotype.7; 8; 13 Both CC004731 and CC04733 caused growth inhibition of the osteosarcoma cell line U2OS (GI50 = 11.0 and 10.6 μM, respectively; Table 2), concentrations ~2-4 fold greater than the IC50 concentration for Nek2 kinase inhibition. In contrast, U2OS cells were relatively resistant to the effects of CC004730, a viridin compound which did not inhibit Nek2 biochemically. The growth of the HeLa cervical cancer cell line was also inhibited by CC004731 and CC004733 (Table 2) suggesting that the antiproliferative effects of these compounds are unlikely to be cell line specific.
Cell-based assay for Nek2 inhibition
The most well-characterized role of Nek2 is in promoting centrosome separation at the onset of mitosis.2 Under normal cell cycle conditions, the activation of Nek2 at the G2/M transition triggers the loss of cohesion between the duplicated interphase centrosomes by phosphorylating and displacing the intercentriolar linker proteins, C-Nap1 and rootletin.3 Ectopic overexpression of wild-type Nek2A in interphase cells induces premature splitting of centrosomes providing a defined phenotype by which Nek2A activity in cells can be measured independently from other mitotic kinases. Here, we made specific use of a U2OS cell line with tetracycline-inducible Nek2A overexpression, U2OS:GFP-Nek2A, that had been generated previously in our laboratory.11
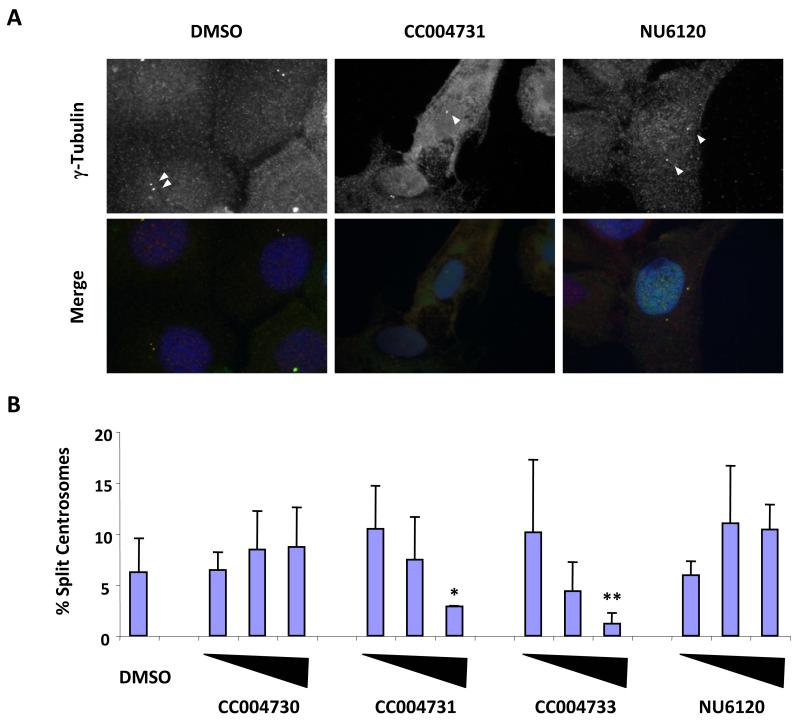
Asynchronous U2OS:GFP-Nek2A cells were either uninduced or induced with doxycycline alone or together with DMSO, CC004731 or CC004733 as active viridin-like compounds, or CC04730, a structurally-related but inactive viridin-like compound. After 24 hours, cells were fixed, stained for γ-tubulin to detect centrosomes and analyzed by immunofluorescence microscopy (Figure 4A). Centrosomes were scored as split when separated by >2 μm. Addition of DMSO (2%) to the induced cells had no effect on assay performance or centrosome splitting, whereas addition of the active Nek2 inhibitors CC004731 and CC004733 reduced centrosome splitting to background levels in a concentration-dependent manner (Figure 4B). The inactive analog, CC04730, did not reduce centrosome splitting. We also tested the effect of a Cdk2 inhibitor, NU6120 (6-cyclohexylmethoxy-2-(4′-hydroxyanilino)purine,28 as overexpression of Cdk2 with either cyclin A or E is also capable of stimulating premature centrosome splitting.29 However, in these cells, NU6120 had no effect on splitting strongly arguing that the assay measures inhibition of Nek2, and not Cdk2 (Figure 4B). Thus, Nek2A-induced centrosome splitting can be used as a convenient cell-based assay to test the activity of compounds in vivo.
Figure 4. Inhibition of Nek2-induced centrosome splitting by viridin compounds.
A. U2OS:GFP-Nek2A stable cells were induced to express GFP-Nek2A by the addition of doxycycline either alone or in the presence of 20 μM CC004731 or NU6120 for 24 hours. Cells were processed for immunofluorescence microscopy and stained for γ-tubulin (top panels) and DNA. Bottom panels show merged images where γ-tubulin is green and DNA is blue. Centrosomes are indicated with arrowheads. B. The percentage of interphase cells containing centrosomes that were separated by >2 μm were scored in the presence of DMSO, or increasing concentrations (0.5, 5 and 50 μM) of CC004730, CC004731, CC004733 or NU6120. The experiment was repeated three times and error bars represent standard deviations. Significance as compared to DMSO control was determined by Students T-test analysis, *p=0.122; **p=0.028.
Automated phenotypic assay for Nek2 inhibition
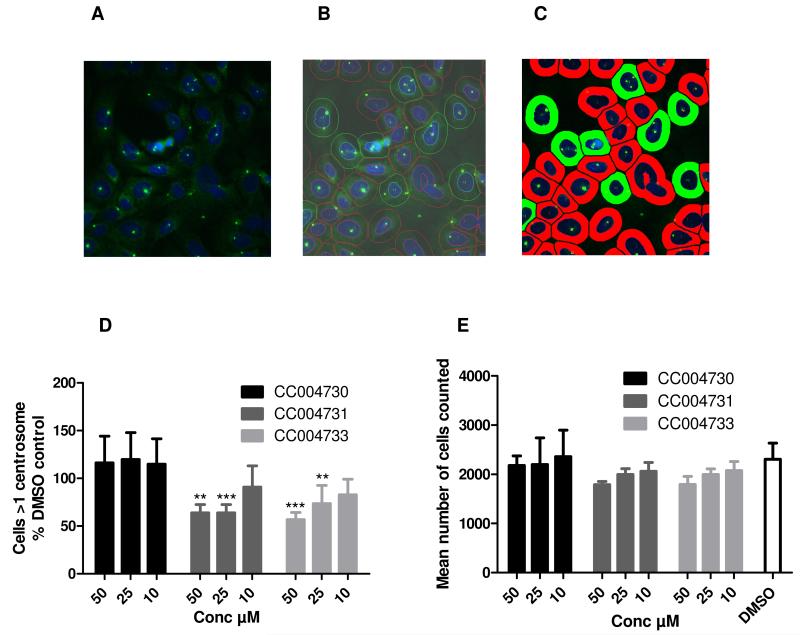
To overcome the laborious process of scoring centrosome splitting manually, the microscopy assay described above was automated. To determine the best fluorescence marker of centrosome localization, a number of antibodies were tested. Centrosomes appeared relatively weak when stained with antibodies against γ-tubulin or C-Nap1 (data not shown). This observation was made under non-induced conditions and may be due to higher than normal levels of Nek2A expression, which may to lead to their displacement from the centrosome.2 In contrast, a strong signal-to-noise ratio for centrosomes was obtained by staining with an antibody against the pericentriolar material marker pericentrin (Figure 5A). To automatically define the approximate perimeter of the cell, cells were co-stained with DAPI to define the nucleus (Figure 5A). Using the IN Cell analysis tools an outline for the cell perimeter was then set arbitrarily to the same shape as the nucleus, but with a radius of 10 μm larger than that of the nucleus (Figure 5B and C) so that the number of cells in each field could be enumerated. We could automatically score whether cells had 1 or 2 pericentrin-stained dots per cell, with 1 representing unsplit centrosomes and 2 representing split centrosomes using this algorithm. The within-plate variation for the percentage of cells with >1 centrosome in the DMSO control wells ranged between 8.8 ± 1.3 and 13.7 ± 2.9 (mean ± sd) with CV ranging from 13-21% in uninduced cells. Using this assay, the two active viridin-like compounds, CC004731 and CC004733, but not the inactive analog, CC004730, reduced centrosome splitting at 10, 25 and 50 μM (Figure 5D) with no significant loss of cell numbers (Figure 5E). Hence, this assay provides an automated approach for assessment of Nek2 inhibition in cells.
Figure 5. Automated cell-based assay for Nek2 inhibition.
A. Cells were processed for immunofluorescence microscopy and stained for pericentrin (green) to identify the centrosomes. B. The cells were also co-stained for DNA (blue). The centrosomes and cell nuclei were discriminated by automated analysis and the cell periphery was determined by setting a uniform distance (10 μm) from the nucleus. Nuclear and cellular areas defined by this algorithm are indicated in red or green where a single (unsplit) or more than one (split) centrosomes are detected per cell respectively. C. An artificial segmentation view of the image. D. The effect of selected viridin compounds (10, 20, and 50 μM for 6h) on centrosome splitting (n=4) in uninduced cells compared to DMSO-treated controls (n=24); Significance was determined by students 2 way ANOVA analysis **p=0.01 and ***p= 0.001. E. The mean number of cells counted under each condition.
DISCUSSION
While cancer drug discovery programs are well advanced for several mitotic kinases e.g. Cdk, Plk and Aurora kinases, interest has only recently turned to the NIMA-related kinases. There is good evidence that several members of this family have mitotic roles, including Nek2, Nek6, Nek7 and Nek9. Expression and depletion studies to date support the notion that Nek2 could indeed be an important anti-cancer target.3; 7; 8 Thus we undertook an unbiased high-throughput screen to identify small molecule inhibitors of Nek2 kinase activity.
Screening assays for Nek2 activity that use either a heterogeneous or homogeneous time-resolved fluorescence endpoint have been described.18 We employed a radiometric proximity assay based on the use of solid scintillant-coated microtitre plates as, when we commenced this work, Nek2 phosphorylation sites had not been identified and non-isotopic assays based on the use of generic serine/threonine kinase substrates and phosphospecific antibodies did not prove successful. The optimized radiometric assay included ATP concentrations below the Km as determined in a gel-based kinase assay and was robust and reproducible with batch variation of ~10% CV, and overall Z′ factors >0.6. The assay was used to screen a compound library with highly acceptable overall performance. Several classes of hit structures were found and these are among the first small molecule inhibitors of Nek2 to be reported.
The hits included non-selective toxoflavin kinase inhibitors and analogs of the natural product viridin. Both the toxoflavins and molecules containing the viridin core are recognized as having the potential to chemically react with proteins. For toxoflavins and other 7-azapteridines, this often involves redox reactions with exposed thiol groups.21; 25; 30 It is interesting to note that the X-ray structure of Nek2 shows two cysteine residues in the ATP-binding site.31; 32 The chemical reactivity of wortmannin with PI3-kinase has been characterized as reaction with Lys802 in the ATP-binding site through nucleophilic attack and ring-opening of the strained furan ring.26; 33 The viridin analogs from this screen may undergo similar covalent binding to an appropriately positioned lysine. Indeed, the Hill slopes for the 2 most active compounds (CC004731 and CC004733) were relatively high (~1.5), whereas those determined for staurosporine approximated to 1.0 (data not shown). This may support the notion that these particular compounds are chemically reactive. The size and calculated physicochemical parameters (MW = 207-350, ClogP = 0-3) of the toxoflavins and viridin-analogs identified here are within the acceptable limits proposed by several groups for the development of targeted molecular therapeutics. However, the possibility of a non-specific mechanism of action through chemical reactivity may increase the risk of off-target toxicities that could frustrate clinical use. The identification in our screen of other drug-like compounds originating from a kinase focused library that do not contain potentially reactive groups demonstrates that more attractive starting points for drug development against Nek2 can be found.
Interestingly, the viridin-like compounds showed a high degree of specificity within the mitotic members of the Nek kinase family with more than 70-fold more activity against Nek2 than Nek6 or Nek7. On the other hand, these compounds generally exhibited similar levels of activity against Plk1, Aurora A and Cdk1 as against Nek2. These data highlight the importance of having structural information on how inhibitors bind to the active sites of different kinases, as such information allows decisions to be made on chemical modifications that would improve selectivity. However, to date, only a limited number of high resolution structures of the Nek2 kinase domain have been solved. The first used a construct containing an inactivating point mutation (T175A) in the T-loop.31 Moreover, this structure was obtained in the presence of the generic kinase inhibitor, SU11652. Further high resolution structures have now been described using both the human wild-type and mutated (T175A) kinase domain of Nek2,32 although it remains to be seen whether these will be representative of full-length Nek2 in the presence of small molecule inhibitors, such as the viridins.
As predicted from Nek2 depletion studies,7; 8; 13 two of the active viridin compounds, but not the inactive analog, showed antiproliferative properties against two human cancer cell lines. However, because of the known promiscuity of this compound class, growth inhibition resulting from non-selective inhibition of other kinases cannot be ruled out as illustrated by the effect of wortmannin on proliferation.
Importantly, the active viridin-like compounds, but not the closely related compound that lacked activity against Nek2, affected premature centrosome splitting in a human tumour cell line. Overexpression of Cdk2/cyclin A or Cdk2/cyclin E, but not Plk1, Aurora A, Cdk1/cyclin B or Cdk4/cyclin D, can also induce significant levels of premature centrosome splitting.29 However, the splitting induced by active Cdk2 occurs as the result of a separate pathway from that induced by Nek2A.29 Furthermore, a compound, previously published as a Cdk2 inhibitor28 but that lacked activity against Nek2, did not block centrosome splitting in the Nek2A-inducible cells. This is consistent with the active viridins acting as direct Nek2 inhibitors in cells.
In view of these promising results, we explored the possibility of automating the manual image-based assay for use as a secondary screen in an ongoing drug discovery project. Antibodies against the PCM marker, pericentrin, gave a single bright spot when centrosomes were unsplit but two distinct dots when centrosomes were split. By additional staining of nuclei and arbitrarily setting an approximate cell perimeter based on the nuclear area, we developed an algorithm that could count the number of pericentrin-stained centrosomes per cell. Automated counting of Nek2A-inducible cells treated with DMSO or the viridin-like compounds demonstrated that this assay was capable of scoring cellular inhibition of Nek2A and would potentially allow more rapid and efficient estimation of cellular potency (EC50) for compounds generated during medicinal chemistry optimization of hit compounds.
In summary, we have successfully completed a small molecule high-throughput screen to identify chemical inhibitors of Nek2 kinase activity. Several series of compounds with activity against Nek2 kinase activity were identified. These included a series of toxoflavins and steroid-like, viridin compounds that demonstrated the validity of the screen. Results with the viridin hits show that pharmacological inhibition of Nek2 kinase results in the expected phenotype of disruption to centrosome function that is associated with growth inhibition and further support Nek2 as a target for cancer drug discovery.
Supplementary Material
ACKNOWLEDGMENTS
We thank all members of our laboratories for useful discussion and are grateful to Prof. R. Griffin (Newcastle) for supply of the NU6120 compound and GE Healthcare for advice in setting up the cell-based assay. This work was supported by grants to AMF, WA and PW from the Discovery Committee of Cancer Research UK. The Cancer Research UK Centre for Cancer Therapeutics is funded primarily by Cancer Research UK Program Grants C309/A2187 and C309/A8274. We acknowledge NHS funding to the NIHR Biomedical Research Centre. PW is a Cancer Research UK Life Fellow.
REFERENCES
- 1.de Carcer G, Perez de Castro I, Malumbres M. Targeting cell cycle kinases for cancer therapy. Curr Med Chem. 2007;14:969–985. doi: 10.2174/092986707780362925. [DOI] [PubMed] [Google Scholar]
- 2.Fry AM. The Nek2 protein kinase: a novel regulator of centrosome structure. Oncogene. 2002;21:6184–6194. doi: 10.1038/sj.onc.1205711. [DOI] [PubMed] [Google Scholar]
- 3.O’Regan L, Blot J, Fry AM. Mitotic regulation by NIMA-related kinases. Cell Div. 2007;2:25. doi: 10.1186/1747-1028-2-25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hayward DG, Fry AM. Nek2 kinase in chromosome instability and cancer. Cancer Lett. 2005;237:155–166. doi: 10.1016/j.canlet.2005.06.017. [DOI] [PubMed] [Google Scholar]
- 5.Barbagallo F, Paronetto MP, Franco R, Chieffi P, Dolci S, Fry AM, Geremia R, Sette C. Increased expression and nuclear localization of the centrosomal kinase Nek2 in human testicular seminomas. J Pathol. 2009;217:431–441. doi: 10.1002/path.2471. [DOI] [PubMed] [Google Scholar]
- 6.Hayward DG, Clarke RB, Faragher AJ, Pillai MR, Hagan IM, Fry AM. The centrosomal kinase Nek2 displays elevated levels of protein expression in human breast cancer. Cancer Res. 2004;64:7370–7376. doi: 10.1158/0008-5472.CAN-04-0960. [DOI] [PubMed] [Google Scholar]
- 7.Kokuryo T, Senga T, Yokoyama Y, Nagino M, Nimura Y, Hamaguchi M. Nek2 as an effective target for inhibition of tumorigenic growth and peritoneal dissemination of cholangiocarcinoma. Cancer Res. 2007;67:9637–9642. doi: 10.1158/0008-5472.CAN-07-1489. [DOI] [PubMed] [Google Scholar]
- 8.Tsunoda N, Kokuryo T, Oda K, Senga T, Yokoyama Y, Nagino M, Nimura Y, Hamaguchi M. Nek2 as a novel molecular target for the treatment of breast carcinoma. Cancer Sci. 2009;100:111–116. doi: 10.1111/j.1349-7006.2008.01007.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Loo LW, Grove DI, Williams EM, Neal CL, Cousens LA, Schubert EL, Holcomb IN, Massa HF, Glogovac J, Li CI, Malone KE, Daling JR, Delrow JJ, Trask BJ, Hsu L, Porter PL. Array comparative genomic hybridization analysis of genomic alterations in breast cancer subtypes. Cancer Res. 2004;64:8541–8549. doi: 10.1158/0008-5472.CAN-04-1992. [DOI] [PubMed] [Google Scholar]
- 10.Weiss MM, Kuipers EJ, Postma C, Snijders AM, Pinkel D, Meuwissen SG, Albertson D, Meijer GA. Genomic alterations in primary gastric adenocarcinomas correlate with clinicopathological characteristics and survival. Cell Oncol. 2004;26:307–317. doi: 10.1155/2004/454238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Faragher AJ, Fry AM. Nek2 kinase stimulates centrosome disjunction and is required for formation of bipolar mitotic spindles. Mol Biol Cell. 2003;14:2876–2889. doi: 10.1091/mbc.E03-02-0108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Fletcher L, Cerniglia GJ, Yen TJ, Muschel RJ. Live cell imaging reveals distinct roles in cell cycle regulation for Nek2A and Nek2B. Biochem Biophys Acta. 2005;1744:89–92. doi: 10.1016/j.bbamcr.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 13.Fletcher L, Cerniglia GJ, Nigg EA, Yend TJ, Muschel RJ. Inhibition of centrosome separation after DNA damage: a role for Nek2. Radiat Res. 2004;162:128–135. doi: 10.1667/rr3211. [DOI] [PubMed] [Google Scholar]
- 14.Qiu XL, Li G, Wu G, Zhu J, Zhou L, Chen PL, Chamberlin AR, Lee WH. Synthesis and biological evaluation of a series of novel inhibitor of Nek2/Hec1 analogues. J Med Chem. 2009;52:1757–1767. doi: 10.1021/jm8015969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Zhang JH, Chung TD, Oldenburg KR. A Simple Statistical Parameter for Use in Evaluation and Validation of High Throughput Screening Assays. J Biomol Screen. 1999;4:67–73. doi: 10.1177/108705719900400206. [DOI] [PubMed] [Google Scholar]
- 16.Asensio CJ, Garcia RC. Determination of a large number of kinase activities using peptide substrates, P81 phosphocellulose paper arrays and phosphor imaging. Anal Biochem. 2003;319:21–33. doi: 10.1016/s0003-2697(03)00282-3. [DOI] [PubMed] [Google Scholar]
- 17.Fry AM, Schultz SJ, Bartek J, Nigg EA. Substrate specificity and cell cycle regulation of the Nek2 protein kinase, a potential human homolog of the mitotic regulator NIMA of Aspergillus nidulans. J Biol Chem. 1995;270:12899–12905. doi: 10.1074/jbc.270.21.12899. [DOI] [PubMed] [Google Scholar]
- 18.Jin G, Aulabaugh A, Pocas J, Liu H, Kriz R, Sampath D. Development and comparison of nonradioactive in vitro kinase assays for NIMA-related kinase 2. Anal Biochem. 2006;358:59–69. doi: 10.1016/j.ab.2006.08.003. [DOI] [PubMed] [Google Scholar]
- 19.Ryan AJ, Gray NM, Lowe PN, Chung CW. Effect of detergent on “promiscuous” inhibitors. J Med Chem. 2003;46:3448–3451. doi: 10.1021/jm0340896. [DOI] [PubMed] [Google Scholar]
- 20.Jadhav A, Ferreira RS, Klumpp C, Mott BT, Austin CP, Inglese J, Thomas CJ, Maloney DJ, Shoichet BK, Simeonov A. Quantitative analyses of aggregation, autofluorescence, and reactivity artifacts in a screen for inhibitors of a thiol protease. J Med Chem. 2010;53:37–51. doi: 10.1021/jm901070c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Goh KC, Wang H, Yu N, Zhou Y, Zheng Y, Lim ZY, Sangthongpitag K, Fang L, Du M, Wang X, Jefferson AB, Rose J, Shampoon B, Reinhard C, Carte B, Entzeroth M, Ni B, Taylor ML, Stunkel W. PLK1 as a potential drug target in cancer therapy. Drug Dev Res. 2004;62:349–361. [Google Scholar]
- 22.Burns S, Travers J, Collins I, Rowlands MG, Newbatt Y, Thompson N, Garrett MD, Workman P, Aherne W. Identification of small-molecule inhibitors of protein kinase B (PKB/AKT) in an AlphaScreenTM high-throughput screen. J Biomol Screen. 2006;11:822–827. doi: 10.1177/1087057106290992. [DOI] [PubMed] [Google Scholar]
- 23.Vlahos CJ, Matter WF, Hui KY, Brown RF. A specific inhibitor of phosphatidylinositol 3-kinase, 2-(4-morpholinyl)-8-phenyl-4H-1-benzopyran-4-one (LY294002) J Biol Chem. 1994;269:5241–5248. [PubMed] [Google Scholar]
- 24.Workman P, Clarke PA, Raynaud FI, van Montfort RL. Drugging the PI3 kinome: from chemical tools to drugs in the clinic. Cancer Res. 2010;70:2146–2157. doi: 10.1158/0008-5472.CAN-09-4355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huth JR, Song D, Mendoza RR, Black-Schaefer CL, Mack JC, Dorwin SA, Ladror US, Severin JM, Walter KA, Bartley DM, Hajduk PJ. Toxicological evaluation of thiol-reactive compounds identified using a la assay to detect reactive molecules by nuclear magnetic resonance. Chemical research in toxicology. 2007;20:1752–1759. doi: 10.1021/tx700319t. [DOI] [PubMed] [Google Scholar]
- 26.Wymann MP, Bulgarelli-Leva G, Zvelebil MJ, Pirola L, Vanhaesebroeck B, Waterfield MD, Panayotou G. Wortmannin inactivates phosphoinositide 3-kinase by covalent modification of Lys-802, a residue involved in the phosphate transfer reaction. Molecular and cellular biology. 1996;16:1722–1733. doi: 10.1128/mcb.16.4.1722. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hilton S, Naud S, Caldwell JJ, Boxall K, Burns S, Anderson VE, Antoni L, Allen CE, Pearl LH, Oliver AW, Wynne Aherne G, Garrett MD, Collins I. Identification and characterisation of 2-aminopyridine inhibitors of checkpoint kinase 2. Bioorg Med Chem. 2010;18:707–718. doi: 10.1016/j.bmc.2009.11.058. [DOI] [PubMed] [Google Scholar]
- 28.Hardcastle IR, Arris CE, Bentley J, Boyle FT, Chen Y, Curtin NJ, Endicott JA, Gibson AE, Golding BT, Griffin RJ, Jewsbury P, Menyerol J, Mesguiche V, Newell DR, Noble ME, Pratt DJ, Wang LZ, Whitfield HJ. N2-substituted O6-cyclohexylmethylguanine derivatives: potent inhibitors of cyclin-dependent kinases 1 and 2. J Med Chem. 2004;47:3710–3722. doi: 10.1021/jm0311442. [DOI] [PubMed] [Google Scholar]
- 29.Meraldi P, Nigg EA. Centrosome cohesion is regulated by a balance of kinase and phosphatase activities. J Cell Sci. 2001;114:3749–3757. doi: 10.1242/jcs.114.20.3749. [DOI] [PubMed] [Google Scholar]
- 30.Guertin KR, Setti L, Qi L, Dunsdon RM, Dymock BW, Jones PS, Overton H, Taylor M, Williams G, Sergi JA, Wang K, Peng Y, Renzetti M, Boyce R, Falcioni F, Garippa R, Olivier AR. Identification of a novel class of orally active pyrimido[5,4-3][1,2,4]triazine-5,7-diamine-based hypoglycemic agents with protein tyrosine phosphatase inhibitory activity. Bioorganic and medicinal chemistry letters. 2003;13:2895–2898. doi: 10.1016/s0960-894x(03)00623-1. [DOI] [PubMed] [Google Scholar]
- 31.Rellos P, Ivins FJ, Baxter JE, Pike A, Nott TJ, Parkinson DM, Das S, Howell S, Fedorov O, Shen QY, Fry AM, Knapp S, Smerdon SJ. Structure and regulation of the human Nek2 centrosomal kinase. J Biol Chem. 2007;282:6833–6842. doi: 10.1074/jbc.M609721200. [DOI] [PubMed] [Google Scholar]
- 32.Westwood I, Cheary DM, Baxter JE, Richards MW, van Montfort RL, Fry AM, Bayliss R. Insights into the conformational variability and regulation of human Nek2 kinase. J Mol Biol. 2009;386:476–485. doi: 10.1016/j.jmb.2008.12.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Walker EH, Pacold ME, Perisic O, Stephens L, Hawkins PT, Wymann MP, Williams RL. Structural determinants of phosphoinositide 3-kinase inhibition by wortmannin, LY294002, quercetin, myricetin, and staurosporine. Molecular cell. 2000;6:909–919. doi: 10.1016/s1097-2765(05)00089-4. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.