Abstract
Interactions between functionalized noble-metal particles in aqueous solution are central to applications relying on controlled equilibrium association. Herein we obtain the potentials of mean force (PMF) for pair-interactions between functionalized gold nanoparticles (AuNPs) in physiological saline, based upon > 1000-ns experiments in silico of all-atom model systems under equilibrium and nonequilibrium conditions. Four types of functionalization are built by coating the globular Au144 cluster each with 60 thiolate groups: GS-AuNP (glutathionate), PhS-AuNP (thiophenol), CyS-AuNP (cysteinyl), and p-APhS-AuNP (para-aminothiophenol), which are, respectively, negatively charged, hydrophobic (neutral-nonpolar), hydrophilic (neutral-polar), and positively charged at neutral pH. The results confirm the behaviour expected of neutral (hydrophilic or hydrophobic) particles in dilute aqueous environment, but the PMF curves demonstrate that the charged AuNPs interact with one another in a unique way — mediated by H2O molecules and electrolyte (Na+,Cl−) — in a physiological environment. In the case of two GS-AuNPs, the excess, neutralizing Na+ ions form a mobile (or ‘dynamic’) cloud of enhanced concentration between like-charged GS-AuNPs, inducing a moderate attraction (~ 25-kT) between them. And, to a lesser degree, for a pair of p-APhS-AuNPs, the excess, neutralizing Cl− ions (less mobile than Na+) also form a cloud of higher concentration between the two like-charged p-APhS-AuNPs, inducing weaker yet significant attractions (~ 12-kT). In the combination of one GS- and one p-APhS-AuNP, the direct, attractive Coulombic force is completely screened out while the solvation effects give rise to moderate repulsion between the two unlike-charged AuNPs.
Due to their intermediacy in size, nanoparticles (NPs) possess unique mesoscopic properties that neither microscopic nor macroscopic bodies have. They are therefore widely investigated for various purposes. For examples, some NPs have been studied in the pursuit of nanomedicine.1–7 In particular, gold nanoparticles (AuNPs) are being exploited for drug delivery and bioimaging,8–15 which can involve solutions at high AuNP concentrations and controlled, reversible association of NPs.16 In light of the current studies, it is of essential relevance to accurately determine the interactions between functionalized AuNPs in aqueous environment approximating physiological conditions. While in vitro and in vivo experiments have the final say, in silico studies, complementing in vitro/vivo experiments, can give atomistic insights of details that cannot be gained by in vitro/vivo means alone. For example, in silico experiments17–20 have been employed to study the diffusion of functionalized AuNPs and to study how an AuNP interacts with biomolecules and cell membranes.21–28 In this paper, we present in silico studies of AuNPs coated with hydrophobic, neutral hydrophilic, positively charged, and negatively charged organic/biological groups that lead to the determination of AuNP-AuNP interactions in physiological saline. The selected AuNP size is one that has been in continuous use in biological investigations over the past decade.29, 30 Our main objective is to elucidate atomistic details of how functionalized AuNPs interact with one another under near-physiological conditions.
It is interesting to note that our in silico study of all-atom models concludes that like-charged AuNPs in physiological saline attract one another significantly over a distance a few times the NP diameter. This counter-intuitive behavior is in line with the long-studied attractions between like-charged colloids in electrolyte solutions.31–57 However, oppositely charged colloids have been rarely studied.58
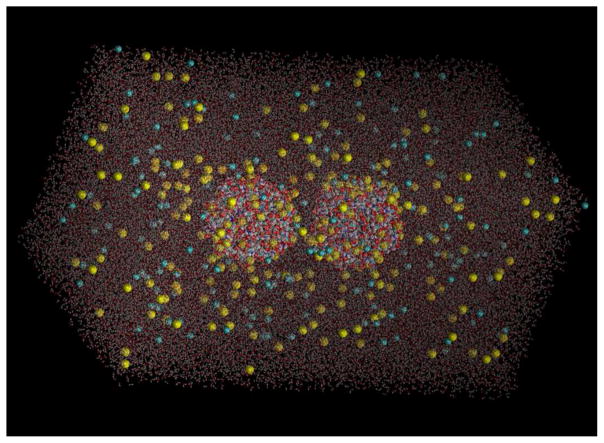
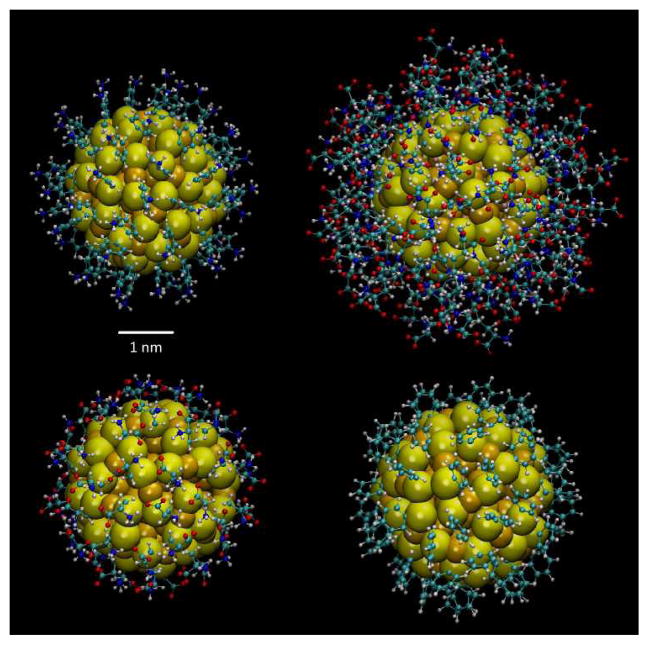
We study five atomistic model systems (see Table 1) each having a pair of AuNPs in a box of physiological saline (150-mM NaCl; 1-bar; 298-K). An example is shown in Fig. 1. A single globular AuNP comprises a compact 144-atom metal core protected by a surface layer of 60 thiolate groups (shown in Fig. 2) either the negatively charged tripeptide L-glutathionates (GS-); the hydrophobic thiophenols (PhS-), the hydrophilic (neutral but polar) amino-acid L-cysteinates (CyS-), or the positively charged aminothiophenols (p-APhS-). The ligand structures are illustrated in the supplemental information (Fig. S1). The atomic structure of the inorganic core, including the sulfur-gold bonding, was taken from Refs. 59–61. System I consists of two GS-AuNPs; System II, two PhS-AuNPs; System III; two CyS-AuNPs; System IV, two p-APhS-AuNPs; System V, one GS-AuNP and one p-APhS-AuNP. All share a common dimension of 2.0-nm diameter (enclosing the inorganic core Au144S60).
Table 1.
Details of systems considered in this study.
| System | NP1 Charge |
NP2 Charge |
Interaction | Range of attraction |
|---|---|---|---|---|
| I | GS-AuNP −60e |
GS-AuNP −60e |
−25 kT | 3.7 to 4 nm |
| II | PhS-AuNP 0e |
PhS-AuNP 0e |
−15 kT | 2.7 to 4 nm |
| III | CyS-AuNP 0e |
CyS-AuNP 0e |
repulsive | |
| IV |
p-APhSAuNP +60e |
p-APhSAuNP +60e |
−12 kT | 3 to 3.7 nm |
| V |
p-APhSAuNP +60e |
GS-AuNP −60e |
repulsive |
Figure 1.
In silico box of physiological saline with two AuNPs each coated with 60 glutathiones. The system dimensions are 10nm×10nm×20nm, consisting of 191355 atoms. Waters are in licorice (sticks) and the NPs and ions in vdw (balls) representations respectively. All colored by element names (Na+, yellow, Cl−, grayish blue, O, red, C, cyan, N, blue, and H white). Au and S are not visible in this graph. All graphics in this paper were rendered with VMD69.
Figure 2.
144-AuNPs coated with 60 aminothiophenols (top left), glutathiones (top right), cysteines (bottom left), and thiophenols (bottom right). Gold and sulphur atoms are in VDW representation (large spheres) in gold and light yellow colors respectively. The ligands are in CPK (ball-and-stick) representations colored with element names (O, red, C, cyan, N, blue, and H white).
All the inter- and intra-molecular (other than Au-Au and S-Au) interactions were represented with the CHARMM36 force field,62, 63 to which we add the following van der Waals (vdW) parameters for Au: σ = 1.66 Å, ε = −0.10585kcal/mol. Water was represented with the TIP3P model.64 Parameters for the interactions involving sulfur and gold atoms were taken from Ref. 65. The in silico experiments, namely, molecular dynamics (MD) simulations, were implemented using NAMD66 as adapted for steered molecular dynamics (SMD).67, 68 Periodic boundary conditions were applied in all dimensions. The cut-off distance applied to the vdW interactions is 1.2 nm, with a switching distance of 1.0 nm and a pair list distance of 1.4 nm. Particle-mesh Ewald was employed to compute the electrostatic interactions in an exact manner. Langevin MD was implemented with a time-step of 1.0 fs for short range interactions and 4.0 fs for long range interactions. The Langevin damping was chosen as 5.0/ps. The temperature was maintained at T=298-K and pressure at 1.0-bar.
The building procedure of System I is summarized as follows: (1) Starting from the widely accepted59–61 structure of Au144(SR)60 cluster, [R = H or CH3] we build a GS-AuNP by bonding one moiety (HS-less glutathione) to each of the 60 S atoms on the surface of the cluster. The mutual spatial arrangements of the 60 glutathionate groups were attained by minimizing the interaction energy of the GS-AuNP while fixing the coordinates of all the Au and S atoms to their initial values. Then we equilibrate the GS-AuNP for 100-ns at 298 K in vacuum, first with the Au and S constrained, and then for another 100-ns, without constraints on any of the atoms. It should be noted that the Au-S core was found to be stable, preserving the icosahedral symmetry, which indicates the validity of the interaction parameters we used. (2) We replicate the GS-AuNP and place two of them in a box of water. We then neutralize the system with 120 Na+ ions and salinate the system with 150 mM NaCl. Thus we have System I. The procedures for building Systems II–V are essentially identical to those for System I and, therefore, not explicitly stated here. Some details are provided in the supplementary information (Figs. S2 and S3).
Equilibrium molecular dynamics
Following initial optimizations, we executed 100-ns equilibrium MD runs for each system. During the last 50-ns of the MD run, we computed the root-mean-square-deviation of the 144-Au atoms from their initial structure (excluding the overall diffusion and rotation of the NP) to check the validity of the interaction parameters we used. Note that none of the atoms were fixed or constrained during the equilibrium MD run. All atoms were freely subject to the stochastic dynamics on the basis of atomistic interactions of the system modeled by the CHARMM36 force field.
Nonequilibrium steered molecular dynamics
We followed the multi-sectional scheme of Ref. 70, conducting SMD runs for each of the Systems I–V in order to compute the PMF71–76 as a function of the distance r between the centres-of-mass of the pairs of AuNPs. In each case, we steered (pulled) the pair of AuNPs toward each other at the same pulling speed vd = 1.0nm/ns in every case. The pulling was along the z-axis and over sections of 0.1-nm each in width. Namely, the center-of-mass (COM) coordinates of the innermost 12-Au core (three degrees of freedom) were controlled as functions (0, 0, zi ± vdt) of time t along a forward-reverse pulling path in the ith section while all other 0.57 million degrees of freedom were left freely determined by the stochastic dynamics of the system. Here zi and zi+1 are, respectively, the z-coordinates of the two end points of the ith section over which we pulled the centre-of-mass of the inner Au core forward and backward to sample four (eight in some cases) forward-reverse pulling paths in each given section. The work done to the system, along these pulling paths (shown in supplementary Figs. S5 to S9), were used in the Brownian-dynamics fluctuation-dissipation theorem77 to compute the free-energy difference (reversible work or PMF) as a function of the distance, i.e. the pairwise separation of AuNPs. Namely, the free-energy difference between two states:
| (1) |
Here WA→B is the work done to the system along a forward path when the NP pair was steered from A to Z. WZ→A = WB→A − WB→Z is the work Z for the part of a reverse path when the NP pair was pulled from Z to A. kB is the Boltzmann constant and T is the absolute temperature. zA and zB are the z-coordinates of the COM of one of the two NPs of the pair at the end states A and B of the system, respectively. Note that the pulling was symmetrical between the two NPs of each pair.
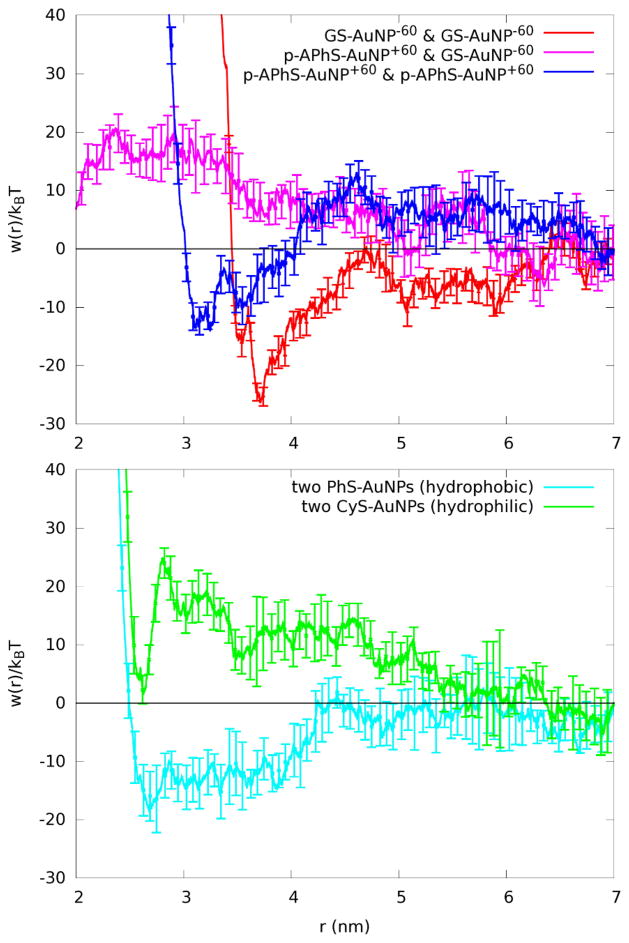
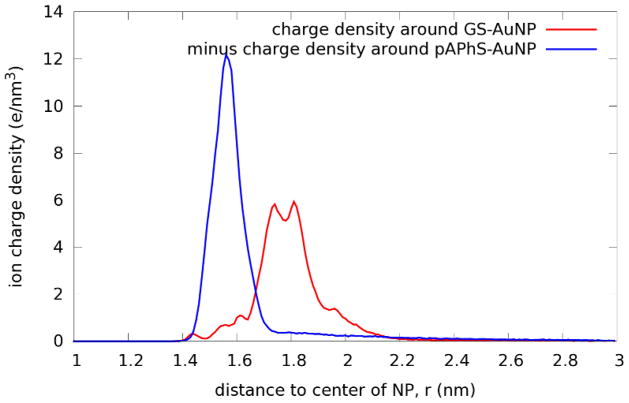
Stability of the globular inorganic Au144S60 core was clearly established in all five cases. None of the four different surfactants has a substantial effect on the arrangement, as the icosahedral symmetry is maintained, including the 30-fold Au-S-Au-S-Au staple geometry. Figure S4 shows quantitatively that the rms-deviation (from the initial Au-atom coordinates) remains within 0.02-nm, confirming the validity of the parameters used to characterize the Au-S and Au-Au interactions, which are not part of the standard CHARMM force field. Our main finding is that like-charged AuNPs in physiological saline do not repel but rather attract one another, via electrolyte ion-mediated attractions, starting from distances around a few times the AuNP diameter. Fig. 3 shows the PMF between two negatively charged GS-AuNPs (−60e charge each), and similarly the PMF between two positively charged p-APhS-AuNPs (+60e charge each). These PMF curves establish that the direct Coulombic repulsion between two like-charged AuNPs are very effectively screened by the physiological saline that provides 120 extra counter ions (120 Na+ ions, for GS-AuNPs; 120 Cl− ions, for pAPhS-AuNPs). Some of the counter ions reside at the surface of these AuNPs, as illustrated in Fig. 4, reducing its effective charge to a small fraction of 60e. The free counter-ions (those not residing at the AuNP surfaces) gather in between the two like-charged AuNPs, as much as electrons gather in between the two protons of an H2 molecule (on the Å length-scale), mediating attraction between two like-charged AuNPs.
Figure 3.
Reversible work for bringing two NPs together. Vertical axis: Potential of mean force. Horizontal axis: Distance between the centers of mass of the two NPs involved.
Figure 4.
The ion-charge density distributions around isolated AuNPs: the positively charged p-APhS-AuNP (blue, sign reversed) and the negatively charged GS-AuNP (red). Each curve includes contribution from all the Cl− and Na+ ions.
The origin of the difference between the GS-AuNP pair and the p-APhS-AuNP pair, as indicated in Fig. 4, is that the Cl− ions are less mobile than Na+ ions and, therefore, are more effective in neutralizing the positively charged pAPhS-AuNP. This implies a reduced effective charge in p-APhS-AuNP, versus GS-AuNP. Consequently, the attraction between p-APhS-AuNPs is weaker than that between GS-AuNPs. Note that the distances corresponding to the minima of the PMFs for both like-charged particle pairs in Fig. 3 coincide with 2 times the distances corresponding to the maxima of the surrounding charge densities in Fig. 4.
By contrast, oppositely-charged AuNPs in physiological saline do not attract one another. The PMF for the unlike-charged pair of AuNPs is also shown in Fig. 3, which clearly indicates a small but significant effective repulsion between the two. In this case, the direct Coulombic attraction, between a positively charged p-APhS-AuNP and a negatively charged GS-AuNP (see supplementary Fig. S10), is totally screened out. Instead, the long-range interaction between them is repulsive due to the solvation effects. 78
Fig. 3 further shows that the neutral hydrophilic AuNPs — the PMF is for a pair of such AuNPs, CyS-AuNPs — tend to separate from one another. This behaviour is easily rationalized in terms of the drive to maximize the total area exposed to aqueous solution.
Finally, the neutral hydrophobic AuNPs tends to coagulate together. Fig. 3 also shows the PMF for a pair of (neutral) thiophenol-coated AuNPs, indicating the long range and deep, broad free-energy minimum associated with the classic hydrophobic effect, i.e. the reduction in the total (non-polar) surface-area exposed to aqueous solution.78
Conclusions
Extensive atomistic in silico experiments (equilibrium MD and nonequilibrium SMD simulations), based on realistic molecular structures for all solution components, lead to the conclusion that the interactions between functionalized AuNPs in physiological saline are fundamentally distinct from the case of vacuum and other environments. On the length-scale of a few times the AuNP diameter, the direct Coulombic interactions are completely screened out. Like-charged AuNPs do not repel and opposite-charged AuNPs do not attract one another. The water molecules and the Na+ and Cl− ions play essential roles to mediate effective attractions between a pair of like-charged AuNPs (positive or negative) as well as a pair of hydrophobic AuNPs. This fact also explains the experimental observation that Thiol passivated AuNPs are stable for long time in sharp contrast with nonpassivated particles. Solvation effects cause both the opposite-charged pair and the neutral-hydrophilic pair to dissociate. These predicted characteristics of various functionalized AuNPs are expected to impact further research of AuNPs in biological applications.
Supplementary Material
Acknowledgments
The authors are indebted for support to the following funding agencies: the NIH (Grants #GM084834 and #G12RR013646), the Welch Foundation (Project AX-1615), and the NSF (Grants #DMR-0934218 and #1103730). They also acknowledge computational support from the Texas Advanced Computing Centre and the NSF XSEDE (TG-MCB130070).
Footnotes
Electronic Supplementary Information (ESI) available: Computational details. See DOI: 10.1039/c000000x/
References
- 1.Ackerson CJ, Jadzinsky PD, Sexton JZ, Bushnell DA, Kornberg RD. Bioconjugate Chemistry. 2010;21:214–218. doi: 10.1021/bc900135d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Antosova A, Gazova Z, Fedunova D, Valusova E, Bystrenova E, Valle F, Daxnerova Z, Biscarini F, Antalik M. Materials Science and Engineering: C. 2012;32:2529–2535. [Google Scholar]
- 3.AshaRani PV, Low Kah Mun G, Hande MP, Valiyaveettil S. ACS Nano. 2008;3:279–290. doi: 10.1021/nn800596w. [DOI] [PubMed] [Google Scholar]
- 4.Bowman MC, Ballard TE, Ackerson CJ, Feldheim DL, Margolis DM, Melander C. Journal of the American Chemical Society. 2008;130:6896–6897. doi: 10.1021/ja710321g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Chung TH, Wu SH, Yao M, Lu CW, Lin YS, Hung Y, Mou CY, Chen YC, Huang DM. Biomaterials. 2007;28:2959–2966. doi: 10.1016/j.biomaterials.2007.03.006. [DOI] [PubMed] [Google Scholar]
- 6.Lin CAJ, Yang TY, Lee CH, Huang SH, Sperling RA, Zanella M, Li JK, Shen JL, Wang HH, Yeh HI, Parak WJ, Chang WH. ACS Nano. 2009;3:395–401. doi: 10.1021/nn800632j. [DOI] [PubMed] [Google Scholar]
- 7.Zhang Y, Yang M, Park JH, Singelyn J, Ma H, Sailor MJ, Ruoslahti E, Ozkan M, Ozkan C. Small. 2009;5:1990–1996. doi: 10.1002/smll.200900520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartczak D, Muskens OL, Sanchez-Elsner T, Kanaras AG, Millar TM. ACS Nano. 2013;7:5628–5636. doi: 10.1021/nn402111z. [DOI] [PubMed] [Google Scholar]
- 9.Bresee J, Maier KE, Boncella AE, Melander C, Feldheim DL. Small. 2011;7:2027–2031. doi: 10.1002/smll.201100420. [DOI] [PubMed] [Google Scholar]
- 10.Chen R, Ratnikova TA, Stone MB, Lin S, Lard M, Huang G, Hudson JS, Ke PC. Small. 2010;6:612–617. doi: 10.1002/smll.200901911. [DOI] [PubMed] [Google Scholar]
- 11.Cho EC, Au L, Zhang Q, Xia Y. Small. 2010;6:517–522. doi: 10.1002/smll.200901622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cho EC, Xie J, Wurm PA, Xia Y. Nano Lett. 2009;9:1080–1084. doi: 10.1021/nl803487r. [DOI] [PubMed] [Google Scholar]
- 13.Kim C, Agasti SS, Zhu Z, Isaacs L, Rotello VM. Nat Chem. 2010;2:962–966. doi: 10.1038/nchem.858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leroueil PR, Berry SA, Duthie K, Han G, Rotello VM, McNerny DQ, Baker JR, Orr BG, Banaszak Holl MM. Nano Lett. 2008;8:420–424. doi: 10.1021/nl0722929. [DOI] [PubMed] [Google Scholar]
- 15.Verma A, Uzun O, Hu Y, Hu Y, Han HS, Watson N, Chen S, Irvine DJ, Stellacci F. Nat Mater. 2008;7:588–595. doi: 10.1038/nmat2202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Murthy AK, Stover RJ, Borwankar AU, Nie GD, Gourisankar S, Truskett TM, Sokolov KV, Johnston KP. ACS Nano. 2012;7:239–251. doi: 10.1021/nn303937k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Heikkilä E, Gurtovenko AA, Martinez-Seara H, Häkkinen H, Vattulainen I, Akola J. The Journal of Physical Chemistry C. 2012;116:9805–9815. [Google Scholar]
- 18.Lee OS, Schatz GC. The Journal of Physical Chemistry C. 2009;113:2316–2321. [Google Scholar]
- 19.Popov KI, Nap RJ, Szleifer I, de la Cruz MO. Journal of Polymer Science Part B: Polymer Physics. 2012;50:852–862. [Google Scholar]
- 20.Djebaili T, Richardi J, Abel S, Marchi M. The Journal of Physical Chemistry C. 2013;117:17791–17800. [Google Scholar]
- 21.Chithrani BD, Ghazani AA, Chan WCW. Nano Lett. 2006;6:662–668. doi: 10.1021/nl052396o. [DOI] [PubMed] [Google Scholar]
- 22.Roiter Y, Ornatska M, Rammohan AR, Balakrishnan J, Heine DR, Minko S. Nano Lett. 2008;8:941–944. doi: 10.1021/nl080080l. [DOI] [PubMed] [Google Scholar]
- 23.Zhang S, Li J, Lykotrafitis G, Bao G, Suresh S. Advanced Materials. 2009;21:419–424. doi: 10.1002/adma.200801393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lin J, Zhang H, Chen Z, Zheng Y. ACS Nano. 2010;4:5421–5429. doi: 10.1021/nn1010792. [DOI] [PubMed] [Google Scholar]
- 25.Verma A, Stellacci F. Small. 2010;6:12–21. doi: 10.1002/smll.200901158. [DOI] [PubMed] [Google Scholar]
- 26.Yang AC, Weng CI. The Journal of Physical Chemistry C. 2010;114:8697–8709. [Google Scholar]
- 27.Lin JQ, Zheng YG, Zhang HW, Chen Z. Langmuir. 2011;27:8323–8332. doi: 10.1021/la201086u. [DOI] [PubMed] [Google Scholar]
- 28.Rocha ELd, Caramori GF, Rambo CR. Physical Chemistry Chemical Physics. 2013;15:2282–2290. doi: 10.1039/c2cp44035k. [DOI] [PubMed] [Google Scholar]
- 29.Hainfeld JF, Liu W, Halsey CMR, Freimuth P, Powell RD. Journal of Structural Biology. 1999;127:185–198. doi: 10.1006/jsbi.1999.4149. [DOI] [PubMed] [Google Scholar]
- 30.Ackerson CJ, Powell RD, Hainfeld JF. In: Methods in Enzymology. Grant JJ, editor. Vol. 481. Academic Press; 2010. pp. 195–230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Langmuir I. The Journal of Chemical Physics. 1938;6:873–896. [Google Scholar]
- 32.Oosawa F. Biopolymers. 1968;6:145–158. [Google Scholar]
- 33.Ise N, Okubo T. Accounts of Chemical Research. 1980;13:303–309. [Google Scholar]
- 34.Patey GN. The Journal of Chemical Physics. 1980;72:5763–5771. [Google Scholar]
- 35.Wu J, Bratko D, Prausnitz JM. Proceedings of the National Academy of Sciences. 1998;95:15169–15172. doi: 10.1073/pnas.95.26.15169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Netz RR, Orland H. Eur Phys J E. 2000;1:203–214. [Google Scholar]
- 37.Otto F, Patey GN. The Journal of Chemical Physics. 2000;112:8939–8949. [Google Scholar]
- 38.Boström M, Williams DRM, Ninham BW. Physical Review Letters. 2001;87:168103. doi: 10.1103/PhysRevLett.87.168103. [DOI] [PubMed] [Google Scholar]
- 39.Gómez-Guzmán O, Ruiz-Garcia J. Journal of Colloid and Interface Science. 2005;291:1–6. doi: 10.1016/j.jcis.2005.04.100. [DOI] [PubMed] [Google Scholar]
- 40.Dahirel V, Jardat M, Dufreche JF, Turq P. Physical Chemistry Chemical Physics. 2008;10:5147–5155. doi: 10.1039/b806315j. [DOI] [PubMed] [Google Scholar]
- 41.Dai L, Mu Y, Nordenskiöld L, van der Maarel JRC. Physical Review Letters. 2008;100:118301. doi: 10.1103/PhysRevLett.100.118301. [DOI] [PubMed] [Google Scholar]
- 42.Tan ZJ, Chen SJ. Biophysical Journal. 2008;95:738–752. doi: 10.1529/biophysj.108.131524. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Tata BVR, Mohanty PS, Valsakumar MC. Solid State Communications. 2008;147:360–365. [Google Scholar]
- 44.Nagornyak E, Yoo H, Pollack GH. Soft Matter. 2009;5:3850–3857. [Google Scholar]
- 45.Dahirel V, Jardat M. Current Opinion in Colloid & Interface Science. 2010;15:2–7. [Google Scholar]
- 46.Ibarra-Armenta JG, Martin-Molina A, Quesada-Perez M. Physical Chemistry Chemical Physics. 2011;13:13349–13357. doi: 10.1039/c1cp21162e. [DOI] [PubMed] [Google Scholar]
- 47.Israelachvili JN. Intermolecular and Surface Forces. 3. Academic Press; Boston: 2011. [Google Scholar]
- 48.Manning GS. Eur Phys J E. 2011;34:1–18. doi: 10.1140/epje/i2011-11132-6. [DOI] [PubMed] [Google Scholar]
- 49.Paillusson F, Dahirel V, Jardat M, Victor JM, Barbi M. Physical Chemistry Chemical Physics. 2011;13:12603–12613. doi: 10.1039/c1cp20324j. [DOI] [PubMed] [Google Scholar]
- 50.Buyukdagli S, Achim CV, Ala-Nissila T. The Journal of Chemical Physics. 2012;137:104902–104918. doi: 10.1063/1.4750044. [DOI] [PubMed] [Google Scholar]
- 51.Jho YS, Safran SA, In M, Pincus PA. Langmuir. 2012;28:8329–8336. doi: 10.1021/la3009943. [DOI] [PubMed] [Google Scholar]
- 52.Turesson M, Jönsson B, Labbez C. Langmuir. 2012;28:4926–4930. doi: 10.1021/la3005008. [DOI] [PubMed] [Google Scholar]
- 53.Ghanbarian S, Rottler J. The Journal of Chemical Physics. 2013;138:084901–084906. doi: 10.1063/1.4793471. [DOI] [PubMed] [Google Scholar]
- 54.Van Lehn RC, Alexander-Katz A. Langmuir. 2013;29:8788–8798. doi: 10.1021/la400756z. [DOI] [PubMed] [Google Scholar]
- 55.Wang LF, Wang LL, Ye XD, Li WW, Ren XM, Sheng GP, Yu HQ, Wang XK. Environmental Science & Technology. 2013;47:5042–5049. doi: 10.1021/es304993j. [DOI] [PubMed] [Google Scholar]
- 56.Wu YY, Wang FH, Tan ZJ. Physics Letters A. 2013;377:1911–1919. [Google Scholar]
- 57.Zhou S. Langmuir. 2013;29:12490–12501. doi: 10.1021/la402860r. [DOI] [PubMed] [Google Scholar]
- 58.Dahirel V, Hansen JP. The Journal of Chemical Physics. 2009;131:084902–084910. doi: 10.1063/1.3193556. [DOI] [PubMed] [Google Scholar]
- 59.Lopez-Acevedo O, Akola J, Whetten RL, Grönbeck H, Häkkinen H. The Journal of Physical Chemistry C. 2009;113:5035–5038. [Google Scholar]
- 60.Bahena D, Bhattarai N, Santiago U, Tlahuice A, Ponce A, Bach SBH, Yoon B, Whetten RL, Landman U, Jose-Yacaman M. The Journal of Physical Chemistry Letters. 2013;4:975–981. doi: 10.1021/jz400111d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Tlahuice-Flores A, Black DM, Bach SBH, Jose-Yacaman M, Whetten RL. Physical Chemistry Chemical Physics. 2013;15:19191–19195. doi: 10.1039/c3cp53902d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Brooks BR, Brooks CL, Mackerell AD, Nilsson L, Petrella RJ, Roux B, Won Y, Archontis G, Bartels C, Boresch S, Caflisch A, Caves L, Cui Q, Dinner AR, Feig M, Fischer S, Gao J, Hodoscek M, Im W, Kuczera K, Lazaridis T, Ma J, Ovchinnikov V, Paci E, Pastor RW, Post CB, Pu JZ, Schaefer M, Tidor B, Venable RM, Woodcock HL, Wu X, Yang W, York DM, Karplus M. Journal of Computational Chemistry. 2009;30:1545–1614. doi: 10.1002/jcc.21287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vanommeslaeghe K, Hatcher E, Acharya C, Kundu S, Zhong S, Shim J, Darian E, Guvench O, Lopes P, Vorobyov I, Mackerell AD. Journal of Computational Chemistry. 2010;31:671–690. doi: 10.1002/jcc.21367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jorgensen WL, Chandrasekhar J, Madura JD, Impey RW, Klein ML. The Journal of Chemical Physics. 1983;79:926–935. [Google Scholar]
- 65.Hautman J, Klein ML. The Journal of Chemical Physics. 1989;91:4994–5001. [Google Scholar]
- 66.Phillips JC, Braun R, Wang W, Gumbart J, Tajkhorshid E, Villa E, Chipot C, Skeel RD, Kalé L, Schulten K. Journal of Computational Chemistry. 2005;26:1781–1802. doi: 10.1002/jcc.20289. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chen LY, Bastien DA, Espejel HE. Physical Chemistry Chemical Physics. 2010;12:6579–6582. doi: 10.1039/b926889h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Chen LY. Physical Chemistry Chemical Physics. 2011;13:6176–6183. doi: 10.1039/c0cp02799e. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Humphrey W, Dalke A, Schulten K. Journal of Molecular Graphics. 1996;14:33–38. doi: 10.1016/0263-7855(96)00018-5. [DOI] [PubMed] [Google Scholar]
- 70.Chen LY. Biochimica et Biophysica Acta (BBA) - Biomembranes. 2013;1828:1786–1793. doi: 10.1016/j.bbamem.2013.03.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Kirkwood JG. The Journal of Chemical Physics. 1935;3:300–313. [Google Scholar]
- 72.Chandler D. The Journal of Chemical Physics. 1978;68:2959–2970. [Google Scholar]
- 73.Pratt LR, Hummer G, Garciá AE. Biophysical Chemistry. 1994;51:147–165. doi: 10.1016/s0301-4622(99)00018-6. [DOI] [PubMed] [Google Scholar]
- 74.Roux B. Computer Physics Communications. 1995;91:275–282. [Google Scholar]
- 75.Allen TW, Andersen OS, Roux B. Biophysical Chemistry. 2006;124:251–267. doi: 10.1016/j.bpc.2006.04.015. [DOI] [PubMed] [Google Scholar]
- 76.Wu J. AIChE Journal. 2006;52:1169–1193. [Google Scholar]
- 77.Chen LY. The Journal of Chemical Physics. 2008;129:144113–144114. doi: 10.1063/1.2992153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ben-Amotz D, Underwood R. Accounts of Chemical Research. 2008;41:957–967. doi: 10.1021/ar7001478. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.