Abstract
Lead toxicity is associated with various human diseases. While Ca2+ binding proteins such as calmodulin (CaM) are often reported to be molecular targets for Pb2+-binding and lead toxicity, the effect of Pb2+ on the Ca2+/CaM regulated biological activities cannot be described by the primary mechanism of ionic displacement (e.g., ionic mimicry). The focus of this study was to investigate the mechanism of lead toxicity through binding differences between Ca2+ and Pb2+ for CaM, an essential intracellular trigger protein with two EF-Hand Ca2+-binding sites in each of its two domains that regulates many molecular targets via Ca2+-induced conformational change. Fluorescence changes in phenylalanine indicated that Pb2+ binds with 8-fold higher affinity than Ca2+ in the N-terminal domain. Additionally, NMR chemical shift changes and an unusual biphasic response observed in tyrosine fluorescence associated with C-terminal domain sites EF-III and EF-IV suggest a single higher affinity Pb2+-binding site with a 3-fold higher affinity than Ca2+, coupled with a second site exhibiting affinity nearly equivalent to that of the N-terminal domain sites. Our results further indicate that Pb2+ displaces Ca2+ only in the N-terminal domain, with minimal perturbation of the C-terminal domain, however significant structural/dynamic changes are observed in the trans-domain linker region which appear to be due to Pb2+-binding outside of the known calcium-binding sites. These data suggest that opportunistic Pb2+-binding in Ca2+/CaM has a profound impact on the conformation and dynamics of the essential molecular recognition sites of the central helix, and provides insight into the molecular toxicity of non-essential metal ions.
Keywords: Calcium, calmodulin, lead, toxicity, opportunistic binding
1 Introduction
Lead toxicity is a persistent global health problem. General physiological and biochemical problems associated with lead toxicity include neurological disorders related to the central and peripheral nervous systems [1–3], interference with heme biosynthesis [4], anemia [5], nephrotoxicity [6], hypertension [7] and reproductive disorders [8–10]. Potential carcinogenic and genetic effects related to lead toxicity have been reviewed [11].
Ionic displacement (e.g., ionic mimicry [12]) is believed to be the primary mechanism associated with several types of Pb2+-induced anemia, first identified almost a century ago [5]. Pb2+ has been found to displace Mg2+ in pyrimidine 5'-nucleotidase type 1 [13], inhibiting the activity of the enzyme. This decreased activity results in increased concentrations of pyrimidines with an increased rate of destruction of red blood cells leading to anemia [14]. Pb2+ has also been shown to replace Zn2+ in 5-aminolevulinic acid dehydratase (ALAD) [15, 16], an important enzyme in heme synthesis, resulting in iron-deficiency anemia. Iron is another important metal which may be a target for Pb2+ displacement. Iron plays important roles in heme biosynthesis, including the formation of the heme precursor protoporphyrin, and in the function of ribonucleotide reductase (RNR) which catalyzes the formation of deoxyribonucleotides through a free radical mechanism. The extent to which Pb2+ may be able to directly interfere with the biological roles of iron is not known, but Pb2+ has been found to displace Fe2+ in divalent cation transporter-1 [17] which may be involved in transport of Pb2+ and cellular uptake, and in the crystal structure of RNR.
Ca2+ binding proteins have also been identified as molecular targets for Pb2+ binding and lead toxicity. Pb2+ can enter cells through Ca2+ channels [18, 19], and can displace Ca2+ in different Ca2+-binding proteins (CaBPs) [20–24]. Protein kinase C (PKC), a family of proteins that mediate various cellular processes including cell proliferation and central nervous system (CNS) development, can be activated by Pb2+ at subnanomolar concentrations [25, 26] which may result in Pb2+-induced neurotoxicity. Calmodulin (CaM) is another Ca2+-binding protein that has previously been identified as playing a potential role in lead toxicity [24, 27], and ionic displacement was assumed to be the mechanism associated with toxicity as Pb2+ will occupy the Ca2+ binding sites in CaM with higher relative affinity than Ca2+ [28, 29]. CaM is a 148 residue, predominantly helical intracellular trigger protein (Fig. S1a) that responds to changes in cytosolic Ca2+ by binding up to four Ca2+ ions in two pairs of cooperative EF-hand sites [30]. The highly-conserved EF-Hand sites are comprised of a helix-loop-helix structure. Residues within the loop are identified by a relative position number 1–12, with binding of Ca2+ typically coordinated by residues in positions 1, 3, 5, 7, 9 and 12, where the coordinating ligand from position 7 is usually a carbonyl oxygen. CaM is divided into two structurally similar domains, each containing a pair of EF-Hand sites, separated by a trans-domain linker region comprising residues 76–84 (Fig. S1b). This region appears helical (Fig. S1c) in X-ray structures [31, 32], and as a flexible loop in NMR solution structures [33, 34]. The intrinsic flexibility of this region (Fig. S1d) allows the two domains to adopt a closer conformation to one another in solution [35] and is believed to be functionally important to CaM’s ability to bind peptides and interact with enzymes as a secondary messenger controlling multiple biological functions [34]. Therefore, activation of CaM-mediated target proteins is largely dependent on the Ca2+ binding process and Ca2+-induced conformational change.
CaM is also one of the only proteins with crystal structure data available showing both the Ca2+- and Pb2+-bound states (Fig. S1e and Fig. S1f) [36, 37]. Previous studies related to lead toxicity have reported that Pb2+ binding to CaM resulted in drastically altered downstream activity, displaying an initial increase (i.e., activation) and subsequent decrease (i.e., inhibition) for both CaM-sensitive phosphodiesterase (PDE) [22, 38] and myosin light-chain kinase (MLCK) [39]. However, it was not clear how Pb2+ or other toxic metals could activate and then subsequently deactivate these processes solely by ionic mimicry, indicating gaps in our understanding of the mechanism associated with lead molecular toxicity. Additionally, the presence of secondary cationic binding sites in CaM [40, 41] has not received detailed attention in discussions regarding lead toxicity, although a statistical analysis completed in our laboratory of Pb2+-bound protein structures in the Protein DataBank (PDB) suggested that Pb2+ exhibits more flexible requirements for binding than Ca2+ and will bind proteins outside of the known Ca2+-binding sites. These apparent inconsistencies led us to hypothesis that lead molecular toxicity may be related to opportunistic binding of Pb2+ in secondary sites, such as the central helix of CaM, resulting in conformational changes that alter or inhibit protein activity [42]. Such an opportunistic binding model would have important human health implications as it would indicate a much broader pool of potential molecular targets for Pb2+ that would better explain the systemic effects observed with lead toxicity.
To test this hypothesis, CaM binding with Ca2+ and Pb2+ was evaluated using fluorescence spectroscopy and multidimensional high resolution NMR with 15N-labeled protein. The effect of Pb2+ on Ca2+/CaM was further examined by titrating Pb2+ into Ca2+-saturated CaM using heteronuclear single quantum coherence (HSQC) NMR, heteronuclear {1H, 15N} nuclear Overhauser effect (NOE) and NMR relaxation studies.
The results of this study demonstrated that Pb2+ alters the conformation of CaM in the Ca2+-bound state, especially at the molecular recognition site, and provides evidence that molecular toxicity may be induced in CaM or other proteins as a result of binding opportunistically in secondary sites. This allosteric mechanism suggests that the promiscuous nature of Pb2+ allows for multiple molecular targets and by extension offers a comprehensive explanation for the resulting systemic pathology of lead toxicity.
2 Materials and Methods
2.1 Metal Standards
Ca2+ was obtained from Ionplus calcium standard (Orion Research Inc., Beverly, MA). Pb2+ was purchased as Pb(NO3)2 (Fisher Scientific, Fair Lawn, NJ) and prepared in ultra milli-Q double deionized water (ddH2O, resistivity = 18.0 MΩ·cm).
2.2 Expression and purification of wild-type calmodulin (wt-CaM)
Recombinant rat wt-CaM (hereafter referred to as CaM) was expressed in Escherichia coli strain BL21(DE3)pLysS. CaM from rat was selected as it shares 99% sequence identity with human CaM. Expression and purification for homonuclear and 15N-labelled proteins in either LB or minimal media followed previously reported protocols [43]. Final concentrations of the proteins were determined by measuring absorbance at 277 nm, and calculating concentration based on the Beer-Lambert Law where the molar absorptivity (ε) for CaM = 3030 M−1 cm−1 [44].
Buffers were treated with Analytical Grade Chelex 100 resin, 100–200 mesh Sodium Form (Bio-Rad Laboratories, Hercule, CA), to selectively remove divalent cations (e.g., Ca2+). Protein samples were treated by passage through a small column packed with 2g polystyrene BAPTA (Invitrogen Molecular Probes, Eugene, OR), commercially referred to as Calcium SpongeTM S, which has a reported approximate Kd for Ca2+ of 300 nM. Unless otherwise specified, all protein samples were prepared in high salt (100 mM KCl) environments, which were found in preliminary studies to have only minimal effect on Pb2+-binding, and experiments were conducted at 37 °C in an effort to mimic physiological conditions as closely as possible.
2.3 Fluorescence studies
Fluorometric spectral analyses were conducted in triplicate using a PTI (Photon Technology International, Birmingham, NJ) spectrofluorometer equipped with a 75 W xenon arc lamp and a model 814 photomultiplier tube (PMT) detector. Samples were evaluated in 1 cm path length quartz cuvettes at 21 °C. Protein sample concentrations were 10 µM in 800 µL, prepared in Chelex-treated buffers of 10 mM Tris pH 7.4, 100 mM KCl.
Previous work with CaM has demonstrated that the distributions of phenylalanine in the N-terminal domain and tyrosine in the C-terminal domain (Fig. S1a) provides a means to monitor binding of Ca2+ ions based on changes in the intrinsic fluorescence of these residues [45, 46]. While this approach cannot provide quantitative data for individual binding sites, it can provide macromolecular dissociation constants (Kd) for the individual domains.
Emission spectra for phenylalanine fluorescence were collected from 265–285 nm (3 nm excitation (Ex) slit widths, 4 nm emission slit widths, λEx = 250 nm, integration 0.2 s, stepsize 1 nm). Emission spectra for tyrosine fluorescence were collected from 290–350 nm (2 nm excitation slit widths, 3 nm emission slit widths, λEx = 277 nm, integration 0.2 s, stepsize 1 nm).
In this study, Kd values based on total metal concentration for direct titrations were calculated from the mean and standard deviation of triplicate experiments using Eq. 1, where [P]T is total protein concentration, [M]T is total metal concentration, and F is the fluorescence intensity.
| (Eq. 1) |
| (Eq. 2) |
| (Eq. 3) |
For competitive titrations involving pre-equilibration of the protein with one metal followed by titration with a second metal, the Kd in Eq. 1 becomes an apparent Kd (Kapp). In Eq. 2, the Kd for a titrant metal (KdM2) can be obtained based on Kapp from Eq. 1, the known Kd for the pre-equilibrated metal ion (KdM1) and the fixed, total concentration of the pre-equilibrated metal ion [M1]T [47]. For titrations where the Kd of the titrant metal is known but not the pre-equilibrated metal, Eq. 2 is rearranged as Eq. 3. Alternatively, for titrations of Ca2+ to CaM which involve cooperative binding, data were fit using the Hill equation (Eq. 4), where n is the Hill coefficient. The free Ca2+ concentration was determined by competitive titration with Oregon Green indicator dye using a calcium buffering system (ethylene glycol tetraacetic acid (EGTA) combined with nitrilotriacetic acid (NTA)) [48].
| (Eq. 4) |
2.4 NMR Analysis
All spectra were acquired on a 600 MHz Varian NMR spectrometer. Conversions of free induction decay (FID) files from Varian to Sparky formats were completed using NMRPipe [49] software. Peak assignment and area integration for 2D and 3D spectra were processed using Sparky software (T.D. Goddard, University of California, San Francisco, CA). Data were analyzed or compiled in Microsoft Excel (Microsoft, Redmond, WA), while curve-fitting was completed using Kaleidagraph software (Synergy Software, Reading, PA). NMR sample tubes were purchased from Wilmad-Labglass (Vineland, NJ). D2O (99.96%) was purchased from Cambridge Isotope Laboratories (Andover, MA).
2.5 2D NMR
The 500 µL samples for evaluation of both Ca2+ and Pb2+ by HSQC NMR were comprised of 253–400 µM 15N-labeled CaM in 10 mM Bis-TRIS pH 6.5, with 5 mM 2-(N-morpholino)ethanesulfonic acid (MES) buffer, 10% D2O, and 0.1 mM NaN3 to inhibit bacterial growth. Samples were analyzed on a 600 MHz Varian NMR spectrometer using the N15 gradient enhanced heteronuclear single quantum coherence pulse sequence (gNHsqc) at 37 °C. Typically a total of 32 dummy scans and 32 acquisition scans were collected across a spectral width of 8384.9 Hz in the proton dimension, with 128 increments across a spectral width of 2000 Hz in the nitrogen dimension. Reference spectra for Ca2+-free or Ca2+-loaded CaM were acquired by treating samples first either with 10 mM EGTA or 20 mM Ca2+. Three titration experiments were completed to monitor structural changes in CaM associated with metal binding. First, HSQC spectra were acquired for CaM with 0, 1, 2, 3, 4 and 6 molar equivalents (MEs) of Ca2+. Similarly, HSQC spectra were acquired for CaM with 0, 1, 2, 3, 4, 5 and 6 MEs Pb2+. Finally, HSQC spectra were obtained for the addition of 0.5, 1.0, 1.5, 2.0, 2.5 and 3.0 MEs Pb2+ to CaM presaturated with 6 MEs Ca2+ to evaluate structural changes associated with competitive binding. Due to sample precipitation problems, we were unable to add more than 3.0 MEs of Pb2+ to CaM samples presaturated with Ca2+. Total chemical shift changes (Δδ) across both dimensions (15N and 1H) were weight-averaged based on Eq. 5. Peaks in the spectra for Ca2+-free and Ca2+-loaded CaM were assigned based on 3D HNCA assignment at 37 °C with previous assignments for Ca2+-free CaM at 23 °C [50], and Ca2+-loaded CaM at 37 °C [51].
| (Eq. 5) |
2.6 Protein Dynamics
To compare dynamic properties of CaM complexed with either Ca2+ or both Ca2+ and Pb2+, heteronuclear NOE data were collected and analyzed following the approach described by Seifert [52]. Samples consisted of 1.14 mM 15N-labeled CaM prepared in 10 mM Tris pH 6.6, 100 mM KCl, 100 µM NaN3, and 10% D2O. For analysis of Ca2+-loaded CaM, 20 mM Ca2+ was added to the sample. For analysis of Pb2+, 2 MEs of Pb2+ was added to CaM pre-loaded with 6 MEs Ca2+. Both analyses were run at 37 °C.
For NOE data, two experiments were conducted with 1H saturation times of 0.0 and 4.0 s using pulse sequence gNnoe. Data for NOE were processed as a ratio (Eq. 6) where t4 and t0 are the integrated peak areas for relaxation times of 4s and 0s respectively.
| (Eq. 6) |
3 Results
3.1 Domain-specific binding affinities of CaM determined by intrinsic fluorescence
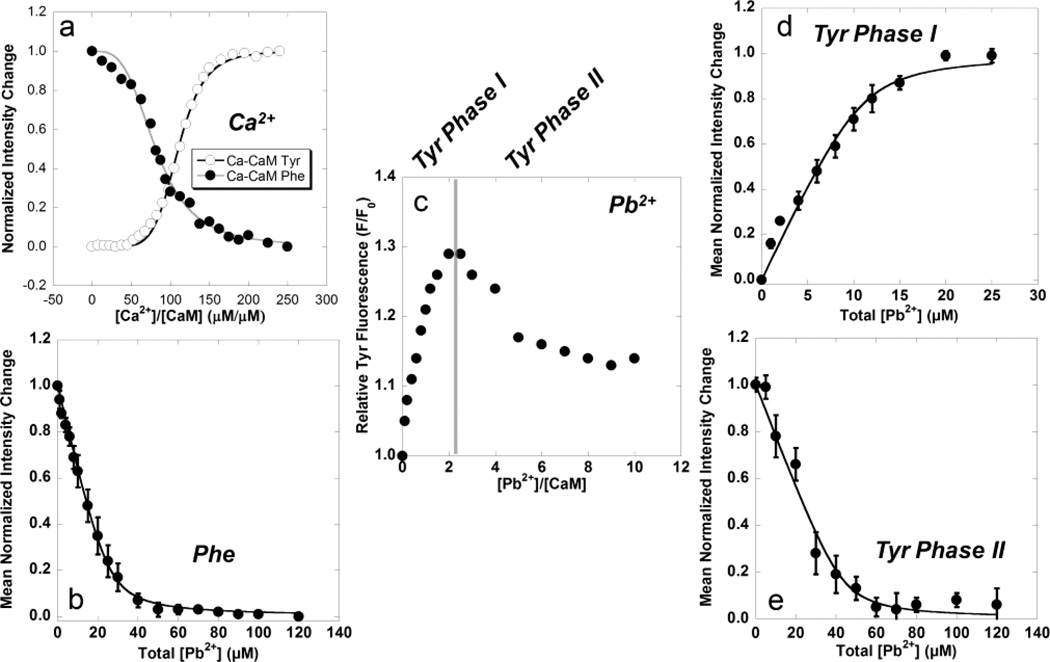
Changes in fluorescent intensities for Phe at 265–285 nm (excited at 250 nm) and Tyr at 290–350 nm (excited at 277 nm) as functions of metal concentration for both Ca2+ and Pb2+ binding with CaM are presented in Fig. 1, and the calculated dissociation constants are summarized in Table 1.
Fig. 1.
(a) Mean normalized Tyr (○) and Phe (●) fluorescence changes as a function of total [Ca2+] using Eq. 4. (b) Mean normalized Phe fluorescence change as a function of total [Pb2+] for the N-terminal domain with calculated Kd of 1.40 ± 0.30 µM. (c) Relative Tyr fluorescence as a function of Pb2+:CaM complex formation. The observed biphasic fluorescent response is divided into (d) Phase 1 with a calculated Kd of 0.73 ± 0.10 µM and (e) Phase 2 with a calculated Kd of 1.93 ± 0.32 µM. The calculated Kd for Phase 2 of the Tyr fluorescence falls within the standard deviation calculated for Phe fluorescence in the N-terminal domain.
Table 1.
Domain-specific binding dissociation constants for CaM.
| Kd (µM) | ||
|---|---|---|
| Domain (Site) | Ca(II) | Pb(II) |
| aND (I, II) | 11.50±0.68 | 1.40±0.30 |
| bCD (III, IV) | 2.04±0.02 | c0.73±0.10 |
| CD (III, IV) | -- | d0.67±0.06 |
| CD (Secondary) | -- | 1.93±0.32 |
N-terminal domain
C-terminal domain
Direct titration
Competitive titration with Ca(II)
Calculated Kd values for Ca2+ for both the N-terminal (11.50±0.68 µM) and C-terminal (2.04±0.02 µM) domains were consistent both with reported values [48] and with known Ca2+ intracellular concentrations in the µM range.
Direct titration of Pb2+ to CaM produced a decrease in Phe fluorescence (Fig. 1b), similar to the response observed with Ca2+ (Fig. 1a). Curve-fitting of data using Eq. 1 produced a calculated Kd of 1.40±0.30 µM for binding of Pb2+ in the N-terminal domain which was as much as 8-fold higher than Ca2+ (Table 1).
Unlike the increase in Tyr fluorescence observed in Ca2+ titrations (Fig. 1a), the direct titration of Pb2+ produced a biphasic response characterized by a rapid initial increase in fluorescence intensity up to ~2:1 MEs of Pb2+/Protein, followed by a hyperbolic decrease in intensity reaching a minimum below 10 MEs of Pb2+ (Fig. 1c). The initial increase (Phase 1, Fig. 1d) which mimics the Ca2+ response and peaks at ~2 MEs of Pb2+, was interpreted as binding of Pb2+ in one of the two binding sites EF-III or EF-IV, while the subsequent decrease (Phase 2, Fig. 1e) was interpreted as binding in the other C-terminal domain site. Curve-fitting of data, based on Eq. 1, produced a calculated Kd of 0.73 ± 0.10 µM for Phase 1 and a Kd of 1.93 ± 0.32 µM for Phase 2 (Table 1). Interestingly, this value and the associated curve of the second phase in the tyrosine titration (Fig. 1e) are nearly identical to the corresponding curve and calculated Kd observed for the Phe signal change for the N-terminal domain. These results suggest a single higher affinity Pb2+-binding site in the C-terminal domain and nearly equivalent affinity for the three remaining sites.
Competitive titrations to analyze changes in Tyr fluorescence were also conducted by presaturating CaM with Ca2+ followed by titration with Pb2+, however, no change in fluorescence intensity was observed using this approach (data not shown), suggesting that Pb2+ does not displace Ca2+ in the C-terminal domain sites, or may do so with slow kinetics. Based on these results, 10 µM samples of Ca2+-free CaM were pre-equilibrated with 20 µM Pb2+, assuming that all Pb2+ was binding to the high affinity C-terminal domain sites, followed by titration of Ca2+. The resulting data is still fit with Eq. 1, but the Kd value returned for Pb2+ is calculated by rearranging Eq. 2 into Eq. 3, and solving for Kdm1 based on the known Kd for Ca2+ (Kdm2), Kd from Eq. 1 which becomes Kapp, and the total, fixed concentration of Pb2+ pre-equilibrated with the protein [M1]T. Results indicated a Kd (Kdm1) of 0.67±0.06 µM (Table 1), which overlaps the standard deviation reported for results obtained by direct titration of Pb2+ (Fig. 1d).
3.2 Monitoring CaM binding with Pb2+ by NMR
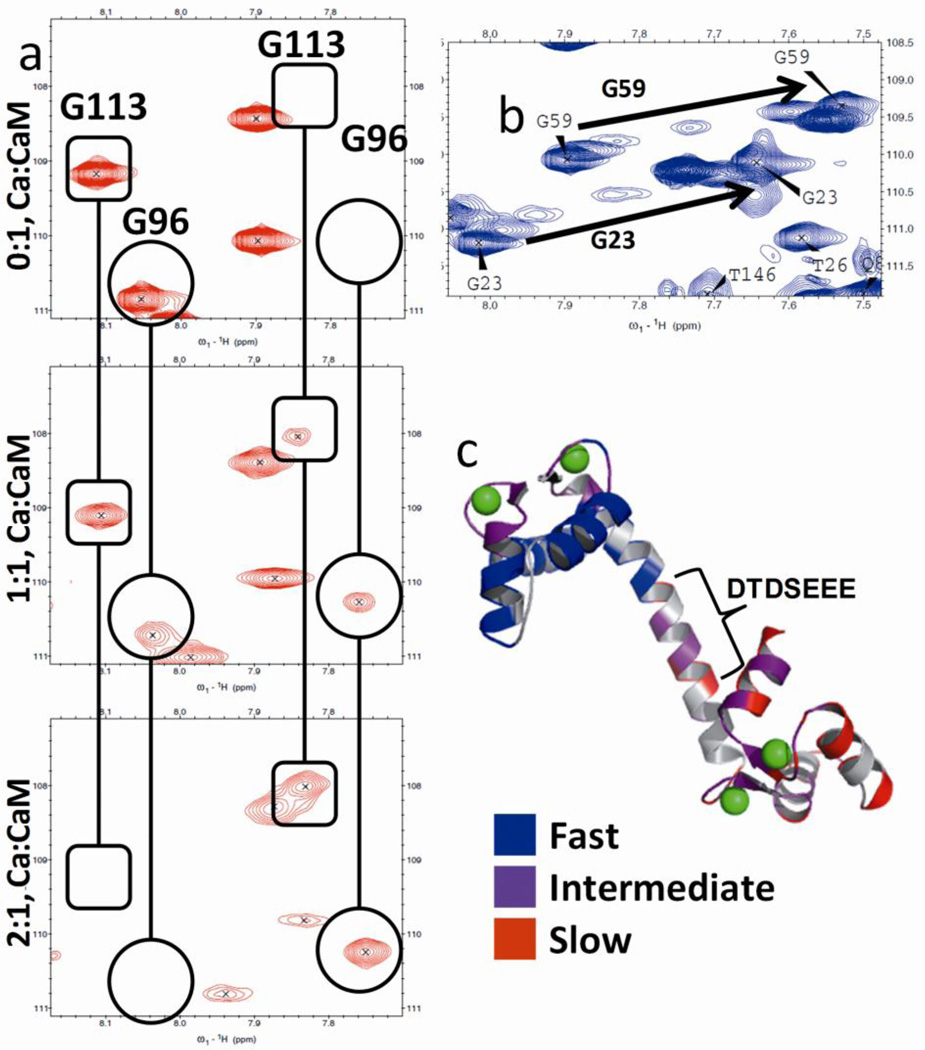
Analyses of spectra for the titration of Ca2+ to apo-CaM indicates a domain-specific pattern of chemical exchange for rat CaM, consistent with results published by Jaren et al. for paramecium CaM [33]. At low concentrations of Ca2+ we observe peak loss due to broadening associated with intermediate chemical exchange, and the appearance of new peaks as a result of slow chemical exchange (Fig. 2a). These changes in the spectra contrast sharply with fast chemical exchange as seen with G59 and G23 (Fig. 2b) indicating single, averaged peaks transient in the spectra in response to Ca2+-binding. In Fig. 2c we color-labeled the residues based on fast (blue), intermediate (purple) or slow (red) chemical exchange, which demonstrates that slow and intermediate exchange occur almost exclusively in the C-terminal domain, with fast exchange observed primarily in the N-terminal domain. A summary of residues and their associated chemical exchange can be found in Table S1.
Fig. 2.
(a) Slow chemical exchange for G113 (in rounded rectangle) and G96 (in circle). Both residues display a single peak at 0 MEs Ca2+, followed by the emergence of a second peak at 1 ME Ca2+. At 2 MEs Ca2+, the original peak in each pair is undetectable, leaving only the second peak. (b) Fast chemical exchange for G23 and G59. (c) CaM (3cln.pdb) with residues color-labeled to indicate fast (blue), intermediate (purple) and slow (red) chemical exchange. The carboxyl-rich central linker is highlighted in the bracket.
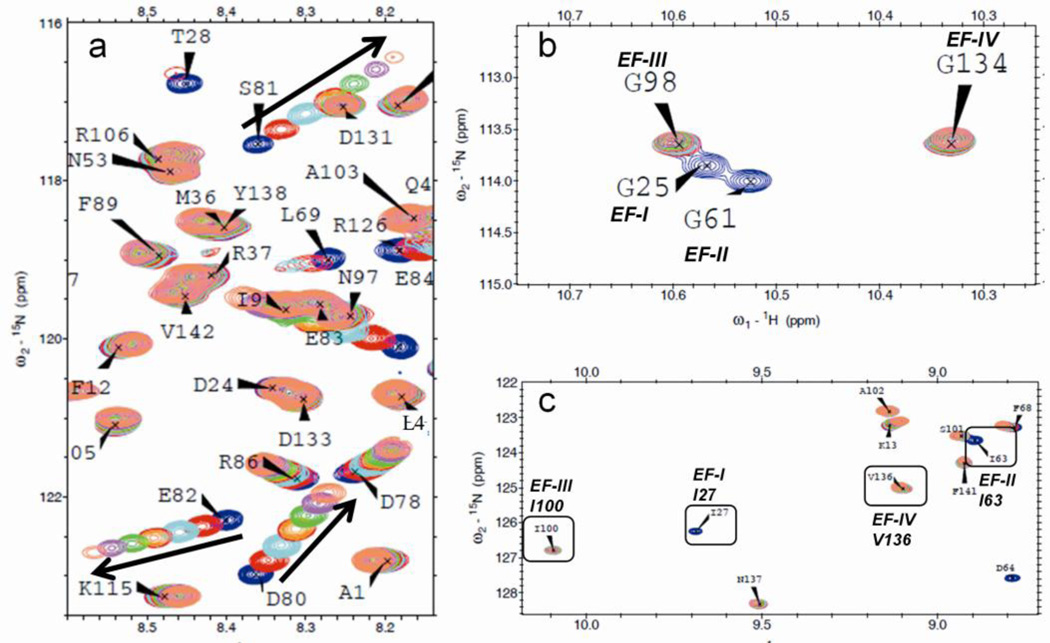
By comparing the spectra from the Ca2+-free to the Ca2+-loaded states, the magnitude of the absolute δ change (i.e., change in chemical shift across both the 1H and 15N dimensions from the initial δ values) reveals that the most significant changes occur for residues within the Ca2+-binding sites (Fig. 3a), while comparatively small changes are observed in the linker region. For the titration of Pb2+ to CaM, some loss of signal is apparent as a number of peaks observed in the Ca2+ spectrum fail to reappear following addition of Pb2+. However, from the peaks that are assigned, it is clear that the same trend is observed with addition of Pb2+, with the most significant changes observed in the canonical EF-Hand sites (Fig. 3b).
Fig. 3.
Weight-averaged chemical shift change (Δδ) in 15N HSQC spectra for titration of (a) Ca2+ and (b) Pb2+ complexed with CaM, and (c) Pb2+ added to Ca2+/CaM complex. The EF-Hand sites and the linker region in the sequence are highlighted in gray. Some loss of data is observed in (b) for addition of Pb2+, however, in both graphs the highest magnitude Δδ is clearly observed for residues within the four EF-Hand Ca2+-binding sites, with minimal change observed in the linker region. In (c) Pb2+ displaces Ca2+ in sites EF-I and EF-II, while the most significant structural changes occur in the linker.
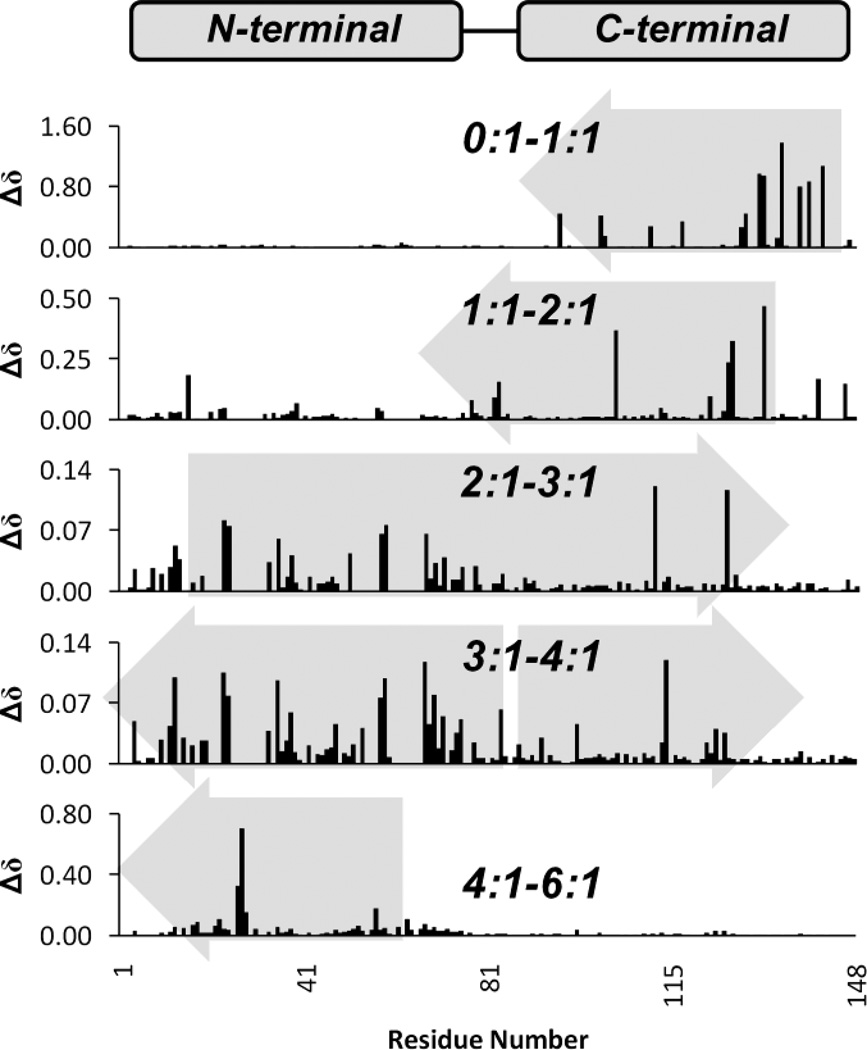
We can also establish a relative order of occupancy for Ca2+ by (1) comparing total Δδ across both dimensions for successive points in the titration (Fig. 4) or (2) plotting the order in which signals disappear relative to MEs of Ca2+ added (Fig. S2). Based on the relative magnitude of Δδ between points in the titration in Fig. 4, we can observe that Ca2+ first binds in the C-terminal domain followed by the N-terminal domain sites. Moreover, binding of Ca2+ in one domain is accompanied by structural changes in the opposite domain. Chemical shift changes from 0–2 MEs Ca2+ (Fig. 4), corresponding to binding in the C-terminal domain, also produce structural changes in the N-terminal domain.
Fig. 4.
Absolute changes in δ between successive points in the titration of Ca2+ to CaM, where changes are expressed in molar ratios of Ca2+:CaM (0:1–6:1). Viewed from top to bottom, Δδ values indicate binding of Ca2+ first in the C-terminal domain, followed by the N-terminal domain. Additionally, binding in one domain affects structural changes in the other. The gray arrows indicate direction of changes.
From 2–3 MEs Ca2+, more restructuring is seen in the N-terminal domain due to binding in either site EF-I or EF-II, but is still accompanied by changes in the C-terminal domain. From 3–4 MEs Ca2+, chemical shift changes indicate restructuring in both domains. The final, Ca2+-saturated state of the protein was not observed until the addition of 6 MEs of Ca2+, as determined by comparison with a reference spectrum obtained for 400 µM CaM in 20 mM Ca2+. Similarly, the disappearance of critical signals in each of the binding sites occurred in a domain-specific order (Fig. S2) with peaks (highlighted in gray) disappearing first in the C-terminal domain followed by the N-terminal domain.
For Pb2+, however, the order of occupancy could not be determined by analysis of Δδ which exhibited simultaneous changes in both domains (data not shown). However, from Fig. S2 we observe the most significant disappearance of peaks first in site EF-IV, followed by nearly-concurrent disappearance of peaks for residues in sites EF-I through EF-III. This is consistent with results of fluorescence analysis suggesting a single higher affinity Pb2+ site in the C-terminal domain with equivalent affinity for the three remaining sites.
The addition of Ca2+ sufficient to saturate CaM produces significant changes in the chemical shifts. Consistent with previous reports, we observed large changes in chemical shifts moving 4–8 ppm downfield in the spectra for I27, I63, I100 and V136 for Ca2+-loaded CaM that were proposed to be related to cooperative binding between the paired binding sites in each domain [53]. Such changes, however, were not observed for binding of Pb2+.
3.3 Titration of Pb2+ to Ca2+-loaded CaM
The titration of Pb2+ to CaM presaturated with 6 MEs of Ca2+ produced interesting variations in the chemical shifts not observed with direct addition of Pb2+ to apo-CaM. Overlaying the HSQC spectra revealed significant movement of signals for key residues in or adjacent to the trans-domain linker region, specifically residues D78, D80, S81, E82, E83 and R86 (Fig. 5a). Analysis of absolute Δδ values (Fig. 3c) indicate displacement of Ca2+ by Pb2+ only in sites EF-I and EF-II, but not sites EF-III and EF-IV, as seen for residues G25 and G61 in Fig. 5b, which disappear following the addition of 0.5 MEs of Pb2+. Also in Fig. 3c, the most significant structural changes occur in the linker region suggesting the presence of at least one additional binding site in this carboxyl-rich region of the sequence that is consistent with the coordination properties of Pb2+ binding [42].
Fig. 5.
(a) Movement of HSQC peaks for CaM bound with 6 MEs Ca2+ followed by titration of Pb2+ from 0–3 MEs in 0.5 ME increments. Residues in the C-terminal domain exhibit stable chemical shifts, while significant changes are observed for residues D78, D80, S81, E82, E83 and R86, which suggests a potential Pb2+-binding site in the linker region (74–82). (b) Residues in sites EF-I and EF-II, but not EF-III and EF-IV, disappear with addition of Pb2+ to Ca2+:CaM complex. (c) Similar results are observed for residues I27 and I63 occupying position 8 in the EF loop regions of sites EF-I and EF-II only.
Additionally, peaks for I27 and I63 disappear following addition of 0.5 MEs Pb2+, which may indicate loss of intradomain cooperativity (Fig. 5c). However, peaks for I100 and V136 remain visible in the spectrum. These results strongly indicate that Pb2+ displaces Ca2+ only in the N-terminal domain sites EF-I and EF-II.
3.4 CaM:Pb dynamics
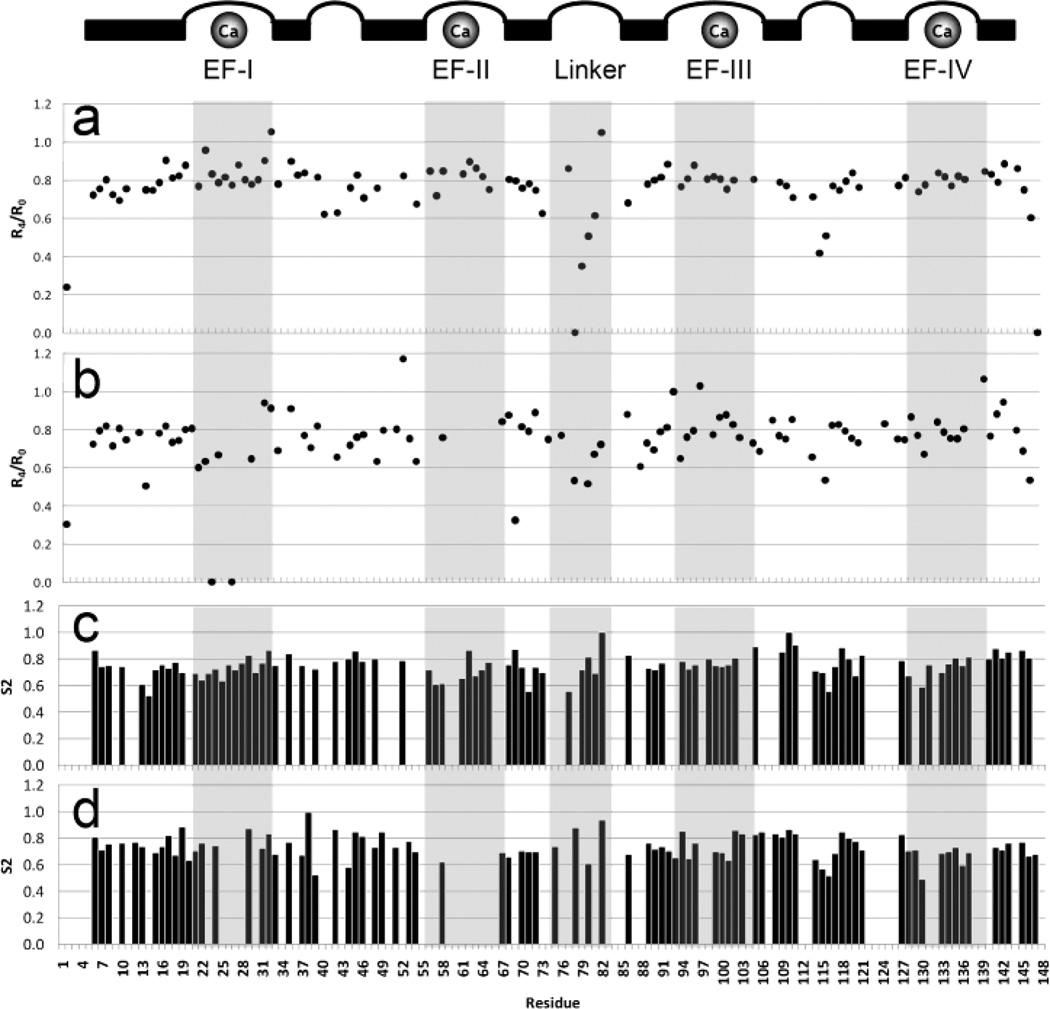
NOE data acquired for Ca2+-saturated CaM (Fig. 6a) followed the same trends reported previously by Barbato [54], with increasing flexibility (i.e., less ordered secondary structure) apparent in the end termini, the central helix, and the small loop region separating sites EF-III and EF-IV.
Fig. 6.
Comparison of NOE data for (a) Ca-CaM and (b) Ca-CaM with the addition of 2 MEs Pb2+. Helices, loop regions and Ca2+-binding sites are identified above the plots. Pb2+ appears to displace Ca2+ in the N-terminal domain but not the C-terminal domain, with additional binding in the linker region.
Comparison of our NOE data between Ca2+-loaded CaM in the absence (Fig. 6a) or presence (Fig. 6b) of 2 MEs Pb2+ suggests increased flexibility in sites EF-I and EF-II, but loss of flexibility in the linker region. Additionally, NOE values for residues in sites EF-III and EF-IV, while exhibiting more variance in the Pb2+-bound protein, do not indicate any significant change in these regions, further indicating that Pb2+ does not displace Ca2+ in these sites.
4 Discussion
In some instances, metal toxicity has been explained by ionic mimicry, the primary mechanism associated with several types of metal toxicity [12]. Since oxygen ligands from proteins for Ca2+ binding can also be used as ligand atoms for different toxic metal ions, it was speculated that the effects of these toxic metal ions are mainly due to displacement of Ca2+ in different CaBPs [20–26] including CaM, CaM-activated skeletal muscle troponin C (TnC) [55], PKC [26], and synaptotagmin [2]. However, there is no direct proof of the theory of ionic mimicry and the observed biphasic contradictory behavior of Pb2+ binding cannot be adequately addressed by this theory [38, 39, 56–59]. Additionally, the presence of secondary cationic binding sites suggests potential alternatives to ionic mimicry as a mechanism of toxicity. To understand the molecular mechanism for lead toxicity, previous studies using different experimental approaches reported that CaM and other Ca2+-binding proteins exhibit relatively higher binding affinities for Pb2+ compared with Ca2+. Fullmer et al., using radioisotopes of Ca2+ and Pb2+ and equilibrium dialysis to facilitate binding, reported that chick CaBP initially binds 3–4 MEs of Pb2+ in preference to Ca2+ with a Kd of 1.1 × 10−6 M, and 1–2 additional MEs of Pb2+ with a Kd of 7 × 10−4 M [21]. Similar higher affinity was reported for binding of Pb2+ to both CaM and troponin C, however, these results were based on determined binding stoichiometries of 4.7 and 4.9 (Pb:Protein) for CaM and troponin C, and were reported as relative affinities rather than calculated dissociation constants. These reported stoichiometries support the existence of an additional Pb2+-binding site for both CaM and troponin C which each have four Ca2+ binding sites. In another study, Aramini et al. [28] used 207Pb NMR to demonstrate binding of Pb2+ in the four EF-hand binding sites of CaM with higher relative affinity than Ca2+. Similarly, NMR studies using 15N labeled Gly residues in CaM by Ouyang and Vogel led them to report concurrent, high-affinity binding of Pb2+ in CaM’s four EF-hand sites [29], however, further site-specific binding information could not be provided by this study due to precipitation of the samples beyond the 2:1 Pb2+:CaM ratio, and dynamic properties were not reported. Nevertheless, these reported studies suggest that an alternative mechanism of toxicity beyond simple ionic displacement exists. Therefore, there is a need to fill in the gaps in our understanding of the molecular mechanism associated with molecular metal toxicity and the structural properties associated with their interactions with proteins.
4.1 Differential domain specific Ca2+ and Pb2+ binding affinities of CaM
The method of determining domain level binding affinities was used in our study and in previous studies because the cooperative binding of Ca2+ in the paired EF-hand sites of CaM precludes a precise determination of affinity in the individual sites. When individual affinity values have been reported, they were obtained either by mutations to deactivate one of the paired sites [60], calculated theoretical affinities based on thermodynamic models [61], or by probing site-specific binding by grafting EF-hand binding sites into a scaffold protein [62]. Alternatively, domain-specific affinity values may be obtained via changes in intrinsic fluorescence [48]. Using intrinsic fluorescence signals from Tyr and Phe, we were able to compare the metal binding affinities for Ca2+ and Pb2+ at the domain level, showing that CaM exhibits a higher relative affinity (~8-fold) for Pb2+ over Ca2+ in the N-terminal domain with a smaller comparative increase (~3-fold) in the C-terminal domain (Table 1). However, the binding affinity for Pb2+ in the N-terminal domain is only ~2-fold higher than that observed for Tyr Phase 1 binding in the C-terminal domain, and equivalent to the calculated Kd for tyrosine Phase II binding (Fig. 1c), which suggests a single, marginally-higher affinity binding site in the C-terminal domain, with the three remaining sites exhibiting approximately equivalent binding affinities. The biphasic response to Pb2+ in the C-terminal domain differs significantly from results reported for binding of Ca2+. We interpreted this biphasic response as two distinct binding events in the paired EF-hand sites, as this was consistent with our NMR data. However, it is also possible that binding of 2 Pb2+ ions is represented only by the initial fluorescence increase up to 2 MEs of Pb2+, and that the decrease observed in the second phase represents fluorescence quenching due to conformational changes induced by binding of Pb2+ in the central helix or elsewhere.
Similarly, analyses of HSQC data using 15N-labelled CaM revealed the disappearance of peaks for residues G61, G132 and G134 at 1 MEs Pb2+, followed by the disappearance of G23, G59, and G96 at 2 MEs Pb2+. Signals for residues G23, G61, and G96 subsequently reappeared in the spectra with increasing Pb2+ concentration (i.e., slow exchange), while signals for residues G25 and G98 remained visible across the spectra. These residues are important because they reside in the loop regions of the EF-hand sites (I through IV). In this study, the initial disappearance of residues G132 and G134 (Fig. S2) at one ME Pb2+ in Ca2+-free CaM suggests binding first in site EF-IV, followed by a concurrent distribution of Pb2+ across sites EF-I through EF-III, consistent with results acquired through fluorescence studies. Also consistent with our findings, Ouyang and Vogel, using 15N-labeled Gly, monitored binding of Pb2+ in the four EF-hand sites based on the disappearance of signals for G23 and G25 (EF-I), G59 and G61 (EF-II), G96 and G98 (EF-III) and G132 and G134 (EF-IV), for one and then two MEs of Pb2+ [29]. However, these studies were limited by sample precipitation reported beyond two MEs of Pb2+, and some of the minor differences reported by Ouyang and our study may be attributed to our use of a higher field strength NMR spectrometer and variations in our experimental approach. Additionally, our results provide quantitative data on binding constant values (Table 1).
4.2 Differential binding effect on EF-hand binding sites
Analysis of the HSQC spectra in this study provides a possible explanation for the similar binding affinities between Pb2+ and the EF-hand CaM Ca2+-binding sites, based on the disappearance of signals for residues I27 and I63 (Fig. 5c). These residues occupy position 8 in the EF-loop sequence. Previous NMR studies reported by Biekofsky et al. [53] indicated that Ca2+ binding with the loop position 7 ligand results in observed deshielding (+4 to +8 ppm) of the mainchain nitrogen in position 8 due to polarization of the O(7)=C(7)-N(8) amido group, which was used to monitor occupancy of Ca2+ in CaM, and provide evidence of cooperativity between the paired EF-Hand sites. It is possible that the disappearance of signals for these residues following addition of Pb2+ may indicate conformational changes resulting in disruption of cooperativity between paired binding sites, particularly in sites EF-I and EF-II. A structural basis for this conformational change may be observed in the crystal structures of Pb2+-bound CaM 1n0y.pdb and 2v01.pdb (Fig. S1e and Fig. S1f), where the T26 Oγ oxygen appears to rotate inward, placing it close enough (~3.5 Å) to the Pb2+ ion to serve as an active coordinating ligand in addition to the carbonyl oxygen utilized in binding of Ca2+ (Fig. S3).
Our results and others suggest only minimal deviation in binding affinities between Pb2+ and the four EF-hand Ca2+-binding sites in CaM, which would imply non-preferential occupancy between equivalent sites. Conversely, Ca2+ first occupies the higher affinity C-terminal domain EF-hand sites in CaM, followed by occupancy of the N-terminal domain sites [63]. Additionally, binding of Ca2+ in each domain involves cooperativity between the paired EF-Hand sites [64, 65] which is believed to be due to the formation of a short β-sheet between residues in position 8 of the paired EF-Loops joining EF-I with EF-II, and EF-III with EF-IV [66, 67]. Loss of cooperativity between the EF-Hand pairs, as suggested by structural differences between the two metals, would provide a plausible explanation for the similar affinities between Pb2+ and the four EF-Hand sites in CaM.
4.3 Differential structural and dynamic changes upon Ca2+ and Pb2+ binding
In this paper, we provide evidence to suggest a unique binding mode for Pb2+ contingent upon Ca2+-induced protein folding. As seen in Fig. 3c, the addition of Pb2+ to Ca2+-loaded CaM results in the disappearance of signals exclusively in sites EF-I (D22, G25 and I27) and EF-II (D56, A57, G61, I63, and E67). Conformational change due to binding is further revealed in the movement of signals in the spectra (Fig. 5), particularly in sites EF-I, EF-II, and the linker region, as plotted in Fig. 3c. These results are closely correlated with our analyses of NOE data which indicate that the addition of Pb2+ to Ca2+-loaded CaM apparently results in loss of flexibility in sites EF-I, EF-II and the linker region (Fig. 6), while residues in sites EF-III and EF-IV appear unperturbed. These data suggest that Pb2+ displaces Ca2+ only in the N-terminal domain sites EF-I and EF-II, and we can speculate that the positive cooperativity associated with Ca2+-binding between the paired sites EF-III and EF-IV [64, 65] in the C-terminal domain is sufficient to inhibit translocation of Pb2+ into the sites, while the 8-fold higher affinity of CaM for Pb2+ compared with Ca2+ in the N-terminal domain is sufficient for Pb2+ to displace Ca2+.
Furthermore, we observed significant δ change due to fast chemical exchange for residues in the linker with the addition of Pb2+ to the Ca2+-bound protein (Fig. 3c), but not with the addition of Pb2+ to Ca2+-free CaM. Moreover, Δδ values for residues in the linker exceed changes observed in sites EF-I and EF-II (Fig. 3c), which suggests that Pb2+ may bind opportunistically in this functionally-important region characterized by high electrostatic potential [42] due to a cluster of oxygen-rich sidechains (DTDSEEE) in positions 78–84. This argues for a unique binding mode observed only when CaM initially adopts an active Ca2+-induced conformer which prevents structural reconfiguration in the C-terminal domain as a consequence of ionic displacement. It should be noted that data fitting of chemical shift perturbations in the central helix (Fig. 5a) could not be performed to determine binding affinity, as the samples were found to precipitate before the endpoint of the titration was reached, reflecting a common problem associated with Pb2+ solubility also reported by Ouyang [29].
The unique Ca2+ potentiated binding mode for Pb2+ with CaM proposed in this paper is supported by several previous studies. Shirran and Barran reported that Pb2+ affinity for CaM increases relative to other divalent cations in the presence of Ca2+ [68]. Mills and Johnson reported that Pb2+ and other metals may bind to Ca2+-bound CaM in secondary sites forming an allosterically potentiated conformer [69], while Raos and Kasprzak suggested the existence of two secondary binding sites occupied by Ni2+ in the Ca2+-bound state [70]. Similarly, a study by Milos et al. [41] presents compelling evidence that Zn2+ interacts with CaM not through the four EF-Hand sites, but by binding in six secondary sites with nearly equivalent affinity. Milos further concluded, based on changes in enthalpy, that binding of Zn2+ and Mg2+ in these secondary sites allosterically antagonizes binding of Ca2+ in the EF-Hand sites, and vice-versa. Binding of Pb2+ in one or more of these secondary binding sites could produce the antagonistic effects described by Milos which could explain the biphasic tyrosine response to Pb2+ (Fig. 1c), Pb2+-induced global conformational changes, and the inhibition of CaM activity at higher concentrations of Pb2+.
It is worth noting that none of these cited studies identified the locations of these secondary sites in CaM, however, strong evidence exists to support our conclusion that at least one of these sites can be found in the central linker of CaM. Bertini et al. [40] reported a potential metal-binding site in the linker region of CaM based on the disappearance of key NMR signals (residues 78–81) following addition of 0.3 MEs of Yb3+, and Kursula and Majava [36] identified a Ca2+-binding site in the linker chelated by residues R74 and D78 in the crystal structure of Pb2+-bound to human CaM.
Binding of Pb2+ in this region is consistent with our previously published study indicating that Pb2+ can bind to carboxyl and hydroxyl groups in regions lacking defined binding geometries yet characterized by high electrostatic potential [42], such as the cluster of oxygen-rich sidechains (DTDSEEE) in positions 78–84.
It is also possible that the observed changes in the linker occur in response to binding in some region of the protein more distant from the linker. The Pb-CaM structure reported by Kursula and Majava [36] depicts binding of Pb2+ by carboxyl groups from D118 and D122, and significant chemical shift changes (>0.05δ) are observed in our data for residues T117 and R126 (Fig. 3c) as a result of Pb2+-binding. The residue sequence 117–123 (Fig. S1a) includes a group of carboxyl-rich sidechains (TDEEVDE) that could potentially bind Pb2+. However, unlike the proposed binding sites in the linker, these residues are all found in an α–helix, and unless the helix itself were to unwind, which was not indicated in the analysis of our dynamic NMR data, this explanation appears less likely as it is not clear how binding in this region would induce major conformational changes in the linker region.
4.4 Implication for activation of CaM
A study by Chao et al. reported that Pb2+ exhibited a biphasic effect on the amount of phosphate transferred from [γ-32P] ATP into MLCK, with stimulation observed at low concentrations followed by inhibition at higher concentrations [56]. Similarly, Habermann observed that Pb2+-bound CaM initially activates PDE with higher potency than Ca2+, but increasing Pb2+ concentration subsequently inhibited CaM-dependent phosphorylation [38].
From these functional assays and structural studies, it is likely that at low concentrations of Pb2+, Pb2+ occupies the Ca2+-binding sites in Ca2+-free CaM. The nearly-equivalent binding affinity of CaM for Pb2+ likely results in multiple complex conformers, one or more resembling the Ca2+/CaM complex in form and function [28, 29]. With increasing Pb2+ concentration, CaM eventually adopts a conformation which inhibits protein function.
Our proposed mechanism addresses the inhibitory effect of Pb2+ binding at higher concentrations as reported by both Chao [56] and Habermann [38]. The introduction of Pb2+ in this environment results in displacement of Ca2+ in sites EF-I and EF-II, coupled with opportunistic binding in the electronegative central helix which alters the conformation of this region, thus inhibiting the ability of CaM to bind other target proteins. Increasing Pb2+ concentration presumably produces a more compact or dynamically-restricted conformer incapable of binding properly with target ligand molecules as was observed with osteocalcin [71]. The loss of functional plasticity in the central helix of CaM interferes with its ability to interact with downstream proteins. Mutating the glutamate cluster of residues 82–84 (EEE) to lysines was found to abolish or greatly impair activation of NAD kinase and MLCK respectively, but had no effect on phosphodiesterase [72]. Conversely, deletion of residues 79–80 (DTD) abolished activation of phosphodiesterase but had no effect on MLCK [73, 74].
5 Conclusions
Our results demonstrated that Pb2+ has higher binding affinity than Ca2+ for both the N- and C-terminal domains. Binding of Pb2+ results in structural changes distinct from the Ca2+-bound state, and may disrupt the cooperativity observed between paired EF-hand sites during binding of Ca2+ [61, 63, 75]. Dynamic NMR studies suggested that Pb2+ displaces Ca2+ only in sites EF-I and EF-II, while the most significant chemical shift changes were observed in the carboxyl-rich linker region (residues 76–84). This provides strong evidence for opportunistic binding of Pb2+ outside of the known Ca2+-binding sites and an alternative mechanism for structural changes in the protein. This mechanism is consistent with the reported concentration-dependent, biphasic activation and inhibition associated with Pb2+-binding of CaM, and may account for the behavior of other toxic metals, such as lanthanides, observed to interact with CaM outside of the known Ca2+-binding sites. Finally, these results suggest that systemic effects of toxicity resulting from exposure to lead or other metals may be due to interactions with multiple molecular targets rather than simple ionic displacement.
Supplementary Material
Highlights.
Pb2+ displaces Ca2+ in the N-terminal, but not the C-terminal domain, of calmodulin.
Pb2+ binding affinity is only 3- to 8-fold higher than Ca2+ in Ca2+-binding sites.
Opportunistic binding of Pb2+ outside of Ca2+ sites alters protein conformation.
Pb2+ toxicity may be due to opportunistic binding, rather than by displacement.
Acknowledgements
The authors would like to thank Natalie White for her critical reading of this paper. Funding for this work was in part provided by NIH Grants EB007268 and GM081749 to JJY, Georgia Research Alliance (Funding to GSU for Varian Inova 600 NMR), and a GSU Molecular Basis of Disease (MBD) fellowship to MK.
Abbreviations
- ALAD
aminolevulinic acid dehydratase
- BAPTA
1,2-bis(o-aminophenoxy)ethane-N,N,N'-N'-tetraacetic acid
- CaBP
calcium binding protein
- CaM
calmodulin
- EGTA
ethylene glycol tetraacetic acid
- HSQC
Heteronuclear Single Quantum Coherence
- LB
lysogeny broth
- ME
Molar Equivalent
- MES
2-(N-morpholino)ethanesulfonic acid
- MLCK
Myosin light-chain kinase
- NTA
nitrilotriacetic acid
- NOE
Nuclear Overhauser Effect
- PDE
phosphodiesterase
- PKC
protein kinase C
- RNR
ribonucleotide reductase
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Bressler J, Kim K-a, Chakraborti T, Goldstein G. Neurochem. Res. 1999;vol. 24:595–600. doi: 10.1023/a:1022596115897. [DOI] [PubMed] [Google Scholar]
- 2.Bouton CM, Frelin LP, Forde CE, Arnold Godwin H, Pevsner J. J. Neurochem. 2001;vol. 76:1724–1735. doi: 10.1046/j.1471-4159.2001.00168.x. [DOI] [PubMed] [Google Scholar]
- 3.Chetty CS, Reddy GR, Murthy KS, Johnson J, Sajwan K, Desaiah D. Int. J. Toxicol. 2001;vol. 20:113–120. doi: 10.1080/109158101317097692. [DOI] [PubMed] [Google Scholar]
- 4.Moore MR, Goldberg A, Yeung-Laiwah AA. Ann. N. Y. Acad. Sci. 1987;vol. 514:191–203. doi: 10.1111/j.1749-6632.1987.tb48774.x. [DOI] [PubMed] [Google Scholar]
- 5.Aub JC, Reznikoff P. J. Exp. Med. 1924;vol. 40:189–208. doi: 10.1084/jem.40.2.189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nolan CV, Shaikh ZA. Toxicology. 1992;vol. 73:127–146. doi: 10.1016/0300-483x(92)90097-x. [DOI] [PubMed] [Google Scholar]
- 7.Khalil-Manesh F, Gonick HC, Weiler EW, Prins B, Weber MA, Purdy RE. Am. J. Hypertens. 1993;vol. 6:723–729. doi: 10.1093/ajh/6.9.723. [DOI] [PubMed] [Google Scholar]
- 8.Apostoli P, Bellini A, Porru S, Bisanti L. Am. J. Ind. Med. 2000;vol. 38:310–315. doi: 10.1002/1097-0274(200009)38:3<310::aid-ajim10>3.0.co;2-9. [DOI] [PubMed] [Google Scholar]
- 9.Ronis MJ, Badger TM, Shema SJ, Roberson PK, Templer L, Ringer D, Thomas PE. J. Toxicol. Environ. Health A. 1998;vol. 54:101–120. doi: 10.1080/009841098158944. [DOI] [PubMed] [Google Scholar]
- 10.Hernandez-Ochoa I, Garcia-Vargas G, Lopez-Carrillo L, Rubio-Andrade M, Moran-Martinez J, Cebrian ME, Quintanilla-Vega B. Reprod. Toxicol. 2005;vol. 20:221–228. doi: 10.1016/j.reprotox.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 11.Johnson FM. Mutat. Res. 1998;vol. 410:123–140. doi: 10.1016/s1383-5742(97)00032-x. [DOI] [PubMed] [Google Scholar]
- 12.Bridges CC, Zalups RK. Toxicol. Appl. Pharmacol. 2005;vol. 204:274–308. doi: 10.1016/j.taap.2004.09.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bitto E, Bingman CA, Wesenberg GE, McCoy JG, Phillips GN., Jr J. Biol. Chem. 2006;vol. 281:20521–20529. doi: 10.1074/jbc.M602000200. [DOI] [PubMed] [Google Scholar]
- 14.Kim Y, Yoo CI, Lee CR, Lee JH, Lee H, Kim SR, Chang SH, Lee WJ, Hwang CH, Lee YH. Ind. Health. 2002;vol. 40:23–27. doi: 10.2486/indhealth.40.23. [DOI] [PubMed] [Google Scholar]
- 15.Bergdahl IA, Grubb A, Schutz A, Desnick RJ, Wetmur JG, Sassa S, Skerfving S. Pharmacol. Toxicol. 1997;vol. 81:153–158. doi: 10.1111/j.1600-0773.1997.tb02061.x. [DOI] [PubMed] [Google Scholar]
- 16.Kelada SN, Shelton E, Kaufmann RB, Khoury MJ. Am. J. Epidemiol. 2001;vol. 154:1–13. doi: 10.1093/aje/154.1.1. [DOI] [PubMed] [Google Scholar]
- 17.Ballatori N. Environ. Health Perspect. 2002;vol. 110(Suppl 5):689–694. doi: 10.1289/ehp.02110s5689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Simons TJ, Pocock G. J. Neurochem. 1987;vol. 48:383–389. doi: 10.1111/j.1471-4159.1987.tb04105.x. [DOI] [PubMed] [Google Scholar]
- 19.Atchison WD. J. Bioenerg. Biomembr. 2003;vol. 35:507–532. doi: 10.1023/b:jobb.0000008023.11211.13. [DOI] [PubMed] [Google Scholar]
- 20.Dowd TL, Rosen JF, Gundberg CM, Gupta RK. Biochim. Biophys. Acta. 1994;vol. 1226:131–137. doi: 10.1016/0925-4439(94)90020-5. [DOI] [PubMed] [Google Scholar]
- 21.Fullmer CS, Edelstein S, Wasserman RH. J. Biol. Chem. 1985;vol. 260:6816–6819. [PubMed] [Google Scholar]
- 22.Goldstein GW, Ar D. Life Sci. 1983;vol. 33:1001–1006. doi: 10.1016/0024-3205(83)90757-9. [DOI] [PubMed] [Google Scholar]
- 23.Godwin HA. Curr. Opin. Chem. Biol. 2001;vol. 5:223–227. doi: 10.1016/s1367-5931(00)00194-0. [DOI] [PubMed] [Google Scholar]
- 24.Goering PL. Neurotoxicology. 1993;vol. 14:45–60. [PubMed] [Google Scholar]
- 25.Long GJ, Rosen JF, Schanne FA. J. Biol. Chem. 1994;vol. 269:834–837. [PubMed] [Google Scholar]
- 26.Markovac J, Goldstein GW. Nature. 1988;vol. 334:71–73. doi: 10.1038/334071a0. [DOI] [PubMed] [Google Scholar]
- 27.Cox JL, Harrison SD., Jr Biochem. Biophys. Res. Commun. 1983;vol. 115:106–111. doi: 10.1016/0006-291x(83)90975-0. [DOI] [PubMed] [Google Scholar]
- 28.Aramini JM, Hiraoki T, Yazawa M, Yuan T, Zhang M, Vogel HJ. J. Biol. Inorg. Chem. 1996;vol. 1:39–48. [Google Scholar]
- 29.Ouyang H, Vogel HJ. Biometals. 1998;vol. 11:213–222. doi: 10.1023/a:1009226215543. [DOI] [PubMed] [Google Scholar]
- 30.Kretsinger RH, Nockolds CE. J. Biol. Chem. 1973;vol. 248:3313–3326. [PubMed] [Google Scholar]
- 31.Chattopadhyaya R, Meador WE, Means AR, Quiocho FA. J. Mol. Biol. 1992;vol. 228:1177–1192. doi: 10.1016/0022-2836(92)90324-d. [DOI] [PubMed] [Google Scholar]
- 32.Wilson MA, Brunger AT. J. Mol. Biol. 2000;vol. 301:1237–1256. doi: 10.1006/jmbi.2000.4029. [DOI] [PubMed] [Google Scholar]
- 33.Jaren OR, Kranz JK, Sorensen BR, Wand AJ, Shea MA. Biochemistry. 2002;vol. 41:14158–14166. doi: 10.1021/bi026340+. [DOI] [PubMed] [Google Scholar]
- 34.Vogel HJ. Biochem. Cell. Biol. 1994;vol. 72:357–376. [PubMed] [Google Scholar]
- 35.Heidorn DB, Trewhella J. Biochemistry. 1988;vol. 27:909–915. doi: 10.1021/bi00403a011. [DOI] [PubMed] [Google Scholar]
- 36.Kursula P, Majava V. Acta Crystallogr. Sect. F Struct. Biol. Cryst. Commun. 2007;vol. 63:653–656. doi: 10.1107/S1744309107034525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wilson MA, Brunger AT. Acta Crystallogr. D Biol. Crystallogr. 2003;vol. 59:1782–1792. doi: 10.1107/s0907444903016846. [DOI] [PubMed] [Google Scholar]
- 38.Habermann E, Crowell K, Janicki P. Arch. Toxicol. 1983;vol. 54:61–70. doi: 10.1007/BF00277816. [DOI] [PubMed] [Google Scholar]
- 39.Chao SH, Bu CH, Cheung WY. Arch. Toxicol. 1995;vol. 69:197–203. doi: 10.1007/s002040050158. [DOI] [PubMed] [Google Scholar]
- 40.Bertini I, Gelis I, Katsaros N, Luchinat C, Provenzani A. Biochemistry. 2003;vol. 42:8011–8021. doi: 10.1021/bi034494z. [DOI] [PubMed] [Google Scholar]
- 41.Milos M, Comte M, Schaer JJ, Cox JA. J. Inorg. Biochem. 1989;vol. 36:11–25. doi: 10.1016/0162-0134(89)80009-1. [DOI] [PubMed] [Google Scholar]
- 42.Kirberger M, Yang JJ. J. Inorg. Biochem. 2008;vol. 102:1901–1909. doi: 10.1016/j.jinorgbio.2008.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhou Y, Yang W, Lurtz MM, Ye Y, Huang Y, Lee HW, Chen Y, Louis CF, Yang JJ. J. Biol. Chem. 2007;vol. 282:35005–35017. doi: 10.1074/jbc.M707728200. [DOI] [PubMed] [Google Scholar]
- 44.Wallace RW, Tallant EA, Cheung WY. Cold Spring Harb. Symp. Quant. Biol. 1982;vol. 46(Pt 2):893–901. doi: 10.1101/sqb.1982.046.01.083. [DOI] [PubMed] [Google Scholar]
- 45.Sorensen BR, Shea MA. Biochemistry. 1998;vol. 37:4244–4253. doi: 10.1021/bi9718200. [DOI] [PubMed] [Google Scholar]
- 46.VanScyoc WS, Sorensen BR, Rusinova E, Laws WR, Ross JB, Shea MA. Biophys. J. 2002;vol. 83:2767–2780. doi: 10.1016/S0006-3495(02)75286-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Yang JJ, Yang J, Wei L, Zurkiya O, Yang W, Li S, Zou J, Zhou Y, Maniccia AL, Mao H, Zhao F, Malchow R, Zhao S, Johnson J, Hu X, Krogstad E, Liu ZR. J. Am. Chem. Soc. 2008;vol. 130:9260–9267. doi: 10.1021/ja800736h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Jiang J, Zhou Y, Zou J, Chen Y, Patel P, Yang JJ, Balog EM. Biochem. J. 2010;vol. 432:89–99. doi: 10.1042/BJ20100505. [DOI] [PubMed] [Google Scholar]
- 49.Delaglio F, Grzesiek S, Vuister GW, Zhu G, Pfeifer J, Bax A. J. Biomol. NMR. 1995;vol. 6:277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 50.Kuboniwa H, Tjandra N, Grzesiek S, Ren H, Klee CB, Bax A. Nat. Struct. Biol. 1995;vol. 2:768–776. doi: 10.1038/nsb0995-768. [DOI] [PubMed] [Google Scholar]
- 51.Torizawa T, Shimizu M, Taoka M, Miyano H, Kainosho M. J. Biomol. NMR. 2004;vol. 30:311–325. doi: 10.1007/s10858-004-3534-2. [DOI] [PubMed] [Google Scholar]
- 52.Seifert MH, Georgescu J, Ksiazek D, Smialowski P, Rehm T, Steipe B, Holak TA. Biochemistry. 2003;vol. 42:2500–2512. doi: 10.1021/bi026481b. [DOI] [PubMed] [Google Scholar]
- 53.Biekofsky RR, Martin SR, Browne JP, Bayley PM, Feeney J. Biochemistry. 1998;vol. 37:7617–7629. doi: 10.1021/bi9800449. [DOI] [PubMed] [Google Scholar]
- 54.Barbato G, Ikura M, Kay LE, Pastor RW, Bax A. Biochemistry. 1992;vol. 31:5269–5278. doi: 10.1021/bi00138a005. [DOI] [PubMed] [Google Scholar]
- 55.Chao SH, Bu CH, Cheung WY. Arch. Toxicol. 1990;vol. 64:490–496. doi: 10.1007/BF01977632. [DOI] [PubMed] [Google Scholar]
- 56.Chao SH, Suzuki Y, Zysk JR, Cheung WY. Mol. Pharmacol. 1984;vol. 26:75–82. [PubMed] [Google Scholar]
- 57.Kern M, Wisniewski M, Cabell L, Audesirk G. Neurotoxicology. 2000;vol. 21:353–363. [PubMed] [Google Scholar]
- 58.Ferguson C, Kern M, Audesirk G. Neurotoxicology. 2000;vol. 21:365–378. [PubMed] [Google Scholar]
- 59.Suzuki Y, Chao SH, Zysk JR, Cheung WY. Arch. Toxicol. 1985;vol. 57:205–211. doi: 10.1007/BF00290889. [DOI] [PubMed] [Google Scholar]
- 60.Maune JF, Klee CB, Beckingham K. J. Biol. Chem. 1992;vol. 267:5286–5295. [PubMed] [Google Scholar]
- 61.Linse S, Helmersson A, Forsen S. J. Biol. Chem. 1991;vol. 266:8050–8054. [PubMed] [Google Scholar]
- 62.Ye Y, Lee HW, Yang W, Shealy S, Yang JJ. J. Am. Chem. Soc. 2005;vol. 127:3743–3750. doi: 10.1021/ja042786x. [DOI] [PubMed] [Google Scholar]
- 63.Ikura M, Hiraoki T, Hikichi K, Mikuni T, Yazawa M, Yagi K. Biochemistry. 1983;vol. 22:2573–2579. doi: 10.1021/bi00279a039. [DOI] [PubMed] [Google Scholar]
- 64.Forsen S, Linse S, Drakenberg T, Kordel J, Akke M, Sellers P, Johansson C, Thulin E, Andersson I, Brodin P, Grundström T, Skelton N, Chazin W. Ciba Found. Symp. 1991;vol. 161:222–236. doi: 10.1002/9780470514146.ch14. [DOI] [PubMed] [Google Scholar]
- 65.Hiraoki T, Vogel HJ. J. Cardiovasc. Pharmacol. 1987;vol. 10(Suppl 1):S14–S31. doi: 10.1097/00005344-198710001-00004. [DOI] [PubMed] [Google Scholar]
- 66.Biekofsky RR, Turjanski AG, Estrin DA, Feeney J, Pastore A. Biochemistry. 2004;vol. 43:6554–6564. doi: 10.1021/bi0497852. [DOI] [PubMed] [Google Scholar]
- 67.Ishida H, Takahashi K, Nakashima K, Kumaki Y, Nakata M, Hikichi K, Yazawa M. Biochemistry. 2000;vol. 39:13660–13668. doi: 10.1021/bi000582x. [DOI] [PubMed] [Google Scholar]
- 68.Shirran SL, Barran PE. J. Am. Soc. Mass. Spectrom. 2009;vol. 20:1159–1171. doi: 10.1016/j.jasms.2009.02.008. [DOI] [PubMed] [Google Scholar]
- 69.Mills JJ, S J. J. Biol. Chem. 1985;vol. 260:15100–15105. [PubMed] [Google Scholar]
- 70.Raos N, Kasprzak KS. Fundam. Appl. Toxicol. 1989;vol. 13:816–822. doi: 10.1016/0272-0590(89)90336-9. [DOI] [PubMed] [Google Scholar]
- 71.Dowd TL, Li L, Gundberg CM. Biochim. Biophys. Acta. 2008;vol. 1784:1534–1545. doi: 10.1016/j.bbapap.2008.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Craig TA, Watterson DM, Prendergast FG, Haiech J, Roberts DM. J. Biol. Chem. 1987;vol. 262:3278–3284. [PubMed] [Google Scholar]
- 73.Tabernero L, Taylor DA, Chandross RJ, VanBerkum MF, Means AR, Quiocho FA, Sack JS. Structure. 1997;vol. 5:613–622. doi: 10.1016/s0969-2126(97)00217-7. [DOI] [PubMed] [Google Scholar]
- 74.VanBerkum MF, George SE, Means AR. J. Biol. Chem. 1990;vol. 265:3750–3756. [PubMed] [Google Scholar]
- 75.Andersson A, Forsen S, Thulin E, Vogel HJ. Biochemistry. 1983;vol. 22:2309–2313. doi: 10.1021/bi00279a001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.