Abstract
Several studies have shown that IL-13 is induced in the esophageal biopsies of EoE patients and promotes esophageal eosinophilia in mice following an IL-13 challenge. However, the role of IL-13 has not been clearly investigated in allergen-induced EoE. Accordingly, we tested the hypothesis that IL-13 is required in allergen-induced EoE. Mice deficient in IL-13, STAT (signal transducer and activator of transcription)6 and both IL-4/IL-13 genes with their respective controls were challenged with aspergillus extract and IL-5 gene-deficient with their control were challenged with recombinant IL-13, intranasally The lung and esophageal eosinophils, mast cells and collagen accumulation were examined. Herein, we report that intranasal delivery of IL-13 promotes IL-5 dependent esophageal eosinophilia. However, allergen-induced EoE is not impaired in the IL-13 gene-deficient mice. In addition, wild type and IL-13 gene-deficient mice demonstrated a comparable level of mast cells and collagen accumulation in the esophagus following allergen-induced experimental EoE. Similarly, we found that esophageal eosinophilia in IL-4/IL-13 double gene-deficient and STAT6 gene-deficient mice were also not reduced following allergen-induced experimental EoE. In contrast, lung eosinophilia was significantly reduced in mice deficient in IL-13, both IL-4/IL-13 and STAT6 genes following allergen challenge. In conclusion, our data establish that allergen-induced EoE pathogenesis is independent of IL-13; whereas, IL-13 is required for allergen-induced lung eosinophilia.
Introduction
Eosinophilic esophagitis (EoE) is a painful and sometimes devastating inflammatory disease of the esophagus, that often leads to swallowing problems, food refusal, food intolerance in infants, dysphagia and food impactions in adolescents and adults.1–4 Both pediatric and adult EoE patients develop fibrosis and other anatomical complications including esophageal strictures.4–12 EoE is now considered a global health problem for children in multiple developed and developing countries over the last decade.1, 5, 12–19 EoE is associated with allergic responses; for example, patients with EoE have a high rate of atopy and their clinical symptoms and eosinophilic infiltrations are ameliorated by an elemental diet or by anti-inflammatory glucocorticoid therapy.20, 21 Interestingly, IL-13 appears to be particularly important since it is produced in high quantities by Th2-cells and regulates multiple characteristics of allergic diseases.22 The levels of elevated IL-13 is an important regulator of a number of allergic diseases including asthma,23 eosinophilic esophagitis,24–28 atopic dermatitis,29–31 and allergic rhinitis.32, 33 IL-13 share a common receptor subunit, with the IL-4 Rα, and signals through the signal transducer and activator of transcription (STAT)6.34 Th2 cells produce the cytokine IL-5, which is specific for the growth and survival of eosinophils. We demonstrated earlier that IL-5 is over-expressed in the esophagus of patients with EoE35 and systemic over-expression of IL-5 (via pharmacological or transgenic approaches) promotes EoE in mice.36 It has been previously shown that in vitro IL-13 activates esophageal epithelial cells and induces eosinophil chemokines, eotaxin-1, -2 and -3.37 Additionally, it has also been shown that IL-13 induces IL-5 that may be responsible for IL-13 induced tissue eosinophilia.26 Therefore, it is important to understand the role of IL-13 in promoting esophageal eosinophilia, whether IL-13 directly acts and promote esophageal eosinophilia or esophageal eosinophila is due to the induction of IL-5 and eotaxins.Of note, mice with targeted deletion of IL-13, both IL-4/IL-13, or STAT6 develop attenuation of certain features of allergic disease like asthma.38, 39 Further, we also previously showed that allergen challenge promotes IL-5 mediated experimental asthma and EoE in mice.40 The allergen-induced experimental EoE in mice mimic most of the characteristic features observed in individuals with various forms of EoE, such as intra-epithelial eosinophils, extracellular granule deposition, and epithelial cell hyperplasia.40 Importantly, over-expression of IL-13, by transgenic approaches, induces multiple features of EoE, including eosinophilia, collagen deposition and reduced lumen circumference.25, 41 Therefore, it is rationale to know whether IL-13 is directly responsible for allergen-induced EoE pathogenesis. Accordingly, we tested the hypothesis that IL-13 is critical in the induction and progression of EoE. Therefore, we delivered Aspergillus allergen to the IL-13, both IL-4/IL-13 and their signaling molecule signal transducer and activatior of transcription (STAT)6 gene-deficient mice. The data presented in this manuscript establish that IL-13 intranasal delivery promote IL-5 dependent esophageal eosinophilia; however, IL-13 signalling is not critical in promoting intranasal allergen associated EoE pathogenesis.
Results
Intranasal IL-13 induces IL-5 mediated esophageal eosinophilia
We were first interested in determining if intranasal delivery of IL-13 induces EoE. In order to test this, 10mg of recombinant IL-13 was delivered intranasally to the wild-type mice 3 times on alternate days. Mice received 3 doses of recombinant IL-13 10μg/ 40μl (or 40μl saline) separated by 24 hours and the eosinophil level in the esophagus was determined 24 hours after the last IL-13 delivery.
IL-13 administration to Balb/c mice promotes esophageal eosinophilia
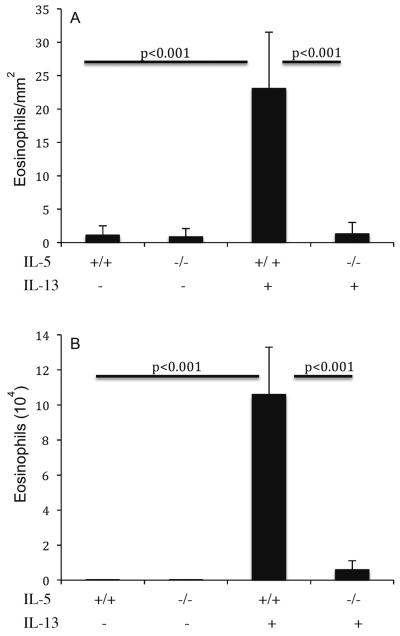
As a control, mice treated with intranasal saline did not have significant levels of esophageal eosinophils. The eosinophil levels in the esophagus of IL-13 and saline treated mice were 24.3 ± 4.6 and 1.1 ± 1.6/mm2 following 3 intranasal IL-13 treatment (mean ± S.D., n = 6; P<0.001), respectively. Next, we tested whether IL-13 induced esophageal eosinophilia is due to IL-5 induction, because it has been shown that IL-13 induces IL-5.26 We addressed this by treating IL-5 gene targeted mice by intranasal IL-13. IL-13 intranasal treatment to IL-5 gene-deficient mice did not develop esophageal eosinophilia; whereas, wild-type control mice developed marked esophageal eosinophilia (Figure 1A). The eosinophil levels in the esophagus of wild-type and IL-5 gene-deficient following 3 doses of 10 mg IL-13 treatment on alternate days were 23.1 ± 8.4 and 1.3 ± 1.7/mm2 (mean ± S.D., n = 4–5; P<0.001), whereas, saline treated wild type and IL-5 gene-deficient mice had 1.1 ± 1.4 and 0.9 ± 1.2/mm2, respectively. For comparison, eosinophil levels in the BALF of wild-type and IL-5 gene targeted mice following 3 doses of 10 mg intranasal IL-13 were 10.6 ± 2.7 × 104 and 0.57 ± 0.48 × 104/lung (mean ± S.D., n = 6–8; P <0.001), whereas, saline treated wild type and IL-5 gene-deficient mice had 0.005 ± 0.01 × 104/ lung and 0.002 ± 0.004 × 104 /Lung, respectively (Figure 1B). Collectively, these results establish an essential role for IL-5 in IL-13-induced esophageal eosinophilia. This data is similar to our earlier reported intratracheal delivered IL-13 findings;25 but to compare aspergillus induced EoE to IL-13 delivered EoE, it was needed to repeat the experiment by delivering IL-13 with a similar route.
Figure 1. IL-13-induced esophageal eosinophilia in wild type and IL-5 gene-targeted mice.
The level of eosinophils in the esophagus and lung of wild type and IL-5 gene targeted mice were analyzed following intranasal 10 mg or rIL-13 challenge. Wild-type (+/+) or IL-5-deficient (−/−) mice were challenged and eosinophils were determined by anti-MBP staining in the esophagus (A) and in the lung fluid (B) by differential counting of cells in BALF. The results are expressed as mean ± S.D. (n = 8 mice/group).
IL-13 gene-deficient mice induce allergen-induced EoE
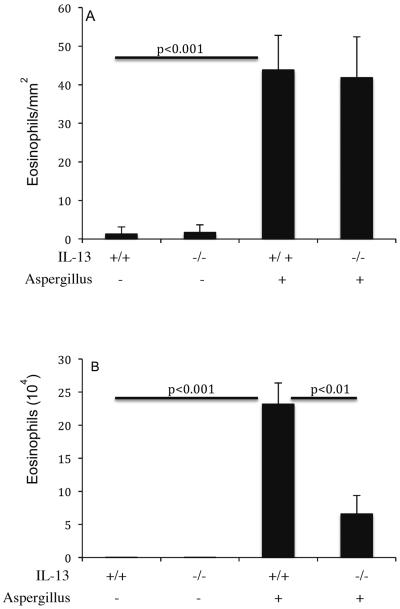
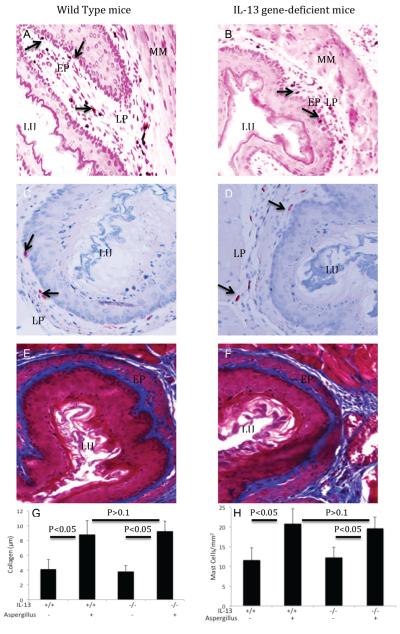
Next, we tested the hypothesis that allergen-induced EoE is IL-13 dependent. Therefore, we induced experimental allergen-induced EoE in IL-13 gene-deficient mice and wild-type control mice were challenged with Aspergillus antigen. As a negative control, both types of mice were challenged with saline. The eosinophil levels in the esophagus of Aspergillus challenged wild-type and IL-13-deficient mice were 43.8 ± 9.1 and 41.8 ± 10.5/mm2 (mean ± S.D., n = 8; P < 0.1), whereas, in saline treated mice they were 1.3 ± 1.8 and 1.2 ± 1.7/mm2 (mean ± S.D., n = 8), respectively. As a comparison, eosinophil levels in the BALF were also examined. The eosinophils in the BALF of Aspergillus challenged wild type mice were 23.2 ± 3.2 × 104/lung (mean ± S.D., n = 8) compared with 6.6 ± 2.8 × 104/lung in IL-13 gene deficient mice (mean ± S.D., n = 8; P<0.01); whereas, saline treated wild type and IL-13 gene-deficient mice had 0.01 ± 0.01 × 104 and 0.01 ± 0.01 × 104/lung (mean ± S.D., n = 8), respectively (Figure 2A, B). Furthermore, most of the EoE characteristics such as the induction of intraepithelial eosinophilia (Figure 3 A, B), esophageal mast cells (Figure 3 C, D) and lamina propria collagen (Figure 3 E, F) was found in comparable levels in both wild type and IL-13 gene-deficient mice following induction of allergen-induced experimental EoE. The quantitation of collagen thickness and mast cell numbers were performed and found comparable in allergen challenged wild type and IL-13 gene-deficient mice (Figure 3,G, H). Collectively, these results establish that IL-13 is not critical in allergen-induced EoE. We also tested the role of IL-4 in allergen-induced EoE by using IL-4Rα gene-deficient mice and similar data to IL-13 gene-deficient mice were found (data not shown). This is not surprising as both, IL-4 and IL-13 are related cytokines that share a common signal transduction mechanism involving the IL-4 receptor alpha chain and STAT6.
Figure 2. Aspergillus-induced eosinophilia in IL-13 gene-targeted mice.
The level of eosinophils in the esophagus and lung of wild type and IL-13 gene targeted mice were analyzed following intranasal nine doses of Aspergillus antigen challenge in 3 weeks of allergen challenge regime. Wild-type (+/+) or IL-13-deficient (−/−) mice were challenged and eosinophils were determined by anti-MBP staining in the esophagus (A) and in the lung fluid (B) by differential counting of cells in BALF. The results are expressed as mean ± S.D. (n = 12 mice/group).
Figure 3. Detection of intraepithelial eosinophils, mast cells and collegen in IL-13 gene-deficient mice following Aspergillus-induced EoE.
The EoE characteristics features were examined in wild type and IL-13 gene-deficient mice following the induction of Aspergillus antigen-induced experimental EoE. Both wild type and IL-13 gene-deficient mice show intraepithelial eosinophils (A, B, original magnification 200×), mast cells (C, D, original magnification 200×) and lamina propria and epithelial mucos collegen accumulation (E, F, original magnification 400×). Arrows indicates intraepithelial eosinophils or lamina propria mast cells in the respective phomicrographs. The quantitation of collagen thickness and mast cells numbers in wild type and IL-13 gen-deficient mice are shown (G, H). The results are expressed as mean ± S.D. (n = 8 mice/group). EP-Epithelium, LP-Lamina propria, LU-Lumen, MM-Muscular Mucosa,
IL-4/IL-13 double gene-deficient mice are not protected from the induction of allergen-induced EoE
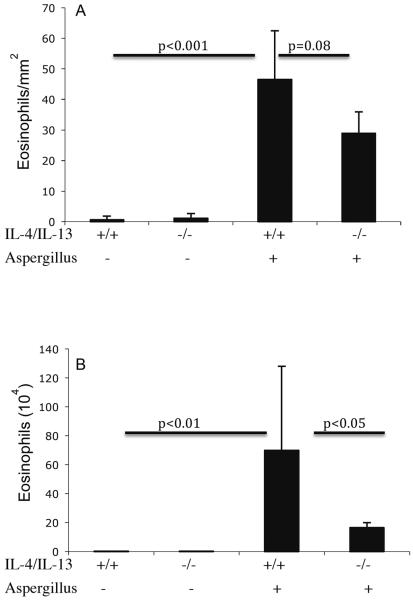
Next, we tested the hypothesis that allergen-induced EoE is IL-4/IL-13 dependent. IL-4/IL-13 has been shown to have a significant role in allergen-induced mouse model of experimental asthma.42, 43 Accordingly, IL-4/IL-13 double gene-deficient mice and strain matched wild-type control mice were challenged with saline or Aspergillus antigen. The eosinophil levels in the esophagus of Aspergillus challenged wild-type and IL-4/IL-13 double gene-deficient mice were 46.5 ± 16.4 and 28.9 ± 7.3/mm2 (mean ± S.D., n = 8; P < 0.08), whereas, in saline treated mice they were 0.6 ± 1.3 and 1.1 ± 1.5/mm2 (mean ± S.D., n = 8), respectively. As a comparison, eosinophil levels in the BALF were also examined. The eosinophils in the BALF of Aspergillus challenged wild type mice were 63.9 ± 51.5 × 104/lung (mean ± S.D., n = 8) compared with 12.8 ± 0.5 × 104/lung in IL-4/IL-13 double gene-deficient mice (mean ± S.D., n = 8; P<0.001); whereas, saline treated wild type and IL-4/IL-13 double gene-deficient mice had 0.01 ± 0.1 × 104 and 0.01 ± 0.1 × 104/lung (mean ± S.D., n = 8), respectively (Figure 4 A, B). Collectively, these results establish that allergen-induced EoE is not dependent on IL-4/IL-13.
Figure 4. Aspergillus-induced eosinophilia in IL-4/IL-13 double gene-targeted mice.
The level of eosinophils in the esophagus and lung of wild type and IL-4/IL-13 gene targeted mice were analyzed following intranasal nine doses of Aspergillus antigen challenge in 3 weeks of allergen challenge regime. Wild-type (+/+) or IL-4/IL-13 double gene-deficient (−/−) mice were challenged and eosinophils were determined by anti-MBP staining in the esophagus (A) and in the lung fluid (B) by differential counting of cells in BALF. The results are expressed as mean ± S.D. (n = 9 mice/group).
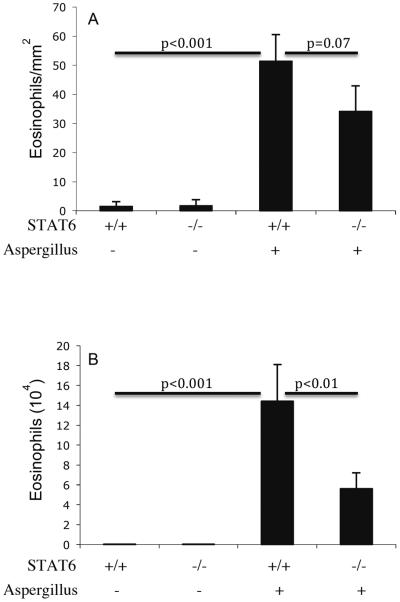
Aspergillus-induced eosinophilic esophagitis is STAT6 independent
We were next interested in determining the role of STAT6 in regulating EoE. STAT6 has been shown to be important in regulating allergen-induced IL-4 and IL-13 mediated eosinophils in the lung, but its effect in the gastrointestinal tract has only been studied in the intestine following parasitic infection.44 Accordingly, we tested the role of STAT6 in the development of allergen-induced EoE, STAT6 gene-deficient mice and strain matched wild-type control mice were challenged with saline or Aspergillus antigen. The eosinophil levels in the esophagus of Aspergillus challenged wild-type and STAT6-deficient mice were 51.4 ± 9.1 and 34.1 ± 8.8/mm2 (mean ± S.D., n = 8; P < 0.07), whereas, in saline treated mice they were 1.4 ± 1.7 and 1.7 ± 2.1/mm2 (mean ± S.D., n = 8), respectively. As a comparison, eosinophil levels in the BALF were also examined. The eosinophils in the BALF of Aspergillus challenged wild type mice were 14.4 ± 3.7 × 104/lung (mean ± S.D., n = 8) compared with 5.6 ± 1.6 × 104/lung in STAT6 gene deficient mice (mean ± S.D., n = 8; P<0.01); whereas, saline treated wild type and STAT6 gene-deficient mice had 0.01 ± 0.01 × 104 and 0.01 ± 0.01 × 104/lung (mean ± S.D., n = 8), respectively (Figure 5 A–B). Collectively, these results establish that Aspergillus-induced EoE is not dependent on STAT6.
Figure 5. Aspergillus-induced eosinophilia in STAT6 gene-targeted mice.
The level of eosinophils in the esophagus and lung of wild type and STAT6 gene targeted mice were analyzed following intranasal nine doses of Aspergillus antigen challenge in 3 weeks of allergen challenge regime. Wild-type (+/+) or STAT6-deficient (−/−) mice were challenged and eosinophils were determined by anti-MBP staining in the esophagus (A) and in the lung fluid (B) by differential counting of cells in BALF. The results are expressed as mean ± S.D. (n = 9 mice/group).
Analysis of eosinophil active cytokine, IL-5 and chemokines Eotaxin-1 and Eotaxin-2 transcript levels in wild type, IL-4/IL-13 double gene-deficient and STAT6 gene-deficient mice
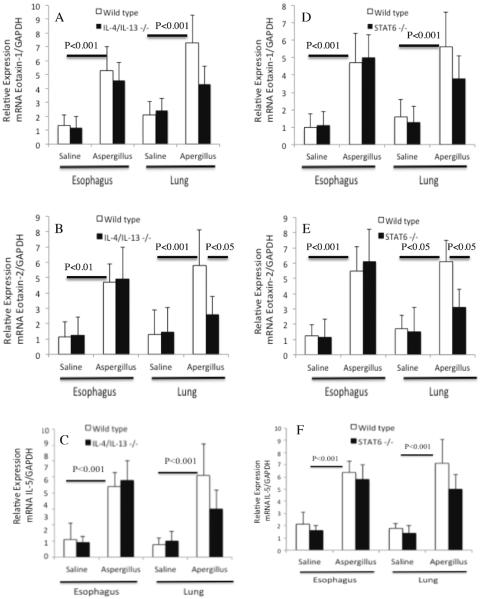
Next, we examined eotaxin-1, eotaxin-2, IL-5 and GADPH mRNA expression in the lung and esophagus of saline- and Aspergillus-challenged wild type, IL-4/IL-13 double gene-deficient and STAT6 gene-deficient mice. Our quantitative PCR analysis showed that Aspergillus-challenged mice had a significant increase of the relative mRNA expression of esophageal and lung eotaxin-1, eotaxin-2 and IL-5 compared to the saline-challenged wild type mice (Figure 6 A–F). However, a comparable transcript expression level of eotaxin-1 and IL-5 were found in allergen challenged wild type and IL-4/IL-13 double gene-deficient mice (Figure 6 A, C) and allergen challenged wild type and STAT6 gene-deficient mice (Figure 6 D, F). The transcript expression level of eotaxin-2 was significantly reduced in the lungs, but not in the esophagus of allergen challenged IL-4/IL-13 double gene-deficient and STAT6 gene-deficient mice compared to their respective wild type mice (Figure 2 B, E). These data indicates that reduced eotaxin-2 transcript in the lung may be responsible for significantly reduced lung eosinophilia in IL-4/IL-13 double gene-deficient and STAT6 gene-deficient mice.
Figure 6. Aspergillus induced mRNA expression of eotaxin-1, eotaxin-2 and IL-5 in the lung and esophagus.
The quantitative real-time PCR analyses of eotaxin-1, eotaxin-2, and IL-5 mRNA levels following saline and Aspergillus challenged in wild type and IL-4/IL-13 double gene-deficient mice are shown (A–C) and wild type and STAT6 gene-deficient mice are shown (D–F). The data are expressed as mean ± S.D, n= 8 mice/group.
Discussion
Eosinophil accumulation in the esophagus is characteristic of a variety of clinical disorders including GERD, eosinophilic gastroenteritis, and EoE.2, 45–49 Recent clinical studies have suggested that the prevalence of these disorders, especially EoE, is increasing frequency.48, 50 It has been reported that IL-13 overexpression is associated with the induction of esophageal eosinophilia and fibrosis, which is commonly observed in the human and experimental model of EoE.40 Herein, we first confirm our previous findings that intranasal IL-13 also promote a similar levels of eosinophil accumulation in the esophagus like intratracheally delivered IL-13.25, 41 However, the signficance of IL-13 in allergen-induced EoE was not earlier proven. Notably, EoE is an allergen-induced esophageal disorder.20, 21 Therefore, the present study was designed to tested the hypothesis that IL-13 is critical in allergen-induced EoE pathogenesis. We show for the first time that intranasal IL-13 induces IL-5 dependent esophageal eosinophilia. These findings are in accordance with the earlier report that indicate IL-5 is required in IL-13 induce eosinophilia in the lung.26 In this report, we first time show that allergen-induced EoE is independent to IL-13 in experimental model, because IL-13 gene-deficient mice are not protected from the development of allergen-induced EoE. Both wild type and IL-13 gene-deficient mice show comparable levels of esophageal eosinophils and induced mast cells. Additionally, we show that both wild type and IL-13 gene-deficient mice have most of the EoE characteristics feature like comparable intraepithelial eosinophils, induced and lamina propria collagen accumulation following allergen challenge. In contrast, the allergen-induced lung eosinophilia was found dependent to IL-13. Our present data indicates that two different mechanism is operational in IL-13 delivered lung eosinophilia and in allergen-induced EoE. We believe that the earlier reported coexpression of IL-5 and IL-13 including eosinophil active chemokine eotaxin-3 in EoE patients35 may be a co-incidence and IL-13 may not be a major contributor in promoting human EoE. The earlier in vitro studies also show that IL-13 induces IL-5 and and eosinophil active chemokines eotaxin-1, -2 and -3 in esophageal epithelial cells,37 which also indicates that IL-13 induced IL-5 and eotaxins may be the primary cause of promoting esophageal eosinophilia. Furthermore, It has been also shown that IL-5 primes eosinophils to respond to chemoattractants and to induce eosinophil adhesion molecule expression and activation.51 Taken together, all these previous reports and our present data suggest that IL-13 induced eosinophil accumulation in the esophagus is eotaxin and IL-5 mediated and not a direct response of IL-13 that has been previously thought and reported.25, 28, 52 IL-13 is a pleotropic cytokine and has been implicated in the variety of diseases. A number of reports indicate that IL-13 levels are increased in patients suffering from chronic asthma and EoE and affect a variety of immune cells mediated functions in several allergic diseases. Therefore, the presented experimental data indicates that IL-13 may not be critical in EoE pathogenesis.
This has been further confirmed by challenging the IL-13, both IL-4/IL-13 and STAT6 gene-deficient mice to the Aspergillus extract that IL-13 signaling is not required in promoting allergen-induced experimental EoE. These studies are important because a clinical report demonstrating that levels of IL-4 secreting T cells in the esophageal lesions along with IL-13 is elevated in patients with EoE.38 Both, IL-4 and IL-13 are related cytokines that share a common signal transduction mechanism involving the IL-4 receptor alpha chain and STAT6.16 Our experimentation showed that neither IL-4 receptor alpha chain deficient mice (data not shown), nor IL-4/IL-13 double gene-deficient mice or the STAT6 gene-deficient mice were protected from allergen-induced EoE. Of note, both IL-4 and IL-13 are regulated by STAT6 and it is noteworthy that antigen-induced lung inflammation is largely dependent upon these signaling molecules. This we further confirmed by showing that Aspergillus challenged IL-4/IL-13 double deficient and STAT6 deficient mice still had higher lung eosinophils compared with saline challenged wild-type mice. In addition, we show that transcript expression levels of eosinophil active cytokine IL-5 and chemokine eotaxin-1 is comparable in the allergen challenged lung and esophagus of wild type, STAT6 gene-deficient and IL-4/IL-13 double gene-deficient mice. However, the eotaxin-2 transcript expression was significantly reduced in the lungs along with partial reduction of lung eosinophilia in IL-4/IL-13 double gene-deficient and STAT6 gene-deficient mice; but no reduction was observed in the esophageal eosinophilia or transcript expression. Therefore, it might be possible that eotaxin-2 may have a role in partial eosinophil reduction in the lung and not in the esophagus. These presented transcript expression data further indicates that two very different mechanisms are operational in the recruitment of eosinophils in lung and esophagus. We provide the first evidence that allergen-induced EoE, is not dependent upon classic Th2 cytokine, IL-13 signaling and draws attention to the importance of IL-5 (not IL-13) in disease pathogenesis. We clearly demonstrated that allergen challenged IL-13 gene-deficient mice not only showing comparable level of eosinophils but also show similar levels of mast cells, intraepithelial esophageal eosinophils and submucosal collagen in experimental EoE. These classical symptoms observed in IL-13 gene-deficient mice and wild type mice following allergen challenges are the characteristic features of experimental and human EoE.40, 53, 54 These presented data further confirm the earlier published reports that IL-5 is critical in EoE and esophageal remodeling in EoE is largely associated with the local expression of IL-5.40,55
In summary, our present investigations dissect the cellular and molecular mechanisms involved in eosinophil accumulation in the esophagus and demonstrate that allergen-induced eosinophilic esophageal disorder is independent to IL-4, IL-13 and its signaling factor STAT6 and further confirm the significance of our most recent findings that allergen-induced EoE is STAT6 independent.56 Taken together, these investigations indicate that IL-13 induced esophageal eosinophilia may be due to the induction of eotaxins and IL-5 and IL-13 has no direct role in allergen-induced eosinophilic esophageal disorder. We conclude our study by stating that the role of IL-13 should be carefully interpretated in promoting EoE or suggesting any therapeutic interventions based on IL-13 in allergen-induced eosinophilic esophageal disorders.
Materials and Methods
Mice
Eight week-old BALB/c mice, STAT6 deficient and IL-5 deficient with matched control mice (Jackson Laboratory, Bar Harbor, ME), IL-13 deficient, IL-4/IL-13 double deficient and their littermate BALB/c controls (originally obtained from A. MacKenzie of Medical Research Council Laboratory of Molecular Biology, Cambridge, United Kingdom) were kept and used at CCHMC, Cincinnati. Partial tissue analysis was performed at Case Western University, Cleveland, Ohio. All procedures were performed in accordance with the ethical guidelines in the Guide for the Care and Use of Laboratory Animals of the Institutional Animal Care and Use Committee approved protocol.
Induction of experimental EoE in mice
Experimental EoE was induced in mice following an established protocol described previously.40 In brief, mice were lightly anesthetized with isoflurane inhalation (methoxy-fluorane; Pittman-Moore, Mundelein, IL) and 100 μg (50 μl) of Aspergillus fumigatus (Greer Laboratories, Lenoir, NC) or 50 μl of normal saline alone was applied to the nares using a micropipette with the mouse held in the supine position. After instillation, mice were held upright until alert. After three treatments per week for three weeks, mice were sacrificed between 18 and 20 hours after the last intranasal challenge. Additionally, we also challenged the mice with 3 doses of recombinant IL-13 10μg/ 40μl (or 40μl saline) separated by 24 hours and the eosinophil level in the esophagus was determined 24 hours after the last IL-13 delivery.
Bronchoalveolar lavage fluid (BALF) collection and analysis
The mice were euthanized by CO2 inhalation. Immediately thereafter a midline neck incision was made and the trachea was cannulated. The lungs were lavaged three times with 1.0 ml PBS containing 1% FCS and 0.5 mM EDTA. The recovered BALF was centrifuged at 400× g for 5 minutes at 4°C, and resuspended in 200 μl PBS containing 1% FCS and 0.5 mM EDTA. Lysis of red blood cells was carried out utilizing RBC lysis buffer (Sigma, St. Louis, MO) according to the manufacturer's recommendations. Total cell numbers were counted with a hemacytometer. Cytospin preparations of 5 × 104 cells were stained with Giemsa-Diff-Quick (Dade Diagnostics of P.R., Inc., Aguada, PR) and differential cell counts were determined for eosinophils percent and absolute numbers.
Eosinophil analysis in the esophagus
The esophagus of adult mice was fixed in 4% paraformaldehyde in phosphate buffer pH 7.4, embedded in paraffin, cut into 5mm sections, fixed to positive charged slides, and immunostained with antiserum against mouse eosinophil major basic protein (anti-MBP), a kind gift of Drs. James and Nancy Lee (Mayo Clinic, Scottsdale, AZ), as described,40 In brief, endogenous peroxidase in the tissues was quenched with 0.3% hydrogen peroxide in methanol followed by nonspecific protein blocking with normal goat serum. Tissue sections were then incubated with rabbit anti-MBP (1:16,000) overnight at 4°C, followed by 1:200 dilution of biotinylated goat anti-rabbit IgG secondary antibody and avidin-peroxidase complex (Vector Laboratories, Burlingame, CA) for 30 minutes each. These slides were further developed with nickel diaminobenzidine-cobalt chloride solution to form a black precipitate, and counterstained with nuclear fast red. Negative controls include replacing the primary antibody with normal rabbit serum to check endogenous biotin and peroxidase activity. Quantification of the immunoreactive cells was carried out by using a video-assistant integrated computer software program Image Pro software analyzer (Media Cybenetics, Warrendale, PA). The eosinophil levels are expressed as cells/mm2.
Mast cell analysis
The 5-μm esophageal paraffin tissue sections were de-paraffinized, stained with a chloroacetate staining and detected by performing histological analysis using light microscopy. The pink color mast cells were quantified by counting stained cells in each tissue section with the assistance of digital morphometry using the Image Pro software analyzer (Media Cybenetics, Warrendale, PA) and expressed as mast cells/mm2 tissue area as described earlier.25, 36
Tissue collagen staining
Esophageal tissue samples were fixed with 4% paraformaldehyde, embedded in paraffin, cut into 5μm sections, and fixed to positively charge slides. Collagen staining was then performed on the tissue sections by Masson's trichrome (Poly Scientific R&D Corp.) method for the detection of collagen fibers according to the manufacturer's recommendations. The collagen thickness was measured using the Image Pro software analyzer (Media Cybenetics, Warrendale, PA) and is expressed as collagen thickness in μm.
Real-time polymerase chain reaction (PCR) analysis
The RNA samples (500 ng) were subjected to reverse transcription using iScript reverse transcriptase (Bio-Rad, Hercules, CA) according to the manufacturer's instructions. The cytokine and chemokine mRNA levels were quantified by real-time PCR using the LightCycler instrument and LightCycler FastStart DNA master SYBR green I as a ready-to-use reaction mix (Roche, Indianapolis, IN). Results were then normalized using previously published human GAPDH primers28,37,52,55 to amplified mouse GAPDH from the same cDNA mix and expressed as fold induction compared to the controls. cDNA were amplified using the primers described earlier. The primers used to perform qPCR are listed in Table 1.
Table 1.
Primers used to quantitate IL-5, eotaxin-1 and eotaxin-2
| Genes | Sense and anti-sense primer sequence |
|---|---|
|
| |
| mIL-5 | 5'-TCCCATGAGCACAGTGGTGAAAG |
| 5'-CACAGTACCCCCACGGACAGTTT | |
|
| |
| mEotaxin-1 | 5'-GGCTCACCCAGGCTCCATCC |
| 5'-TTTTGGTCCAGGTGCTTTGTGG | |
|
| |
| mEotaxin-2 | 5'-CTCCTTCTCCTGGTAGCCTGC |
| 5'-GTGATGAAGATGACCCCTGCCTT | |
|
| |
| GAPDH | 5'-TGGAAATCCCATCACCATCT |
| 5'GTCTTCTGGGTGGCAGTGAT | |
Statistical analysis
Data are expressed as mean ± Standard Deviation (S.D.). Statistical significance comparing different set of mice was determined by unpaired InStat GraphPad t-test.
Acknowledgements
This work was supported by in part by NIH R01 DK067255 (AM), NIH R01 AI080581 (AM). We thank Drs. James and Nancy Lee (Mayo Clinic, Scottsdale, AZ) for the generous supply of anti-MBP, and Marc Rothenberg, MD., PhD (Cincinnati Childrens, Cincinnati, Ohio) for his support in providing the gene-deficient mice at Cincinnati Childrens Hospital, Cincinnati, OH. We also acknowledge Dr. Fabio Cominelli, MD., PhD, Professor and Chief, Division of Gastroenterology and Liver Disease for providing the facility at Case Western Reserve University, Cleveland to continue our EoE research.
Abbreviations used in this paper
- BALF
bronchoalveolar lavage fluid
- EoE
eosinophilic esophagitis
- MBP
major basic protein
- STAT
signal-transducer-and-activator-of-transcription
References
- 1.Orenstein SR, Shalaby TM, Di Lorenzo C, Putnam PE, Sigurdsson L, Kocoshis SA. The spectrum of pediatric eosinophilic esophagitis beyond infancy: a clinical series of 30 children. Am J Gastroenterol. 2000;95(6):1422–1430. doi: 10.1111/j.1572-0241.2000.02073.x. [DOI] [PubMed] [Google Scholar]
- 2.Walsh SV, Antonioli DA, Goldman H, Fox VL, Bousvaros A, Leichtner AM, et al. Allergic esophagitis in children: a clinicopathological entity. Am J Surg Pathol. 1999;23(4):390–396. doi: 10.1097/00000478-199904000-00003. [DOI] [PubMed] [Google Scholar]
- 3.Liacouras CA, Ruchelli E. Eosinophilic esophagitis. Curr Opin Pediatr. 2004;16(5):560–566. doi: 10.1097/01.mop.0000141071.47572.eb. [DOI] [PubMed] [Google Scholar]
- 4.Sant'Anna AM, Rolland S, Fournet JC, Yazbeck S, Drouin E. Eosinophilic Esophagitis in Children: Symptoms, Histology and pH Probe Results. Journal of pediatric gastroenterology and nutrition. 2004;39(4):373–377. doi: 10.1097/00005176-200410000-00013. [DOI] [PubMed] [Google Scholar]
- 5.Croese J, Fairley SK, Masson JW, Chong AK, Whitaker DA, Kanowski PA, et al. Clinical and endoscopic features of eosinophilic esophagitis in adults. Gastrointest Endosc. 2003;58(4):516–522. doi: 10.1067/s0016-5107(03)01870-4. [DOI] [PubMed] [Google Scholar]
- 6.Dahshan A, Rabah R. Correlation of endoscopy and histology in the gastroesophageal mucosa in children: are routine biopsies justified? J Clin Gastroenterol. 2000;31(3):213–216. doi: 10.1097/00004836-200010000-00005. [DOI] [PubMed] [Google Scholar]
- 7.Ruigomez A, Alberto Garcia Rodriguez L, Wallander MA, Johansson S, Eklund S. Esophageal stricture: incidence, treatment patterns, and recurrence rate. Am J Gastroenterol. 2006;101(12):2685–2692. doi: 10.1111/j.1572-0241.2006.00828.x. [DOI] [PubMed] [Google Scholar]
- 8.Zimmerman SL, Levine MS, Rubesin SE, Mitre MC, Furth EE, Laufer I, et al. Idiopathic eosinophilic esophagitis in adults: the ringed esophagus. Radiology. 2005;236(1):159–165. doi: 10.1148/radiol.2361041100. [DOI] [PubMed] [Google Scholar]
- 9.White RJ, Zhang Y, Morris GP, Paterson WG. Esophagitis-related esophageal shortening in opossum is associated with longitudinal muscle hyperresponsiveness. Am J Physiol Gastrointest Liver Physiol. 2001;280(3):G463–469. doi: 10.1152/ajpgi.2001.280.3.G463. [DOI] [PubMed] [Google Scholar]
- 10.Paterson WG, Kolyn DM. Esophageal shortening induced by short-term intraluminal acid perfusion in opossum: a cause for hiatus hernia? Gastroenterology. 1994;107(6):1736–1740. doi: 10.1016/0016-5085(94)90814-1. [DOI] [PubMed] [Google Scholar]
- 11.Martin-Munoz MF, Lucendo AJ, Navarro M, Letran A, Martin-Chavarri S, Burgos E, et al. Food allergies and eosinophilic esophagitis--two case studies. Digestion. 2006;74(1):49–54. doi: 10.1159/000096595. [DOI] [PubMed] [Google Scholar]
- 12.Attwood SE, Smyrk TC, Demeester TR, Jones JB. Esophageal eosinophilia with dysphagia. A distinct clinicopathologic syndrome. Dig Dis Sci. 1993;38(1):109–116. doi: 10.1007/BF01296781. [DOI] [PubMed] [Google Scholar]
- 13.Cury EK, Schraibman V, Faintuch S. Eosinophilic infiltration of the esophagus: gastroesophageal reflux versus eosinophilic esophagitis in children--discussion on daily practice. J Pediatr Surg. 2004;39(2):e4–7. doi: 10.1016/j.jpedsurg.2003.10.028. [DOI] [PubMed] [Google Scholar]
- 14.Cantu P, Velio P, Prada A, Penagini R. Ringed oesophagus and idiopathic eosinophilic oesophagitis in adults: an association in two cases. Dig Liver Dis. 2005;37(2):129–134. doi: 10.1016/j.dld.2004.04.008. [DOI] [PubMed] [Google Scholar]
- 15.Fujiwara H, Morita A, Kobayashi H, Hamano K, Fujiwara Y, Hirai K, et al. Infiltrating eosinophils and eotaxin: their association with idiopathic eosinophilic esophagitis. Ann Allergy Asthma Immunol. 2002;89(4):429–432. doi: 10.1016/S1081-1206(10)62047-9. [DOI] [PubMed] [Google Scholar]
- 16.Munitiz V, Martinez de Haro LF, Ortiz A, Pons JA, Bermejo J, Serrano A, et al. Primary eosinophilic esophagitis. Dis Esophagus. 2003;16(2):165–168. doi: 10.1046/j.1442-2050.2003.00319.x. [DOI] [PubMed] [Google Scholar]
- 17.Lucendo AJ, Carrion G, Navarro M, Pascual JM, Gonzalez P, Castillo P, et al. Eosinophilic esophagitis in adults: an emerging disease. Dig Dis Sci. 2004;49(11–12):1884–1888. doi: 10.1007/s10620-004-9588-x. [DOI] [PubMed] [Google Scholar]
- 18.Straumann A, Spichtin HP, Bucher KA, Heer P, Simon HU. Eosinophilic esophagitis: red on microscopy, white on endoscopy. Digestion. 2004;70(2):109–116. doi: 10.1159/000080934. [DOI] [PubMed] [Google Scholar]
- 19.Straumann A. What is your diagnosis? Primary eosinophilic esophagitis. Schweiz Rundsch Med Prax. 2004;93(19):795–796. doi: 10.1024/0369-8394.93.19.795. [DOI] [PubMed] [Google Scholar]
- 20.Kelly KJ, Lazenby AJ, Rowe PC, Yardley JH, Perman JA, Sampson HA. Eosinophilic esophagitis attributed to gastroesophageal reflux: improvement with an amino acid-based formula. Gastroenterology. 1995;109(5):1503–1512. doi: 10.1016/0016-5085(95)90637-1. [DOI] [PubMed] [Google Scholar]
- 21.Liacouras CA, Wenner WJ, Brown K, Ruchelli E. Primary eosinophilic esophagitis in children: successful treatment with oral corticosteroids. Journal of pediatric gastroenterology and nutrition. 1998;26(4):380–385. doi: 10.1097/00005176-199804000-00004. [DOI] [PubMed] [Google Scholar]
- 22.Wills-Karp M. IL-12/IL-13 axis in allergic asthma. J Allergy Clin Immunol. 2001;107(1):9–18. doi: 10.1067/mai.2001.112265. [DOI] [PubMed] [Google Scholar]
- 23.Humbert M, Durham SR, Kimmitt P, Powell N, Assoufi B, Pfister R, et al. Elevated expression of messenger ribonucleic acid encoding IL-13 in the bronchial mucosa of atopic and nonatopic subjects with asthma. J Allergy Clin Immunol. 1997;99(5):657–665. doi: 10.1016/s0091-6749(97)70028-9. [DOI] [PubMed] [Google Scholar]
- 24.Blanchard C, Mishra A, Saito-Akei H, Monk P, Anderson I, Rothenberg ME. Inhibition of human interleukin-13-induced respiratory and oesophageal inflammation by anti-human-interleukin-13 antibody (CAT-354) Clin Exp Allergy. 2005;35(8):1096–1103. doi: 10.1111/j.1365-2222.2005.02299.x. [DOI] [PubMed] [Google Scholar]
- 25.Mishra A, Rothenberg ME. Intratracheal IL-13 induces eosinophilic esophagitis by an IL-5, eotaxin-1, and STAT6-dependent mechanism. Gastroenterology. 2003;125(5):1419–1427. doi: 10.1016/j.gastro.2003.07.007. [DOI] [PubMed] [Google Scholar]
- 26.Pope SM, Brandt EB, Mishra A, Hogan SP, Zimmermann N, Matthaei KI, et al. IL-13 induces eosinophil recruitment into the lung by an IL-5- and eotaxin-dependent mechanism. J Allergy Clin Immunol. 2001;108(4):594–601. doi: 10.1067/mai.2001.118600. [DOI] [PubMed] [Google Scholar]
- 27.Fulkerson PC, Fischetti CA, Hassman LM, Nikolaidis NM. Rothenberg ME. Persistent effects induced by IL-13 in the lung. Am J Respir Cell Mol Biol. 2006;35(3):337–346. doi: 10.1165/rcmb.2005-0474OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Blanchard C, Stucke EM, Rodriguez-Jimenez B, Burwinkel K, Collins MH, Ahrens A, et al. A striking local esophageal cytokine expression profile in eosinophilic esophagitis. J Allergy Clin Immunol. 2011;127(1):208–217. 217, e201–207. doi: 10.1016/j.jaci.2010.10.039. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Akdis M, Akdis CA, Weigl L, Disch R, Blaser K. Skin-homing, CLA+ memory T cells are activated in atopic dermatitis and regulate IgE by an IL-13-dominated cytokine pattern: IgG4 counter-regulation by CLA- memory T cells. J Immunol. 1997;159(9):4611–4619. [PubMed] [Google Scholar]
- 30.Katagiri K, Itami S, Hatano Y, Takayasu S. Increased levels of IL-13 mRNA, but not IL-4 mRNA, are found in vivo in peripheral blood mononuclear cells (PBMC) of patients with atopic dermatitis (AD) Clin Exp Immunol. 1997;108(2):289–294. doi: 10.1046/j.1365-2249.1997.d01-1015.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ying S, Meng Q, Barata LT, Robinson DS, Durham SR, Kay AB. Associations between IL-13 and IL-4 (mRNA and protein), vascular cell adhesion molecule-1 expression, and the infiltration of eosinophils, macrophages, and T cells in allergen-induced late-phase cutaneous reactions in atopic subjects. J Immunol. 1997;158(10):5050–5057. [PubMed] [Google Scholar]
- 32.Pawankar RU, Okuda M, Hasegawa S, Suzuki K, Yssel H, Okubo K, et al. Interleukin-13 expression in the nasal mucosa of perennial allergic rhinitis. Am J Respir Crit Care Med. 1995;152(6 Pt 1):2059–2067. doi: 10.1164/ajrccm.152.6.8520776. [DOI] [PubMed] [Google Scholar]
- 33.Pawankar R, Okuda M, Yssel H, Okumura K, Ra C. Nasal mast cells in perennial allergic rhinitics exhibit increased expression of the Fc epsilonRI, CD40L, IL-4, and IL-13, and can induce IgE synthesis in B cells. J Clin Invest. 1997;99(7):1492–1499. doi: 10.1172/JCI119311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jiang H, Harris MB, Rothman P. IL-4/IL-13 signaling beyond JAK/STAT. J Allergy Clin Immunol. 2000;105(6 Pt 1):1063–1070. doi: 10.1067/mai.2000.107604. [DOI] [PubMed] [Google Scholar]
- 35.Straumann A, Bauer M, Fischer B, Blaser K, Simon HU. Idiopathic eosinophilic esophagitis is associated with a T(H)2-type allergic inflammatory response. J Allergy Clin Immunol. 2001;108(6):954–961. doi: 10.1067/mai.2001.119917. [DOI] [PubMed] [Google Scholar]
- 36.Mishra A, Hogan SP, Brandt EB. Rothenberg ME. IL-5 promotes eosinophil trafficking to the esophagus. J Immunol. 2002;168(5):2464–2469. doi: 10.4049/jimmunol.168.5.2464. [DOI] [PubMed] [Google Scholar]
- 37.Blanchard C, Stucke EM, Burwinkel K, Caldwell JM, Collins MH, Ahrens A, et al. Coordinate interaction between IL-13 and epithelial differentiation cluster genes in eosinophilic esophagitis. J Immunol. 184(7):4033–4041. doi: 10.4049/jimmunol.0903069. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Blease K, Schuh JM, Jakubzick C, Lukacs NW, Kunkel SL, Joshi BH, et al. Stat6-deficient mice develop airway hyperresponsiveness and peribronchial fibrosis during chronic fungal asthma. Am J Pathol. 2002;160(2):481–490. doi: 10.1016/S0002-9440(10)64867-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Venkayya R, Lam M, Willkom M, Grunig G, Corry DB, Erle DJ. The Th2 lymphocyte products IL-4 and IL-13 rapidly induce airway hyperresponsiveness through direct effects on resident airway cells. Am J Respir Cell Mol Biol. 2002;26(2):202–208. doi: 10.1165/ajrcmb.26.2.4600. [DOI] [PubMed] [Google Scholar]
- 40.Mishra A, Hogan SP, Brandt EB, Rothenberg ME. An etiological role for aeroallergens and eosinophils in experimental esophagitis. J Clin Invest. 2001;107(1):83–90. doi: 10.1172/JCI10224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Zuo L, Fulkerson PC, Finkelman FD, Mingler M, Fischetti CA, Blanchard C, et al. IL-13 induces esophageal remodeling and gene expression by an eosinophil-independent, IL-13R alpha 2-inhibited pathway. J Immunol. 2010;185(1):660–669. doi: 10.4049/jimmunol.1000471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Devouassoux G, Foster B, Scott LM, Metcalfe DD, Prussin C. Frequency and characterization of antigen-specific IL-4- and IL-13- producing basophils and T cells in peripheral blood of healthy and asthmatic subjects. J Allergy Clin Immunol. 1999;104(4 Pt 1):811–819. doi: 10.1016/s0091-6749(99)70292-7. [DOI] [PubMed] [Google Scholar]
- 43.Luttmann W, Matthiesen T, Matthys H, Virchow J. Synergistic effects of interleukin-4 or interleukin-13 and tumor necrosis factor-alpha on eosinophil activation in vitro. Am J Respir Cell Mol Biol. 1999;20(3):474–480. doi: 10.1165/ajrcmb.20.3.3326. [DOI] [PubMed] [Google Scholar]
- 44.Urban JF, Jr., Noben-Trauth N, Donaldson DD, Madden KB, Morris SC, Collins M, et al. IL-13, IL-4Ralpha, and Stat6 are required for the expulsion of the gastrointestinal nematode parasite Nippostrongylus brasiliensis. Immunity. 1998;8(2):255–264. doi: 10.1016/s1074-7613(00)80477-x. [DOI] [PubMed] [Google Scholar]
- 45.Furuta GT, Ackerman SJ, Wershil BK. The role of the eosinophil in gastrointestinal diseases. Current Opinions in Gastroenterology. 1995;11:541–547. [Google Scholar]
- 46.Sampson HA. Food allergy. Part 1: immunopathogenesis and clinical disorders. J Allergy Clin Immunol. 1999;103(5 Pt 1):717–728. doi: 10.1016/s0091-6749(99)70411-2. [DOI] [PubMed] [Google Scholar]
- 47.Kelly KJ. Eosinophilic gastroenteritis. Journal of pediatric gastroenterology and nutrition. 2000;30(Suppl):S28–35. doi: 10.1097/00005176-200001001-00005. [DOI] [PubMed] [Google Scholar]
- 48.Fox VL, Nurko S, Furuta GT. Eosinophilic esophagitis: It's not just kid's stuff. Gastrointest Endosc. 2002;56(2):260–270. doi: 10.1016/s0016-5107(02)70188-0. [DOI] [PubMed] [Google Scholar]
- 49.Sampson HA, Anderson JA. Summary and recommendations: Classification of gastrointestinal manifestations due to immunologic reactions to foods in infants and young children. Journal of pediatric gastroenterology and nutrition. 2000;30(Suppl):S87–94. doi: 10.1097/00005176-200001001-00013. [DOI] [PubMed] [Google Scholar]
- 50.Rothenberg ME, Mishra A, Collins MH, Putnam PE. Pathogenesis and clinical features of eosinophilic esophagitis. J Allergy Clin Immunol. 2001;108(6):891–894. doi: 10.1067/mai.2001.120095. [DOI] [PubMed] [Google Scholar]
- 51.Barnes PJ. Therapeutic strategies for allergic diseases. Nature. 1999;402(6760 Suppl):B31–38. doi: 10.1038/35037026. [DOI] [PubMed] [Google Scholar]
- 52.Blanchard C, Mingler MK, Vicario M, Abonia JP, Wu YY, Lu TX, et al. IL-13 involvement in eosinophilic esophagitis: transcriptome analysis and reversibility with glucocorticoids. J Allergy Clin Immunol. 2007;120(6):1292–1300. doi: 10.1016/j.jaci.2007.10.024. [DOI] [PubMed] [Google Scholar]
- 53.Rayapudi M, Mavi P, Zhu X, Pandey AK, Abonia JP, Rothenberg ME, et al. Indoor insect allergens are potent inducers of experimental eosinophilic esophagitis in mice. J Leukoc Biol. 88(2):337–346. doi: 10.1189/jlb.0110025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Rajavelu P, Rayapudi M, Moffitt M, Mishra A, Mishra A. Significance of para esophageal lymph nodes in food or aeroallergen-induced iNKT cell-mediated experimental eosinophilic esophagitis. Am J Physiol Gastrointest Liver Physiol. 2012;302(7):G645–654. doi: 10.1152/ajpgi.00223.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mishra A, Wang M, Pemmaraju VR, Collins MH, Fulkerson PC, Abonia JP, et al. Esophageal Remodeling Develops as a Consequence of Tissue Specific IL-5-Induced Eosinophilia. Gastroenterology. 2008;134(1):204–214. doi: 10.1053/j.gastro.2007.10.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zhu X, Wang M, Mavi P, Rayapudi M, Pandey AK, Kaul A, et al. Interleukin-15 expression is increased in human eosinophilic esophagitis and mediates pathogenesis in mice. Gastroenterology. 139(1):182–193. e187. doi: 10.1053/j.gastro.2010.03.057. [DOI] [PMC free article] [PubMed] [Google Scholar]