Abstract
This review will discuss the existing literature that has examined the role of calcineurin (CnA) in the regulation of skeletal muscle mass in conditions associated with hypertrophic growth or atrophy. Muscle mass is determined by the balance between protein synthesis and degradation which is controlled by a number of intracellular signaling pathways, most notably the insulin/IGF/phosphatidylinositol 3-kinase (PI3K)/Akt system. Despite being activated by IGF-1 and having well-described functions in the determination of muscle fiber phenotypes, calcineurin (CnA), a Ca2+-activated serine/threonine phosphatase, and its downstream signaling partners have garnered little attention as a regulator of muscle mass. Compared to other signaling pathways, the relatively few studies that have examined the role of CnA in the regulation of muscle size have produced discordant results. The reasons for these differences is not obvious but may be due to the selective nature of the genetic models studied, fluctuations in the endogenous level of CnA activity in various muscles, and the variable use of CnA inhibitors to inhibit CnA signaling. Despite the inconsistent nature of the outcomes, there is sufficient direct and indirect evidence to conclude that CnA plays a role in the regulation of skeletal muscle mass.
Keywords: calcineurin, skeletal muscle, atrophy, hypertrophy, cell signaling
INTRODUCTION
Skeletal muscle hypertrophy is an adaptive process that results largely from stimulation of mTOR and protein synthesis pathways by exercise, growth factors and other anabolic hormones, and neuronal input. In contrast, muscle atrophy is a debilitating consequence of a range of pathologic and metabolic conditions including disuse/inactivity, cancer, renal failure, diabetes, AIDS, and sepsis. It is widely accepted that skeletal muscle atrophy is a combinatorial process that involves both a decrease in muscle protein synthesis and an increase in protein degradation (Eley and Tisdale, 2007, Jackman and Kandarian, 2004, Powers et al., 2007). Given the physiological complexity of maintaining muscle mass, it is important to understand the underlying mechanisms that lead to changes in skeletal muscle protein synthesis and degradation during growth and atrophy.
Over the past decade, a number of studies have examined the role of calcineurin (CnA) in the regulation of skeletal muscle mass. CnA, also known as protein phosphatase-3, is a serine/threonine phosphatase that is activated by sustained increased levels of intracellular Ca2+ (Crabtree, 1999). CnA is a heterodimeric complex composed of a catalytic A subunit and smaller calcium-binding regulatory B subunit (Crabtree, 1999, Rusnak and Mertz, 2000). Three 59–62 kDa isoforms of the catalytic CnA subunit have been identified that arise from separate genes: CnAα, CnAβ, and CnAγ (Klee et al., 1998, Rusnak and Mertz, 2000). CnAα and CnAβ are abundant in a wide variety of tissues including skeletal muscle. In contrast, CnAγ is predominately located in the testis and brain as a result of differential gene expression (Klee, Ren, 1998, Rusnak and Mertz, 2000). The presence of microRNAs that specifically target CnAγ may also contribute to its tissue expression pattern (Chhabra et al., 2011).
Although the downstream effectors of CnA have not been fully elucidated, CnA signaling is known to be important for proper function of both cardiac and skeletal muscle. In skeletal muscle, CnA participates in a variety of processes including myoblast recruitment, myotube differentiation, fiber type specification, and recovery from muscle injury and dystrophic muscle damage (Friday et al., 2000, Horsley et al., 2001, Horsley et al., 2003, Naya et al., 2000, Parsons et al., 2003, Stupka et al., 2006). There are several excellent reviews of the central role of CnA signaling in cardiac muscle and specification of fiber types in skeletal muscle (Fiedler and Wollert, 2005, Houser and Molkentin, 2008, Liu et al., 2010, Mallinson et al., 2009, Molkentin, 2004, Schiaffino and Serrano, 2002, Wilkins and Molkentin, 2004), but only a few have detailed the regulation of skeletal muscle size by CnA (Mitchell and Pavlath, 2002, Schiaffino and Serrano, 2002). This report will focus on the role of CnA in the control of muscle mass and discuss how manipulation of CnA signaling pathways may have potential applications for treating skeletal muscle wasting.
ROLE OF CALCINEURIN IN MUSCLE GROWTH AND HYPERTROPHY
Protein synthesis is the major mechanism responsible for muscle growth. Generally speaking, protein synthesis is thought to be regulated primarily through a highly conserved signaling pathway involving insulin and IGF-1 in skeletal muscle (Schiaffino and Mammucari, 2011). These hormones activate a common signaling cascade involving PI3K, Akt, and mTOR that mediates skeletal muscle hypertrophy. Papers from the late 1990s indicate that IGF-1-mediated growth requires CnA-NFAT signaling in addition to the PI3K/Akt/mTOR signaling. Musaro et al. (Musaro et al., 1999) demonstrated that IGF-1 activates CnA and its downstream targets, the NFAT transcription factors, but the activation mechanism was not described. In similar studies, treatment of cultured myotubes with the CnA inhibitor cyclosporine A (CsA) prevented IGF-1-induced hypertrophy (Musaro, McCullagh, 1999, Semsarian et al., 1999). These early reports consistently show that activation of CnA is necessary for muscle growth in response to IGF-1. Subsequent studies using various in vitro and in vivo models of muscle growth produced inconsistent outcomes. Some conclude that CnA is required for skeletal muscle hypertrophy whereas others exclude a role for CnA signaling. Several observations may explain some of these disparities and will be discussed in detail in the next two sections.
Genetic models that evaluate the effect of CnA on muscle size
A number of groups have examined the impact of CnA signaling using genetically modified animal models (Kegley et al., 2001, Parsons et al., 2004, Parsons, Wilkins, 2003). Parsons et al. (Parsons, Wilkins, 2003) studied both CnAα−/− and CnAβ−/− mice at 8–10 weeks of age and reported that neither had smaller muscle sizes relative to wild-type controls. This conclusion was based on muscle weight data that was normalized to body weight and importantly, both knockout strains of mice had overall diminished body size. Thus, it is likely that the absolute mass of the muscles were smaller in mice with reduced CnA activity. In later studies, the same group observed that deletion of CnAβ failed to block IGF-1-induced or mechanical overload-induced hypertrophy (Parsons, Millay, 2004). They commented that the numbers of muscle fibers in hindlimb muscles of the CnAβ−/− mice were reduced suggesting the initial size of the muscles might have been smaller.
In related studies, Kegley and colleagues (Kegley, Gephart, 2001) evaluated muscle mass in NFATc3−/− transgenic mice and found that they had smaller soleus and EDL muscles. The decrease in size was attributed to a reduction in the number of muscle fibers. Fiber size and muscle organization were unaltered. Importantly, they noted the levels of other CnA regulated NFAT isoforms were unaltered in the model. Thus, the possibility cannot be excluded that other active NFATs present in muscle were sufficient to maintain adequate CnA signaling.
In a gain of function study, Naya et al (Naya, Mercer, 2000) found no evidence of muscle hypertrophy in hindlimb muscles of 3-month old transgenic mice overexpressing CnAα under the control of the muscle creatine kinase (MCK) promoter (MCK-Cn*); activated CnA is expressed in all skeletal muscles in these mice. Strangely, the Naya group (Talmadge et al., 2004) reported in a later study with MCK-Cn* mice that predominantly slow fiber muscles underwent hypertrophy whereas predominantly fast fiber muscles had impaired growth at 3 months of age. In yet another study using MCK-Cn* mice, Jiang et al. (Jiang et al., 2010) reported no change in the size of soleus muscles whereas the EDL was smaller. The authors concluded that MCK-Cn* mice undergo adaptive changes in muscle fiber proportions and mitochondrial content that “mimic the response to chronic resistance training”.
Dunn et al. (Dunn et al., 2000) studied both MCK-Cn* mice and transgenic mice that overexpress activated CnAα under the control of the fast MLC (myosin light chain) promoter which is active only in fast fibers. In both cases, there was no increase in size of either predominantly fast or slow muscles. Furthermore, responses to mechanical overload were similar in muscles of wild type, MCK-Cn* and MLC-Cn* mice. In contrast, they reported that overexpression of a peptide inhibitor of calmodulin, which is immediately upstream of CnA and required for its activation, blocks overload-induced hypertrophy.
Oh and colleagues (Oh et al., 2005) utilized a muscle-specific, inducible Flox-On transgenic mouse model to examine the role of CnA signaling in muscle. They characterized mice that overexpress an endogenous CnA inhibitor protein known as MCIP1 (also called RCAN1 (Davies et al., 2007)) in skeletal muscle and found they had a significant reduction in the size of the soleus but not other muscles.
What are potential explanations for the disagreement in findings among these various gain or loss of function models? One possibility is that variable levels of basal and stimulated CnA activity are achieved in the different models. For example, the level of basal CnA activity in the hindlimb of mice developed by Dunn and colleagues (Dunn, Chin, 2000) was several times lower than in the mice studied by Talmadge et al. (Dunn, Chin, 2000, Talmadge, Otis, 2004). Dunn and colleagues (Dunn, Chin, 2000) interestingly concluded that endogenous CnA is able to accommodate large changes in upstream Ca2+-activated signals. Another possibility is that CnAα and CnAβ have both overlapping and uniquely different functions (Williams and Gooch, 2012). The same appears to be true for the NFAT proteins (Mancini and Toker, 2009). Therefore, it is difficult to compare data from models in which different proteins are manipulated. Finally, hypertrophy in response to growth factors and other physiological stimuli (e.g., overload or regrowth after injury) involves a variety of CnA-dependent and independent signaling pathways. Pertubations in one CnA pathway component may not be sufficient to impact the overall growth process.
Pharmacological models that evaluate the effect of CnA on muscle size
A second strategy that is frequently employed to study the role of CnA in growth and hypertrophy involves the use of pharmacological inhibitors of CnA (e.g., cyclosporine; FK506 (also known as tacrolimus)). In contrast to genetic knockout models, these compounds affect both CnAα and CnAβ and presumably result in a more complete inhibition of CnA signaling.
Bodine and colleagues (Bodine et al., 2001) administered either 15mg/kg of CsA or 3 mg/kg of FK506 once daily for 30 days to rats following mechanical overload and found no differences between muscles of treated and untreated animals. A number of other groups reported that administration of 25mg/kg of CsA at least once daily for two or more weeks prevents hypertrophy in both hindlimb suspension/reloading and mechanical overload models of growth (Dunn et al., 1999, Mitchell et al., 2002, Oishi et al., 2008, Sakuma et al., 2008). Similarly, Miyazaki and colleagues (Miyazaki et al., 2006) found that FK506 (5 mg/kg daily) blocks the regrowth of the soleus muscle following hindlimb suspension.
These data suggest that inconsistent levels of CnA suppression may have been achieved with different doses of inhibitors, thus resulting in different conclusions about the role of CnA in the regulation of muscle growth. At higher doses, CnA inhibitors prevent regrowth or hypertrophy although as in the genetic models, the effects seem to be more evident in predominantly slow-fiber muscles like soleus. This is logical since CnA activity is higher in slow than fast muscles.
ROLE OF CALCINEURIN IN SKELETAL MUSCLE PROTEIN DEGRADATION
The role of CnA in the maintenance of muscle size during atrophic conditions has only recently been investigated in detail. Muscle atrophy largely results from activation of intracellular proteolytic systems including the ubiquitin-proteasome pathway (Hudson et al., 2012, Lecker et al., 2006, Lecker et al., 2004, Sacheck et al., 2007), the autophagy-lysosome system (Mammucari et al., 2007), and select members of the calpain and caspase families (Franch and Price, 2005, Lecker and Mitch, 2011, Powers et al., 2012, Smuder et al., 2012, Whidden et al., 2010, Zheng et al., 2010). The cellular responses giving rise to increased activities of these systems include a coordinated increase in gene expression for key components of multiple proteolytic systems (i.e., atrophy-inducing genes or atrogenes). Details of these processes can be found in a number of excellent reviews (Glass, 2005, Jackman and Kandarian, 2004, Lecker, Jagoe, 2004, Powers et al., 2005, Schiaffino et al., 2013, Ventadour and Attaix, 2006, Workeneh and Mitch, 2010). To briefly summarize, the PI3K/Akt signaling system is the central pathway that controls proteolysis (as well as protein synthesis). Inhibition of the pathway, genetically or pharmacologically, results in the same atrophy-related responses that occur in conditions associated with muscle wasting (Price et al., 2010, Sacheck et al., 2004, Schiaffino and Mammucari, 2011). When activated, Akt phosphorylates the FOXO transcription factors that inhibit the expression of the muscle-specific E3 ubiquitin ligases, atrogin-1/MAFbx and MuRF1, as well as other proteolytic genes. When insulin or IGF-1 signaling is low or impaired (e.g., diabetes, kidney disease), the activities of the FOXOs increase, resulting in upregulation of the ubiquitin-proteasome and autophagy-lysosome systems (Mammucari, Milan, 2007, Sandri et al., 2004, Zhao et al., 2007, Zheng, Ohkawa, 2010). Reduction of FOXO3 by siRNA or overexpression of a dominant-negative plasmid attenuates muscle atrophy and in some cases actually induces muscle growth (Reed et al., 2012, Sandri, Sandri, 2004).
Interestingly, PGC-1α, a transcriptional coactivator that regulates various aspects of skeletal muscle metabolism, is reduced in skeletal muscle during conditions associated with FOXO activation and atrophy (Arany, 2008, Mootha et al., 2003, Patti et al., 2003, Roberts-Wilson et al., 2010, Sandri et al., 2006). This is notable because Sandri et al. (Sandri, Lin, 2006) reported that overexpression of PGC-1α attenuates FOXO activity; this effect decreased the expression of atrogenes and protected muscles from denervation or starvation-induced atrophy. Similarly, Wenz and colleagues (Wenz et al., 2009) found that overexpression of PGC-1α prevented sarcopenia and muscle wasting associated with metabolic disorders. The animals also had improved insulin signaling.
Recently, our lab identified an interaction between CnA activity and PGC-1α expression in muscle during atrophy induced by streptozotocin-induced diabetes (Roberts-Wilson, Reddy, 2010). PGC-1α expression and CnA signaling were decreased in diabetic rat muscle. Although CnA activity was not directly measured, several findings indicate that CnA signaling was suppressed. First, luciferase activity in hindlimb muscles of NFAT-luciferase transgenic mice was lower in diabetic than control mice. Second, the level of mRNA for RCAN1-4, a gene target of NFAT, was also decreased in muscle by diabetes; RCAN1-4 are a generally accepted surrogate of CnA activity (Ni et al., 2006, Ni et al., 2007). Last, a direct link between PGC-1α expression and CnA activity is indicated by the reduced level of PGC-1α mRNA measured in the hindlimb muscle of both CnAα−/− and CnAβ−/− mice versus wild type controls. Others have reported that overexpression of CnA in skeletal muscle results in increased PGC-1α expression (Guerfali et al., 2007, Jiang, Garcia-Roves, 2010, Long et al., 2007). In a recent study, Banzet and colleagues (Banzet et al., 2012) reported that CsA treatment does not change PGC-1α mRNA expression in rat muscle. The caveat to these experiments is that CsA was administered at a low dose (12.5mg/kg) for a short time (2.5 days) and CnA activity was not consistently suppressed in all muscles tested.
There are several other less conventional mechanisms by which CnA may affect protein degradation. Knockdown of a naturally occurring constitutively active variant of CnA, CnAβ1, activates FOXO3 (Lara-Pezzi et al., 2007). Ectopic overexpression of CnAβ1 inhibits FOXO3 and prevents atrophy while transgenic overexpression of CnAβ1 in muscle enhances muscle regeneration following damage (Lara-Pezzi, Winn, 2007). Despite the mechanism of CnAβ1 action being unknown, the study suggests that selective activation of specific CnA isoforms could provide a novel approach for treating muscle atrophy.
CnA could also indirectly protect muscle fibers from atrophy by raising the proportion of slow fibers in muscles. For reasons that are not well understood, slow or Type I muscle fibers are more resistant to atrophy than fast or Type II fibers (Sandri, 2008, Schiaffino et al., 2007). CnA supports the slow fiber phenotype, in part, by maintaining PGC-1α through activation of the MEF2 and NFAT transcription factors (Lin et al., 2002). In many diseases (e.g., diabetes, cancer, kidney disease), fiber-type switching from slow to fast fibers occurs (Stevenson et al., 2003). Atrogin-1, a muscle-specific E3 ubiquitin ligase that is upregulated during atrophy, may contribute to this process because it initiates CnA degradation in cardiomyocytes (Li et al., 2004). Therefore, a potential mechanism underlying the disease-related fiber switching process is a reduction in both CnA and PGC-1α.
MicroRNAs provide yet another novel mechanism by which CnA may influence muscle atrophy. In recent years, microRNAs have been suggested to have a therapeutic potential for treating pathologic conditions (Quiat and Olson, 2013). MicroRNAs provide a post-transcriptional mechanism by which the levels of specific proteins can be controlled in cells (Allen and Loh, 2011, McCarthy et al., 2009, Wada et al., 2011). Although the regulation of microRNAs is poorly understood in general, a few recent papers implicate CnA-related signaling in the regulation of at least one microRNA that targets proteins involved in muscle wasting.
In 2011, Wada and colleagues (Wada, Kato, 2011) reported that microRNA-23a (miR-23a) suppresses the translation of both atrogin-1 and MuRF1 in skeletal muscle. The potential physiological importance of miR-23a during atrophy was demonstrated when ectopic expression of miR-23a protected muscle from atrophy in vivo and in vitro. In the same study, NFATc3 induced the activity of a miR-23a-responsive reporter gene in C2C12 muscle cells; curiously, activated CnAα did not induce the same response (Wada, Kato, 2011). It remains unclear why activated CnAα did not increase miR-23a reporter gene expression because both CnA and NFAT up-regulate miR-23a in cardiomyocytes (Lin et al., 2009). Moreover, the miR-23a-27a-24-2 microRNA cluster in which miR-23a is located is controlled by a single promoter that is responsive to CnA signaling in C2C12 myotubes (Allen and Loh, 2011). Additional studies are needed to extensively evaluate the relationship between CnA signaling, miR-23a expression, and the control of muscle size.
CONCLUSIONS
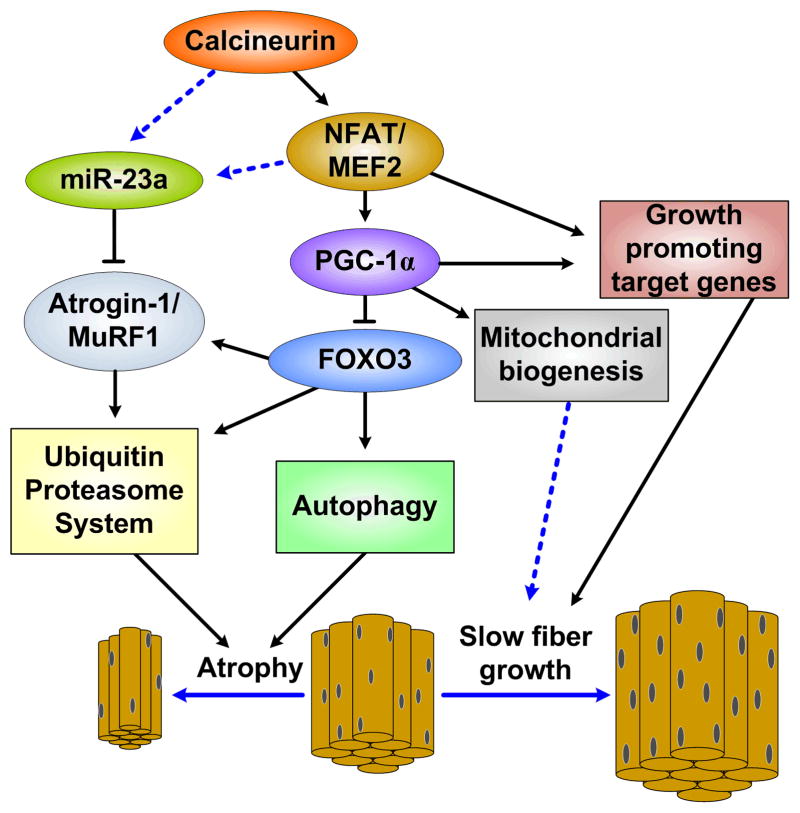
The maintenance of skeletal muscle mass requires a delicate balance between a variety of positive and negative biological inputs. Most inputs achieve their results by activating signaling pathways that coordinate a complex interplay between transcriptional, post-transcriptional, and post-translation responses that fine tune protein synthesis and degradation. A growing number of publications indicate that CnA is an integral part of some of these regulatory mechanisms. The preponderance of evidence indicates that CnA is involved in both muscle growth and wasting (Fig. 1). Details about how these responses are achieved remain largely unknown; however, research during the last few years has uncovered new, unexpected ways by which CnA seems to regulate protein turnover.
Figure 1. A basic overview of the regulation of muscle size by CnA.
Solid lines represent established pathways while dotted lines represent potential or unverified interactions. CnA’s activation of PGC-1α via MEF2 and NFAT is currently thought to be the primary signaling cascade by which CnA prevents muscle wasting. However, CnA also potentially activates miR-23a, thus inhibiting Atrogin-1 and MuRF1, either by direct activation or by first activating MEF2 and/or NFAT, representing an additional mechanism whereby CnA may prevent muscle loss.
CnA inhibitors, including cyclosporine and tacrolimus (FK506), are often prescribed to organ transplant patients as part of their immunosuppressive drug regimen (Masuda and Inui, 2006). Most take CnA inhibitors along with other medications known to alter skeletal muscle signaling including glucocorticoids, statins, and/or colchicine(Masuda and Inui, 2006). Moreover, well known side-effects associated with sustained use of CnA inhibitors include nephrotoxicity, new onset diabetes mellitis, and peripheral neuropathy (English et al., 2002, Heisel et al., 2004) which put patients at higher risks of developing muscle atrophy. These complications make it difficult to ascertain the CnA inhibitor-related effect on skeletal muscle in these patients (Breil and Chariot, 1999). These are important questions to sort out in future studies.
In summary, current evidence supports both correlative and causal links between CnA and muscle fiber size. The use of various genetic models to address questions about whether CnA is required for muscle growth and hypertrophy have yielded inconsistent results. Outcomes of similar studies performed using CnA have been more consistent when higher doses of the compounds were used and indicate that intact CnA signaling is necessary to maintain muscle size, especially in slow muscles. Recent evidence indicates that CnA signaling is suppressed by atrophy-inducing conditions. This response causes a reduction in a variety of mediators that act to antagonize key components of the protein degradation machinery in muscle. Future examination of the signaling pathways regulating CnA as well as its downstream mediators will be important areas of future research. Such studies could provide novel insights about the regulation of muscle size in response to diverse anabolic and catabolic conditions.
Acknowledgments
Our work has been supported by grants from the National Institute of Diabetes, Digestive and Kidney Disease (RO1 DK-95610, S.R.P.) (T32 DK007656, M.B.H), the American Heart Association (GRNT7660020, S.R.P.), and the Department of Veterans Affairs (I01 BX001456, S.R.P.).
Abbreviations
- CnA
calcineurin
- CnAα
α isoform of the CnA catalytic subunit
- CnAβ
β isoform of the CnA catalytic subunit
- CsA
cyclosporine
- MEF2
myocyte enhancing factor 2
- NFAT
nuclear factor of activated T cells
- phosphatidylinositol 3-kinase
PI3K
- FOXO
Forkhead box O
- IGF-1
insulin-like growth factor-1
- MCK
muscle creatine kinase
- MLC
myosin light chain
Footnotes
Conflicts of interest: The authors have no conflicts of interest.
References
- Allen DL, Loh AS. Posttranscriptional mechanisms involving microRNA-27a and b contribute to fast-specific and glucocorticoid-mediated myostatin expression in skeletal muscle. Am J Physiol Cell Physiol. 2011;300:C124–37. doi: 10.1152/ajpcell.00142.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arany Z. PGC-1 coactivators and skeletal muscle adaptations in health and disease. Curr Opin Genet Dev. 2008;18:426–34. doi: 10.1016/j.gde.2008.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Banzet S, Sanchez H, Chapot R, Peinnequin A, Bigard X, Koulmann N. Basal peroxisome proliferator activated receptor gamma coactivator 1alpha expression is independent of calcineurin in skeletal muscle. Metabolism. 2012;61:389–94. doi: 10.1016/j.metabol.2011.07.016. [DOI] [PubMed] [Google Scholar]
- Bodine SC, Stitt TN, Gonzalez M, Kline WO, Stover GL, Bauerlein R, et al. Akt/mTOR pathway is a crucial regulator of skeletal muscle hypertrophy and can prevent muscle atrophy in vivo. Nat Cell Biol. 2001;3:1014–9. doi: 10.1038/ncb1101-1014. [DOI] [PubMed] [Google Scholar]
- Breil M, Chariot P. Muscle disorders associated with cyclosporine treatment. Muscle Nerve. 1999;22:1631–6. doi: 10.1002/(sici)1097-4598(199912)22:12<1631::aid-mus3>3.0.co;2-v. [DOI] [PubMed] [Google Scholar]
- Chhabra R, Dubey R, Saini N. Gene expression profiling indicate role of ER stress in miR-23a~27a~24–2 cluster induced apoptosis in HEK293T cells. RNA Biol. 2011;8:648–64. doi: 10.4161/rna.8.4.15583. [DOI] [PubMed] [Google Scholar]
- Crabtree GR. Generic signals and specific outcomes: signaling through Ca2+, calcineurin, and NF-AT. Cell. 1999;96:611–4. doi: 10.1016/s0092-8674(00)80571-1. [DOI] [PubMed] [Google Scholar]
- Davies KJ, Ermak G, Rothermel BA, Pritchard M, Heitman J, Ahnn J, et al. Renaming the DSCR1/Adapt78 gene family as RCAN: regulators of calcineurin. FASEB J. 2007;21:3023–8. doi: 10.1096/fj.06-7246com. [DOI] [PubMed] [Google Scholar]
- Dunn SE, Burns JL, Michel RN. Calcineurin is required for skeletal muscle hypertrophy. J Biol Chem. 1999;274:21908–12. doi: 10.1074/jbc.274.31.21908. [DOI] [PubMed] [Google Scholar]
- Dunn SE, Chin ER, Michel RN. Matching of calcineurin activity to upstream effectors is critical for skeletal muscle fiber growth. J Cell Biol. 2000;151:663–72. doi: 10.1083/jcb.151.3.663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eley HL, Tisdale MJ. Skeletal muscle atrophy, a link between depression of protein synthesis and increase in degradation. J Biol Chem. 2007;282:7087–97. doi: 10.1074/jbc.M610378200. [DOI] [PubMed] [Google Scholar]
- English RF, Pophal SA, Bacanu SA, Fricker J, Boyle GJ, Ellis D, et al. Long-term comparison of tacrolimus- and cyclosporine-induced nephrotoxicity in pediatric heart-transplant recipients. Am J Transplant. 2002;2:769–73. doi: 10.1034/j.1600-6143.2002.20811.x. [DOI] [PubMed] [Google Scholar]
- Fiedler B, Wollert KC. Targeting calcineurin and associated pathways in cardiac hypertrophy and failure. Expert Opin Ther Targets. 2005;9:963–73. doi: 10.1517/14728222.9.5.963. [DOI] [PubMed] [Google Scholar]
- Franch HA, Price SR. Molecular signaling pathways regulating muscle proteolysis during atrophy. Curr Opin Clin Nutr Metab Care. 2005;8:271–5. doi: 10.1097/01.mco.0000165005.01331.45. [DOI] [PubMed] [Google Scholar]
- Friday BB, Horsley V, Pavlath GK. Calcineurin activity is required for the initiation of skeletal muscle differentiation. J Cell Biol. 2000;149:657–66. doi: 10.1083/jcb.149.3.657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glass DJ. Skeletal muscle hypertrophy and atrophy signaling pathways. Int J Biochem Cell Biol. 2005;37:1974–84. doi: 10.1016/j.biocel.2005.04.018. [DOI] [PubMed] [Google Scholar]
- Guerfali I, Manissolle C, Durieux AC, Bonnefoy R, Bartegi A, Freyssenet D. Calcineurin A and CaMKIV transactivate PGC-1alpha promoter, but differentially regulate cytochrome c promoter in rat skeletal muscle. Pflugers Arch. 2007;454:297–305. doi: 10.1007/s00424-007-0206-6. [DOI] [PubMed] [Google Scholar]
- Heisel O, Heisel R, Balshaw R, Keown P. New onset diabetes mellitus in patients receiving calcineurin inhibitors: a systematic review and meta-analysis. Am J Transplant. 2004;4:583–95. doi: 10.1046/j.1600-6143.2003.00372.x. [DOI] [PubMed] [Google Scholar]
- Horsley V, Friday BB, Matteson S, Kegley KM, Gephart J, Pavlath GK. Regulation of the growth of multinucleated muscle cells by an NFATC2-dependent pathway. J Cell Biol. 2001;153:329–38. doi: 10.1083/jcb.153.2.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsley V, Jansen KM, Mills ST, Pavlath GK. IL-4 acts as a myoblast recruitment factor during mammalian muscle growth. Cell. 2003;113:483–94. doi: 10.1016/s0092-8674(03)00319-2. [DOI] [PubMed] [Google Scholar]
- Houser SR, Molkentin JD. Does contractile Ca2+ control calcineurin-NFAT signaling and pathological hypertrophy in cardiac myocytes? Sci Signal. 2008;1:pe31. doi: 10.1126/scisignal.125pe31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hudson MB, Smuder AJ, Nelson WB, Bruells CS, Levine S, Powers SK. Both high level pressure support ventilation and controlled mechanical ventilation induce diaphragm dysfunction and atrophy. Crit Care Med. 2012;40:1254–60. doi: 10.1097/CCM.0b013e31823c8cc9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackman RW, Kandarian SC. The molecular basis of skeletal muscle atrophy. Am J Physiol Cell Physiol. 2004;287:C834–43. doi: 10.1152/ajpcell.00579.2003. [DOI] [PubMed] [Google Scholar]
- Jiang LQ, Garcia-Roves PM, de Castro Barbosa T, Zierath JR. Constitutively active calcineurin in skeletal muscle increases endurance performance and mitochondrial respiratory capacity. Am J Physiol Endocrinol Metab. 2010;298:E8–E16. doi: 10.1152/ajpendo.00403.2009. [DOI] [PubMed] [Google Scholar]
- Kegley KM, Gephart J, Warren GL, Pavlath GK. Altered primary myogenesis in NFATC3(−/−) mice leads to decreased muscle size in the adult. Dev Biol. 2001;232:115–26. doi: 10.1006/dbio.2001.0179. [DOI] [PubMed] [Google Scholar]
- Klee CB, Ren H, Wang X. Regulation of the calmodulin-stimulated protein phosphatase, calcineurin. J Biol Chem. 1998;273:13367–70. doi: 10.1074/jbc.273.22.13367. [DOI] [PubMed] [Google Scholar]
- Lara-Pezzi E, Winn N, Paul A, McCullagh K, Slominsky E, Santini MP, et al. A naturally occurring calcineurin variant inhibits FoxO activity and enhances skeletal muscle regeneration. J Cell Biol. 2007;179:1205–18. doi: 10.1083/jcb.200704179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lecker SH, Goldberg AL, Mitch WE. Protein degradation by the ubiquitin-proteasome pathway in normal and disease states. J Am Soc Nephrol. 2006;17:1807–19. doi: 10.1681/ASN.2006010083. [DOI] [PubMed] [Google Scholar]
- Lecker SH, Jagoe RT, Gilbert A, Gomes M, Baracos V, Bailey J, et al. Multiple types of skeletal muscle atrophy involve a common program of changes in gene expression. FASEB J. 2004;18:39–51. doi: 10.1096/fj.03-0610com. [DOI] [PubMed] [Google Scholar]
- Lecker SH, Mitch WE. Proteolysis by the ubiquitin-proteasome system and kidney disease. J Am Soc Nephrol. 2011;22:821–4. doi: 10.1681/ASN.2010090958. [DOI] [PubMed] [Google Scholar]
- Li HH, Kedar V, Zhang C, McDonough H, Arya R, Wang DZ, et al. Atrogin-1/muscle atrophy F-box inhibits calcineurin-dependent cardiac hypertrophy by participating in an SCF ubiquitin ligase complex. J Clin Invest. 2004;114:1058–71. doi: 10.1172/JCI22220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin J, Wu H, Tarr PT, Zhang CY, Wu Z, Boss O, et al. Transcriptional co-activator PGC-1 alpha drives the formation of slow-twitch muscle fibres. Nature. 2002;418:797–801. doi: 10.1038/nature00904. [DOI] [PubMed] [Google Scholar]
- Lin Z, Murtaza I, Wang K, Jiao J, Gao J, Li PF. miR-23a functions downstream of NFATc3 to regulate cardiac hypertrophy. Proc Natl Acad Sci U S A. 2009;106:12103–8. doi: 10.1073/pnas.0811371106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu HB, Yang BF, Dong DL. Calcineurin and electrical remodeling in pathologic cardiac hypertrophy. Trends Cardiovasc Med. 2010;20:148–53. doi: 10.1016/j.tcm.2010.12.003. [DOI] [PubMed] [Google Scholar]
- Long YC, Glund S, Garcia-Roves PM, Zierath JR. Calcineurin regulates skeletal muscle metabolism via coordinated changes in gene expression. J Biol Chem. 2007;282:1607–14. doi: 10.1074/jbc.M609208200. [DOI] [PubMed] [Google Scholar]
- Mallinson J, Meissner J, Chang KC. Chapter 2. Calcineurin signaling and the slow oxidative skeletal muscle fiber type. Int Rev Cell Mol Biol. 2009;277:67–101. doi: 10.1016/S1937-6448(09)77002-9. [DOI] [PubMed] [Google Scholar]
- Mammucari C, Milan G, Romanello V, Masiero E, Rudolf R, Del Piccolo P, et al. FoxO3 controls autophagy in skeletal muscle in vivo. Cell Metab. 2007;6:458–71. doi: 10.1016/j.cmet.2007.11.001. [DOI] [PubMed] [Google Scholar]
- Mancini M, Toker A. NFAT proteins: emerging roles in cancer progression. Nat Rev Cancer. 2009;9:810–20. doi: 10.1038/nrc2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masuda S, Inui K. An up-date review on individualized dosage adjustment of calcineurin inhibitors in organ transplant patients. Pharmacol Ther. 2006;112:184–98. doi: 10.1016/j.pharmthera.2006.04.006. [DOI] [PubMed] [Google Scholar]
- McCarthy JJ, Esser KA, Peterson CA, Dupont-Versteegden EE. Evidence of MyomiR network regulation of beta-myosin heavy chain gene expression during skeletal muscle atrophy. Physiol Genomics. 2009;39:219–26. doi: 10.1152/physiolgenomics.00042.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell PO, Mills ST, Pavlath GK. Calcineurin differentially regulates maintenance and growth of phenotypically distinct muscles. Am J Physiol Cell Physiol. 2002;282:C984–92. doi: 10.1152/ajpcell.00483.2001. [DOI] [PubMed] [Google Scholar]
- Mitchell PO, Pavlath GK. Multiple roles of calcineurin in skeletal muscle growth. Clin Orthop Relat Res. 2002:S197–202. doi: 10.1097/00003086-200210001-00023. [DOI] [PubMed] [Google Scholar]
- Miyazaki M, Hitomi Y, Kizaki T, Ohno H, Katsumura T, Haga S, et al. Calcineurin-mediated slow-type fiber expression and growth in reloading condition. Med Sci Sports Exerc. 2006;38:1065–72. doi: 10.1249/01.mss.0000222833.43520.6e. [DOI] [PubMed] [Google Scholar]
- Molkentin JD. Calcineurin-NFAT signaling regulates the cardiac hypertrophic response in coordination with the MAPKs. Cardiovasc Res. 2004;63:467–75. doi: 10.1016/j.cardiores.2004.01.021. [DOI] [PubMed] [Google Scholar]
- Mootha VK, Lindgren CM, Eriksson KF, Subramanian A, Sihag S, Lehar J, et al. PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat Genet. 2003;34:267–73. doi: 10.1038/ng1180. [DOI] [PubMed] [Google Scholar]
- Musaro A, McCullagh KJ, Naya FJ, Olson EN, Rosenthal N. IGF-1 induces skeletal myocyte hypertrophy through calcineurin in association with GATA-2 and NF-ATc1. Nature. 1999;400:581–5. doi: 10.1038/23060. [DOI] [PubMed] [Google Scholar]
- Naya FJ, Mercer B, Shelton J, Richardson JA, Williams RS, Olson EN. Stimulation of slow skeletal muscle fiber gene expression by calcineurin in vivo. J Biol Chem. 2000;275:4545–8. doi: 10.1074/jbc.275.7.4545. [DOI] [PubMed] [Google Scholar]
- Ni YG, Berenji K, Wang N, Oh M, Sachan N, Dey A, et al. Foxo transcription factors blunt cardiac hypertrophy by inhibiting calcineurin signaling. Circulation. 2006;114:1159–68. doi: 10.1161/CIRCULATIONAHA.106.637124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni YG, Wang N, Cao DJ, Sachan N, Morris DJ, Gerard RD, et al. FoxO transcription factors activate Akt and attenuate insulin signaling in heart by inhibiting protein phosphatases. Proc Natl Acad Sci U S A. 2007;104:20517–22. doi: 10.1073/pnas.0610290104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oh M, Rybkin, Copeland V, Czubryt MP, Shelton JM, van Rooij E, et al. Calcineurin is necessary for the maintenance but not embryonic development of slow muscle fibers. Mol Cell Biol. 2005;25:6629–38. doi: 10.1128/MCB.25.15.6629-6638.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Oishi Y, Ogata T, Yamamoto KI, Terada M, Ohira T, Ohira Y, et al. Cellular adaptations in soleus muscle during recovery after hindlimb unloading. Acta Physiol (Oxf) 2008;192:381–95. doi: 10.1111/j.1748-1716.2007.01747.x. [DOI] [PubMed] [Google Scholar]
- Parsons SA, Millay DP, Wilkins BJ, Bueno OF, Tsika GL, Neilson JR, et al. Genetic loss of calcineurin blocks mechanical overload-induced skeletal muscle fiber type switching but not hypertrophy. J Biol Chem. 2004;279:26192–200. doi: 10.1074/jbc.M313800200. [DOI] [PubMed] [Google Scholar]
- Parsons SA, Wilkins BJ, Bueno OF, Molkentin JD. Altered skeletal muscle phenotypes in calcineurin Aalpha and Abeta gene-targeted mice. Mol Cell Biol. 2003;23:4331–43. doi: 10.1128/MCB.23.12.4331-4343.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Patti ME, Butte AJ, Crunkhorn S, Cusi K, Berria R, Kashyap S, et al. Coordinated reduction of genes of oxidative metabolism in humans with insulin resistance and diabetes: Potential role of PGC1 and NRF1. Proc Natl Acad Sci U S A. 2003;100:8466–71. doi: 10.1073/pnas.1032913100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Powers SK, Kavazis AN, DeRuisseau KC. Mechanisms of disuse muscle atrophy: role of oxidative stress. Am J Physiol Regul Integr Comp Physiol. 2005;288:R337–44. doi: 10.1152/ajpregu.00469.2004. [DOI] [PubMed] [Google Scholar]
- Powers SK, Kavazis AN, McClung JM. Oxidative stress and disuse muscle atrophy. J Appl Physiol. 2007;102:2389–97. doi: 10.1152/japplphysiol.01202.2006. [DOI] [PubMed] [Google Scholar]
- Powers SK, Smuder AJ, Judge AR. Oxidative stress and disuse muscle atrophy: cause or consequence? Curr Opin Clin Nutr Metab Care. 2012;15:240–5. doi: 10.1097/MCO.0b013e328352b4c2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Price SR, Gooch JL, Donaldson SK, Roberts-Wilson TK. Muscle atrophy in chronic kidney disease results from abnormalities in insulin signaling. J Ren Nutr. 2010;20:S24–8. doi: 10.1053/j.jrn.2010.05.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Quiat D, Olson EN. MicroRNAs in cardiovascular disease: from pathogenesis to prevention and treatment. J Clin Invest. 2013;123:11–8. doi: 10.1172/JCI62876. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed SA, Sandesara PB, Senf SM, Judge AR. Inhibition of FoxO transcriptional activity prevents muscle fiber atrophy during cachexia and induces hypertrophy. FASEB J. 2012;26:987–1000. doi: 10.1096/fj.11-189977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts-Wilson TK, Reddy RN, Bailey JL, Zheng B, Ordas R, Gooch JL, et al. Calcineurin signaling and PGC-1alpha expression are suppressed during muscle atrophy due to diabetes. Biochim Biophys Acta. 2010;1803:960–7. doi: 10.1016/j.bbamcr.2010.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rusnak F, Mertz P. Calcineurin: form and function. Physiol Rev. 2000;80:1483–521. doi: 10.1152/physrev.2000.80.4.1483. [DOI] [PubMed] [Google Scholar]
- Sacheck JM, Hyatt JP, Raffaello A, Jagoe RT, Roy RR, Edgerton VR, et al. Rapid disuse and denervation atrophy involve transcriptional changes similar to those of muscle wasting during systemic diseases. FASEB J. 2007;21:140–55. doi: 10.1096/fj.06-6604com. [DOI] [PubMed] [Google Scholar]
- Sacheck JM, Ohtsuka A, McLary SC, Goldberg AL. IGF-I stimulates muscle growth by suppressing protein breakdown and expression of atrophy-related ubiquitin ligases, atrogin-1 and MuRF1. Am J Physiol Endocrinol Metab. 2004;287:E591–601. doi: 10.1152/ajpendo.00073.2004. [DOI] [PubMed] [Google Scholar]
- Sakuma K, Akiho M, Nakashima H, Nakao R, Hirata M, Inashima S, et al. Cyclosporin A modulates cellular localization of MEF2C protein and blocks fiber hypertrophy in the overloaded soleus muscle of mice. Acta Neuropathol. 2008;115:663–74. doi: 10.1007/s00401-008-0371-5. [DOI] [PubMed] [Google Scholar]
- Sandri M. Signaling in muscle atrophy and hypertrophy. Physiology (Bethesda) 2008;23:160–70. doi: 10.1152/physiol.00041.2007. [DOI] [PubMed] [Google Scholar]
- Sandri M, Lin J, Handschin C, Yang W, Arany ZP, Lecker SH, et al. PGC-1alpha protects skeletal muscle from atrophy by suppressing FoxO3 action and atrophy-specific gene transcription. Proc Natl Acad Sci U S A. 2006;103:16260–5. doi: 10.1073/pnas.0607795103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sandri M, Sandri C, Gilbert A, Skurk C, Calabria E, Picard A, et al. Foxo transcription factors induce the atrophy-related ubiquitin ligase atrogin-1 and cause skeletal muscle atrophy. Cell. 2004;117:399–412. doi: 10.1016/s0092-8674(04)00400-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiaffino S, Dyar KA, Ciciliot S, Blaauw B, Sandri M. Mechanisms regulating skeletal muscle growth and atrophy. FEBS J. 2013 doi: 10.1111/febs.12253. [DOI] [PubMed] [Google Scholar]
- Schiaffino S, Mammucari C. Regulation of skeletal muscle growth by the IGF1-Akt/PKB pathway: insights from genetic models. Skelet Muscle. 2011;1:4. doi: 10.1186/2044-5040-1-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schiaffino S, Sandri M, Murgia M. Activity-dependent signaling pathways controlling muscle diversity and plasticity. Physiology (Bethesda) 2007;22:269–78. doi: 10.1152/physiol.00009.2007. [DOI] [PubMed] [Google Scholar]
- Schiaffino S, Serrano A. Calcineurin signaling and neural control of skeletal muscle fiber type and size. Trends Pharmacol Sci. 2002;23:569–75. doi: 10.1016/s0165-6147(02)02111-9. [DOI] [PubMed] [Google Scholar]
- Semsarian C, Wu MJ, Ju YK, Marciniec T, Yeoh T, Allen DG, et al. Skeletal muscle hypertrophy is mediated by a Ca2+-dependent calcineurin signalling pathway. Nature. 1999;400:576–81. doi: 10.1038/23054. [DOI] [PubMed] [Google Scholar]
- Smuder AJ, Hudson MB, Nelson WB, Kavazis AN, Powers SK. Nuclear factor-kappaB signaling contributes to mechanical ventilation-induced diaphragm weakness*. Crit Care Med. 2012;40:927–34. doi: 10.1097/CCM.0b013e3182374a84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stevenson EJ, Giresi PG, Koncarevic A, Kandarian SC. Global analysis of gene expression patterns during disuse atrophy in rat skeletal muscle. J Physiol. 2003;551:33–48. doi: 10.1113/jphysiol.2003.044701. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stupka N, Plant DR, Schertzer JD, Emerson TM, Bassel-Duby R, Olson EN, et al. Activated calcineurin ameliorates contraction-induced injury to skeletal muscles of mdx dystrophic mice. J Physiol. 2006;575:645–56. doi: 10.1113/jphysiol.2006.108472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Talmadge RJ, Otis JS, Rittler MR, Garcia ND, Spencer SR, Lees SJ, et al. Calcineurin activation influences muscle phenotype in a muscle-specific fashion. BMC Cell Biol. 2004;5:28. doi: 10.1186/1471-2121-5-28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ventadour S, Attaix D. Mechanisms of skeletal muscle atrophy. Curr Opin Rheumatol. 2006;18:631–5. doi: 10.1097/01.bor.0000245731.25383.de. [DOI] [PubMed] [Google Scholar]
- Wada S, Kato Y, Okutsu M, Miyaki S, Suzuki K, Yan Z, et al. Translational suppression of atrophic regulators by microRNA-23a integrates resistance to skeletal muscle atrophy. J Biol Chem. 2011;286:38456–65. doi: 10.1074/jbc.M111.271270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wenz T, Rossi SG, Rotundo RL, Spiegelman BM, Moraes CT. Increased muscle PGC-1alpha expression protects from sarcopenia and metabolic disease during aging. Proc Natl Acad Sci U S A. 2009;106:20405–10. doi: 10.1073/pnas.0911570106. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- Whidden MA, Smuder AJ, Wu M, Hudson MB, Nelson WB, Powers SK. Oxidative stress is required for mechanical ventilation-induced protease activation in the diaphragm. J Appl Physiol. 2010;108:1376–82. doi: 10.1152/japplphysiol.00098.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilkins BJ, Molkentin JD. Calcium-calcineurin signaling in the regulation of cardiac hypertrophy. Biochem Biophys Res Commun. 2004;322:1178–91. doi: 10.1016/j.bbrc.2004.07.121. [DOI] [PubMed] [Google Scholar]
- Williams CR, Gooch JL. Calcineurin inhibitors and immunosuppression - a tale of two isoforms. Expert Rev Mol Med. 2012;14:e14. doi: 10.1017/erm.2012.8. [DOI] [PubMed] [Google Scholar]
- Workeneh BT, Mitch WE. Review of muscle wasting associated with chronic kidney disease. Am J Clin Nutr. 2010;91:1128S–32S. doi: 10.3945/ajcn.2010.28608B. [DOI] [PubMed] [Google Scholar]
- Zhao J, Brault JJ, Schild A, Cao P, Sandri M, Schiaffino S, et al. FoxO3 coordinately activates protein degradation by the autophagic/lysosomal and proteasomal pathways in atrophying muscle cells. Cell Metab. 2007;6:472–83. doi: 10.1016/j.cmet.2007.11.004. [DOI] [PubMed] [Google Scholar]
- Zheng B, Ohkawa S, Li H, Roberts-Wilson TK, Price SR. FOXO3a mediates signaling crosstalk that coordinates ubiquitin and atrogin-1/MAFbx expression during glucocorticoid-induced skeletal muscle atrophy. FASEB J. 2010;24:2660–9. doi: 10.1096/fj.09-151480. [DOI] [PMC free article] [PubMed] [Google Scholar]