Abstract
Cell biology and microbiology are some of the oldest areas of scientific inquiry. Despite the depth of knowledge we now have in these respective fields, much remains unclear about how microorganisms interact with host intracellular organelles. Perhaps nowhere is this statement more accurate than in the role of peroxisomes in microbial infections. Peroxisomes were one of the first organelles discovered by Christian De Duve over 50 years ago (de Duve, 1982). These organelles are ubiquitously found in eukaryotic cells, where they serve several well-defined functions in lipid and oxygen homeostasis (Waterham and Wanders, 2012). This chapter will discuss the emerging evidence that indicates that in addition to their functions in cellular metabolism, peroxisomes play an important role in viral infections.
Keywords: Peroxisomes, Mitochondria, MAM, MAVS, RLRs, antiviral immunity
4.1 Peroxisomes and their interactions with intracellular pathogens
Peroxisomes interact functionally and morphologically with other organelles, such as the endoplasmic reticulum (ER), mitochondria and lipid droplets (Hettema and Motley, 2009; Schrader, 2006; Waterham and Wanders, 2012). All of these organelles are involved in interactions between the host cell and virus (Fig. 1). For example, poliovirus infection results in massive reorganization of intracellular membranes, mainly ER membranes, into vesicles that harbor replication complexes at their surface (Bienz et al., 1987). These vesicles are found in close proximity to remnants of the ER (Bienz et al., 1987) and the poliovirus viroporin 2B was shown to localize to the ER. Similarly, the polymerase (Protein A) of the insect pathogen flock house virus (FHV) associates with the mitochondrial outer membrane (Miller et al., 2001). This results in viral replication on mitochondria, which serve as important sites of innate immune signal transduction to fight viral infections (Seth et al., 2005). Finally, proteins derived from Hepatitis C virus (HCV), rotavirus or the C protein of Dengue virus (DENV) are located on lipid droplets (Cheung et al., 2010; Moradpour et al., 1996; Samsa et al., 2009). Thus, organelles that interact with peroxisomes play important roles in the lifecycles of diverse viruses.
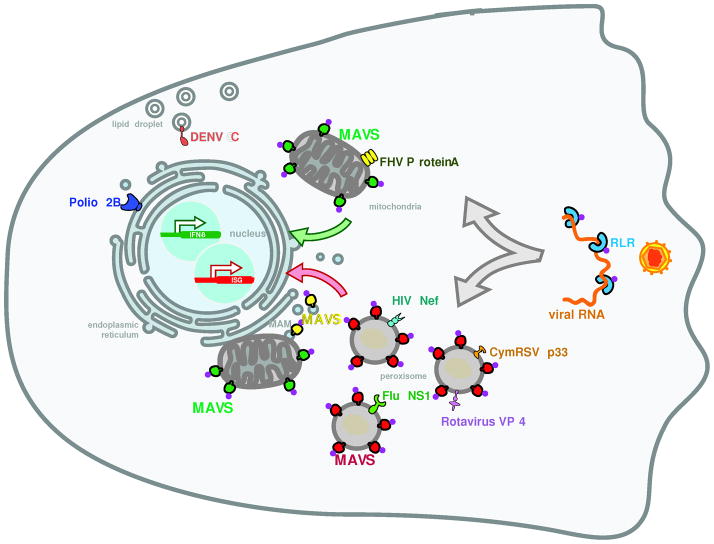
Figure 1. Localization of viral proteins and components of the antiviral innate immune machinery.
Proteins encoded by viruses localize to the organelles that interact with peroxisomes. Poliovirus 2B is localized on the endoplasmic reticulum, Flock House virus (FHV) protein A is found on mitochondria and Dengue virus (DENV) C protein is on lipid droplets. Peroxisomes are also a localization site of viral proteins: VP4 from Rotavirus, NS1 from Influenza (Flu), HIV Nef and Cymbidium ringspot virus (CymRSV) p33 are found on peroxisomes. The antiviral adaptor MAVS also localizes to mitochondria, peroxisomes and mitochondria-associated membranes (MAM). From these locations, MAVS transduces signals that originate from the recognition of viral RNA by RIG-I like receptors (RLR).
Peroxisomes themselves were shown to be sites of viral protein localization as well as assembly of replication complexes. A member of the replicase complex of some viruses of the tombusvirus family, p33, was shown to associate with plant peroxisomes, and viral replication was shown to occur on the peroxisomal membrane (McCartney et al., 2005; Panavas et al., 2005). A member of this virus family, Cymbidium ringspot virus (CymRSV) induces profound changes to peroxisome morphology, forming small vesicles in the periphery of peroxisomes (Russo et al., 1983). As the infection proceeds, these vesicles fill the peroxisomes, leading to the disappearance of the matrix. Strikingly, these intra-peroxisomal vesicles were shown to contain dsRNA, most probably replicative forms of viral RNA (Russo et al., 1983). In mammals, the Nef protein from HIV, the VP4 protein from rotavirus and the NS1 protein from influenza have been detected on peroxisomes (Cohen et al., 2000; Lazarow, 2011; Mohan et al., 2002). Bioinformatic approaches have identified several other viral proteins with putative peroxisomal targeting sequences (Mohan and Atreya, 2003), but most of these predicted targeting sequences have yet to be examined experimentally. Overall, these data indicate that like the organelles they interact with, peroxisomes interact with viral components. While the precise role of peroxisomes in the life cycle of most viruses is unclear, recent data on the innate immune responses that fight these infections suggests a critical role for these organelles in host defense.
4.2 Subcellular localization of mammalian sensors of microbial infection
The innate immune system of mammals is comprised of a series of structurally diverse but functionally related families of receptors that detect the presence of microorganisms (Akira et al., 2006). These receptor families are the Toll-like Receptors (TLRs), NOD-like Receptors (NLRs), RIG-I like Receptors (RLRs) and the C-type Lectin Receptors of the Dectin family (Brennan and Bowie, 2010; Goodridge et al., 2012). These receptors are classically called Pattern Recognition Receptors (PRRs), because they evolved to recognize Pathogen Associated Molecular Patterns (PAMPs) (Janeway, 1989), such as bacterial lipopolysaccharides, flagellin, lipoproteins and double-stranded RNA, among others. Microbial detection by PRRs leads to the activation of signal transduction pathways that activate several transcription factors (Medzhitov and Horng, 2009). These factors then induce major changes in the host transcriptional response, as hundreds of proinflammatory and immunomodulatory factors are expressed, including the cytokines interleukin-1, TNF and Type I and III interferons (IFNs) (Medzhitov and Horng, 2009).
Cell biological analyses of PRRs revealed that each receptors family surveys a distinct subcellular compartment for the presence of microbial products (Kagan, 2012). For example, the TLRs and the Dectin family are type I transmembrane proteins that contain an extracellular ligand-binding domain and an intracellular signaling domain (Akira et al., 2006). These receptors survey the extracellular and lumenal compartments of endosomes, and as such are located in these locations. In contrast, the NLRs and the RLRs do not contain transmembrane domains and are rather found in the cytosol (Fig. 1), where they survey this compartment for the presence of microbes (Kagan, 2012).
During a host-microbe encounter, pathogenic and non-pathogenic microbes will be found in the extracellular space. Therefore, PRRs located at the cell surface (TLRs and Dectins) have the ability to detect all microbes. Microbial pathogens encode sophisticated activities that manipulate the function of host cells, often enabling them to survive and replicate intracellularly (Vance et al., 2009). The outcome of these encounters will almost always results in interactions between the pathogen and components of the host cell cytosol (Vance et al., 2009). For example, bacteria encode secretion systems or secreted toxins that can deliver proteins to the cytosol (Cambronne and Roy, 2006). Likewise, a necessary step in the pathogenesis of all viruses is the delivery of their genetic material and proteins in the cytosol of host cells, where they can access the various metabolic activities necessary to complete their lifecycles. Therefore, since the cytosol is accessed only by pathogens, the PRRs located in the cytosol (RLRs and NLRs) can be considered legitimate pathogen detection receptors.
4.3 Activation of antiviral innate immune responses by RLRs
With the exception of the NLRs, all other PRR families enlist the aid of a transmembrane protein at the receptor-proximal level to activate innate immune responses upon microbial infection. In the case of the plasma membrane localized PRRs (TLRs and Dectins), the transmembrane proteins are the receptors themselves. By contrast, RLRs are cytoplasmic receptors. Upon viral infection, they engage a downstream transmembrane domain-containing adaptor protein to induce innate immune responses. This adaptor is called MAVS (also known as IPS-1, VISA or Cardif, Fig. 1) (Kawai et al., 2005; Meylan et al., 2005; Seth et al., 2005; Xu et al., 2005). MAVS contains a C-terminal “tail anchor” transmembrane domain that was originally identified to target this protein to the mitochondrial outer membrane (Seth et al., 2005). In resting cells, the best characterized RLR, RIG-I, is phosphorylated (Nistal-Villan et al., 2010). During an infection, RIG-I binds to viral RNA that displays one or more features, such as a short double-stranded region, a polyU-rich 3′ end, and a 5′ end with a triphosphate group (Kowalinski et al., 2011). RIG-I binding to viral RNA results in its dephosphorylation by unknown phosphatases and its ubiquitination by the E3 ubiquitin ligase TRIM25 (Gack et al., 2007; Nistal-Villan et al., 2010). A chaperone called 14-3-3ε is then able to engage this modified form of RIG-I and together they translocate to the MAVS adaptor on mitochondria (Liu et al., 2012). MAVS binding by active RIG-I results in the oligomerization of this adaptor into a prion-like state, the signaling competent form of this protein (Liu et al., 2012). Oligomerized MAVS then activates a series of signal transduction pathways that involve ubiquitin ligases (e.g. TRAF3 and TRAF6) and kinases (e.g. TBK1, IKKi) to activate NF-κB and IRF3 transcription factors (Liu et al., 2012). These transcription factors are responsible for upregulating the expression of numerous genes involved in antiviral immunity such as type I and III IFNs, chemokines and IFN-stimulated genes (ISGs) (Belgnaoui et al., 2011). The importance of MAVS localization to mitochondria for RLR signaling was originally revealed through the study of the HCV protease NS3/4a. When overexpressed in mammalian cells, NS3/4a cleaves MAVS near its transmembrane domain, resulting in its release from membranes (Li et al., 2005; Meylan et al., 2005). The release of MAVS into the cytosol renders the RLR signaling pathway inactive. Thus, membrane localization of MAVS is important for its signaling functions.
4.4 Role of peroxisomes in RLR signal transduction
Detailed cell biological analysis of the MAVS protein revealed a more complex integration of the RLR signaling pathway into the infrastructure of the cell. As discussed above, MAVS contains a C-terminal transmembrane domain that anchors it to the mitochondrial outer membrane. This type of localization motif is found in other outer membrane proteins of the mitochondria, such as Fis1 and Mff that control the dynamic changes that occur in mitochondrial morphology during cellular homeostasis (Gandre-Babbe and van der Bliek, 2008; Koch and Brocard, 2012). Subsequent work revealed that in addition to localizing to mitochondria, Fis1 and Mff are located on peroxisomes and that these proteins regulate the morphology of both organelles (Gandre-Babbe and van der Bliek, 2008; Koch and Brocard, 2012).
Based on the similarities between the localization domains of MAVS, Fis1 and Mff, we recently examined the localization of MAVS to peroxisomes (Dixit et al., 2010). MAVS is localized to peroxisomes in human hepatocytes, murine fibroblasts and macrophages (Fig. 1). Like the mitochondria, the localization of MAVS to peroxisomes is dependent on the C-terminal tail anchor transmembrane domain (Dixit et al., 2010). Using mutant alleles of MAVS that encode variants that target this protein to either mitochondria, peroxisomes or the cytosol, we found that RLR signaling can occur from multiple subcellular locations (Dixit et al., 2010). For example, during infection of fibroblasts with mammalian reovirus, RLR signaling through mitochondrial MAVS induces the expression of all the genes that have been identified as targets of this signaling pathway (e.g. type I IFNs, ISGs and chemokines), albeit with delayed kinetics when compared to wild-type MAVS. In contrast, peroxisomal MAVS induces chemokines and ISGs, but not Type I IFNs. Also in contrast to mitochondrial MAVS, peroxisomal MAVS induces antiviral gene expression with kinetics similar to that observed in cells expressing wild-type MAVS. As expected from prior studies on MAVS cleavage from membranes by the HCV NS3/4a protease (Li et al., 2005; Loo et al., 2006), MAVS engineered to localize to the cytosol is incapable of participating in RLR signaling (Dixit et al., 2010). Perhaps most notably, the signaling outputs from MAVS located in distinct subcellular compartments correlates with the cell’s ability to control viral infections. Fibroblasts expressing wild-type MAVS readily control the replication of vesicular stomatitis virus (VSV) while MAVS-deficient cells or cells expressing cytosolic MAVS cannot (Dixit et al., 2010). Cells expressing MAVS located on peroxisomes also control VSV replication, but not as efficiently as wild type MAVS. In contrast mitochondrial MAVS cannot control VSV replication, despite the fact that cells expressing mitochondrial MAVS induce IFNs and ISGs. These data suggest that the speed at which antiviral responses are induced during an infection is just as important as the type of response that is generated. Thus, the delay in IFN and ISG expression observed in cells expressing mitochondrial MAVS may be responsible for the inability of these cells to control infections.
4.5 Coordinating the role of peroxisomal and mitochondrial MAVS in RLR signal transduction around mitochondria-associated membranes
Recent work has revealed that MAVS is also located on a subdomain of the ER called mitochondria-associated membranes (MAM, Fig. 1) (Horner et al., 2011). The MAM are defined as contact sites between the ER and mitochondria, and are thought to play important roles in lipid metabolism and calcium signaling (Bononi et al., 2012; Vance and Shiao, 1996). A role for the MAM in activation of the inflammasome, a protein complex that regulates the secretion of a subset of inflammatory cytokines, has also been reported (Raturi and Simmen, 2012). Detailed subcellular fractionation studies revealed that in addition to peroxisomes and mitochondria, MAVS is located on the MAM (Horner et al., 2011). A striking observation was made by the Gale group. They found that within hepatocytes encoding HCV replicons, the viral NS4/4a protease preferentially cleaves MAVS from the MAM, as opposed to mitochondria (Horner et al., 2011). Whether NS3/4a cleaves MAVS from peroxisomes is unknown. Gale and colleagues also found that during viral infections of hepatocytes, peroxisomes and mitochondria interact with one another at the MAM (Horner et al., 2011). These contacts have been proposed to form an “innate immune signaling synapse”, which coordinates the signaling functions of MAVS on peroxisomes and mitochondria (Horner et al., 2011). These studies also revealed an intriguing link between the MAM and MAVS distribution on mitochondria and peroxisomes. Cells deficient for mitofusin-2, which cannot form mitochondria-ER contacts efficiently, exhibit higher levels of MAVS on peroxisomes than wild-type cells (Horner et al., 2011). These results suggest that MAVS is likely “cycling” between the MAM, mitochondria and peroxisomes. This cycling may be altered to enrich this protein in one or more compartments by altering the cell biological interactions between these organelles.
4.6 Perspectives on the future of peroxisome research
For many years, the study of basic cell biological processes has occurred separately from the study of host-pathogen interactions and innate immunity. Despite this statement, it is very clear that intracellular pathogens are excellent cell biologists. In fact, in several areas of research, our knowledge of given biological processes has been greatly influenced by the study of pathogens that manipulate said process (Mostowy and Cossart, 2009). As such, it is imperative that we continue to consider all aspects of the host-pathogen interaction when studies of pathogenesis are performed. Similarly, it is imperative that basic cell biologists consider infectious models of other disease models to better inform us of the importance of the process they are studying. An excellent example of this statement comes from the recent work (highlighted above) on the role of peroxisomes in antiviral immunity. For many years, peroxisomes were considered metabolic organelles, with no other cell function. Many other organelles (e.g. plasma membrane, endosomes, lysosomes, mitochondria, ER) were also first defined through their metabolic activities, but have since emerged as critical regulators of innate immune signal transduction. Peroxisomes therefore are not unique in their dual role in metabolism and infection. Rather, a common theme is emerging whereby most (perhaps all) organelles will have a metabolic and immune function. A challenge that now faces the community is understanding how the dynamics of peroxisome morphology and inter-organelle interactions influences the operation of the antiviral pathways that operates from these locations. Human patients with peroxisomal biogenesis disorders such as Zellweger Syndrome may prove useful in this regard, as would further characterization of cells deficient in mitochondria/peroxisome/MAM interactions. Additionally, although many viral proteins are predicted to be localized to peroxisomes, whether they are actually present on these organelles is unclear. If they are present, we need to determine the role of peroxisomes, and peroxisomal localization in the infectious cycle of these viruses. One possibility is that like the HCV protease NS3/4a, which cleaves and inactivates MAVS, some of these purported viral peroxisomal proteins may act to interfere with RLR signaling from this organelle. Finally, it remains unclear if the RLR network is the only signaling pathway that operates from peroxisomes. Other signaling pathways (involved in immunity or otherwise) may also operate from this location.
In summary, the years of work invested in understanding basic properties of peroxisomes and antiviral immunity has created an opportunity where these two areas can be integrated rapidly. The peroxisome may therefore emerge as the model of choice to understand the coordination of protein localization and signal transduction. This work may also have the wonderful outcome as revealing novel means of diagnosing and/or treating patients with peroxisomal disorders.
References
- Akira S, Uematsu S, Takeuchi O. Pathogen recognition and innate immunity. Cell. 2006;124:783–801. doi: 10.1016/j.cell.2006.02.015. [DOI] [PubMed] [Google Scholar]
- Belgnaoui SM, Paz S, Hiscott J. Orchestrating the interferon antiviral response through the mitochondrial antiviral signaling (MAVS) adapter. Curr Opin Immunol. 2011;23:564–572. doi: 10.1016/j.coi.2011.08.001. [DOI] [PubMed] [Google Scholar]
- Bienz K, Egger D, Pasamontes L. Association of polioviral proteins of the P2 genomic region with the viral replication complex and virus-induced membrane synthesis as visualized by electron microscopic immunocytochemistry and autoradiography. Virology. 1987;160:220–226. doi: 10.1016/0042-6822(87)90063-8. [DOI] [PubMed] [Google Scholar]
- Bononi A, Missiroli S, Poletti F, Suski JM, Agnoletto C, Bonora M, De Marchi E, Giorgi C, Marchi S, Patergnani S, et al. Mitochondria-associated membranes (MAMs) as hotspot Ca(2+) signaling units. Adv Exp Med Biol. 2012;740:411–437. doi: 10.1007/978-94-007-2888-2_17. [DOI] [PubMed] [Google Scholar]
- Brennan K, Bowie AG. Activation of host pattern recognition receptors by viruses. Curr Opin Microbiol. 2010;13:503–507. doi: 10.1016/j.mib.2010.05.007. [DOI] [PubMed] [Google Scholar]
- Cambronne ED, Roy CR. Recognition and delivery of effector proteins into eukaryotic cells by bacterial secretion systems. Traffic. 2006;7:929–939. doi: 10.1111/j.1600-0854.2006.00446.x. [DOI] [PubMed] [Google Scholar]
- Cheung W, Gill M, Esposito A, Kaminski CF, Courousse N, Chwetzoff S, Trugnan G, Keshavan N, Lever A, Desselberger U. Rotaviruses associate with cellular lipid droplet components to replicate in viroplasms, and compounds disrupting or blocking lipid droplets inhibit viroplasm formation and viral replication. J Virol. 2010;84:6782–6798. doi: 10.1128/JVI.01757-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen GB, Rangan VS, Chen BK, Smith S, Baltimore D. The human thioesterase II protein binds to a site on HIV-1 Nef critical for CD4 down-regulation. J Biol Chem. 2000;275:23097–23105. doi: 10.1074/jbc.M000536200. [DOI] [PubMed] [Google Scholar]
- de Duve C. Peroxisomes and related particles in historical perspective. Ann N Y Acad Sci. 1982;386:1–4. doi: 10.1111/j.1749-6632.1982.tb21402.x. [DOI] [PubMed] [Google Scholar]
- Dixit E, Boulant S, Zhang Y, Lee AS, Odendall C, Shum B, Hacohen N, Chen ZJ, Whelan SP, Fransen M, et al. Peroxisomes are signaling platforms for antiviral innate immunity. Cell. 2010;141:668–681. doi: 10.1016/j.cell.2010.04.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gack MU, Shin YC, Joo CH, Urano T, Liang C, Sun L, Takeuchi O, Akira S, Chen Z, Inoue S, Jung JU. TRIM25 RING-finger E3 ubiquitin ligase is essential for RIG-I-mediated antiviral activity. Nature. 2007;446:916–920. doi: 10.1038/nature05732. [DOI] [PubMed] [Google Scholar]
- Gandre-Babbe S, van der Bliek AM. The novel tail-anchored membrane protein Mff controls mitochondrial and peroxisomal fission in mammalian cells. Mol Biol Cell. 2008;19:2402–2412. doi: 10.1091/mbc.E07-12-1287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goodridge HS, Underhill DM, Touret N. Mechanisms of fc receptor and dectin-1 activation for phagocytosis. Traffic. 2012;13:1062–1071. doi: 10.1111/j.1600-0854.2012.01382.x. [DOI] [PubMed] [Google Scholar]
- Hettema EH, Motley AM. How peroxisomes multiply. J Cell Sci. 2009;122:2331–2336. doi: 10.1242/jcs.034363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horner SM, Liu HM, Park HS, Briley J, Gale M., Jr Mitochondrial-associated endoplasmic reticulum membranes (MAM) form innate immune synapses and are targeted by hepatitis C virus. Proc Natl Acad Sci U S A. 2011;108:14590–14595. doi: 10.1073/pnas.1110133108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Janeway CA., Jr Approaching the asymptote? Evolution and revolution in immunology. Cold Spring Harb Symp Quant Biol. 1989;54(Pt 1):1–13. doi: 10.1101/sqb.1989.054.01.003. [DOI] [PubMed] [Google Scholar]
- Kagan JC. Defining the subcellular sites of innate immune signal transduction. Trends Immunol. 2012;33:442–448. doi: 10.1016/j.it.2012.06.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kawai T, Takahashi K, Sato S, Coban C, Kumar H, Kato H, Ishii KJ, Takeuchi O, Akira S. IPS-1, an adaptor triggering RIG-I- and Mda5-mediated type I interferon induction. Nat Immunol. 2005;6:981–988. doi: 10.1038/ni1243. [DOI] [PubMed] [Google Scholar]
- Koch J, Brocard C. PEX11 proteins attract Mff and human Fis1 to coordinate peroxisomal fission. J Cell Sci. 2012;125:3813–3826. doi: 10.1242/jcs.102178. [DOI] [PubMed] [Google Scholar]
- Kowalinski E, Lunardi T, McCarthy AA, Louber J, Brunel J, Grigorov B, Gerlier D, Cusack S. Structural basis for the activation of innate immune pattern-recognition receptor RIG-I by viral RNA. Cell. 2011;147:423–435. doi: 10.1016/j.cell.2011.09.039. [DOI] [PubMed] [Google Scholar]
- Lazarow PB. Viruses exploiting peroxisomes. Curr Opin Microbiol. 2011;14:458–469. doi: 10.1016/j.mib.2011.07.009. [DOI] [PubMed] [Google Scholar]
- Li XD, Sun L, Seth RB, Pineda G, Chen ZJ. Hepatitis C virus protease NS3/4A cleaves mitochondrial antiviral signaling protein off the mitochondria to evade innate immunity. Proc Natl Acad Sci U S A. 2005;102:17717–17722. doi: 10.1073/pnas.0508531102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HM, Loo YM, Horner SM, Zornetzer GA, Katze MG, Gale M., Jr The mitochondrial targeting chaperone 14-3-3epsilon regulates a RIG-I translocon that mediates membrane association and innate antiviral immunity. Cell Host Microbe. 2012;11:528–537. doi: 10.1016/j.chom.2012.04.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo YM, Owen DM, Li K, Erickson AK, Johnson CL, Fish PM, Carney DS, Wang T, Ishida H, Yoneyama M, et al. Viral and therapeutic control of IFN-beta promoter stimulator 1 during hepatitis C virus infection. Proc Natl Acad Sci U S A. 2006;103:6001–6006. doi: 10.1073/pnas.0601523103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCartney AW, Greenwood JS, Fabian MR, White KA, Mullen RT. Localization of the tomato bushy stunt virus replication protein p33 reveals a peroxisome-to-endoplasmic reticulum sorting pathway. Plant Cell. 2005;17:3513–3531. doi: 10.1105/tpc.105.036350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Medzhitov R, Horng T. Transcriptional control of the inflammatory response. Nat Rev Immunol. 2009;9:692–703. doi: 10.1038/nri2634. [DOI] [PubMed] [Google Scholar]
- Meylan E, Curran J, Hofmann K, Moradpour D, Binder M, Bartenschlager R, Tschopp J. Cardif is an adaptor protein in the RIG-I antiviral pathway and is targeted by hepatitis C virus. Nature. 2005;437:1167–1172. doi: 10.1038/nature04193. [DOI] [PubMed] [Google Scholar]
- Miller DJ, Schwartz MD, Ahlquist P. Flock house virus RNA replicates on outer mitochondrial membranes in Drosophila cells. J Virol. 2001;75:11664–11676. doi: 10.1128/JVI.75.23.11664-11676.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan KV, Atreya CD. Novel organelle-targeting signals in viral proteins. Bioinformatics. 2003;19:10–13. doi: 10.1093/bioinformatics/19.1.10. [DOI] [PubMed] [Google Scholar]
- Mohan KV, Som I, Atreya CD. Identification of a type 1 peroxisomal targeting signal in a viral protein and demonstration of its targeting to the organelle. J Virol. 2002;76:2543–2547. doi: 10.1128/jvi.76.5.2543-2547.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moradpour D, Englert C, Wakita T, Wands JR. Characterization of cell lines allowing tightly regulated expression of hepatitis C virus core protein. Virology. 1996;222:51–63. doi: 10.1006/viro.1996.0397. [DOI] [PubMed] [Google Scholar]
- Mostowy S, Cossart P. From pathogenesis to cell biology and back. Cell Host Microbe. 2009;5:510–513. doi: 10.1016/j.chom.2009.06.002. [DOI] [PubMed] [Google Scholar]
- Nistal-Villan E, Gack MU, Martinez-Delgado G, Maharaj NP, Inn KS, Yang H, Wang R, Aggarwal AK, Jung JU, Garcia-Sastre A. Negative role of RIG-I serine 8 phosphorylation in the regulation of interferon-beta production. J Biol Chem. 2010;285:20252–20261. doi: 10.1074/jbc.M109.089912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panavas T, Hawkins CM, Panaviene Z, Nagy PD. The role of the p33:p33/p92 interaction domain in RNA replication and intracellular localization of p33 and p92 proteins of Cucumber necrosis tombusvirus. Virology. 2005;338:81–95. doi: 10.1016/j.virol.2005.04.025. [DOI] [PubMed] [Google Scholar]
- Raturi A, Simmen T. Where the endoplasmic reticulum and the mitochondrion tie the knot: The mitochondria-associated membrane (MAM) Biochim Biophys Acta. 2012 doi: 10.1016/j.bbamcr.2012.04.013. [DOI] [PubMed] [Google Scholar]
- Russo M, Di Franco A, Martelli GP. The fine structure of Cymbidium ringspot virus infections in host tissues. III. Role of peroxisomes in the genesis of multivesicular bodies. J Ultrastruct Res. 1983;82:52–63. doi: 10.1016/s0022-5320(83)90096-5. [DOI] [PubMed] [Google Scholar]
- Samsa MM, Mondotte JA, Iglesias NG, Assuncao-Miranda I, Barbosa-Lima G, Da Poian AT, Bozza PT, Gamarnik AV. Dengue virus capsid protein usurps lipid droplets for viral particle formation. PLoS Pathog. 2009;5:e1000632. doi: 10.1371/journal.ppat.1000632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schrader M. Shared components of mitochondrial and peroxisomal division. Biochim Biophys Acta. 2006;1763:531–541. doi: 10.1016/j.bbamcr.2006.01.004. [DOI] [PubMed] [Google Scholar]
- Seth RB, Sun L, Ea CK, Chen ZJ. Identification and characterization of MAVS, a mitochondrial antiviral signaling protein that activates NF-kappaB and IRF 3. Cell. 2005;122:669–682. doi: 10.1016/j.cell.2005.08.012. [DOI] [PubMed] [Google Scholar]
- Vance JE, Shiao YJ. Intracellular trafficking of phospholipids: import of phosphatidylserine into mitochondria. Anticancer Res. 1996;16:1333–1339. [PubMed] [Google Scholar]
- Vance RE, Isberg RR, Portnoy DA. Patterns of pathogenesis: discrimination of pathogenic and nonpathogenic microbes by the innate immune system. Cell Host Microbe. 2009;6:10–21. doi: 10.1016/j.chom.2009.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterham HR, Wanders RJ. Metabolic functions and biogenesis of peroxisomes in health and disease. Biochim Biophys Acta. 2012;1822:1325. doi: 10.1016/j.bbadis.2012.06.001. [DOI] [PubMed] [Google Scholar]
- Xu LG, Wang YY, Han KJ, Li LY, Zhai Z, Shu HB. VISA is an adapter protein required for virus-triggered IFN-beta signaling. Mol Cell. 2005;19:727–740. doi: 10.1016/j.molcel.2005.08.014. [DOI] [PubMed] [Google Scholar]