SUMMARY
Androgen receptor (AR) is a ligand-dependent transcription factor that plays a key role in prostate cancer. Little is known about the nature of AR cis-regulatory sites in the human genome. We have mapped the AR binding regions on two chromosomes in human prostate cancer cells by combining chromatin immunoprecipitation (ChIP) with tiled oligonucleotide microarrays. We find that the majority of AR binding regions contain noncanonical AR-responsive elements (AREs). Importantly, we identify a noncanonical ARE as a cis-regulatory target of AR action in TMPRSS2, a gene fused to ETS transcription factors in the majority of prostate cancers. In addition, through the presence of enriched DNA-binding motifs, we find other transcription factors including GATA2 and Oct1 that cooperate in mediating the androgen response. These collaborating factors, together with AR, form a regulatory hierarchy that governs androgen-dependent gene expression and prostate cancer growth and offer potential new opportunities for therapeutic intervention.
INTRODUCTION
The androgen receptor (AR) plays a key role in the growth and maintenance of the normal prostate and the onset and progression of prostate cancer (Heinlein and Chang, 2004). AR is a ligand-dependent transcription factor belonging to the nuclear hormone receptor (NR) superfamily (Mangelsdorf et al., 1995). NRs regulate transcription through recruitment of a number of non-DNA-binding coregulatory factors including coactivators and corepressors to the cis-regulatory regions of target genes. Although NRs share common coregulatory complexes, the temporal, spatial, and functional binding pattern of each receptor such as AR and its coregulatory factors to genuine chromatin targets is distinct. Previous chromatin immunoprecipitation (ChIP) studies focus primarily on AR regulation of only a single target gene, prostate-specific antigen (PSA), which is commonly used for prostate cancer diagnosis and for monitoring therapeutic response (Balk et al., 2003). In the presence of androgens, AR and its coactivators increasingly bind to PSA-regulatory regions over time, loading primarily on the PSA enhancer. Following enhancer occupancy, the AR coactivator complex communicates with the PSA promoter through chromosomal looping and RNA polymerase II (Pol II) tracking (Jia et al., 2004; Louie et al., 2003; Wang et al., 2005). By contrast, antiandrogens facilitate AR and corepressor recruitment primarily to the PSA promoter (Kang et al., 2004; Shang et al., 2002).
Despite the well-characterized dynamics of AR transcription complex loading on PSA-regulatory regions, few other AR cis-regulatory sites across the genome have been analyzed in detail. Furthermore, while there has been considerable progress in describing the role of AR coregulators in androgen-dependent gene regulation, little is known concerning the roles of other DNA-binding transcription factors that may collaborate with AR in mediating the androgen response. Addressing these issues is critical to the elucidation of the mechanism of androgen-stimulated prostate cancer growth, to understanding so-called “androgen-independent” prostate cancer, and for the identification of new therapeutic targets. For example, the androgen-regulated gene TMPRSS2 has been recently reported to fuse with oncogenes ERG or ETV1 (or other ETS transcription factors) and mediate androgen-responsive overexpression of ERG or ETV1 in 80% of prostate tumors (Tomlins et al., 2005). However, how AR and other potential cooperating factors regulate the TMPRSS2 or the TMPRSS2-ETS fusion genes is unknown.
ChIP combined with tiled DNA oligonucleotide microarray analysis (ChIP-on-chip) has recently evolved as an unbiased technique for mapping the in vivo binding sites of transcription factors in the genome (Carroll et al., 2005, 2006). Here we used this approach to map AR binding sites on chromosomes 21 and 22 in a prostate cancer cell line. We identify 90 AR binding sites on chromosomes 21 and 22. Interestingly, the majority of these sites contain noncanonical AREs. Importantly, we identified a noncanonical ARE as a previously unknown site of AR binding upstream of the TMPRSS2 coding region that serves as an androgen-stimulated enhancer and regulates the expression of TMPRSS2 and the TMPRSS2-ETS fusion genes. We further find that three transcription factor DNA recognition motifs are significantly enriched within the AR binding regions. By combining the ChIP-on-chip data with gene expression profiles from prostate cancer, we show that three specific transcription factors, namely FoxA1, GATA2, and Oct1, recognize the motifs found by ChIP-on-chip. Finally, we demonstrate that GATA2 and Oct1, together with AR, form a regulatory hierarchy that controls androgen-mediated transcription and prostate cancer cell growth.
RESULTS
Identification of 90 AR Binding Sites on Chromosomes 21 and 22
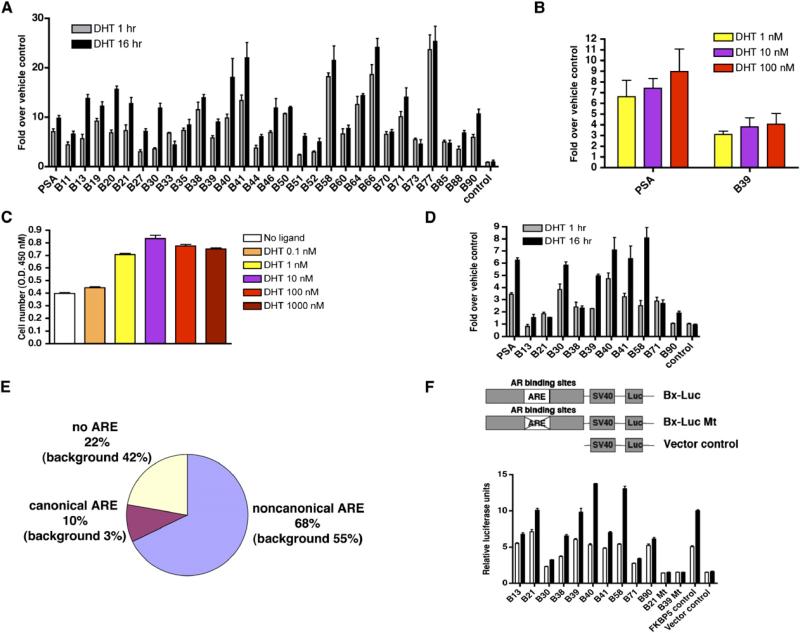
We performed AR ChIP-on-chip assays in the prostate cancer cell line LNCaP that expresses endogenous AR protein (Horoszewicz et al., 1980). Because the occupancy of AR on PSA-regulatory regions increases gradually following androgen exposure and peaks at 16 hr (Jia et al., 2004; Louie et al., 2003; Wang et al., 2005), we treated LNCaP cells for a brief (1 hr) or prolonged (16 hr) period of time with saturating levels of the physiological androgen 5α-dihydrotestosterone (DHT) and performed ChIP with an anti-AR antibody. The ChIP-enriched, AR-associated DNA was amplified, labeled, and hybridized to Affymetrix-tiled oligonucleotide microarrays that cover the entire nonrepetitive regions of human chromosomes 21 and 22 (Carroll et al., 2005). Genomic regions enriched for AR binding were identified by an analysis using generalized Mann-Whitney U test (see the Supplemental Experimental Procedures in the Supplemental Data available with this article online). Based on a stringent threshold (p value < 1E-05), we identified 90 AR binding sites on chromosomes 21 and 22 (Figure 1). We then carried out quantitative anti-AR ChIP followed by real-time PCR to validate a subset of these sites including a majority that were randomly selected. The PSA enhancer region (chromosome 19) was used as a positive control (Wang et al., 2005), and the XBP-1 promoter region (chromosome 22) served as a negative control (Carroll et al., 2005). Androgen induced significant enrichment of AR binding to all of the 29 tested binding regions (≥5-fold) (Figures 2A). In addition, the fold enrichment determined by real-time PCR is highly correlated with ChIP-on-chip p-value (DHT 1 hr r = 0.83, p value 9.51E-08; DHT 16 hr r = 0.77, p value 2.92E-06), suggesting that very few of the identified AR binding sites are false positives. Interestingly, the occupancy of AR on most AR binding sites increases after longer androgen exposure, generalizing our long-term AR recruitment model developed for the PSA gene (Wang et al., 2005).
Figure 1. Map of AR Binding Sites on Human Chromosomes 21 and 22.
RefSeq genes are shown in dark bars, and AR binding regions are shown as blue bars. Ten binding regions for further detailed analyses are indicated.
Figure 2. Characterization of AR Binding Sites on Chromosomes 21 and 22.
(A) Standard ChIP assays of AR recruitment to various potential AR binding regions. LNCaP cells were treated with 100 nM DHT for 1 or 16 hr. ChIP assays were performed with anti-AR antibodies. Immunoprecipitated DNA was quantified by real-time PCR using primers (Table S1) spanning various potential AR binding regions.
(B and C) Effects of DHT concentration on AR binding to chromatin and cell proliferation. (B) Androgen-depleted LNCaP cells were treated with 1, 10, or 100 nM DHT for 1 hr. ChIP assays were performed with an anti-AR antibody, and DNA precipitates were measured with real-time PCR using primers spanning the PSA enhancer and the B39 site. (C) LNCaP cells were cultured in hormone-depleted medium for 3 days and then treated with vehicle or DHT from 0.1 to 1000 nM for another 3 days. Cell proliferation was measured using the WST-1 assay.
(D) Standard ChIP assays of Pol II recruitment to various potential AR binding regions. ChIP assays were performed with anti-Pol II antibodies.
(E) Distribution of the types of AR binding motifs within AR binding sites and the chromosome 21 and 22 genomic background.
(F) AR binding regions function as enhancers. Ten selected AR binding regions, mutant AR binding regions with deleted AREs (B21 and B39), and the FKBP5 enhancer were subcloned into the pGL3-promoter vector and transfected into androgen-depleted LNCaP cells. Empty pGL3 promoter vector was used as a negative control. Cells were stimulated with 100 nM DHT or vehicle for 24 hr. The data were presented as the mean ± SE of two to three replicates, (A)–(D) and (F).
In order to determine the dose response of AR recruitment, we conducted directed AR ChIP to both the PSA enhancer and the newly identified B39 site over a range of DHT concentrations. We found that AR recruitment was little changed between 1 and 100 nM DHT (the saturating level used for ChIP-on-chip) (Figure 2B). In addition, consistent with a previous report (Lin et al., 1998), we found that, while 10 nM DHT is the optimal concentration for the stimulation of LNCaP cell proliferation, concentrations as high as 1000 nM induce significant cell growth (Figure 2C).
Selected AR Binding Sites Play Transcriptional Roles
We and others have previously found that RNA Pol II associates with the AR binding regions of the PSA enhancer and promoter, which is critical for PSA transcription (Louie et al., 2003; Shang et al., 2002; Wang et al., 2005). To address whether Pol II is recruited to the AR binding sites on chromosomes 21 and 22, we performed Pol II ChIP on several AR binding sites. Interestingly, Pol II was significantly recruited to 50% of binding sites tested in an androgen-dependent manner (≥3-fold) (Figure 2D), supporting the assertion that a significant number of binding sites play transcriptional regulatory roles. We further performed a comparative genomic analysis of 90 AR binding sites between genomes of seven vertebrate species (chimp, dog, mouse, rat, chicken, fugu, and zebrafish). The results showed a strong conservation at the center of the AR binding sites (Figure S1), consistent with the notion that noncoding sequences that have survived selection play important roles in gene regulation (Hardison, 2000).
Most AR Binding Sites Are Far from the Promoters of Androgen-Regulated Genes
To correlate AR binding with androgen-dependent gene expression, we examined the gene expression profile in LNCaP cells in response to DHT using an expression microarray. LNCaP cells were deprived of hormone for 72 hr and then treated with vehicle or DHT for 4 and 16 hr. Total RNA was extracted and then hybridized to Affymetrix U133 plus 2.0 expression array. We found 62 transcripts on chromosomes 21 and 22 that were differentially expressed in response to androgen (Table S2). Consistent with previous studies (Nelson et al., 2002; Velasco et al., 2004), we find that the expression of only a small number of transcripts on chromosomes 21 and 22 is very highly changed by androgens. In order to avoid a bias toward this small number of highly differentially expressed genes, we have included all transcripts that were differentially expressed by at least 1.1-fold. Real-time RT-PCR validation of five selected genes and PSA as a positive control confirmed the expression array results (Figure S2). Interestingly, on chromosomes 21 and 22, most AR binding sites are far from transcription start sites of androgen-regulated genes (>10 kb) (Table S3). In total, only 34 out of 90 binding sites (38%) were located within 500 kb of transcriptional start sites of genes regulated by androgens under these conditions in LNCaP cells.
Noncanonical AR-Responsive Elements Mediate AR-Dependent Transcription
We next performed a DNA-binding motif search on the 90 AR binding regions. Surprisingly, we found that, although 78% of the binding regions contain the AR binding half-site motif TGTTCT, only 10% of the binding regions have a canonical class I NR (AGAACAnnnTGTTCT) binding motif when we allow up to two positions to vary from the palindromic consensus with 3 nt spacing (for detail, see “Sequence Analysis” in the Supplemental Experimental Procedures) (Verrijdt et al., 2003) (Figure 2E and Table S4). Of the 90 binding sites, 68% have one or more noncanonical AREs including isolated ARE half-sites, head-to-head AREs (AGAACA [0–8 n] TGTTCT, n ≠ 3), tail-to-tail AREs (TGTTCT [0–8 n] AGAACA), and ARE direct repeats (AGAACA [0–8 n] AGAACA) (Table S4 and “Sequence Analysis” in the Supplemental Experimental Procedures).
To further address whether AR binding regions containing noncanonical AREs can function as enhancers, we subcloned 10 AR binding sites and the recently characterized enhancer of the androgen-regulated gene FKBP5 (Magee et al., 2006) as a positive control, upstream of an SV40 promoter in the pGL3 luciferase vector. The pGL3 luciferase vector with the minimal SV40 promoter alone served as a negative control. These constructs were transfected into androgen-depleted LNCaP cells, and luciferase activity was measured after 24 hr treatment with vehicle alone or in the presence of saturating concentrations of DHT. As shown in Figure 2F, all AR binding regions tested demonstrate enhancer activity (≥2-fold) with varying degrees of androgen dependence comparable to the activity of the FKBP5 enhancer. To confirm that the enhancer characteristics of the AR binding sites require AREs, we deleted the canonical ARE within the B21 and the noncanonical ARE within the B39 enhancer constructs and transfected these mutated vectors into LNCaP cells. As expected, deletion of the single typical ARE within B21 completely abolished its enhancer activity (Figure 2F). More importantly, deletion of the single noncanonical (head-to-head, 8 nt spacing) ARE from B39 also completely abolished the enhancer activity, thus demonstrating that the noncanonical AREs within AR binding regions are required for their enhancer function (Figure 2F).
Characterization of the TMPRSS2-Regulatory Region
As mentioned above, most prostate cancers have recently been shown to harbor either TMPRSS2:ERG or TMPRSS2: ETV1 fusions (Tomlins et al., 2005). However, how the TMPRSS2 gene or the TMPRSS2-ETS fusions are regulated by AR is unknown. Using quantitative ChIP, we confirmed AR binding to site B39, which is ~13.5 kb upstream of the TMPRSS2 mRNA start site both in LNCaP cells that do not harbor a rearranged TMPRSS2 gene (Figures 2A) and in VCaP cells that contain the TMPRSS2:ERG fusion (Figure 3A). In order to test whether this AR binding site functions as an AR-dependent enhancer and communicates with the TMPRSS2 promoter, we performed a ChIP combined with chromosome conformation capture (5C) assay in order to detect looped chromatin structures in vivo (Wang et al., 2005). Hormone-depleted cells were stimulated with vehicle or androgen. The fixed chromatin was digested with EcoRI followed by anti-AR ChIP and ligation. After reversing the crosslinks, one primer in the TMPRSS2 enhancer and one primer in the promoter region were used for PCR amplification of equal amounts of eluted DNA. As shown in Figure 3B, the specific PCR product is formed only in the presence of ligase and is significantly enhanced by androgen stimulation, while control PCR for the level of input chromatin was equivalent under all conditions (Figure 3B). The 383 bp PCR amplified band was sequenced to confirm that it indeed represents the product of the correct ligation of the TMPRSS2 enhancer and promoter. This result demonstrates that the TMPRSS2 enhancer is in close proximity to the promoter in vivo. As negative controls, the interaction of the PSA enhancer with the TMPRSS2 promoter and the TMPRSS2 enhancer with the PSA promoter was tested by 5C. In neither case was a specific PCR product formed (data not shown), supporting the specificity of the 5C product formed between the TMPRSS2 enhancer and promoter. In addition, a control region 9 kb upstream of the TMPRSS2 transcription start site that contains two putative AR-responsive element (ARE) motifs that were not found to bind AR did not contact the TMPRSS2 promoter by 5C (Figure S3). Interestingly, an AR binding site 462 kb downstream of TMPRSS2 (B38) also failed to form a loop with the TMPRSS2 promoter (Figure 3B). This result serves as another negative control and is consistent with the conclusion that regions upstream of TMPRSS2 that are retained in the TMPRSS2-ETS fusions rather than those downstream harbor the important functional elements. These data demonstrate that the –13.5 kb AR binding site specifically interacts with the TMPRSS2 promoter region, supporting the formation of a chromosomal loop between this upstream enhancer and the proximal promoter. The identification of this upstream TMPRSS2 enhancer also provides a molecular explanation of how expression TMPRSS2-ETS factor fusions are driven in an androgen-dependent manner. Interestingly, a newly identified prostate cancer oncogene resulting from the fusion of TMPRSS2 and the ETS transcription factor ETV4 does not include any of the TMPRSS2 coding exons, as the breakpoint is ~8 kb upstream of the mRNA start site though ~5.5 kb downstream of this newly identified TMPRSS2 enhancer (Tomlins et al., 2006), suggesting that this enhancer acts to drive ETV4 expression in this setting.
Figure 3. Characterization of TMPRSS2-Regulatory Regions.
(A) ChIP analyses of AR occupancy on the PSA and B39 in VCaP cells. VCaP cells (a prostate cancer cell line that expresses TMPRSS2:ERG fusion gene) were treated with 100 nM DHT for 1 hr. ChIP assays were performed with an anti-AR antibody.
(B) Spatial communication between the TMPRSS2 enhancer and promoter. 5C was performed using fixed or EcoRI- or BtgI-digested chromatin from vehicle- or DHT-treated LNCaP cells. Primers (Table S1) flanking the –13.5 kb or 462 kb AR binding region and –700 bp promoter region were used to PCR amplify DNA after ligation. Control PCR was performed using chromatin before restriction enzyme digestion.
(C) Schematic representation of the TMPRSS2 14 kb upstream regulatory region. Potential five ARE clusters and fourteen 1 kb fragments are shown.
(D) ChIP analyses of AR recruitment to five potential ARE regions. ChIP assays were performed with anti-AR antibodies in LNCaP cells treated with vehicle or 100 nM DHT for 4 or 16 hr.
(E) Systematic mapping of enhancer elements within the TMPRSS2 14 kb upstream regulatory region. Fourteen 1 kb TMPRSS2 upstream sequences were subcloned into the pGL3-promoter vector and transfected into LNCaP cells. Cells were treated with vehicle or 100 nM DHT for 24 hr. The data are presented as the mean ± SE of two to three replicates, (A), (D), and (E).
Although we found that the –13.5 kb B39 binding site in the TMPRSS2 gene is a functional androgen-regulated enhancer (Figure 2F) and is in close proximity to the TMPRSS2 promoter in vivo (Figure 3B), we investigated the possibility that other functionally important AR binding sites located between –13.5 kb and the transcription start site may have been missed by the ChIP-on-chip assay. We therefore performed an AR motif search within the 14 kb region upstream of the TMPRSS2 gene and found five clusters of potential AREs (Figure 3C). Despite the presence of AREs, quantitative AR ChIP analyses found only significant AR binding to ARE region V corresponding exactly to the region identified by ChIP-on-chip (–13.5 kb) (Figure 3D). In addition, to further address whether other regions within the 14 kb upstream sequence might play transcriptional roles, we systematically subcloned fourteen 1 kb regions (Figure 3C) upstream of an SV40 promoter in the pGL3 luciferase vector and transfected these into androgen-depleted LNCaP cells. As shown in Figure 3E, construct A (–14 to –13 kb) corresponding to the B39 element had the most significant enhancer activity. In addition, construct N (–1 kb to +24 bp) that contains the TMPRSS2 promoter also demonstrated weak enhancer activity in this reporter assay. Significantly, quantitative AR ChIP showed that AR was only weakly recruited to the endogenous TMPRSS2 promoter region. Therefore the B39 binding site is the primary site of AR binding and functional enhancer activity within the 14 kb upstream of the TMPRSS2 gene. Of course, we cannot rule out the possibility that other cis-regulatory regions outside of the 14 kb region (Figure S4) may also play a role in TMPRSS2 expression.
Identification of AR-Collaborating Transcription Factor Complexes
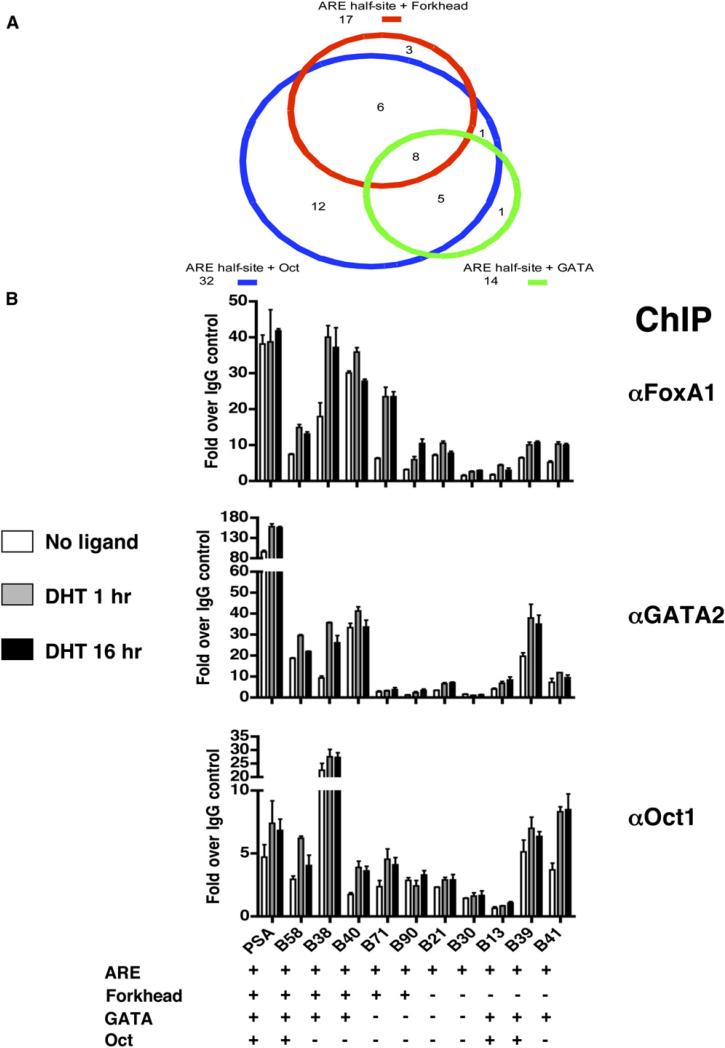
We hypothesized that other transcription factors might play important collaborative roles in AR function. To search for these AR partners, we mapped all of the mammalian transcription factor motifs in the TRANSFAC database (Matys et al., 2003) to the 90 AR binding regions to identify motifs significantly enriched compared to the chromosomes 21 and 22 genomic background. While there was only a moderate enrichment (1.55-fold, p value 3.00E-3) for the AR half-site, the co-occurrence of an AR half-site motif with each of three DNA motifs, Forkhead, GATA, and Oct, was significantly enriched compared with the genomic background (AR half-site+ Forkhead, 5.33-fold, p value 1.63E-08; AR half-site + GATA, 3.19-fold, p value 1.16E-04; AR half-site + Oct, 2.03-fold, p value 3.09E-05) (Figure 4A). These associations strongly suggested a functional interaction between AR and sequence-specific DNA-binding transcription factors that recognize each of these motifs.
Figure 4. Collaborative Transcription Factors Are Recruited to AR Binding Regions.
(A) Schematic representation of association of AR half-site and transcription factor motifs within AR binding regions. Each pairwise association is designated by a circle with different color. The area of the circle is proportional to the frequency of the association.
(B) The recruitment of collaborative factors to AR binding regions. LNCaP cells were cultured in hormone-depleted medium for 3 days and then treated with vehicle or DHT for 1 and 16 hr. ChIP analyses were then performed with indicated antibodies or control IgG. DNA precipitates were measured by real-time PCR using primers spanning the PSA enhancer and nine AR binding regions on chromosomes 21 and 22. The results are shown as the average of two to three replicates ± SE. The occurrence of ARE, Forkhead, GATA, and Oct motifs in each region is also shown.
Given the three significantly enriched DNA motifs, we were faced with the question of which of the large number of transcription factors that can potentially bind the motifs were in fact playing a functional role. To generate hypotheses on which factors might be responsible, we searched gene expression profiles from several human prostate cancer studies available on the Oncomine database (Rhodes et al., 2004) and found the three transcription factors FoxA1, GATA2, and Oct1 to be of potential interest on the basis of their high expression in prostate cancers. FoxA1 and GATA2 have been shown to be required for androgen-induced PSA gene transcription in reporter gene assays (Gao et al., 2003; Perez-Stable et al., 2000). Moreover, FoxA1 and Oct1 are known to interact with AR in vivo (Gao et al., 2003; Gonzalez and Robins, 2001). To examine whether the three collaborating transcription factors are actually recruited to AR binding regions on chromosomes 21 and 22, we performed quantitative ChIP assays using antibodies against each of the three factors or IgG (control) using primers spanning ten of the validated AR binding regions and the well-characterized PSA enhancer as a control. As shown in Figure 4B, the three collaborating factors are recruited (≥2-fold) to 80% of the AR binding sites tested. Significant binding of each of the three factors to the AR binding regions was observed in the absence of androgen while androgen did increase specific factor binding at specific sites. These results demonstrate that AR and various combinations of each of the three potential collaborating transcription factors are recruited to specific regions of the genome. Given the complex transcriptional response to androgen, it is not surprising that there is a wide variation in the degree of factor occupancy and androgen dependence seen at these sites.
To explore the physical association of AR and the three transcription factors, we first performed coimmunoprecipitations of the endogenous proteins in vivo. Each of the three transcription factors was immunoprecipitated from DHT-treated or untreated LNCaP cells and subsequently probed by western blot with anti-AR antibodies. As shown in Figure 5A, AR interacts with GATA2 and Oct1 in a hormone-dependent manner, but its interaction with FoxA1 is not affected by androgen. To address whether AR forms a single complex with all three transcription factors, we immunoprecipitated GATA2 from LNCaP cells followed by western blotting with anti-Oct1 antibodies. The results showed that GATA2 does not interact with Oct1 (data not shown), suggesting that AR may form independent complexes with each of the collaborating factors.
Figure 5. AR Interacts with Collaborative Transcription Factors In Vivo.
(A) AR coimmunopreciptates with its collaborating factors in vivo. LNCaP cells were grown in the presence or absence of DHT for 24 hr. Whole-cell lysates were immunoprecipitated (IP) with indicated antibodies or control IgG. Western blot (WB) was performed with indicated antibodies.
(B) AR-collaborating factor complexes form on chromatin. LNCaP cells were treated with or without DHT for 16 hr. ChIP assays were performed with anti-AR antibodies. The immunoprecipitated chromatin was eluted, reimmunoprecipitated with the indicated antibodies or control IgG, and amplified by PCR using primers flanking the PSA and B38 enhancer regions.
(C and D) Colocalized cis-active elements are required for AR-dependent transcription. A wild-type PSA reporter construct and PSA reporter construct with deleted GATA or Oct motifs within the PSA enhancer or promoter regions (C) or B39 reporter wild-type construct and B39 reporters with GATA or Oct motif deletions (D) were transfected into LNCaP cells treated with vehicle or 100 nM DHT for 24 hr. Luciferase activities were determined, and results are presented as the mean ± SE of the triplicate experiments.
To demonstrate that AR and the three transcription factors form complexes on chromatin, we performed serial ChIP experiments (re-ChIP). The crosslinked chromatin was first immunoprecipitated with anti-AR antibodies and reimmunoprecipitated with either control IgG or antibodies against each of the three transcription factors. The PSA enhancer and B38 regions were enriched after re-ChIP in a ligand-dependent manner (Figure 5B), demonstrating that AR can act together on chromatin with FoxA1, GATA2, or Oct1.
While the above ChIP, coimmunoprecipitation, and re-ChIP assays demonstrated the protein-protein interaction between AR and potential collaborating transcription factors, it is not clear if these interactions are based on functional interactions between colocalized cis-active motifs with the AR binding regions. To address this issue, we generated GATA and Oct motif deletion mutants in the PSA enhancer within the full-length 6 kb PSA-regulatory region upstream of a luciferase gene. As a control, we also generated an Oct motif deletion mutant in the PSA promoter region. We transiently transfected these mutated constructs along with the wild-type PSA construct into hormone-depleted LNCaP cells, and luciferase was measured after 24 hr DHT stimulation. Consistent with a previous report (Perez-Stable et al., 2000), deletion of the GATA motif within the PSA enhancer region dramatically decreased androgen-stimulated transcriptional activities (Figure 5C). Interestingly, deletion of the Oct motif within the PSA enhancer, but not the promoter region, also significantly attenuated androgen-induced transcriptional activity (Figure 5C). These data suggest a functional interaction among colocalized cis-active elements within the PSA enhancer region. Significantly, both the Oct and GATA motifs were also required for the activity of the TMPRSS2 enhancer (Figure 5D).
AR-Collaborating Transcription Factors Regulate AR-Mediated Transcription and Prostate Cancer Cell Growth
To address the functional roles of the three collaborating transcription factors in mediating AR-dependent transcription, we reduced expression levels of the three factors by RNA interference (Figure 6A). We then investigated the effect of silencing of the three factors on androgen-stimulated expression of two previously reported androgen-regulated genes PSA and TMPRSS2. We transiently transfected siRNA targeting each of the three factors into LNCaP cells. Forty-eight hours after transfection, cells were treated with DHT (1 or 100 nM) for 4 or 16 hr. Quantitative RT-PCR was then performed to measure androgen-regulated mRNA levels. As shown in Figure 6B, reduction of GATA2 or Oct1 levels significantly decreased PSA at 16 hr and TMPRSS2 mRNA expression at 4 hr while not changing control GADPH expression levels. Interestingly, TMPRSS2 expression returns to control levels at 16 hr, suggesting the existence of gene-specific bypass pathways. In contrast, FoxA1 silencing had no effect on the expression of these two genes (Figures 6B). While this result may suggest that FoxA1 does not play a critical role in androgen signaling, it is also possible that FoxA1 function is required for only a subset of AR targets or is redundant with another Forkhead family member.
Figure 6. Functional Analyses of Collaborating Transcription Factors in Mediating AR-Dependent Transcription of the PSA and TMPRSS2 Genes.
(A) Suppression of AR-collaborating factor levels by RNAi. LNCaP cells were transfected with siRNA targeting each factor and a control siRNA. Forty-eight hours posttransfection, cells were treated with or without 100 nM DHT for 16 hr, and western blots were performed using the antibodies indicated.
(B) Effects of siRNA on PSA, TMPRSS2, and GADPH gene expression. Forty-eight hours after siRNA transfection, cells were treated with or without 1 or 100 nM DHT for 4 and 16 hr. Total RNA was isolated and amplified by real-time RT-PCR using transcript-specific primers (Table S1). The no-ligand control was measured at 4 hr.
(C) Effects of silencing GATA2 and Oct1 on AR, Pol II, Oct1, and GATA2 recruitment to the PSA and TMPRSS2 enhancers. AR, Pol II, Oct1, and GATA2 ChIPs were performed after vehicle or 4 hr 100 nM DHT treatment of siLuc, siGATA2, or siOct1-transfected cells. Graphical representations of the mean ± SE of two to three independent experiments are shown in (B) and (C).
To investigate the mechanism of GATA2 and Oct1 action on PSA and TMPRSS2 expression, we performed ChIP assays to study the effect of GATA2 and Oct1 silencing on AR and Pol II recruitment to the PSA and TMPRSS2 enhancers. LNCaP cells were transiently transfected with siGATA2, siOct1, or control siLuc; cultured for 48 hr; and then stimulated with 100 nM DHT for 4 hr. As shown in Figure 6C, recruitment of AR and Pol II to the PSA and TMPRSS2 enhancers was significantly decreased in si-GATA2 transfected cells as compared with the siLuc control, both in the absence of ligand and, to a greater extent, in the presence of hormone. In contrast, silencing of Oct1 did not affect AR binding but greatly reduced Pol II loading on the PSA and TMPRSS2 enhancers, both in the absence and presence of hormone (Figure 6C). Interestingly, Oct1 recruitment to the PSA and TMPRSS2 enhancers was similarly attenuated when GATA2 was silenced, whereas GATA2 binding to these enhancers was not affected by Oct1 silencing (Figure 6C). Western blot analyses showed that silencing of GATA2 and Oct1 had no obvious effect on the level of AR or Pol II expression (Figure 6A). This suggests that GATA2 and Oct1 play necessary roles both in the basal recruitment of AR and/or Pol II to chromatin and in their recruitment following androgen stimulation. More interestingly, these data suggest that GATA2 and Oct1 act at distinct steps in AR signaling, with GATA2 acting upstream of Oct1 recruitment.
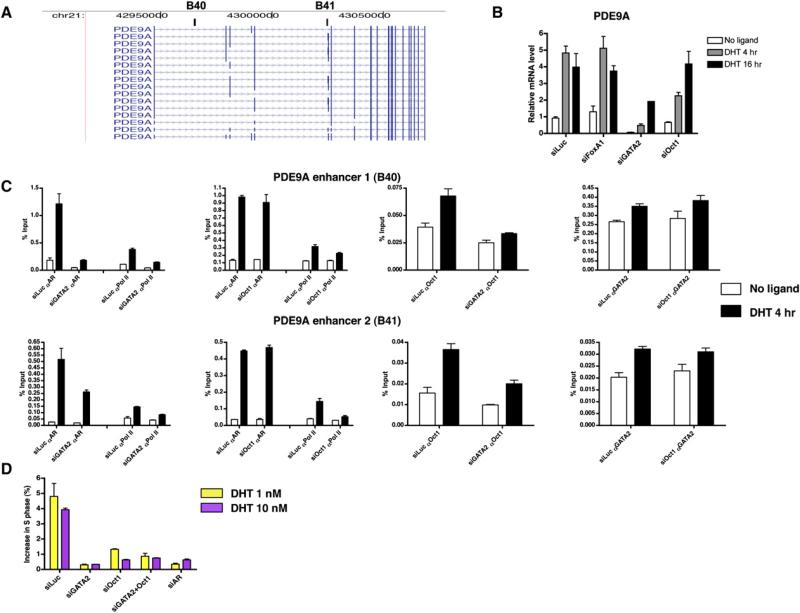
In order to extend these findings to a gene not previously known to be androgen regulated, we analyzed the role of GATA2, Oct1, and FoxA1 in the expression of the PDE9A gene. PDE9A is a member of the cyclic nucleotide phosphodiesterase (PDE) family that plays a critical role in regulating intracellular concentrations of cyclic nucleotides and is highly expressed in prostate (Fisher et al., 1998; Guipponi et al., 1998). Our ChIP-on-chip analysis identified two intronic AR binding sites (B40 and B41) that are 18.4 kb and 77.8 kb downstream of the PDE9A transcription start site, respectively (Figure 7A). In addition, our gene expression studies identified PDE9A as a gene significantly upregulated by androgen in LNCaP cells (Table S2 and Figure S2). Similar to our findings with the PSA and TMPRSS2 genes, we find that GATA2 and Oct1, but not FoxA1, play necessary collaborative and hierarchical roles in AR-mediated PDE9A gene expression (Figures 7B and 7C).
Figure 7. Functional Roles of Collaborating Transcription Factors in Mediating the PDE9A Gene Transcription and Prostate Cancer Cell Proliferation.
(A) AR binding sites relative to the PDE9A gene. The black blocks represent AR binding sites. The PDE9A gene is shown in its 5′-3′ orientation, and the blue arrows indicate the direction of the gene (June 05, University of California, Santa Cruz [UCSC], known genes).
(B) Effects of FoxA1, GATA2, and Oct1 silencing on PDE9A mRNA expression. siRNA-RTPCR analyses were performed as described in Figure 6B.
(C) Effects of silencing GATA2 and Oct1 on AR, Pol II, Oct1, and GATA2 recruitment to the PDE9A enhancers. siRNA-ChIP analyses were performed as described in Figure 6C.
(D) Effects of AR and cofactor silencing on androgen-stimulated cell cycle entry. Forty-eight hours after siRNA transfection, cells were treated with or without 1 or 10 nM DHT for 24 hr. Cells were then fixed, stained with propidium iodide, and analyzed by flow cytometry. Values represent the mean ± SE of two to three independent experiments (B) to (D).
As AR is essential for the growth of prostate cancer cells (Heinlein and Chang, 2004) and previous studies have shown AR silencing inhibits prostate cancer cell proliferation (Wright et al., 2003; Zegarra-Moro et al., 2002), we next tested the effects of silencing individually or in combination the collaborating transcription factors on cell cycle progression. Forty-eight hours after siRNA transfection, LNCaP cells were treated with or without 1 or 10 nM DHT for 24 hr. Cells were then stained with propidium iodide for DNA content. Silencing of GATA2 and Oct1 alone or in combination significantly decreased androgen-induced cell cycle progression and to the same extent as silencing AR itself (Figures 7D). To control for potential off-target effects, silencing GATA2 and Oct1 with an independent set of siRNAs also inhibited androgen-dependent cell cycle progression (Figure S5). In addition, the response to the silencing of GATA2 and Oct1 was specific, as their silencing had no effect on cell cycle progression in HeLa cells (Figure S6). Interestingly, consistent with its nonessential role in androgen-stimulated gene expression, silencing of FoxA1, a chromatin “pioneer” factor (Cirillo et al., 2002), did not affect androgen-stimulated cell cycle progression (Figure S7). Thus, GATA2 and Oct 1, but not FoxA1, play essential roles in androgen-stimulated cell cycle progression in prostate cancer cells.
DISCUSSION
Whereas previous ChIP studies have provided useful information pertaining to AR transcription complex assembly (Jia et al., 2004; Louie et al., 2003; Kang et al., 2004; Shang et al., 2002; Wang et al., 2005), the data are mainly restricted to the PSA gene. In this study, we used ChIP-on-chip to map 90 previously unknown AR binding sites on chromosomes 21 and 22. As chromosomes 21 and 22 represent approximately 2% of the entire nonrepetitive human genome, these results predict ~4500 AR binding sites across the entire human genome.
Interestingly, most of the binding sites identified on chromosomes 21 and 22 are far from androgen-regulated genes. We previously have found that most estrogen receptor (ER) binding sites are also distant from estrogen-regulated genes, consistent with their functioning as enhancers rather than promoters (Carroll et al., 2005, 2006). Whether distance-independent binding is a general rule for NR action awaits similar studies of other NRs. While it is likely that many of these sites act as transcriptional enhancers of neighboring genes, other functions may also exist. In addition, it is likely that only a subset of the binding sites are functional in LNCaP under the specific experimental conditions tested and may be functional in different cell types and/or under different conditions, as previously suggested (van Steensel, 2005). It is also possible that some AR binding sites are indeed nonfunctional. Finally, while a proportion of androgen-regulated genes have AR binding sites associated with the gene, androgen-stimulated transcription may in some cases be independent of AR binding to chromatin.
Interestingly, we found that the majority of AR binding sites on chromosomes 21 and 22 contain noncanonical AREs and that a significant proportion of those tested can function as AR-dependent enhancers and that the noncanonical AREs are necessary for this activity. Although in vitro DNA-binding assays demonstrated AR binds to canonical AREs with highest affinity (Kallio et al., 1994), a small number of noncanonical AREs such as ARE direct repeats have been previously reported (Verrijdt et al., 2003). In the genuine chromatin environment, it is likely that collaborating factors may assist AR in binding to non-canonical AREs. Our finding of a preponderance of atypical modes of AR binding suggests that transcription factor binding motifs defined in the pre-ChIP era need to be readdressed in the face of the ability to find large numbers of additional binding sites using ChIP-on-chip.
In contrast to NR coregulatory factors identified by biochemical methods (McKenna and O'Malley, 2002; Perissi and Rosenfeld, 2005), we used a systems approach uniting diverse data types combining chromosome-scale ChIP-on-chip analysis and gene expression profiling to identify a network of AR-collaborating transcription factors. Our data demonstrate a physical interaction between AR and these transcription factors and that the collaborating partners have distinct functional roles in androgen-dependent gene transcription and cell proliferation. Interestingly, we found that GATA2 and Oct1 hierarchically regulate androgen-dependent transcription. Previous studies have found that other GATA family members can open the compact chromatin through their conserved zinc finger motifs (Boyes et al., 1998; Cirillo et al., 2002). Given our finding that GATA2 is required for AR binding, it may function similarly in this case as a pioneer factor that alters chromatin to allow AR binding. In contrast, Oct1 is not required for AR to bind the PSA, TMPRSS2, and PDE9A enhancers, suggesting it functions at a step subsequent to GATA2 action and in conjunction with AR. Consistent with the roles of GATA2 and Oct1 in androgen target gene expression, silencing of these two collaborating transcription factors significantly decreases androgen-induced cell cycle progression. In other systems it has been reported that GATA2 and Oct1 promote cell proliferation through mechanisms including a p53-dependent pathway in the case of GATA2 (Tsai and Orkin, 1997) and activation of histone H2B transcription in the case of Oct1 (Ewen, 2000).
There is abundant evidence that AR plays a critical role both in early hormone-dependent prostate cancer and in so-called advanced androgen-independent prostate cancer (Debes and Tindall, 2004) where AR can be amplified. The elucidation of the authentic AR cis-regulatory elements allows an understanding of how critical androgen-stimulated targets such as TMPRSS2-ETS fusions are regulated. Finally, the identification of the critical AR-cooperating transcription factors provides potential new targets for therapeutic intervention.
EXPERIMENTAL PROCEDURES
ChIP-on-Chip Assay
ChIP was performed with an anti-AR antibody (N20, Santa Cruz Biotechnology, CA) as previously described (Wang et al., 2005) except that 100 μg/ml yeast tRNA was used as blocking reagent instead of 2 μg salmon sperm DNA. Ligation-mediated PCR and labeling were performed as previously described (Carroll et al., 2005). Microarrays used were Affymetrix Genechip chromosome 21/22 tiling set P/N 900545. Biological triplicates were performed. The ChIP-on-chip raw data are accessible at http://chip.dfci.harvard.edu/~wli/Androgen.Receptor.ChIP2/. Details for ChIP-on-chip data and sequence analyses are available in the Supplemental Experimental Procedures.
ChIP and Re-ChIP
ChIP and re-ChIP were performed as previously described (Shang et al., 2000; Wang et al., 2005). Details are available in the Supplemental Experimental Procedures.
Cell Proliferation Assay
Cell proliferation was measured using a WST-1 kit (Roche, Indianapolis, IN).
Expression Array Experiments
Hormone-depleted LNCaP cells were grown in the absence or presence of 100 nM DHT for 4 and 16 hr. Total RNA was isolated using an RNeasy kit (QIAGEN, Valencia, CA). Total RNA from three biological replicates was hybridized at the Dana-Farber Cancer Institute Microarray Core Facility to human U133 plus 2.0 expression array. The data were analyzed by using dChip microarray software (Li and Wong, 2001).
ChIP Combined Chromosome Conformation Capture Assay
5C was performed as previously described (Wang et al., 2005). The primers used are listed in Table S1. The PCR amplified band was sequence verified.
Construction of Plasmids
AR binding sites were PCR amplified from LNCaP genomic DNA. Fourteen 1 kb TMPRSS2 upstream sequences were PCR amplified from BAC RP11-814F13. These PCR products were subcloned into pGL3-promoter vector (Promega, Madison, WI). ARE, GATA, and Oct motif deletion mutants were sequence verified. The PCR primers are listed in Table S1.
Reporter Gene Assays
Hormone-depleted LNCaP cells were transfected using Lipofectamine 2000 (Invitrogen, Carlsbad, CA). Details are available in the Supplemental Experimental Procedures.
Coimmunoprecipitation and Western Blotting
Coimmunoprecipitation and western blotting were performed as previously described (Wang et al., 2002) with a few modifications. Details are available in the Supplemental Experimental Procedures.
Small Interfering RNA
Small interfering RNA (siRNA) duplexes were transfected into LNCaP cells using Lipofectamine 2000 (Invitrogen). Forty-eight hours after siRNA transfection, cells were treated with DHT or vehicle and then harvested. The siRNA sequences are listed in Table S1.
Real-Time RT-PCR
Real-time RT-PCR was performed as previously described (Wang et al., 2005). Primers for RT-PCR are listed in Table S1.
Flow Cytometry
siRNA-transfected LNCaP cells were treated with DHT for 24 hr. Cells were collected and stained with propidium iodide. DNA contents were analyzed by Dana-Farber Cancer Institute Cytometry Core Facility.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported by the Claudia Adams Barr Program in Innovative Basic Cancer Research (M.B.), Prostate Cancer SPORE Grants P50 CA90381 (M.B.) and P50 CA69568 (K.J.P.), Department of Defense award W81XWH-05-01-0023 (Q.W.), and European Union contract LSHM-CT-2005-018652 (O.A.J.). K.J.P. is an American Cancer Society Clinical Research Professor. The authors also thank Drs. Jérôme Eeckhoute and Kirsten Fertuck for helpful discussions.
Footnotes
Supplemental Data
Supplemental Data include Supplemental Experimental Procedures, seven figures, and four tables and can be found with this article online at http://www.molecule.org/cgi/content/full/27/3/380/DC1/.
Accession Numbers
The microarray data in this study have been deposited in NCBI's Gene Expression Omnibus (GEO, http://www.ncbi.nlm.nih.gov/geo/) and are accessible under GEO Series accession number GSE7868.
REFERENCES
- Balk SP, Ko YJ, Bubley GJ. Biology of prostate-specific antigen. J. Clin. Oncol. 2003;21:383–391. doi: 10.1200/JCO.2003.02.083. [DOI] [PubMed] [Google Scholar]
- Boyes J, Byfield P, Nakatani Y, Ogryzko V. Regulation of activity of the transcription factor GATA-1 by acetylation. Nature. 1998;396:594–598. doi: 10.1038/25166. [DOI] [PubMed] [Google Scholar]
- Carroll JS, Liu XS, Brodsky AS, Li W, Meyer CA, Szary AJ, Eeckhoute J, Shao W, Hestermann EV, Geistlinger TR, et al. Chromosome-wide mapping of estrogen receptor binding reveals long-range regulation requiring the forkhead protein FoxA1. Cell. 2005;122:33–43. doi: 10.1016/j.cell.2005.05.008. [DOI] [PubMed] [Google Scholar]
- Carroll JS, Meyer CA, Song J, Li W, Geistlinger TR, Eeckhoute J, Brodsky AS, Keeton EK, Fertuck KC, Hall GF, et al. Genome-wide analysis of estrogen receptor binding sites. Nat. Genet. 2006;38:1289–1297. doi: 10.1038/ng1901. [DOI] [PubMed] [Google Scholar]
- Cirillo LA, Lin FR, Cuesta I, Friedman D, Jarnik M, Zaret KS. Opening of compacted chromatin by early developmental transcription factors HNF3 (FoxA) and GATA-4. Mol. Cell. 2002;9:279–289. doi: 10.1016/s1097-2765(02)00459-8. [DOI] [PubMed] [Google Scholar]
- Debes JD, Tindall DJ. Mechanisms of androgen-refractory prostate cancer. N. Engl. J. Med. 2004;351:1488–1490. doi: 10.1056/NEJMp048178. [DOI] [PubMed] [Google Scholar]
- Ewen ME. Where the cell cycle and histones meet. Genes Dev. 2000;14:2265–2270. doi: 10.1101/gad.842100. [DOI] [PubMed] [Google Scholar]
- Fisher DA, Smith JF, Pillar JS, St Denis SH, Cheng JB. Isolation and characterization of PDE9A, a novel human cGMP-specific phosphodiesterase. J. Biol. Chem. 1998;273:15559–15564. doi: 10.1074/jbc.273.25.15559. [DOI] [PubMed] [Google Scholar]
- Gao N, Zhang J, Rao MA, Case TC, Mirosevich J, Wang Y, Jin R, Gupta A, Rennie PS, Matusik RJ. The role of hepatocyte nuclear factor-3 alpha (Forkhead Box A1) and androgen receptor in transcriptional regulation of prostatic genes. Mol. Endocrinol. 2003;17:1484–1507. doi: 10.1210/me.2003-0020. [DOI] [PubMed] [Google Scholar]
- Gonzalez MI, Robins DM. Oct-1 preferentially interacts with androgen receptor in a DNA-dependent manner that facilitates recruitment of SRC-1. J. Biol. Chem. 2001;276:6420–6428. doi: 10.1074/jbc.M008689200. [DOI] [PubMed] [Google Scholar]
- Guipponi M, Scott HS, Kudoh J, Kawasaki K, Shibuya K, Shin-tani A, Asakawa S, Chen H, Lalioti MD, Rossier C, et al. Identification and characterization of a novel cyclic nucleotide phosphodiesterase gene (PDE9A) that maps to 21q22.3: alternative splicing of mRNA transcripts, genomic structure and sequence. Hum. Genet. 1998;103:386–392. doi: 10.1007/s004390050838. [DOI] [PubMed] [Google Scholar]
- Hardison RC. Conserved noncoding sequences are reliable guides to regulatory elements. Trends Genet. 2000;16:369–372. doi: 10.1016/s0168-9525(00)02081-3. [DOI] [PubMed] [Google Scholar]
- Heinlein CA, Chang C. Androgen receptor in prostate cancer. Endocr. Rev. 2004;25:276–308. doi: 10.1210/er.2002-0032. [DOI] [PubMed] [Google Scholar]
- Horoszewicz JS, Leong SS, Chu TM, Wajsman ZL, Friedman M, Papsidero L, Kim U, Chai LS, Kakati S, Arya SK, Sand-berg AA. The LNCaP cell line—a new model for studies on human prostatic carcinoma. Prog. Clin. Biol. Res. 1980;37:115–132. [PubMed] [Google Scholar]
- Jia L, Choong CS, Ricciardelli C, Kim J, Tilley WD, Coetzee GA. Androgen receptor signaling: mechanism of interleukin-6 inhibition. Cancer Res. 2004;64:2619–2626. doi: 10.1158/0008-5472.can-03-3486. [DOI] [PubMed] [Google Scholar]
- Kallio PJ, Palvimo JJ, Mehto M, Janne OA. Analysis of androgen receptor-DNA interactions with receptor proteins produced in insect cells. J. Biol. Chem. 1994;269:11514–11522. [PubMed] [Google Scholar]
- Kang Z, Janne OA, Palvimo JJ. Coregulator recruitment and histone modifications in transcriptional regulation by the androgen receptor. Mol. Endocrinol. 2004;18:2633–2648. doi: 10.1210/me.2004-0245. [DOI] [PubMed] [Google Scholar]
- Li C, Wong WH. Model-based analysis of oligonucleo-tide arrays: expression index computation and outlier detection. Proc. Natl. Acad. Sci. USA. 2001;98:31–36. doi: 10.1073/pnas.011404098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MF, Meng TC, Rao PS, Chang C, Schonthal AH, Lin FF. Expression of human prostatic acid phosphatase correlates with androgen-stimulated cell proliferation in prostate cancer cell lines. J. Biol. Chem. 1998;273:5939–5947. doi: 10.1074/jbc.273.10.5939. [DOI] [PubMed] [Google Scholar]
- Louie MC, Yang HQ, Ma AH, Xu W, Zou JX, Kung HJ, Chen HW. Androgen-induced recruitment of RNA polymerase II to a nuclear receptor-p160 coactivator complex. Proc. Natl. Acad. Sci. USA. 2003;100:2226–2230. doi: 10.1073/pnas.0437824100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Magee JA, Chang LW, Stormo GD, Milbrandt J. Direct, androgen receptor-mediated regulation of the FKBP5 gene via a distal enhancer element. Endocrinology. 2006;147:590–598. doi: 10.1210/en.2005-1001. [DOI] [PubMed] [Google Scholar]
- Mangelsdorf DJ, Thummel C, Beato M, Herrlich P, Schutz G, Umesono K, Blumberg B, Kastner P, Mark M, Chambon P, et al. The nuclear receptor superfamily: the second decade. Cell. 1995;83:835–839. doi: 10.1016/0092-8674(95)90199-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matys V, Fricke E, Geffers R, Gossling E, Haubrock M, Hehl R, Hornischer K, Karas D, Kel AE, Kel-Margoulis OV, et al. TRANSFAC: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McKenna NJ, O'Malley BW. Combinatorial control of gene expression by nuclear receptors and coregulators. Cell. 2002;108:465–474. doi: 10.1016/s0092-8674(02)00641-4. [DOI] [PubMed] [Google Scholar]
- Nelson PS, Clegg N, Arnold H, Ferguson C, Bonham M, White J, Hood L, Lin B. The program of androgen-responsive genes in neoplastic prostate epithelium. Proc. Natl. Acad. Sci. USA. 2002;99:11890–11895. doi: 10.1073/pnas.182376299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perez-Stable CM, Pozas A, Roos BA. A role for GATA transcription factors in the androgen regulation of the prostate-specific antigen gene enhancer. Mol. Cell. Endocrinol. 2000;167:43–53. doi: 10.1016/s0303-7207(00)00300-2. [DOI] [PubMed] [Google Scholar]
- Perissi V, Rosenfeld MG. Controlling nuclear receptors: the circular logic of cofactor cycles. Nat. Rev. Mol. Cell Biol. 2005;6:542–554. doi: 10.1038/nrm1680. [DOI] [PubMed] [Google Scholar]
- Rhodes DR, Yu J, Shanker K, Deshpande N, Varambally R, Ghosh D, Barrette T, Pandey A, Chinnaiyan AM. ONCOMINE: a cancer microarray database and integrated data-mining platform. Neoplasia. 2004;6:1–6. doi: 10.1016/s1476-5586(04)80047-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shang Y, Hu X, DiRenzo J, Lazar MA, Brown M. Cofactor dynamics and sufficiency in estrogen receptor-regulated transcription. Cell. 2000;103:843–852. doi: 10.1016/s0092-8674(00)00188-4. [DOI] [PubMed] [Google Scholar]
- Shang Y, Myers M, Brown M. Formation of the androgen receptor transcription complex. Mol. Cell. 2002;9:601–610. doi: 10.1016/s1097-2765(02)00471-9. [DOI] [PubMed] [Google Scholar]
- Tomlins SA, Rhodes DR, Perner S, Dhanasekaran SM, Mehra R, Sun XW, Varambally S, Cao X, Tchinda J, Kuefer R, et al. Recurrent fusion of TMPRSS2 and ETS transcription factor genes in prostate cancer. Science. 2005;310:644–648. doi: 10.1126/science.1117679. [DOI] [PubMed] [Google Scholar]
- Tomlins SA, Mehra R, Rhodes DR, Smith LR, Roulston D, Helgeson BE, Cao X, Wei JT, Rubin MA, Shah RB, Chin-naiyan AM. TMPRSS2:ETV4 gene fusions define a third molecular subtype of prostate cancer. Cancer Res. 2006;66:3396–3400. doi: 10.1158/0008-5472.CAN-06-0168. [DOI] [PubMed] [Google Scholar]
- Tsai FY, Orkin SH. Transcription factor GATA-2 is required for proliferation/survival of early hematopoietic cells and mast cell formation, but not for erythroid and myeloid terminal differentiation. Blood. 1997;89:3636–3643. [PubMed] [Google Scholar]
- van Steensel B. Mapping of genetic and epigenetic regulatory networks using microarrays. Nat. Genet. Suppl. 2005;37:S18–S24. doi: 10.1038/ng1559. [DOI] [PubMed] [Google Scholar]
- Velasco AM, Gillis KA, Li Y, Brown EL, Sadler TM, Achilleos M, Greenberger LM, Frost P, Bai W, Zhang Y. Identification and validation of novel androgen-regulated genes in prostate cancer. Endocrinology. 2004;145:3913–3924. doi: 10.1210/en.2004-0311. [DOI] [PubMed] [Google Scholar]
- Verrijdt G, Haelens A, Claessens F. Selective DNA recognition by the androgen receptor as a mechanism for hormone-specific regulation of gene expression. Mol. Genet. Metab. 2003;78:175–185. doi: 10.1016/s1096-7192(03)00003-9. [DOI] [PubMed] [Google Scholar]
- Wang Q, Sharma D, Ren Y, Fondell JD. A coregulatory role for the TRAP-mediator complex in androgen receptor-mediated gene expression. J. Biol. Chem. 2002;277:42852–42858. doi: 10.1074/jbc.M206061200. [DOI] [PubMed] [Google Scholar]
- Wang Q, Carroll JS, Brown M. Spatial and temporal recruitment of androgen receptor and its coactivators involves chromosomal looping and polymerase tracking. Mol. Cell. 2005;19:631–642. doi: 10.1016/j.molcel.2005.07.018. [DOI] [PubMed] [Google Scholar]
- Wright ME, Tsai MJ, Aebersold R. Androgen receptor represses the neuroendocrine transdifferentiation process in prostate cancer cells. Mol. Endocrinol. 2003;17:1726–1737. doi: 10.1210/me.2003-0031. [DOI] [PubMed] [Google Scholar]
- Zegarra-Moro OL, Schmidt LJ, Huang H, Tindall DJ. Disruption of androgen receptor function inhibits proliferation of androgen-refractory prostate cancer cells. Cancer Res. 2002;62:1008–1013. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.