Abstract
Introduction
Aberrant promoter DNA methylation is a well-described mechanism of leukemogenesis within hematologic malignancies, including acute lymphoblastic leukemia (ALL). However, the importance of methylation patterns among the adolescent and young adult (AYA) ALL population has not been well established.
Methods
DNA methylation of 18 candidate genes in 33 AYA ALL patients was analyzed at diagnosis and during treatment, to evaluate the frequency and clinical relevance of aberrant methylation in an AYA population treated on a uniform therapeutic regimen.
Results
Of 16 informative genes, there was a median of 6 methylated genes per AYA ALL patient. Correlations were identified between increasing number of methylated genes with male sex (p=0.04), increased white blood cell (WBC) count (p=0.04) and increased bone-marrow blast percentage (p=0.04). Increasing age was associated with EPHA5 methylation (p=0.05). Overall, patients experienced favorable outcomes with median survival that was not reached. On univariate analysis, methylation of CYP1B1 was associated with worse overall survival (HR 10.7, 95% CI 1.3–87.6, p=0.03), disease-free survival (HR 3.7, 95% CI 1.1–9.2, p=0.04) and correlated with decreased CYP1B1 gene expression.
Conclusions
A significant incidence of methylation within the AYA ALL population was identified, with increased methylation associated with distinct clinicopathologic features including male gender and elevated WBC count. Our results suggest aberrant methylation among AYA patients is frequent, and may provide a common pathogenic mechanism. The inferior outcome identified with methylation of the cytochrome p450 gene CYP1B1, an enzyme involved in drug metabolism and steroid synthesis, warrants further investigation.
Keywords: acute lymphoblastic leukemia, DNA methylation, adolescent young adult
Introduction
Acute lymphoblastic leukemia (ALL) is a heterogeneous disease affecting both pediatric and adult patient populations. With the advent of intensified combination chemotherapy regimens, children with ALL now enjoy a greater than 80% disease-free long-term survival1–3, whereas adults unfortunately continue to experience long-term survival rates closer to 40–50%4–6. In addition to age, several factors have been associated with ALL clinical outcome such as cytogenetic aberrations including mixed lineage leukemia (MLL) rearrangements and the Philadelphia (Ph) chromosome, specific leukemia-associated immunophenotypes, patient performance status, and presenting white blood cell count3,7–10.
In ALL, the adolescent and young adult (AYA) population comprises a unique cohort of patients. These patients may be treated by either pediatric or adult oncologists, and treatment regimens have historically varied based on the training background of the treating physician. The AYA cohort has received much recent attention, as retrospective reviews have suggested that AYA patients have intermediate survival compared to children or adults, but that AYA patients treated on pediatric regimens have improved outcome compared to patients treated on adult protocols11–18. Many different factors for the differences in outcome in AYA patients have been proposed, including treatment intensity, psychosocial factors, and variations in the disease biology itself including cytogenetic and immunophenotypic variations.19–22
In addition to standard prognostic factors in ALL, the importance of recurrent abnormalities within epigenetic patterning, such as aberrant promoter DNA methylation, is also of interest with respect to prognostication within ALL and within the AYA cohort in particular. DNA methylation at promoter cytosine residues within conserved CpG islands is an essential component of regulated gene expression, and aberrant methylation can lead to gene silencing of critical genes and has been a well described phenomenon in hematologic malignancies including ALL23–26. Within ALL, abnormal methylation of various genes appears to cluster within specific molecular functional pathways, including Wnt-related genes, the glucocorticoid pathway, Ephrin family kinases, and mitogen-activated protein kinase (MAPK) pathways27–29.
Correlations between the presence and number of detectable methylated genes in ALL have been associated with various clinicopathologic features and in some studies have been linked with poor prognosis. For example, increased methylation of the p73/p15/p57KIP2 cell cycle regulatory pathway members was found to occur rarely in pediatrics but frequently in adults, and was associated with worse outcome30. Within the pediatric population, published reports suggest a correlation between methylation and prognosis31,32, with increased methylation frequencies particularly identified in T-cell phenotype ALL patients33, and a globally hyper-methylated phenotype associated with infants with mixed lineage leukemia (MLL) rearrangements, a high-risk pediatric group with particularly poor outcome34.
There is preliminary information to suggest that prognostic differences between and within age groups could be related in part to differences in methylation patterns, and that identification of aberrantly methylated subsets may improve risk-based treatment decisions. In light of the differences in ALL survival between children, AYAs and adults, we have investigated DNA methylation patterns of 16 informative genes in ALL patients treated on a uniform treatment regimen, to evaluate the frequency and clinical relevance of aberrant methylation in an AYA population.
Methods
Patients and Treatment
Primary DNA samples from 33 adolescent and young adult patients with newly diagnosed ALL, age 13 to 37, who received treatment on our augmented Berlin-Frankfurt-Munster (BFM) therapeutic protocol at the University of Texas M.D. Anderson Cancer Center were enrolled from 2006 to 2009. Clinical information of all patients in this study is provided in Table 1, including patient age, sex, ALL subtype, white blood cell (WBC) count at diagnosis, platelet (PLT) count at diagnosis, karyotype, LDH, bone marrow percentage, and overall patient outcome. All patients were negative for the Philadelphia chromosome rearrangement by virtue of a negative BCR/ABL quantitative PCR. In addition to the pretreatment samples, there were 24 patients with at least one repeat sample obtained during induction treatment that were analyzed for changes in methylation over time. All samples were collected following institutional guidelines with informed consent in accordance with the Declaration of Helsinki. Evaluation of promoter methylation levels was a pre-specified aim of the augmented BFM protocol.
Table 1.
Cohort Patient Characteristics
| Total Cohort (n=33) | |
|---|---|
| Age (yrs) | |
| Median | 20 |
| Range | 13–37 |
| Male Sex | 23 (70%) |
| ALL Subtype | |
| Pre-B ALL | 27 (82%) |
| Pre-T ALL | 4 (12%) |
| Pre-B LL | 1 (3%) |
| Pre-T LL | 1 (3%) |
| WBC count | |
| Median | 5.2 |
| Range | 0.4 – 170.1 |
| PLT count | |
| Median | 61 |
| Range | 4 – 274 |
| BM Blast % | |
| Median | 88 |
| Range | 0–98 |
| LDH | |
| Median | 1040 |
| Range | 284 – 9784 |
| Karyotype | |
| Diploid | 17 (40%) |
| Hyperdiploid | 7 (17%) |
| Hypodiploid | 4 (10%) |
| Other | 6 (14%) |
| Unavailable | 8 (19%) |
| CR Rate | 33 (100%) |
| Relapse | 12 (36%) |
| Survival | 24 (73%) |
Gene Selection, DNA Extraction and Methylation Analysis
A total of 18 genes, including those within the Notch pathway (Notch3, Jag1), Ephrin pathway (EPHA2, EPHA4 – EPHA7, EPHA10), and glucocorticoid pathway (HSPAL4, PTPRM, CYP1B1) were analyzed. The selected genes were chosen based on our results of a previous genome-wide analysis, identifying 26 candidate genes that were subsequently validated and confirmed as hyper-methylated in ALL cell lines.27 Genes with a high likelihood of importance in ALL pathogenesis by their location in top canonical pathways such as cellular growth and proliferation, gene expression and cell death, using Ingenuity Pathway Analysis (IPA) software were analyzed. Candidate genes from top canonical pathways such as the Ephrin receptor signaling pathway were similarly chosen. Our methods of DNA extraction, bisulfite modification and methylation analysis have been previously described26,35,36 and the specific primers used are available in Supplementary Table 1. GAPDH was used as the reference control gene. Evaluation of promoter methylation was performed by PCR amplification of peripheral blood followed by pyrosequencing of CpG sites within target genes. A candidate gene was considered methylated if the methylation density was ≥10% as previously established37.
Statistical Analysis
Associations between methylation of single genes or groups of genes were assessed using the Spearman rank correlation coefficient. Categorical variables were compared using the χ2 or Fisher’s exact test, and continuous variables using the Wilcoxon rank-sum test. The number of methylated genes was analyzed as both a continuous and categorical variable. When indicated, p-values to indicate statistical significance were adjusted for testing of multiple comparisons using the Bonferonni adjustment.
Disease-free survival (DFS) and overall survival (OS) were based on the Kaplan-Meier method, with differences tested using the log-rank test. OS was measured as the time from ALL diagnosis to death or date of last follow-up (censored). Disease-free survival was defined as the time from complete remission (CR) to treatment failure including relapse, death, or date at last follow-up (censored). All analyses were performed using STATA software, version 12.0 (College Station, TX).
Results
Clinical and disease-specific patient characteristics of samples are detailed in Table 1. Methylation density results of each of the 18 genes analyzed (TP73, SKP2, HOXD13, Olig2, Jag1, Notch3, EPHA2, EPHA4, EPHA5, EPHA6, EPHA7, EPHA10, EPHB3, ENFA5, GNA14, HSPAL4, PTPRM and CYP1B1) are presented in Table 2. Of note, SKP2 was methylated in all analyzed samples and TP73 in all but three samples, with minimal variation in methylation levels of these two genes, and thus TP73 and SKP2 were not evaluated for further correlations given the near-universal methylation observed in our AYA cohort. All other genes were methylated in a proportion of patients; the frequency of methylated genes ranged from EPHB3 which was methylated in only 2 samples (6%), to EPHA5 which was methylated in 30 (91%) of samples (Table 2). Seven genes (Olig2, EPHA5, EPHA10, ENFA5, GNA14, HSPAL4 and PTPRM) were methylated in more than 50% of the samples analyzed.
Table 2.
Methylation Densities of Analyzed Genes
| # Samples with gene promoter ≥10% methylated | Median methylation % (range) | |
|---|---|---|
| TP73 | 25 (89%) | 12% (7–18%) |
| SKP2 | 29 (100%) | 28% (25–32%) |
| HOXD13 | 14 (42%) | 8% (2–47%) |
| Olig2 | 22 (67%) | 13% (4–74%) |
| Jag1 | 12 (41%) | 8% (3–50%) |
| Notch3 | 14 (44%) | 8% (3–66%) |
| EPHA2 | 6 (18%) | 7% (4–52%) |
| EPHA4 | 5 (16%) | 4% (2–41%) |
| EPHA5 | 30 (91%) | 18% (8–47%) |
| EPHA6 | 12 (41%) | 6% (2–58%) |
| EPHA7 | 2 (6%) | 3% (2–33%) |
| EPHA10 | 25 (76%) | 21% (5–74%) |
| EPHB3 | 2 (6%) | 6% (0–11%) |
| ENFA5 | 18 (62%) | 11% (4–39%) |
| GNA14 | 18 (62%) | 10% (3–54%) |
| HSPAL4 | 19 (70%) | 11% (3–39%) |
| PTPRM | 16 (59%) | 10% (4–70%) |
| CYP1B1 | 14 (47%) | 9% (3–66%) |
note not all columns have n=33 samples
Using a standard methylation cut-point of 10% or more, only one sample (3%) was without any observed methylation of the n=16 genes. 9 (27%) had methylation of 1–4 genes, 11 samples (33%) had methylation of 5–9 genes, and 12 (36%) had methylation of 10 or more genes (Table 3) suggestive of a “hypermethylated phenotype”. In a Spearman’s correlation matrix assessment using the Bonferroni adjustment for multiple comparisons, methylation of certain genes was found to be significantly associated with the presence of methylation of other analyzed genes (Table 4). For instance, methylation of Notch3 was associated with methylation of Jag1 (p=0.009), EPHA10 (p=0.006) and GNA14 (p=0.02).
Table 3.
Number of Methylated Genes per Patient at Diagnosis
| # Methylated Genes | Frequency |
|---|---|
| 0 | 1 (3%) |
| 1 | 1 (3%) |
| 2 | 2 (6%) |
| 3 | 1 (3%) |
| 4 | 5 (15%) |
| 5 | 4 (12%) |
| 6 | 1 (3%) |
| 7 | 2 (6%) |
| 8 | 4 (12%) |
| 10 | 5 (15%) |
| 11 | 5 (15%) |
| 12 | 2 (6%) |
Table 4.
Methylation Correlation Matrix
| HOXD13 | Olig2 | Jag1 | Notch3 | EPHA2 | EPHA4 | EPHA5 | EPHA6 | EPHA7 | EPHA10 | EPHB3 | ENFA5 | GNA14 | HSPAL4 | PTPRM | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| HOXD13 | |||||||||||||||
| Olig2 | 0.08 | ||||||||||||||
| Jag1 | 0.29 | 1.0 | |||||||||||||
| Notch3 | 0.19 | 1.0 | 0.009 | ||||||||||||
| EPHA2 | 1.0 | 0.002 | 1.0 | 1.0 | |||||||||||
| EPHA4 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | ||||||||||
| EPHA5 | 0.54 | 0.002 | 1.0 | 1.0 | 0.36 | 1.0 | |||||||||
| EPHA6 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 0.004 | 1.0 | ||||||||
| EPHA7 | 1.0 | 0.04 | 1.0 | 1.0 | 1.0 | 1.0 | 0.8 | 1.0 | |||||||
| EPHA10 | 0.21 | 0.02 | 1.0 | 0.006 | 1.0 | 1.0 | 0.0001 | 1.0 | 1.0 | ||||||
| EPHB3 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | |||||
| ENFA5 | 0.05 | 1.0 | 0.04 | 0.87 | 1.0 | 1.0 | 0.5 | 1.0 | 1.0 | 1.0 | 1.0 | ||||
| GNA14 | 0.91 | 1.0 | 1.0 | 0.02 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 0.01 | 1.0 | 0.20 | |||
| HSPAL4 | 1.0 | 1.0 | 1.0 | 0.32 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 1.0 | 0.75 | ||
| PTPRM | 0.86 | 0.04 | 1.0 | 1.0 | 0.03 | 1.0 | 0.002 | 1.0 | 0.17 | 0.0001 | 1.0 | 1.0 | 0.02 | 1.0 | |
| CYP1B1 | 1.0 | 1.0 | 1.0 | 0.09 | 1.0 | 1.0 | 0.16 | 1.0 | 1.0 | 0.009 | 1.0 | 0.07 | 0.08 | 1.0 | 1.0 |
reported values are level of significance using Bonferroni adjustment for multiple comparisons
There was no association with the number of methylated genes and either ALL subtype, EGIL classification or patient karyotype. Specifically, when analyzed by ALL subtype, the 27 patients with precursor-B (pre-B) ALL had a median of 7 methylated genes, and the 4 patients with pre-T ALL had a median number of 6 methylated genes. The one patient with pre-B lymphoblastic lymphoma (LL) had 5 methylated genes, and the one patient with pre-T LL had one methylated gene.
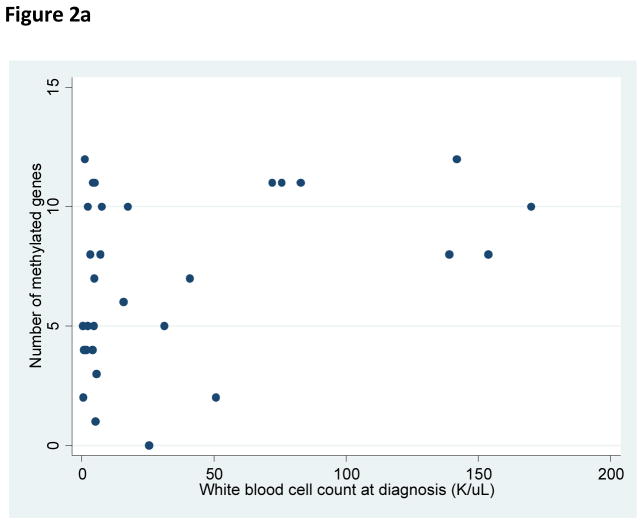
There was no relationship identified between the number of methylated genes with patient age, platelet count at diagnosis, or presenting LDH count. Significant correlations were identified between increasing number of methylated genes and male sex (p=0.02), increased white blood cell count at diagnosis (p=0.04), and an increased BM blast percentage at diagnosis (p=0.04) (Figure 2).
Figure 2.
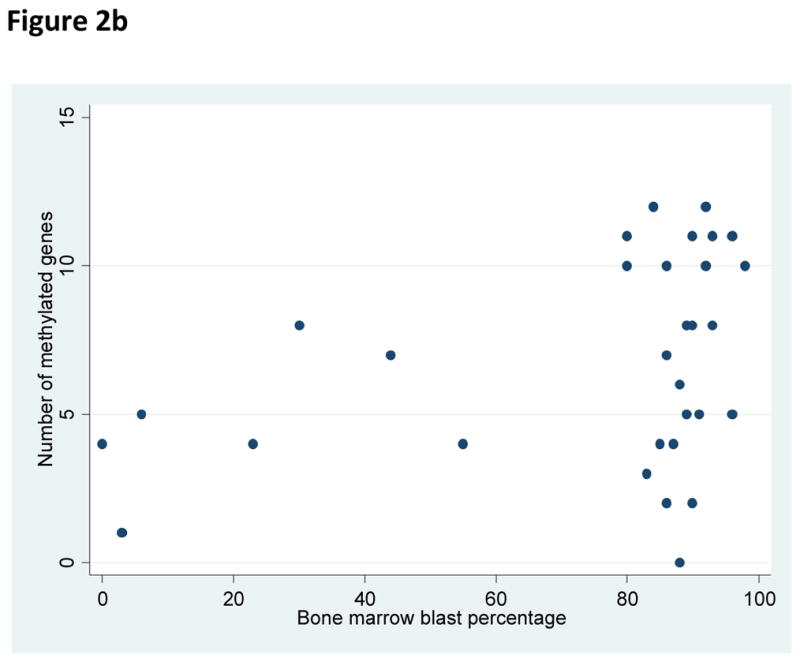
Figure 2a. Relationship of white blood cell count at diagnosis and number of methylated genes per sample at diagnosis.
Figure 2b. Relationship of bone marrow blast percentage at diagnosis and number of methylated genes per sample at diagnosis.
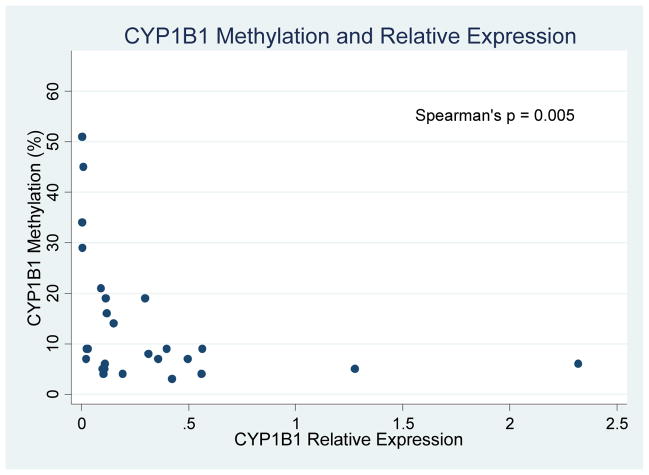
All 33 patients achieved an initial CR on therapy. Overall, patients experienced favorable outcomes with a median survival that was not reached. On univariate survival analysis, the presence of methylated CYP1B1 was associated with worse OS (HR 10.7, 95% CI 1.3 – 87.6, p=0.03) and DFS (HR 3.7, 95% CI 1.1 – 9.2, p=0.04) which retained significance in multivariate analysis (HR 4.4, 95% CI 1.1–18.3, p=0.04). To evaluate CYP1B1 further, we additionally performed quantitative PCR of CYP1B1 in n=27 AYA subjects with remaining samples, as well as in six normal controls. Compared to normal controls, 21 of 27 AYA ALL patients had lower relative CYP1B1 gene expression. We furthermore identified a significant inverse association (Spearman’s rho = −0.54, p = 0.005) between increased CYP1B1 methylation and decreased CYP1B1 expression (Figure 5). In addition, a non-significant trend towards worse survival was identified in patients with methylated ENFA5 (HR 5.5, 95% CI 0.67 – 44.5, p=0.06). No other relationships were identified between single genes or methylation patterns on patient outcome. Specifically, the number of methylated genes did not correlate with prognosis.
Figure 5.
Inverse relation of CYP1B1 methylation % and relative gene expression
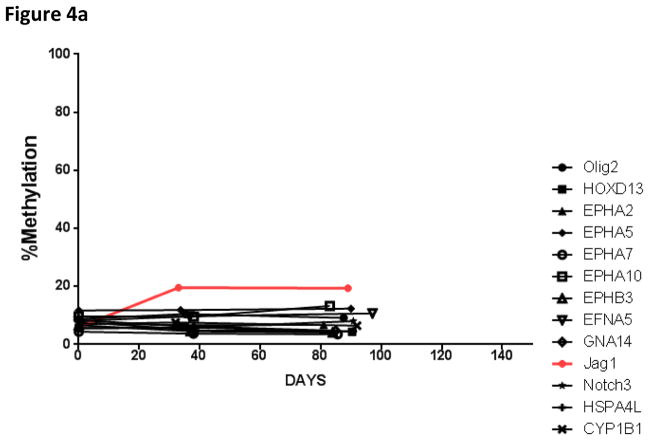
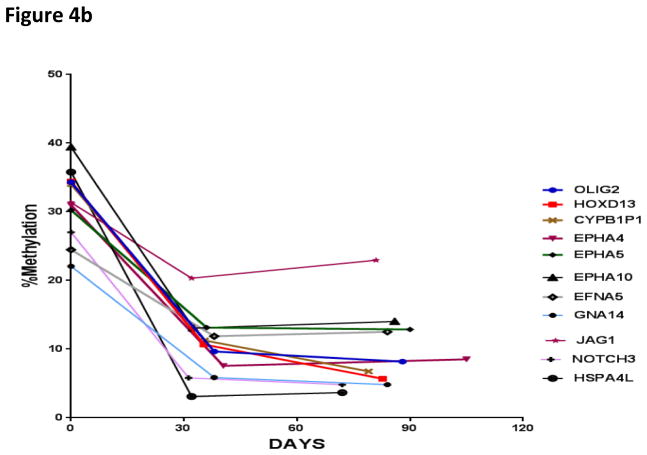
There were 24 patients who had repeat samples obtained for at least one additional time point in addition to the pretreatment baseline sample (repeat samples were obtained from day 28 – to day 110 of therapy). In samples without gene methylation at diagnosis, no significant increase in methylation over time was noted with the exception of Jag1, which experienced increased methylation during the first thirty days of therapy (Figure 3). Importantly, in samples with methylated pretreatment genes, the average methylation score was noted to decrease during the first thirty days of therapy. The change in methylation at the time of relapse was not able to be determined, as only one patient relapsed during the first 150 days; this patient had 10 genes methylated at diagnosis (HOXD13, Olig2, Jag1, EPHA2, EPHA5, EPHA6, EPHA10, GNA14, PTPRM, and CYP1B1).
Figure 3.
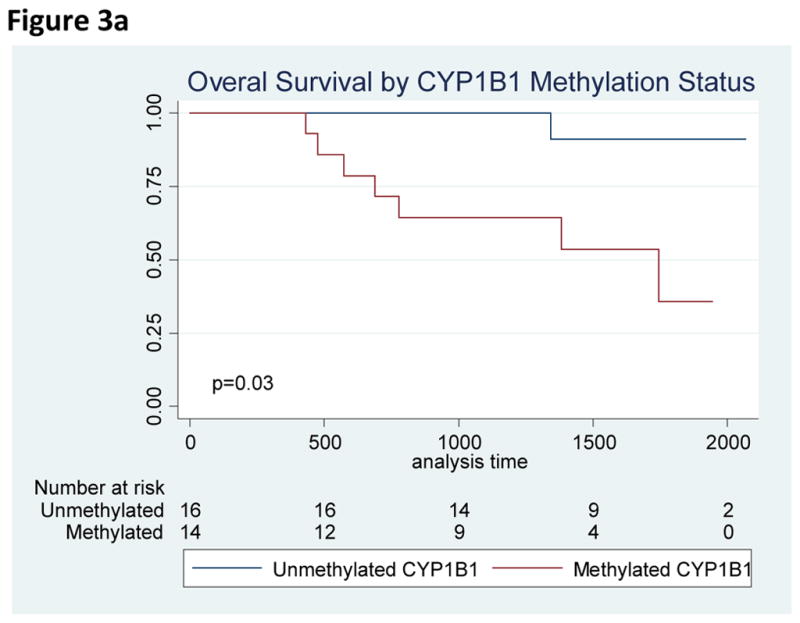
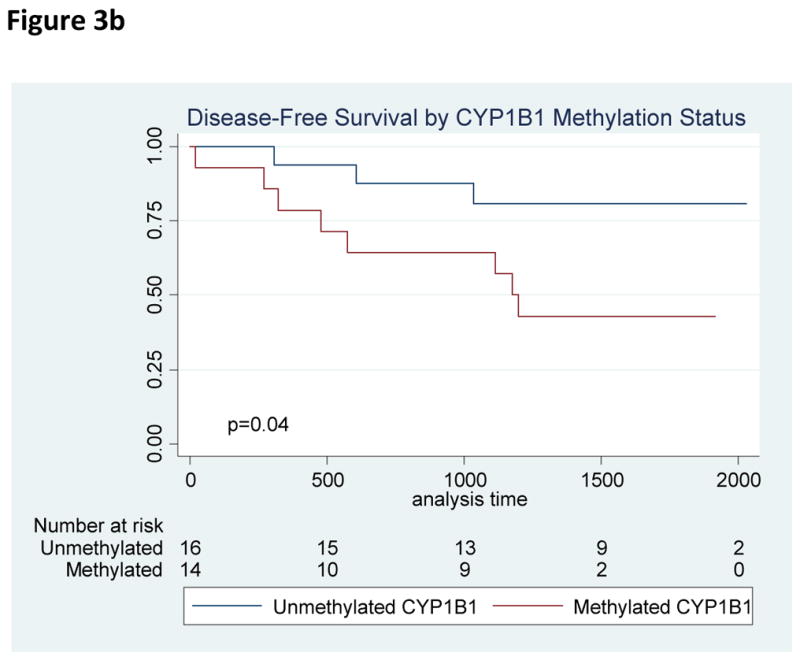
Figure 3a. Overall Survival by CYP1B1 Methylation Status
Figure 3b. Disease-Free Survival by CYP1B1 Methylation Status
Finally, we divided our AYA cohort into a “pediatric cohort” aged 13–19 and an “adult cohort” aged 20–37, to determine whether methylation characteristics were different within these two groups. There were no identified differences in important clinical or biologic characteristics between these age groups (Supplementary Table 2). The adult cohort was more likely to be methylated at EPHA4 (p=0.05); this did not impact patient outcome. Otherwise, no statistically significant differences were observed in terms of gene specific methylation between both groups.
Discussion
The adolescent and young adult population comprises a unique cohort among ALL patients, with outcomes that historically fall in-between the published pediatric and adult ALL outcomes. Multiple different factors have been purported to influence outcomes in AYA patients, however the role of DNA promoter methylation within this population has not been systematically investigated. In our study, we evaluate the methylation profile of 33 AYA patients treated on a uniform protocol, the change in methylation during initial treatment, and investigate the role of methylation on various clinical and biologic ALL characteristics, as well as overall patient outcome.
Previous studies have identified that increased methylation is associated with age and worse overall survival31,32,38,39, while other studies have not identified these correlations. Our analysis identified an increased rate of methylation of EPHA4 in the adult cohort compared to our pediatric cohort within our AYA population, but otherwise we were unable to detect differences between gene-specific methylation based on patient age. However, it is important to note that some of the genes analyzed in our study are different than those previously evaluated, with inclusion of several novel genes in our study due to their relative significance using IPA software, and exclusion of other genes found to be methylated in ALL in prior publications. Interestingly, the p53 homologue p73 and the MYC target gene SKP2 were methylated in nearly all and all AYA patients within our cohort, respectively. Previous studies have identified a low incidence of p73 methylation in pediatric ALL, with an increased frequency of p73 methylation (>20% incidence) in adults with ALL.26,38 While absent or limited expression of p73 as a result of promoter methylation has been a well described phenomena in hematologic malignancies,40,41 universal methylation has not been previously described in other leukemia cohorts to date.
A significant adverse prognosis was identified in patients with methylated CYP1B1, a gene which belongs to the cytochrome p450 enzymatic superfamily and is thought to catalyze reactions involved in drug metabolism and steroid synthesis.42 This novel finding was supported by the additional recognition that increased CYP1B1 methylation inversely correlates with CYP1B1 gene expression, suggesting down-regulation of this gene may decrease glucocorticoid efficacy. The association of CYP1B1 with patient response to glucocorticoids during leukemia treatment, as well as drug metabolism and therapy-related toxicity will be essential future relationships to investigate. While a relationship between CYP1B1 and acute lymphoblastic leukemia has not been previously identified and will require validation in an independent cohort, overexpression of other genes involved in glucocorticoid resistance (i.e. the S100 family members including S100A8 and S100A9) have been recently identified to predict for steroid-resistance in ALL patients, and therapeutic inhibition of this pathway is currently under evaluation.43 In the methylation correlation matrix analysis, methylation of CYP1B1 was significantly associated with EPHA10 methylation. The significance of this correlation is not clear, however the prognostic significance of CYP1B1 was not dependent on methylation status of EPHA10.
We additionally identified several clinical-biological associations that have not been previously well described, including a correlation between increased methylation scores with male gender, elevated white blood cell count at diagnosis, and an increased bone marrow blast percentage at diagnosis. Whether these findings are due to chance or represent true findings will require additional testing of larger datasets. Given that our study utilized peripheral blood of patients for methylation analysis, it is also possible that these correlations of “tumor burden” with methylation may be related to the dilution of methylated DNA in the periphery. Of note, the majority of the published literature on methylation patterns within ALL exclusively use samples with >80–90% bone marrow blasts, so this relationship may have been masked in previous studies32,44.
The augmented BFM combination chemotherapy regimen in our AYA population provided excellent outcomes, with a median OS and DFS that were not reached. This and additional preliminary data from our ongoing trial helps support the notion that AYA patients treated with intensive pediatric regimens can achieve similarly favorable outcomes as younger pediatric patients. The exceptional survival in this cohort could actually have proven to be a limitation in our study, as the lack of “events” may have prevented the detection of differences in patient outcome, based on relevant methylation characteristics.
Importantly, as opposed to other recurrent genetic changes in leukemias such as mutations and cytogenetic abnormalities, DNA methylation is an epigenetic event which does not alter the underlying genetic code, and is thus potentially reversible with therapy. There is only limited experience with epigenetic therapy in ALL, but preliminary studies have identified both histone deacetylase inhibitors and DNA methyltransferase inhibitors can lead to re-expression of genes shown to be methylated and silenced in ALL45. Further evaluation of epigenetic therapy for ALL is warranted, although it is important to note that in our analysis of methylation levels over time, we observed that the majority of patients treated with intensive ALL therapy obtain “reversal” of methylation with standard ALL therapy.
In summary, we identified a significant presence of methylated genes within the AYA ALL population, suggesting a potential epigenetic role of promoter DNA methylation in leukemogenesis. Increased methylation of evaluated genes were associated with distinct clinical and pathologic ALL features including male gender and elevated white blood cell count and bone marrow blast percentage at diagnosis. Finally, given the prognostic importance of the cytochrome p450 gene CYP1B1 in our cohort, further evaluation of CYP1B1 as a prognostic and/or therapeutic marker is justified.
Supplementary Material
Figure 1.
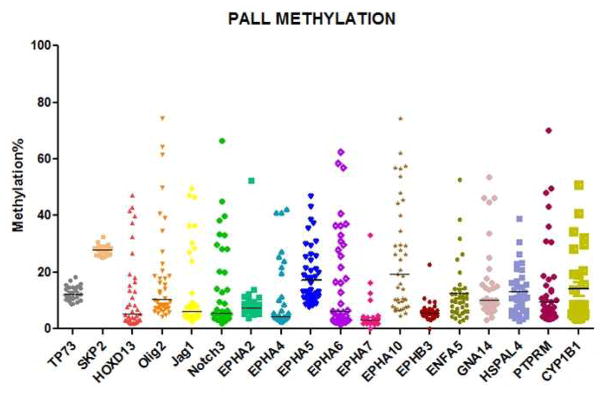
Baseline methylation levels of selected genes (as %) in patient samples (n=33)
Figure 4.
Figure 4a. Change in methylation over time in samples with unmethylated gene status at diagnosis
Figure 4b. Change in methylation over time in samples with methylated gene status at diagnosis
Acknowledgments
This work was supported in part by the MD Anderson Cancer Center Support Grant (CCSG) CA016672.
Footnotes
Conflict of Interest Disclosure: The authors declare no competing financial interests
References
- 1.Pui CH, Evans WE. Acute lymphoblastic leukemia. The New England journal of medicine. 1998;339(9):605–15. doi: 10.1056/NEJM199808273390907. [DOI] [PubMed] [Google Scholar]
- 2.Nachman J. Clinical characteristics, biologic features and outcome for young adult patients with acute lymphoblastic leukaemia. British journal of haematology. 2005;130(2):166–73. doi: 10.1111/j.1365-2141.2005.05544.x. [DOI] [PubMed] [Google Scholar]
- 3.Pulte D, Gondos A, Brenner H. Improvement in survival in younger patients with acute lymphoblastic leukemia from the 1980s to the early 21st century. Blood. 2009;113(7):1408–11. doi: 10.1182/blood-2008-06-164863. [DOI] [PubMed] [Google Scholar]
- 4.Kantarjian HM, Walters RS, Keating MJ, Smith TL, O’Brien S, Estey EH, et al. Results of the vincristine, doxorubicin, and dexamethasone regimen in adults with standard- and high-risk acute lymphocytic leukemia. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 1990;8(6):994–1004. doi: 10.1200/JCO.1990.8.6.994. [DOI] [PubMed] [Google Scholar]
- 5.Rowe JM, Buck G, Burnett AK, Chopra R, Wiernik PH, Richards SM, et al. Induction therapy for adults with acute lymphoblastic leukemia: results of more than 1500 patients from the international ALL trial: MRC UKALL XII/ECOG E2993. Blood. 2005;106(12):3760–7. doi: 10.1182/blood-2005-04-1623. [DOI] [PubMed] [Google Scholar]
- 6.Kantarjian HM, O’Brien S, Smith TL, Cortes J, Giles FJ, Beran M, et al. Results of treatment with hyper-CVAD, a dose-intensive regimen, in adult acute lymphocytic leukemia. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2000;18(3):547–61. doi: 10.1200/JCO.2000.18.3.547. [DOI] [PubMed] [Google Scholar]
- 7.Faderl S, Kantarjian HM, Talpaz M, Estrov Z. Clinical significance of cytogenetic abnormalities in adult acute lymphoblastic leukemia. Blood. 1998;91(11):3995–4019. [PubMed] [Google Scholar]
- 8.Johansson B, Mertens F, Mitelman F. Clinical and biological importance of cytogenetic abnormalities in childhood and adult acute lymphoblastic leukemia. Annals of medicine. 2004;36(7):492–503. doi: 10.1080/07853890410018808. [DOI] [PubMed] [Google Scholar]
- 9.Cytogenetic abnormalities in adult acute lymphoblastic leukemia: correlations with hematologic findings outcome. A Collaborative Study of the Group Francais de Cytogenetique Hematologique. Blood. 1996;87(8):3135–42. [PubMed] [Google Scholar]
- 10.Dreyer ZE, Dinndorf PA, Camitta B, Sather H, La MK, Devidas M, et al. Analysis of the role of hematopoietic stem-cell transplantation in infants with acute lymphoblastic leukemia in first remission and MLL gene rearrangements: a report from the Children’s Oncology Group. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2011;29(2):214–22. doi: 10.1200/JCO.2009.26.8938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Boissel N, Auclerc MF, Lheritier V, Perel Y, Thomas X, Leblanc T, et al. Should adolescents with acute lymphoblastic leukemia be treated as old children or young adults? Comparison of the French FRALLE-93 and LALA-94 trials. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2003;21(5):774–80. doi: 10.1200/JCO.2003.02.053. [DOI] [PubMed] [Google Scholar]
- 12.de Bont JM, Holt B, Dekker AW, van der Does-van den Berg A, Sonneveld P, Pieters R. Significant difference in outcome for adolescents with acute lymphoblastic leukemia treated on pediatric vs adult protocols in the Netherlands. Leukemia: official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2004;18(12):2032–5. doi: 10.1038/sj.leu.2403538. [DOI] [PubMed] [Google Scholar]
- 13.Hallbook H, Gustafsson G, Smedmyr B, Soderhall S, Heyman M Swedish Adult Acute Lymphocytic Leukemia G et al. Treatment outcome in young adults and children >10 years of age with acute lymphoblastic leukemia in Sweden: a comparison between a pediatric protocol and an adult protocol. Cancer. 2006;107(7):1551–61. doi: 10.1002/cncr.22189. [DOI] [PubMed] [Google Scholar]
- 14.Linker C, Damon L, Ries C, Navarro W. Intensified and shortened cyclical chemotherapy for adult acute lymphoblastic leukemia. Journal of clinical oncology: official journal of the American Society of Clinical Oncology. 2002;20(10):2464–71. doi: 10.1200/JCO.2002.07.116. [DOI] [PubMed] [Google Scholar]
- 15.Stock W, La M, Sanford B, Bloomfield CD, Vardiman JW, Gaynon P, et al. What determines the outcomes for adolescents and young adults with acute lymphoblastic leukemia treated on cooperative group protocols? A comparison of Children’s Cancer Group and Cancer and Leukemia Group B studies. Blood. 2008;112(5):1646–54. doi: 10.1182/blood-2008-01-130237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ramanujachar R, Richards S, Hann I, Goldstone A, Mitchell C, Vora A, et al. Adolescents with acute lymphoblastic leukaemia: outcome on UK national paediatric (ALL97) and adult (UKALLXII/E2993) trials. Pediatric blood & cancer. 2007;48(3):254–61. doi: 10.1002/pbc.20749. [DOI] [PubMed] [Google Scholar]
- 17.Pole JD, Alibhai SM, Ethier MC, Teuffel O, Portwine C, Zelcer S, et al. Adolescents with acute lymphoblastic leukemia treated at pediatric versus adult hospitals. Annals of oncology: official journal of the European Society for Medical Oncology/ESMO. 2012 doi: 10.1093/annonc/mds518. [DOI] [PubMed] [Google Scholar]
- 18.Ram R, Wolach O, Vidal L, Gafter-Gvili A, Shpilberg O, Raanani P. Adolescents and young adults with acute lymphoblastic leukemia have a better outcome when treated with pediatric-inspired regimens: systematic review and meta-analysis. American journal of hematology. 2012;87(5):472–8. doi: 10.1002/ajh.23149. [DOI] [PubMed] [Google Scholar]
- 19.Wood WA, Lee SJ. Malignant hematologic diseases in adolescents and young adults. Blood. 2011;117(22):5803–15. doi: 10.1182/blood-2010-12-283093. [DOI] [PubMed] [Google Scholar]
- 20.Ribera JM, Oriol A. Acute lymphoblastic leukemia in adolescents and young adults. Hematology/oncology clinics of North America. 2009;23(5):1033–42. vi. doi: 10.1016/j.hoc.2009.07.002. [DOI] [PubMed] [Google Scholar]
- 21.Ramanujachar R, Richards S, Hann I, Webb D. Adolescents with acute lymphoblastic leukaemia: emerging from the shadow of paediatric and adult treatment protocols. Pediatric blood & cancer. 2006;47(6):748–56. doi: 10.1002/pbc.20776. [DOI] [PubMed] [Google Scholar]
- 22.Nachman JB. Adolescents with acute lymphoblastic leukemia: a new “age”. Reviews in clinical and experimental hematology. 2003;7(3):261–9. [PubMed] [Google Scholar]
- 23.Jones PA, Baylin SB. The fundamental role of epigenetic events in cancer. Nature reviews Genetics. 2002;3(6):415–28. doi: 10.1038/nrg816. [DOI] [PubMed] [Google Scholar]
- 24.Issa JP, Baylin SB, Herman JG. DNA methylation changes in hematologic malignancies: biologic and clinical implications. Leukemia: official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 1997;11(Suppl 1):S7–11. [PubMed] [Google Scholar]
- 25.Garcia-Manero G, Daniel J, Smith TL, Kornblau SM, Lee MS, Kantarjian HM, et al. DNA methylation of multiple promoter-associated CpG islands in adult acute lymphocytic leukemia. Clinical cancer research: an official journal of the American Association for Cancer Research. 2002;8(7):2217–24. [PubMed] [Google Scholar]
- 26.Garcia-Manero G, Jeha S, Daniel J, Williamson J, Albitar M, Kantarjian HM, et al. Aberrant DNA methylation in pediatric patients with acute lymphocytic leukemia. Cancer. 2003;97(3):695–702. doi: 10.1002/cncr.11090. [DOI] [PubMed] [Google Scholar]
- 27.Kuang SQ, Tong WG, Yang H, Lin W, Lee MK, Fang ZH, et al. Genome-wide identification of aberrantly methylated promoter associated CpG islands in acute lymphocytic leukemia. Leukemia: official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2008;22(8):1529–38. doi: 10.1038/leu.2008.130. [DOI] [PubMed] [Google Scholar]
- 28.Hogan LE, Meyer JA, Yang J, Wang J, Wong N, Yang W, et al. Integrated genomic analysis of relapsed childhood acute lymphoblastic leukemia reveals therapeutic strategies. Blood. 2011;118(19):5218–26. doi: 10.1182/blood-2011-04-345595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Ge X, Wang X. Role of Wnt canonical pathway in hematological malignancies. Journal of hematology & oncology. 2010;3:33. doi: 10.1186/1756-8722-3-33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Canalli AA, Yang H, Jeha S, Hoshino K, Sanchez-Gonzalez B, Brandt M, et al. Aberrant DNA methylation of a cell cycle regulatory pathway composed of P73, P15 and P57KIP2 is a rare event in children with acute lymphocytic leukemia. Leukemia research. 2005;29(8):881–5. doi: 10.1016/j.leukres.2004.11.023. [DOI] [PubMed] [Google Scholar]
- 31.Gutierrez MI, Siraj AK, Bhargava M, Ozbek U, Banavali S, Chaudhary MA, et al. Concurrent methylation of multiple genes in childhood ALL: Correlation with phenotype and molecular subgroup. Leukemia: official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2003;17(9):1845–50. doi: 10.1038/sj.leu.2403060. [DOI] [PubMed] [Google Scholar]
- 32.Takeuchi S, Matsushita M, Zimmermann M, Ikezoe T, Komatsu N, Seriu T, et al. Clinical significance of aberrant DNA methylation in childhood acute lymphoblastic leukemia. Leukemia research. 2011;35(10):1345–9. doi: 10.1016/j.leukres.2011.04.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kraszewska MD, Dawidowska M, Larmonie NS, Kosmalska M, Sedek L, Szczepaniak M, et al. DNA methylation pattern is altered in childhood T-cell acute lymphoblastic leukemia patients as compared with normal thymic subsets: insights into CpG island methylator phenotype in T-ALL. Leukemia: official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2012;26(2):367–71. doi: 10.1038/leu.2011.208. [DOI] [PubMed] [Google Scholar]
- 34.Stumpel DJ, Schneider P, van Roon EH, Pieters R, Stam RW. Absence of global hypomethylation in promoter hypermethylated Mixed Lineage Leukaemia-rearranged infant acute lymphoblastic leukaemia. Eur J Cancer. 2012 doi: 10.1016/j.ejca.2012.07.013. [DOI] [PubMed] [Google Scholar]
- 35.Shen L, Toyota M, Kondo Y, Obata T, Daniel S, Pierce S, et al. Aberrant DNA methylation of p57KIP2 identifies a cell-cycle regulatory pathway with prognostic impact in adult acute lymphocytic leukemia. Blood. 2003;101(10):4131–6. doi: 10.1182/blood-2002-08-2466. [DOI] [PubMed] [Google Scholar]
- 36.Clark SJ, Harrison J, Paul CL, Frommer M. High sensitivity mapping of methylated cytosines. Nucleic acids research. 1994;22(15):2990–7. doi: 10.1093/nar/22.15.2990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Toyota M, Kopecky KJ, Toyota MO, Jair KW, Willman CL, Issa JP. Methylation profiling in acute myeloid leukemia. Blood. 2001;97(9):2823–9. doi: 10.1182/blood.v97.9.2823. [DOI] [PubMed] [Google Scholar]
- 38.Gutierrez MI, Siraj AK, Ibrahim MM, Hussain A, Bhatia K. Childhood and adult ALL: differences in epigenetic lesions associated with cell cycle genes. American journal of hematology. 2005;80(2):158–60. doi: 10.1002/ajh.20458. [DOI] [PubMed] [Google Scholar]
- 39.Roman-Gomez J, Castillejo JA, Jimenez A, Gonzalez MG, Moreno F, del Rodriguez MC, et al. 5′ CpG island hypermethylation is associated with transcriptional silencing of the p21(CIP1/WAF1/SDI1) gene and confers poor prognosis in acute lymphoblastic leukemia. Blood. 2002;99(7):2291–6. doi: 10.1182/blood.v99.7.2291. [DOI] [PubMed] [Google Scholar]
- 40.Kawano S, Miller CW, Gombart AF, Bartram CR, Matsuo Y, Asou H, et al. Loss of p73 gene expression in leukemias/lymphomas due to hypermethylation. Blood. 1999;94(3):1113–20. [PubMed] [Google Scholar]
- 41.Pluta A, Nyman U, Joseph B, Robak T, Zhivotovsky B, Smolewski P. The role of p73 in hematological malignancies. Leukemia: official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2006;20(5):757–66. doi: 10.1038/sj.leu.2404166. [DOI] [PubMed] [Google Scholar]
- 42.Tang YM, Wo YY, Stewart J, Hawkins AL, Griffin CA, Sutter TR, et al. Isolation and characterization of the human cytochrome P450 CYP1B1 gene. The Journal of biological chemistry. 1996;271(45):28324–30. doi: 10.1074/jbc.271.45.28324. [DOI] [PubMed] [Google Scholar]
- 43.Spijkers-Hagelstein JA, Schneider P, Hulleman E, de Boer J, Williams O, Pieters R, et al. Elevated S100A8/S100A9 expression causes glucocorticoid resistance in MLL-rearranged infant acute lymphoblastic leukemia. Leukemia: official journal of the Leukemia Society of America, Leukemia Research Fund, UK. 2012;26(6):1255–65. doi: 10.1038/leu.2011.388. [DOI] [PubMed] [Google Scholar]
- 44.Nordlund J, Milani L, Lundmark A, Lonnerholm G, Syvanen AC. DNA methylation analysis of bone marrow cells at diagnosis of acute lymphoblastic leukemia and at remission. PloS one. 2012;7(4):e34513. doi: 10.1371/journal.pone.0034513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Bhatla T, Wang J, Morrison DJ, Raetz EA, Burke MJ, Brown P, et al. Epigenetic reprogramming reverses the relapse-specific gene expression signature and restores chemosensitivity in childhood B-lymphoblastic leukemia. Blood. 2012;119(22):5201–10. doi: 10.1182/blood-2012-01-401687. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.