Abstract
Centrosomes are microtubule-organizing centres of animal cells. They influence the morphology of the microtubule cytoskeleton, function as the base for the primary cilium and serve as a nexus for important signalling pathways. At the core of a typical centrosome are two cylindrical microtubule-based structures termed centrioles, which recruit a matrix of associated pericentriolar material. Cells begin the cell cycle with exactly one centrosome, and the duplication of centrioles is constrained such that it occurs only once per cell cycle and at a specific site in the cell. As a result of this duplication mechanism, the two centrioles differ in age and maturity, and thus have different functions; for example, the older of the two centrioles can initiate the formation of a ciliary axoneme. We discuss spatial aspects of the centrosome duplication cycle, the mechanism of centriole assembly and the possible consequences of the inherent asymmetry of centrioles and centrosomes.
Centrosomes and associated components determine the geometry of microtubule arrays throughout the cell cycle, and thus influence cell shape, polarity and motility, as well as spindle formation, chromosome segregation and cell division1. Importantly, centrioles also function as basal bodies for the formation of cilia and flagella. These in turn play important roles in locomotion, transport and signalling2. Phylogenetic studies indicate that centrioles/basal bodies existed in the last common ancestor of eukaryotes but were lost from specific branches, such as yeasts and vascular plants3. Their presence correlates strictly with the occurrence of cilia, indicating that selective pressure was exerted on basal body functionality. Aberrations in centriole/basal body formation and function are associated with a plethora of human diseases, including ciliopathies, brain diseases and cancer. Accordingly, recent years have seen a surge of interest in the biogenesis and function of these elaborate intracellular structures, as reflected in the number of excellent and comprehensive reviews now published4–8. Here we focus on recent advances and a selection of seminal papers that bear on centriole biogenesis, duplication, function and association with cellular asymmetries.
Centriole biogenesis and the control of centriole number
In cycling cells, exactly one new centriole forms adjacent to each pre-existing centriole, reminiscent of the replication of DNA. In contrast, in differentiating multiciliated epithelial cells, hundreds of centrioles are formed near-simultaneously adjacent to deuterosomes, amorphous proteinaceous structures unique to this cell type. Importantly, it is now recognized that ‘de novo’ formation of centrioles occurs more commonly than previously thought9 and even cycling cells display this ability if the resident centrioles are experimentally removed10. Moreover, recent studies demonstrate that the pathways underlying centriole/basal body formation in eukaryotes share a common set of key regulatory proteins, indicating that they represent variations on a common theme6. One intriguing corollary of this view is that the de novo formation of centrioles is possible in most if not all cell types, unless it is actively suppressed by pre-existing centrioles. In this context, it will be interesting to clarify the role of pericentriolar material (PCM) components associated with those centrioles in the spatial and numerical control of centriole assembly11–13.
Although proteomic analyses of human centrosomes have revealed hundreds of proteins and considerable complexity14,15, genetic and RNAi screens in Caenorhabditis elegans identified just five gene products as strictly required for centriole formation16. These key components have been conserved during evolution and, although additional proteins important for centriole formation have been identified in other species17,18, the core machinery for centriole biogenesis seems to rely on a surprisingly small number of proteins6. Prominent among these are the protein kinase PLK4 (also known as SAK in Drosophila; in C. elegans ZYG-1 is a functional orthologue of PLK4, but is structurally distinct) and the coiled-coil protein SAS-6 (ref. 13). Levels of these proteins are critical for centriole assembly in both invertebrates and vertebrates18–22.
A priori, the maintenance of constant centriole numbers in proliferating cells requires two types of controls: first, every centriole must duplicate once and only once in every cell cycle (cell cycle control) and, second, exactly one new centriole must form next to every pre-existing centriole (copy number control). To what extent the two control mechanisms rely on common molecular components remains to be determined. As summarized in Fig. 1, centriole duplication begins at the G1 to S transition of the cell cycle with the formation of one procentriole adjacent to each pre-existing parental centriole. This event is under the control of PLK4 and one recent study identifies the F-box protein FBXW5 as one critical substrate of this kinase22. The same study shows that SAS-6 is a substrate of the SCF–FBXW5 E3-ubiquitin ligase, leading to an attractive model according to which phosphorylation of FBXW5 by PLK4 results in the stabilization of SAS-6, thus triggering centriole duplication22. However, it is unclear whether this mechanism is relevant to embryonic cells, which also initiate centriole duplication only once per cycle, but where the abundance of centriole components is not limiting. In C. elegans, SAS-6 itself has been identified as a substrate of the putative PLK4 functional orthologue ZYG-1 (ref. 23), but whether human SAS-6 is a direct substrate of PLK4 is unknown. How procentriole formation is normally restricted to exactly one new copy remains to be understood. One critical parameter is the activity of PLK4 itself, as excess PLK4 triggers the formation of supernumerary centrioles, whereas its depletion causes a reduction in centriole number24,25. Levels of PLK4 are tightly controlled through SCFβTrCP/ubiquitin-dependent proteolysis, which in turn depends on PLK4 autophosphorylation and perhaps the action of other kinases26–30. In addition, there is evidence that a type 2 phosphatase31,32 and the amount of PCM surrounding the parental centrioles limits the local availability of essential centriolar components12,33.
Figure 1.
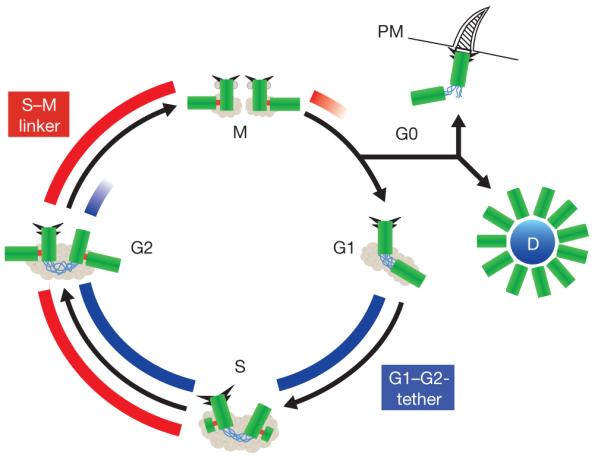
Centriole biogenesis. This schematic representation of the centriole duplication cycle shows centrioles (green) and PCM (grey), with emphasis on two distinct linker structures. The G1–G2 tether (GGT; blue) connects the proximal ends of the two parental centrioles from G1 to late G2; it is important to ensure microtubule nucleation from a single microtubule organizing centre. The S–M Linker (SML; red) forms during S phase and connects the proximal end of the nascent procentriole to the lateral surface of the mother centriole. The removal of this tight connection in late M phase (disengagement) is an important element of cell cycle control of centriole duplication. Both the molecular components of the GGT and SML as well as the regulation of the formation and dissolution of these structures are expected to be distinct, although some PCM components are likely to be important for both GGT and SML. Also depicted are subdistal and distal appendages (triangles); although readily visible in electron micrographs during interphase, these appendages are difficult to visualize during M phase. In quiescent cells, the appendage-bearing centriole associates with the plasma membrane (PM) and acts as a basal body to form a primary cilium. Finally, in multi-ciliated epithelial cells, multiple centrioles form simultaneously from an amorphous structure termed the deuterosome (D).
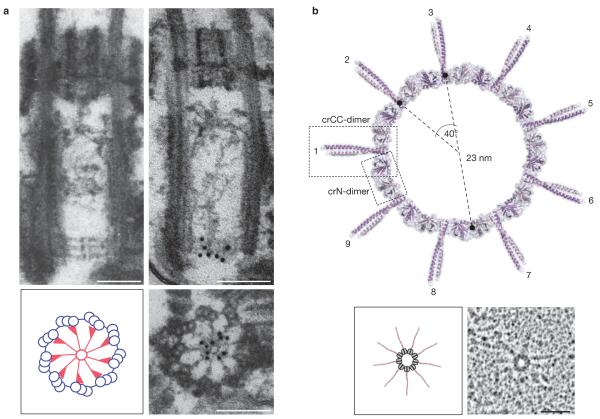
At a morphological level, the initial formation of procentrioles is characterized by the emergence of a cartwheel-shaped structure with ninefold symmetry6 or, in C. elegans, a central tube structure34. Recent studies identify the conserved coiled-coil protein SAS-6 as critical for cartwheel formation. First, in Chlamydomonas, SAS-6 was localized by immuno-electron microscopy to the central part of the cartwheel (Fig. 2a) and SAS-6 null mutants (bld12 mutants) lack this central hub35. Likewise, SAS-6 is important for ninefold symmetry in Drosophila19,36. Second, tubules highly reminiscent of the cartwheel hub could be assembled in Drosophila spermatocytes by the co-expression of SAS-6 with its binding partner ANA2 (ref. 37). Additional co-expression with PLK4 resulted not only in centriole overduplication and enhanced recruitment of SAS-6–ANA2 to the proximal ends of parental centrioles, but also, surprisingly, to increased centriole cohesion37. Third, analysis of the SAS-6 structure by X-ray crystallography revealed that this protein itself displays self-assembly properties that can readily explain the formation of a central cartwheel ring with nine emanating spokes38,39 (Fig. 2b). Taken together, these studies provide an appealing explanation for the evolutionary conservation of centriole structure and its ninefold symmetry and thus represent a major breakthrough in our understanding of centriole biogenesis. In future, it will be interesting to determine how other key centriole proteins, notably CEP135 (putative homologue of Bld10p), and SAS-5, ANA2 and STIL (putative partners of SAS-6), contribute to the assembly and stabilization of the cartwheel structure.
Figure 2.
Identification of SAS-6 as a key element of the centriolar cartwheel. (a) Immunolocalization of the SAS-6 protein (also known as Bld12p) to the central hub of the cartwheel in Chlamydomonas reinhardtii imaged by electron microscopy. Top images show longitudinal sections through wild-type centrioles; note the immunogold-labelling of the carthwheel by anti-SAS-6 antibodies (right). Bottom; immunogold-labelling of centriole in cross-section, showing that SAS-6 localizes to the central part of the cartwheel (right). Schematic representation (left) shows cartwheel in red. Scale bars,100 nm. Reproduced with permission from ref. 35. (b). Structural model of a SAS-6 oligomer (upper panel) and rotary-metalshadowing electron micrographs of the same structure (lower right panel; schematic representation of structure is shown on the left). Both images emphasize the importance of SAS-6 for conferring ninefold rotational symmetry to the centriole. (Reproduced with permission from ref. 38).
Once formed, procentrioles elongate throughout S and G2 phase. This elongation process depends on several proteins, including SAS-4 (also known as CPAP or CENPJ)40–42, POC5 (ref. 43), OFD1 (ref. 44) and CP110 (refs 41,45). Although SAS-4 promotes centriole elongation through the deposition of centriolar microtubules, CP110 localizes to the distal ends of growing centrioles and possibly functions as a cap to limit microtubule extension. Consistent with this interpretation, CP110, along with binding partners CEP76 and CEP290, suppresses ciliogenesis46. Interestingly, the abundance of CP110 is controlled by SCFCyclinF/ubiquitin-dependent proteolysis47, strengthening the evidence that proteolysis makes an important contribution to centriole biogenesis and homeostasis22,26,29,48. After completion of one-and-a-half cell cycles since its birth, a centriole reaches full maturity through the acquisition of distal and subdistal appendages (Figs 1 and 3). Several mammalian appendage proteins have been characterized and the available evidence indicates that these structures are important for microtubule anchoring49 and, as described below, ciliogenesis. Mature centrioles also have associated PCM; much remains to be learned about the assembly and role of individual PCM components, many of which are large coiled-coil proteins14. Some, like CEP152 (asterless in Drosophila50–52) and CEP192 (also known as SPD-2 in C. elegans) are tightly associated with centrioles and implicated in the recruitment of centriole-duplication factors. Others, like CEP215 (CNN in Drosophila), function in PCM recruitment and assembly53.
Figure 3.
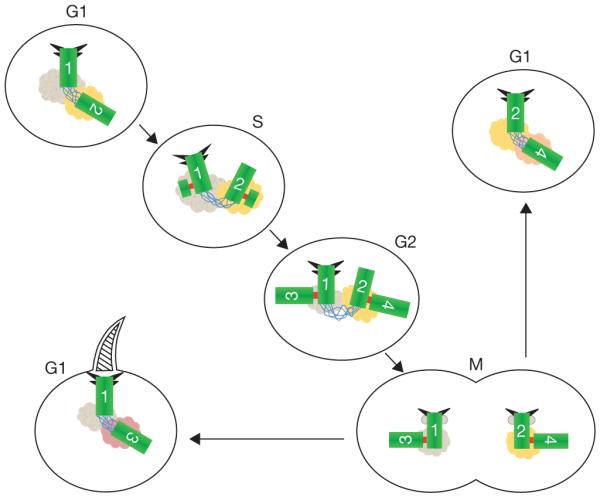
Centriole and centrosome asymmetries. Schematic representation of centrioles (green), with distal and subdistal appendages (triangles), G1–G2 tether (blue), S–M linker (red) and pericentriolar material associated with the base of each centriole. The centrioles are numbered to indicate their origin and age. The centriole marked ‘1’ (centriole 1) is the older of the two centrioles in the G1 cell at the upper left. Centriole 2 formed in the previous cell cycle, as a procentriole adjacent to centriole 1. The centrioles in this cell are disengaged (no S–M linker), but tethered (G1–G2 tether). In S phase new procentrioles grow from each of centrioles 1 and 2 and elongate in G2 phase. These new centrioles (3 and 4) are engaged to their mother centrioles (1 and 2, respectively), but are otherwise equivalent. Centriole 2 acquires appendage proteins at the G2/M transition and appendages proper in the subsequent G1. The two centrosomes segregate at mitosis, with one cell receiving the 1, 3 pair and the other receiving the 2, 4 pair. Although the centriole pairs are morphologically equivalent, there is a functional difference, in that the cell receiving the older mother centriole (centriole 1), is able to form a primary cilium earlier in the cell cycle than the other cell. The pericentriolar material at the base of each centriole is represented in different colours to indicate the possibility that proteins associated with the centrioles might be asymmetrically segregated at mitosis with them.
Progression through the centrosome duplication cycle involves the formation and dissolution of two fundamentally different types of connections between centrioles (Fig. 1). A first type of linker, here termed ‘S–M linker’ (SML), forms during S phase and connects each newly forming procentriole to its adjacent ‘mother’. This linker is tight and persists until ‘disengagement’ of the mother-daughter centriole pair late in cell division (Fig. 1). The composition of the SML remains poorly understood, but recent data from Drosophila suggest that SML functionality depends on the SAS-6–ANA2 complex54. This notion is supported by the cell-cycle-dependent association of SAS-6 with the mother-daughter centriole pair13,18. In addition to providing an attachment between centrioles, a key function of the SML relates to the cell cycle control of centriole duplication. In particular, an attractive model posits that centriole re-duplication is prevented as long as procentriole and mother centriole remain connected through the SML (that is, ‘engaged’) and that dissolution of the SML (‘disengagement’) constitutes a critical step for the licensing of a subsequent round of centriole duplication55. Disengagement requires the action of PLK1, a mitotic kinase distantly related to PLK4 (ref. 56), as well as separase, the protease responsible for sister chromatid separation at the metaphase-to-anaphase transition57. As PLK1 and separase are normally activated only in mitosis, this licensing model offers an appealing explanation for the coupling of centriole duplication to the traverse of the cell cycle55,58. It also ensures that disengagement of centriole pairs is coordinated with chromatid separation; effectively preventing one potential source of multipolar spindles. Sister chromatid cohesion is maintained by the cohesin protein complex, which is cleaved by separase at the metaphase-to-anaphase transition. Remarkably, cleavage of cohesin is also required for centriole disengagement59, suggesting that cohesin might make up part of the SML. It is still unclear how the same protein complex could be involved in maintaining the association of two very different structures before mitosis, but such a mechanistic linkage highlights the importance of faithfully segregating both the genetic material (chromosomes) and the potential to make a cilium (centrioles) at mitosis.
The second type of connection, here termed the ‘G1–G2 tether’ (GGT) provides a loose connection between the proximal ends of the two parental centrioles and thereby helps to focus the microtubule-organizing activity of the centrosome (Fig. 1). Assembly of the GGT occurs in G1 cells concomitant with (or shortly after) the dissolution of the SML (disengagement), suggesting that GGT assembly probably depends on disengagement. Proteins implicated in GGT function include C-Nap1 (ref. 60), rootletin61,62, as well as CEP68 and CEP215–CDK5RAP2 (ref. 63). Live-cell imaging shows that parental centrioles have coordinated movements but can transiently separate over distances of micrometres64,65, suggesting that the GGT is best considered as a loose tether. Disassembly of the GGT occurs in G2, when the two centrosomes (each comprised of a pair of engaged centrioles) separate in preparation for assembly of the mitotic spindle. Loss of the GGT is triggered when the activity of the NIMA-family protein kinase NEK2 exceeds that of a counteracting type 1γ phosphatase66,67, which then results in the phosphorylation of at least two GGT components, C-Nap1 and rootletin. Subsequently, the untethered centrosomes are separated through the kinesin-related motor Eg5, which is recruited to centrosomes in response to phosphorylation by multiple kinases68. Although Eg5 is normally sufficient to trigger centrosome separation, the NEK2 pathway becomes essential when Eg5 activity is impaired69. This cooperation has obvious implications for the possible use of corresponding inhibitors as anti-proliferative therapeutics. Interestingly, NEK2 activity at the centrosome depends not only on Mst1/2 kinases69 but also on PLK1, which contributes to control both the Mst1/2–NEK2 pathway and Eg5 recruitment70. Mst1/2 are components of the Hippo pathway, which is best known for its anti-proliferative and pro-apoptotic function71. It will be interesting to explore the precise relationship of Hippo pathway components to centrosome biology, particularly as additional components of this pathway, Ndr kinases, have been implicated in the control of centriole duplication72.
The spatial organization of centrioles relative to each other thus plays a key role in the regulation of both centriole duplication and microtubule organization. As discussed above, the two connecting structures depicted in Fig. 1 play fundamentally different roles and this is reflected in at least partly different compositions and different modes of regulation. Although the SML is important for limiting centriole duplication to once per cell cycle, the GGT allows the duplicated centrosomes to function as a single microtubule-organizing centre during interphase, until GGT disassembly sets the stage for bipolar spindle formation and chromosome segregation during cell division.
Centrosome cycle and genome stability
Centrosomes and chromosomes are both segregated on the mitotic spindle and it was noticed long ago that in cancer cells there is a strong correlation between aberrant centrosome number and aberrant chromosome number73. It is important to note that centrosomes are not strictly required for mitosis in many cell types, even though they are often active participants in the process. As a particularly striking example of this, it has been possible to generate adult flies lacking centrioles74 and the main defects in such flies are related to the absence of cilia in specific neurons and not to any difficulty with adult mitotic divisions (note that centrosomes were still required for the embryonic divisions in this case). Rather, it is the presence of extra centrosomes that potentially presents more serious problems for mitosis75 and recent studies have begun to reveal the mechanisms through which extra centrosomes give rise to chromosome segregation errors.
Some chromosomally unstable cells proliferate with many centrosomes and usually undergo a bipolar mitotic division despite the aberrant centrosome number, with the centrosomes clustered at each pole. More recently it was shown that when multipolar mitosis does occur as the result of having extra centrosomes, the resulting cells are usually not viable76,77 and thus unlikely to contribute to tumour formation. However, these studies revealed another mechanism by which extra centrosomes can contribute to genomic instability, showing that even in cells that undergo bipolar division extra centrosomes increase the chances for single kinetochores to undergo attachments to microtubules emanating from opposite poles — so-called merotelic attachments76,77. Such merotelic attachments, unless resolved, constitute an important cause of aneuploidy. As most multipolar mitoses result in inviable products, it has been proposed that proteins required for centrosome clustering might be attractive targets for the selective elimination of tumour cells with extra centrosomes78,79.
Another link between centrosomes and genome stability relates to the response to DNA damage and incomplete DNA replication. In Drosophila embryos, centrosomes become inactive for microtubule nucleation in response to entering mitosis with such compromised DNA80 and this might represent an alternative mechanism to the DNA-damage checkpoint for preventing mitosis under conditions where segregation of chromosomes would be catastrophic. In cultured mammalian cells, entry into mitosis with compromised DNA can also result in centriole disengagement and formation of multipolar spindles81; as this often results in inviable products, this might also be a similar fail-safe mechanism. Although several proteins involved in the DNA-damage response have been reported to localize to centrosomes, it is still unclear how communication between the centrosome and DNA-damage sensing occurs. For example, the human checkpoint kinase Chk1 has been reported to delay mitotic entry through an association with centrosomes82, but a recent study convincingly demonstrates that this kinase instead acts from within the nucleus83.
The centrosome cycle and the chromosome cycle can become uncoupled during prolonged DNA damage/replication checkpoint arrest in some cell types84. This re-duplication of centrosomes has been confounding because it is at odds with the apparent requirement for passage through mitosis to disengage centrioles as a prerequisite of duplication. Of particular interest in this context is the recent demonstration that centrosome amplification occurs only during such arrest when levels of PLK1 kinase activity are sufficiently high to allow procentriole maturation and centriole disengagement85,86. PLK1 has recently been shown to also be required to convert new centrioles to centrosomes as cells progress from mitosis to interphase87. In the absence of PLK1 activity, daughter centrioles formed in the previous cycle were unable to organize pericentriolar material and unable to duplicate. A recent study proposes that the unmodified daughter centrioles exert a negative effect on the mother to which they are engaged, preventing re-duplication in a single cycle, and that this effect is only relieved by PLK1 activity in mitosis87. Perhaps it is this activity of PLK1, unleashed prematurely in damage-induced S/G2 arrest, that allows re-duplication. Subsequent experiments along these lines may help to answer the long-standing question of why only some transformed cell lines undergo re-duplication during arrest, and how this relates to genome stability.
Centrioles, cilia and cellular asymmetries
The duplication cycle described above results in several asymmetries intrinsic to the structure of the centrioles and centrosome. In Fig. 3 the centrioles are numbered by age to illustrate this point.
One asymmetry that is apparent in this cycle is that the older centriole in a G1 cell (centriole 1) has centriolar appendages and the younger (centriole 2) does not. The subdistal appendages are involved in organizing the interphase microtubule cytoskeleton, whereas the distal appendages allow the older centriole to function as a basal body, nucleating a primary cilium88. The distal appendages have a ninefold symmetry, reflecting the ninefold symmetry of the centriole microtubule structure89 and are defined molecularly by the protein CEP164, which is required for primary cilium formation90. The distal appendages link the distal end of the basal body/centriole to the plasma membrane, thus defining the ciliary compartment. The new mother centriole (centriole 2 in Fig. 3) only acquires appendage proteins at G2/M and defined appendages in G1 of the next cycle and little is known of how assembly of these structures is directed to this centriole. One hint is that in cells depleted of PLK4, and thus unable to grow new centrioles, the new mother centriole fails to acquire appendage proteins at G2/M, suggesting that presence of an engaged daughter centriole is a requirement for recognition for the initiation of appendage assembly91.
A second asymmetry is that the inherent difference in age of the centrioles results in one daughter cell receiving the old mother centriole, formed sometime in the past (Fig. 3; cell in lower left, centriole 1) and the other receiving the new mother, formed in the previous cell cycle (Fig. 3; cell in upper right, centriole 2; centrioles 3 and 4 are both new daughters and are presumed to be equivalent). Is there any functional consequence to receiving the old mother versus the new mother centriole? Anderson and Stearns92 reported that although both daughter cells are competent to make a primary cilium after division, the cell receiving the old mother usually does so first, with this asynchrony persisting for several hours. Interestingly, this resulted in a differential response to Sonic hedgehog, a signalling pathway that requires the cilium.
That this inherent centriole age difference can result in phenotypic differences in the two daughter cells of a division suggests that it could play a part in asymmetric cell divisions. Drosophila provides several excellent examples of just such a connection. In asymmetrically dividing Drosophila larval neuroblasts and male germline stem cells, only one centriole accumulates PCM during interphase93–95 and the PCM-accumulating centriole remains close to the stem cell niche and segregates into the stem cell93–96. Recent experiments by Januschke et al.97 showed that, contrary to expectation, it is the daughter centriole in Drosophila neuroblasts that accumulates PCM and is segregated to the stem cell. When this pattern of centrosome function and segregation is disrupted cell-fate determinants are not accurately segregated75,94,96. Asymmetric centriole segregation is also observed in radial glial stem cells in mice. In this case, the older mother centriole co-segregates with the stem cell98 and depletion of a centriole appendage protein in developing brain resulted in a failure of asymmetric cell divisions. It is possible that cilium asynchrony and the associated signalling phenotypes, as described in vitro92, are responsible for this in vivo effect of centrosome asymmetry, but it should be noted that several centrosomal proteins have now been shown to be required to maintain the radial glial stem cell population and no common mechanism is yet apparent.
Lastly, there is the potential for asymmetry in the make-up of the PCM associated with each of the two centrosomes in a dividing cell. Although some PCM components exchange rapidly with a cytoplasmic pool, it is possible that others are more stably associated with each of the mother centrioles, creating an opportunity for asymmetry. There is evidence for centrosome-based asymmetric segregation of proteins destined for destruction in mammalian cells99 and of specific mRNAs during early embryonic cleavages in mollusc embryo100. Given the large number of currently uncharacterized proteins that continue to be identified as components of the centrosome, it seems likely some will be associated with asymmetric distribution of material, or themselves be so distributed, providing the potential for phenotypic differences. The unique duplication and segregation cycle of centrioles results in an inherent asymmetry, such that every cell division is asymmetric at the level of the centrioles and it will be of great interest to learn more of how this asymmetry is leveraged by cells as part of developmental and differentiation mechanisms.
ACKNOWLEDGEMENTS
We thank the members of the Nigg and Stearns labs for helpful discussion and apologise to our colleagues whose work we were unable to cite for space limitations. E.A.N. was supported by the Swiss National Science Foundation and T.S. was supported by the National Institutes of Health.
Footnotes
COMPETING FINANCIAL INTERESTS The authors declare that they have no competing financial interests.
Contributor Information
Erich A. Nigg, Biozentrum, University of Basel, Klingelbergstrasse 50/70, CH-4056 Basel, Switzerland
Tim Stearns, Department of Biology, Stanford University, Stanford, California 94305 USA.
References
- 1.Luders J, Stearns T. Microtubule-organizing centres: a re-evaluation. Nat. Rev. Mol. Cell Biol. 2007;8:161–167. doi: 10.1038/nrm2100. [DOI] [PubMed] [Google Scholar]
- 2.Goetz SC, Anderson KV. The primary cilium: a signalling centre during vertebrate development. Nat. Rev. Genet. 2010;11:331–344. doi: 10.1038/nrg2774. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carvalho-Santos Z, Azimzadeh J, Pereira-Leal JB, Bettencourt-Dias M. Evolution: Tracing the origins of centrioles, cilia, and flagella. J. Cell Biol. 2011;194:165–175. doi: 10.1083/jcb.201011152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nigg EA, Raff JW. Centrioles, centrosomes, and cilia in health and disease. Cell. 2009;139:663–678. doi: 10.1016/j.cell.2009.10.036. [DOI] [PubMed] [Google Scholar]
- 5.Vaughan S, Dawe HR. Common themes in centriole and centrosome movements. Trends Cell Biol. 2011;21:57–66. doi: 10.1016/j.tcb.2010.09.004. [DOI] [PubMed] [Google Scholar]
- 6.Azimzadeh J, Marshall WF. Building the centriole. Curr. Biol. 2010;20:R816–R825. doi: 10.1016/j.cub.2010.08.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bettencourt-Dias M, Hildebrandt F, Pellman D, Woods G, Godinho SA. Centrosomes and cilia in human disease. Trends Genet. 2011;27:307–315. doi: 10.1016/j.tig.2011.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Avidor-Reiss T. The cellular and developmental program connecting the centrosome and cilium duplication cycle. Semin. Cell Dev. Biol. 2010;21:139–141. doi: 10.1016/j.semcdb.2010.01.014. [DOI] [PubMed] [Google Scholar]
- 9.Marshall WF. Centriole evolution. Curr. Opin. Cell Biol. 2009;21:14–19. doi: 10.1016/j.ceb.2009.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khodjakov A, et al. De novo formation of centrosomes in vertebrate cells arrested during S phase. J. Cell Biol. 2002;158:1171–1181. doi: 10.1083/jcb.200205102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dammermann A, Maddox PS, Desai A, Oegema K. SAS-4 is recruited to a dynamic structure in newly forming centrioles that is stabilized by the γ-tubulin-mediated addition of centriolar microtubules. J. Cell Biol. 2008;180:771–785. doi: 10.1083/jcb.200709102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Loncarek J, Hergert P, Magidson V, Khodjakov A. Control of daughter centriole formation by the pericentriolar material. Nat. Cell Biol. 2008;10:322–328. doi: 10.1038/ncb1694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Strnad P, et al. Regulated HsSAS-6 levels ensure formation of a single procentriole per centriole during the centrosome duplication cycle. Dev. Cell. 2007;13:203–213. doi: 10.1016/j.devcel.2007.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Andersen JS, et al. Proteomic characterization of the human centrosome by protein correlation profiling. Nature. 2003;426:570–574. doi: 10.1038/nature02166. [DOI] [PubMed] [Google Scholar]
- 15.Jakobsen L, et al. Novel asymmetrically localizing components of human centrosomes identified by complementary proteomics methods. EMBO J. 2011;30:1520–1535. doi: 10.1038/emboj.2011.63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Strnad P, Gonczy P. Mechanisms of procentriole formation. Trends Cell Biol. 2008;18:389–396. doi: 10.1016/j.tcb.2008.06.004. [DOI] [PubMed] [Google Scholar]
- 17.Dobbelaere J, et al. A genome-wide RNAi screen to dissect centriole duplication and centrosome maturation in Drosophila. PLoS Biol. 2008;6:e224. doi: 10.1371/journal.pbio.0060224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Kleylein-Sohn J, et al. Plk4-induced centriole biogenesis in human cells. Dev. Cell. 2007;13:190–202. doi: 10.1016/j.devcel.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 19.Rodrigues-Martins A, et al. DSAS-6 organizes a tube-like centriole precursor, and its absence suggests modularity in centriole assembly. Curr. Biol. 2007;17:1465–1472. doi: 10.1016/j.cub.2007.07.034. [DOI] [PubMed] [Google Scholar]
- 20.Rodrigues-Martins A, Riparbelli M, Callaini G, Glover DM, Bettencourt-Dias M. Revisiting the role of the mother centriole in centriole biogenesis. Science. 2007;316:1046–1050. doi: 10.1126/science.1142950. [DOI] [PubMed] [Google Scholar]
- 21.Peel N, Stevens NR, Basto R, Raff JW. Overexpressing centriole-replication proteins in vivo induces centriole overduplication and de novo formation. Curr. Biol. 2007;17:834–843. doi: 10.1016/j.cub.2007.04.036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Puklowski A, et al. The SCF–FBXW5 E3-ubiquitin ligase is regulated by PLK4 and targets HsSAS-6 to control centrosome duplication. Nat. Cell Biol. 2011;13:1004–1009. doi: 10.1038/ncb2282. [DOI] [PubMed] [Google Scholar]
- 23.Kitagawa D, Busso C, Fluckiger I, Gonczy P. Phosphorylation of SAS-6 by ZYG-1 is critical for centriole formation in C. elegans embryos. Dev. Cell. 2009;17:900–907. doi: 10.1016/j.devcel.2009.11.002. [DOI] [PubMed] [Google Scholar]
- 24.Bettencourt-Dias M, et al. SAK/PLK4 is required for centriole duplication and flagella development. Curr. Biol. 2005;15:2199–2207. doi: 10.1016/j.cub.2005.11.042. [DOI] [PubMed] [Google Scholar]
- 25.Habedanck R, Stierhof YD, Wilkinson CJ, Nigg EA. The Polo kinase Plk4 functions in centriole duplication. Nat. Cell Biol. 2005;7:1140–1146. doi: 10.1038/ncb1320. [DOI] [PubMed] [Google Scholar]
- 26.Cunha-Ferreira I, et al. The SCF/Slimb ubiquitin ligase limits centrosome amplification through degradation of SAK/PLK4. Curr. Biol. 2009;19:43–49. doi: 10.1016/j.cub.2008.11.037. [DOI] [PubMed] [Google Scholar]
- 27.Guderian G, Westendorf J, Uldschmid A, Nigg EA. Plk4 trans-autophosphorylation regulates centriole number by controlling βTrCP-mediated degradation. J. Cell Sci. 2010;123:2163–2169. doi: 10.1242/jcs.068502. [DOI] [PubMed] [Google Scholar]
- 28.Holland AJ, Lan W, Niessen S, Hoover H, Cleveland DW. Polo-like kinase 4 kinase activity limits centrosome overduplication by autoregulating its own stability. J. Cell Biol. 2010;188:191–198. doi: 10.1083/jcb.200911102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rogers GC, Rusan NM, Roberts DM, Peifer M, Rogers SL. The SCF Slimb ubiquitin ligase regulates Plk4/Sak levels to block centriole reduplication. J. Cell Biol. 2009;184:225–239. doi: 10.1083/jcb.200808049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sillibourne JE, et al. Autophosphorylation of polo-like kinase 4 and its role in centriole duplication. Mol. Biol. Cell. 2010;21:547–561. doi: 10.1091/mbc.E09-06-0505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kitagawa D, et al. PP2A phosphatase acts upon SAS-5 to ensure centriole formation in C. elegans embryos. Dev. Cell. 2011;20:550–562. doi: 10.1016/j.devcel.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 32.Song MH, Liu Y, Anderson DE, Jahng WJ, O’Connell KF. Protein phosphatase 2A-SUR-6/B55 regulates centriole duplication in C. elegans by controlling the levels of centriole assembly factors. Dev. Cell. 2011;20:563–571. doi: 10.1016/j.devcel.2011.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Dammermann A, et al. Centriole assembly requires both centriolar and pericentriolar material proteins. Dev. Cell. 2004;7:815–829. doi: 10.1016/j.devcel.2004.10.015. [DOI] [PubMed] [Google Scholar]
- 34.Pelletier L, O’Toole E, Schwager A, Hyman AA, Muller-Reichert T. Centriole assembly in Caenorhabditis elegans. Nature. 2006;444:619–623. doi: 10.1038/nature05318. [DOI] [PubMed] [Google Scholar]
- 35.Nakazawa Y, Hiraki M, Kamiya R, Hirono M. SAS-6 is a cartwheel protein that establishes the 9-fold symmetry of the centriole. Curr. Biol. 2007;17:2169–2174. doi: 10.1016/j.cub.2007.11.046. [DOI] [PubMed] [Google Scholar]
- 36.Gopalakrishnan J, et al. Self-assembling SAS-6 multimer is a core centriole building block. J. Biol. Chem. 2010;285:8759–8770. doi: 10.1074/jbc.M109.092627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stevens NR, Dobbelaere J, Brunk K, Franz A, Raff JW. Drosophila Ana2 is a conserved centriole duplication factor. J. Cell Biol. 2010;188:313–323. doi: 10.1083/jcb.200910016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Kitagawa D, et al. Structural basis of the 9-fold symmetry of centrioles. Cell. 2011;144:364–375. doi: 10.1016/j.cell.2011.01.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.van Breugel M, et al. Structures of SAS-6 suggest its organization in centrioles. Science. 2011;331:1196–1199. doi: 10.1126/science.1199325. [DOI] [PubMed] [Google Scholar]
- 40.Kohlmaier G, et al. Overly long centrioles and defective cell division upon excess of the SAS-4-related protein CPAP. Curr. Biol. 2009;19:1012–1018. doi: 10.1016/j.cub.2009.05.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Schmidt TI, et al. Control of centriole length by CPAP and CP110. Curr. Biol. 2009;19:1005–1011. doi: 10.1016/j.cub.2009.05.016. [DOI] [PubMed] [Google Scholar]
- 42.Tang CJ, Fu RH, Wu KS, Hsu WB, Tang TK. CPAP is a cell-cycle regulated protein that controls centriole length. Nat. Cell Biol. 2009;11:825–831. doi: 10.1038/ncb1889. [DOI] [PubMed] [Google Scholar]
- 43.Azimzadeh J, et al. hPOC5 is a centrin-binding protein required for assembly of full-length centrioles. J. Cell Biol. 2009;185:101–114. doi: 10.1083/jcb.200808082. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Singla V, Romaguera-Ros M, Garcia-Verdugo JM, Reiter JF. Ofd1, a human disease gene, regulates the length and distal structure of centrioles. Dev. Cell. 2010;18:410–424. doi: 10.1016/j.devcel.2009.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Spektor A, Tsang WY, Khoo D, Dynlacht BD. Cep97 and CP110 suppress a cilia assembly program. Cell. 2007;130:678–690. doi: 10.1016/j.cell.2007.06.027. [DOI] [PubMed] [Google Scholar]
- 46.Tsang WY, et al. CP110 suppresses primary cilia formation through its interaction with CEP290, a protein deficient in human ciliary disease. Dev. Cell. 2008;15:187–197. doi: 10.1016/j.devcel.2008.07.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.D’Angiolella V, et al. SCF(Cyclin F) controls centrosome homeostasis and mitotic fidelity through CP110 degradation. Nature. 2010;466:138–142. doi: 10.1038/nature09140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Korzeniewski N, Cuevas R, Duensing A, Duensing S. Daughter centriole elongation is controlled by proteolysis. Mol. Biol. Cell. 2010;21:3942–3951. doi: 10.1091/mbc.E09-12-1049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bornens M. Centrosome composition and microtubule anchoring mechanisms. Curr. Opin. Cell Biol. 2002;14:25–34. doi: 10.1016/s0955-0674(01)00290-3. [DOI] [PubMed] [Google Scholar]
- 50.Cizmecioglu O, et al. Cep152 acts as a scaffold for recruitment of Plk4 and CPAP to the centrosome. J. Cell Biol. 2010;191:731–739. doi: 10.1083/jcb.201007107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Dzhindzhev NS, et al. Asterless is a scaffold for the onset of centriole assembly. Nature. 2010;467:714–718. doi: 10.1038/nature09445. [DOI] [PubMed] [Google Scholar]
- 52.Hatch EM, Kulukian A, Holland AJ, Cleveland DW, Stearns T. Cep152 interacts with Plk4 and is required for centriole duplication. J. Cell Biol. 2010;191:721–729. doi: 10.1083/jcb.201006049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Conduit PT, et al. Centrioles regulate centrosome size by controlling the rate of Cnn incorporation into the PCM. Curr. Biol. 2010;20:2178–2186. doi: 10.1016/j.cub.2010.11.011. [DOI] [PubMed] [Google Scholar]
- 54.Stevens NR, Roque H, Raff JW. DSas-6 and Ana2 coassemble into tubules to promote centriole duplication and engagement. Dev. Cell. 2010;19:913–919. doi: 10.1016/j.devcel.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Tsou MF, Stearns T. Mechanism limiting centrosome duplication to once per cell cycle. Nature. 2006;442:947–951. doi: 10.1038/nature04985. [DOI] [PubMed] [Google Scholar]
- 56.Tsou MF, et al. Polo kinase and separase regulate the mitotic licensing of centriole duplication in human cells. Dev. Cell. 2009;17:344–354. doi: 10.1016/j.devcel.2009.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Uhlmann F, Wernic D, Poupart MA, Koonin EV, Nasmyth K. Cleavage of cohesin by the CD clan protease separin triggers anaphase in yeast. Cell. 2000;103:375–386. doi: 10.1016/s0092-8674(00)00130-6. [DOI] [PubMed] [Google Scholar]
- 58.Nigg EA. Centrosome duplication: of rules and licenses. Trends Cell Biol. 2007;17:215–221. doi: 10.1016/j.tcb.2007.03.003. [DOI] [PubMed] [Google Scholar]
- 59.Schöckel L, Möckel M, Mayer B, Boos D, Stemmann O. Cleavage of cohesin rings coordinates the separation of centrioles and chromatids. Nat. Cell Biol. 2011;13:966–972. doi: 10.1038/ncb2280. [DOI] [PubMed] [Google Scholar]
- 60.Mayor T, Stierhof YD, Tanaka K, Fry AM, Nigg EA. The centrosomal protein C-Nap1 is required for cell cycle-regulated centrosome cohesion. J. Cell Biol. 2000;151:837–846. doi: 10.1083/jcb.151.4.837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bahe S, Stierhof YD, Wilkinson CJ, Leiss F, Nigg EA. Rootletin forms centriole-associated filaments and functions in centrosome cohesion. J. Cell Biol. 2005;171:27–33. doi: 10.1083/jcb.200504107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Yang J, Adamian M, Li T. Rootletin interacts with C-Nap1 and may function as a physical linker between the pair of centrioles/basal bodies in cells. Mol. Biol. Cell. 2006;17:1033–1040. doi: 10.1091/mbc.E05-10-0943. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Graser S, Stierhof YD, Nigg EA. Cep68 and Cep215 (Cdk5rap2) are required for centrosome cohesion. J. Cell Sci. 2007;120:4321–4331. doi: 10.1242/jcs.020248. [DOI] [PubMed] [Google Scholar]
- 64.Piel M, Meyer P, Khodjakov A, Rieder CL, Bornens M. The respective contributions of the mother and daughter centrioles to centrosome activity and behavior in vertebrate cells. J. Cell Biol. 2000;149:317–330. doi: 10.1083/jcb.149.2.317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Piel M, Nordberg J, Euteneuer U, Bornens M. Centrosome-dependent exit of cytokinesis in animal cells. Science. 2001;291:1550–1553. doi: 10.1126/science.1057330. [DOI] [PubMed] [Google Scholar]
- 66.Fry AM, et al. C-Nap1, a novel centrosomal coiled-coil protein and candidate substrate of the cell cycle-regulated protein kinase Nek2. J. Cell Biol. 1998;141:1563–1574. doi: 10.1083/jcb.141.7.1563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Helps NR, Luo X, Barker HM, Cohen PT. NIMA-related kinase 2 (Nek2), a cell-cycle-regulated protein kinase localized to centrosomes, is complexed to protein phosphatase 1. Biochem. J. 2000;349:509–518. doi: 10.1042/0264-6021:3490509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Bertran MT, et al. Nek9 is a Plk1-activated kinase that controls early centrosome separation through Nek6/7 and Eg5. EMBO J. 2011;30:2634–2647. doi: 10.1038/emboj.2011.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Mardin BR, et al. Components of the Hippo pathway cooperate with Nek2 kinase to regulate centrosome disjunction. Nat. Cell Biol. 2010;12:1166–1176. doi: 10.1038/ncb2120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mardin BR, Agircan FG, Lange C, Schiebel E. Plk1 Controls the Nek2A-PP1γ antagonism in centrosome disjunction. Curr. Biol. 2011;21:1145–1151. doi: 10.1016/j.cub.2011.05.047. [DOI] [PubMed] [Google Scholar]
- 71.Halder G, Johnson RL. Hippo signaling: growth control and beyond. Development. 2011;138:9–22. doi: 10.1242/dev.045500. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Hergovich A, et al. The MST1 and hMOB1 tumor suppressors control human centrosome duplication by regulating NDR kinase phosphorylation. Curr. Biol. 2009;19:1692–1702. doi: 10.1016/j.cub.2009.09.020. [DOI] [PubMed] [Google Scholar]
- 73.Hardy PA, Zacharias H. Reappraisal of the Hansemann-Boveri hypothesis on the origin of tumors. Cell Biol. Int. 2005;29:983–992. doi: 10.1016/j.cellbi.2005.10.001. [DOI] [PubMed] [Google Scholar]
- 74.Basto R, et al. Flies without centrioles. Cell. 2006;125:1375–1386. doi: 10.1016/j.cell.2006.05.025. [DOI] [PubMed] [Google Scholar]
- 75.Basto R, et al. Centrosome amplification can initiate tumorigenesis in flies. Cell. 2008;133:1032–1042. doi: 10.1016/j.cell.2008.05.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ganem NJ, Godinho SA, Pellman D. A mechanism linking extra centrosomes to chromosomal instability. Nature. 2009;460:278–282. doi: 10.1038/nature08136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Silkworth WT, Nardi IK, Scholl LM, Cimini D. Multipolar spindle pole coalescence is a major source of kinetochore mis-attachment and chromosome mis-segregation in cancer cells. PLoS One. 2009;4:e6564. doi: 10.1371/journal.pone.0006564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Kwon M, et al. Mechanisms to suppress multipolar divisions in cancer cells with extra centrosomes. Genes Dev. 2008;22:2189–2203. doi: 10.1101/gad.1700908. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Nigg EA. Centrosome aberrations: cause or consequence of cancer progression? Nat. Rev. Cancer. 2002;2:815–825. doi: 10.1038/nrc924. [DOI] [PubMed] [Google Scholar]
- 80.Sibon OC, Kelkar A, Lemstra W, Theurkauf WE. DNA-replication/DNA-damage-dependent centrosome inactivation in Drosophila embryos. Nat. Cell Biol. 2000;2:90–95. doi: 10.1038/35000041. [DOI] [PubMed] [Google Scholar]
- 81.Hut HM, et al. Centrosomes split in the presence of impaired DNA integrity during mitosis. Mol. Biol. Cell. 2003;14:1993–2004. doi: 10.1091/mbc.E02-08-0510. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kramer A, et al. Centrosome-associated Chk1 prevents premature activation of cyclin-B–Cdk1 kinase. Nat. Cell Biol. 2004;6:884–891. doi: 10.1038/ncb1165. [DOI] [PubMed] [Google Scholar]
- 83.Matsuyama M, et al. Nuclear Chk1 prevents premature mitotic entry. J. Cell Sci. 2011;124:2113–2119. doi: 10.1242/jcs.086488. [DOI] [PubMed] [Google Scholar]
- 84.Balczon R, et al. Dissociation of centrosome replication events from cycles of DNA synthesis and mitotic division in hydroxyurea-arrested Chinese hamster ovary cells. J. Cell Biol. 1995;130:105–115. doi: 10.1083/jcb.130.1.105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Inanc B, Dodson H, Morrison CG. A centrosome-autonomous signal that involves centriole disengagement permits centrosome duplication in G2 phase after DNA damage. Mol. Biol. Cell. 2010;21:3866–3877. doi: 10.1091/mbc.E10-02-0124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Loncarek J, Hergert P, Khodjakov A. Centriole reduplication during prolonged interphase requires procentriole maturation governed by Plk1. Curr. Biol. 2010;20:1277–1282. doi: 10.1016/j.cub.2010.05.050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Wang WJ, Soni RK, Uryu K, Bryan Tsou MF. The conversion of centrioles to centrosomes: essential coupling of duplication with segregation. J. Cell Biol. 2011;193:727–739. doi: 10.1083/jcb.201101109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Hoyer-Fender S. Centriole maturation and transformation to basal body. Semin. Cell Dev. Biol. 2010;21:142–147. doi: 10.1016/j.semcdb.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 89.Paintrand M, Moudjou M, Delacroix H, Bornens M. Centrosome organization and centriole architecture: their sensitivity to divalent cations. J. Struct. Biol. 1992;108:107–128. doi: 10.1016/1047-8477(92)90011-x. [DOI] [PubMed] [Google Scholar]
- 90.Graser S, et al. Cep164, a novel centriole appendage protein required for primary cilium formation. J. Cell Biol. 2007;179:321–330. doi: 10.1083/jcb.200707181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Mahjoub MR, Xie Z, Stearns T. Cep120 is asymmetrically localized to the daughter centriole and is essential for centriole assembly. J. Cell Biol. 2010;191:331–346. doi: 10.1083/jcb.201003009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Anderson CT, Stearns T. Centriole age underlies asynchronous primary cilium growth in mammalian cells. Curr. Biol. 2009;19:1498–1502. doi: 10.1016/j.cub.2009.07.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Yamashita YM, Jones DL, Fuller MT. Orientation of asymmetric stem cell division by the APC tumor suppressor and centrosome. Science. 2003;301:1547–1550. doi: 10.1126/science.1087795. [DOI] [PubMed] [Google Scholar]
- 94.Rusan NM, Peifer M. A role for a novel centrosome cycle in asymmetric cell division. J. Cell Biol. 2007;177:13–20. doi: 10.1083/jcb.200612140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Rebollo E, et al. Functionally unequal centrosomes drive spindle orientation in asymmetrically dividing Drosophila neural stem cells. Dev. Cell. 2007;12:467–474. doi: 10.1016/j.devcel.2007.01.021. [DOI] [PubMed] [Google Scholar]
- 96.Januschke J, Gonzalez C. The interphase microtubule aster is a determinant of asymmetric division orientation in Drosophila neuroblasts. J. Cell Biol. 2010;188:693–706. doi: 10.1083/jcb.200905024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Januschke J, Llamazares S, Reina J, Gonzalez C. Drosophila neuroblasts retain the daughter centrosome. Nat. Commun. 2011;2:243. doi: 10.1038/ncomms1245. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Wang X, et al. Asymmetric centrosome inheritance maintains neural progenitors in the neocortex. Nature. 2009;461:947–955. doi: 10.1038/nature08435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Fuentealba LC, Eivers E, Geissert D, Taelman V, De Robertis EM. Asymmetric mitosis: Unequal segregation of proteins destined for degradation. Proc. Natl Acad. Sci. USA. 2008;105:7732–7737. doi: 10.1073/pnas.0803027105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Lambert JD, Nagy LM. Asymmetric inheritance of centrosomally localized mRNAs during embryonic cleavages. Nature. 2002;420:682–686. doi: 10.1038/nature01241. [DOI] [PubMed] [Google Scholar]