Abstract
Induced pluripotent stem cells (iPSCs) and embryonic stem cells (ESCs) are two types of pluripotent stem cells that hold great promise for biomedical research and medical applications. iPSCs were initially favorably compared to ESCs. This view was first based on ethical arguments (the generation of iPSCs does not require the destruction of an embryo) and on immunological reasons (it is easier to derive patient HLA-matched iPSCs than ESCs). However, several reports suggest that iPSCs might be characterized by higher occurrence of epigenetic and genetic aberrations than ESCs as a consequence of the reprogramming process. We focus here on the DNA integrity of pluripotent stem cells and examine the three main sources of genomic abnormalities in iPSCs: (1) genomic variety of the parental cells, (2) cell reprogramming, and (3) in vitro cell culture. Recent reports claim that it is possible to generate mouse or human iPSC lines with a mutation level similar to that of the parental cells, suggesting that “genome-friendly” reprogramming techniques can be developed. The issue of iPSC DNA integrity clearly highlights the crucial need of guidelines to define the acceptable level of genomic integrity of pluripotent stem cells for biomedical applications. We discuss here the main issues that such guidelines should address.
Keywords: Animals; Cell Culture Techniques; Cell Differentiation; DNA; genetics; Embryonic Stem Cells; cytology; immunology; Epigenesis, Genetic; Genomic Instability; Humans; Induced Pluripotent Stem Cells; cytology; immunology; Mice
Keywords: Induced pluripotent stem cells, cell reprogramming, genetic abnormalities, pluripotency, DNA damage, genome integrity
INTRODUCTION
Embryonic stem cells (ESCs), which can theoretically differentiate into any cell type, have become a key cellular tool in biology and have paved the way for cell-based treatments of human diseases [1]. However, human ES cells (hESCs) have two major drawbacks. The first one is the ethical issue concerning the destruction of human embryos to generate hESCs, although these embryos were doomed to destruction anyway [2]. The second barrier to their potential medical use is the need of Human Leukocyte Antigen (HLA)-compatible hESC lines. Both issues are circumvented by induced pluripotent stem cells (iPSCs) as they are directly reprogrammed from adult cells [3], thus opening the way to the derivation of patient-specific pluripotent stem cell (PCS) lines [4]. Moreover, by carefully selecting donor cells, for instance from donors who are homozygous for HLA, it will be possible to set up PSC banks that cover a large part of the genetic diversity of a population [5]. As iPSC derivation allows the in vitro production of cells from potentially any tissue in an unlimited supply and from any genetic background, many applications can therefore be foreseen, particularly: (i) in vitro modeling of human development, (ii) in vitro modeling of human genetic diseases, (iii) supply of normal or diseased cells for drug testing, (iv) supply of cells for cell therapies, including patient-specific cells, and (v) rejuvenation of old/senescent cells for regenerative medicine [6]. In addition, the iPS technology can be mastered with a reasonable amount of training and costs and is now successfully used in many laboratories worldwide (for a discussion on the iPS technology and its applications we refer the readers to other recent reviews [3,7]). However, several reports are raising concerns about the potential weaknesses of iPSCs, such as the appearance of genetic or epigenetic abnormalities linked to the process of cell reprogramming. By altering the iPSC biological behavior or threatening their therapeutic safety, these defects could undermine their use for modeling diseases and medical applications. This problem might tip the balance back towards ESCs. In this review, we will focus on the genetic alterations observed in iPSCs and ESCs and discuss the fact that cell reprogramming is only one of the possible sources of genetic variation in PSCs.
TRACKING THE GENETIC DAMAGE PRODUCED BY CELL REPROGRAMMING
The generation of iPS by cell reprogramming opens the way to two potential sources of mutations. First, insertional mutagenesis due to the use of integrative vectors for cell reprogramming is an obvious cause of DNA damage (Figure 1). In the initial reports, Southern blot analyses showed that each iPSC clone was characterized by the presence of more than 20 integration events (about half a dozen integration events for each transcription factor) [8]. Although aberrant transcription of an oncogene following the integration of a retroviral vector in its proximity has been already described in gene therapy trials [9], similar examples of insertional mutagenesis with functional consequences on endogenous genes are not a typical finding in iPSCs. Nonetheless, the definitive insertion of reprogramming vectors in the iPSC genome is a serious concern and the reactivation of the c-MYC transgene might have fatal consequences, as illustrated by the development of tumors in germline-competent adult chimeras, which were obtained using iPSCs generated with retroviral vectors, and in their progeny [10]. Safer reprogramming strategies have been developed and are now available, such as the use of non-integrating viral vectors, or the direct delivery of reprogramming transcription factor proteins or synthetic mRNAs (discussed below).
Figure 1. Causes of genome alterations in iPSCs.
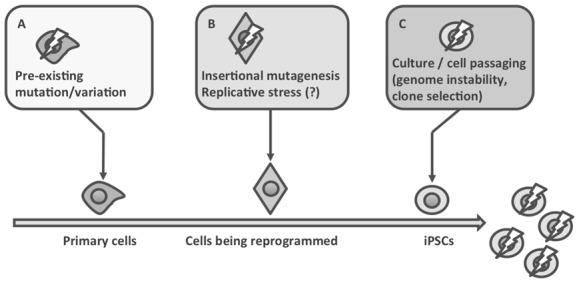
Genome alterations in iPSCs can be the consequence of: (A) variations or mutations that are present at high or low frequency in the parental cells; clonal selection during iPSC generation can select and/or fix these variants; (B) insertional mutagenesis, replicative stress, caused for example by forced expression of an oncogene, can result in DNA damage that might not be properly repaired; (C) iPSCs could be intrinsically genetically unstable, and sub-optimal culture conditions might favor the generation and/or the selection of clones that are fitter for in vitro culture conditions.
Second, the reprogramming process itself could also cause genomic alterations. Although iPSC lines have often normal karyotypes at early passages [11,12], recent reports show that iPSCs, like ESCs, can display chromosomal aberrations [13–16]. The proportion of karyotype abnormalities may vary between iPSCs and ESCs, but most aberrations can be found in both PSC types [14]. In addition, detailed analyses, by using CGH microarrays [17,18], SNP microarrays [19,20] or next-generation sequencing techniques [21,22], suggest that more subtle abnormalities, such as copy number variations (CNV) and mutations, occur in iPSCs at much higher frequency than originally thought. These findings have raised serious concerns about the functional reliability and safety of iPSCs [23,24]. The involved mechanisms are multiple. The reprogramming process, particularly the forced expression of oncogenes, such as c-MYC which is often included in the reprogramming gene cocktail [18], could cause replicative stress and consequently DNA damage. This can be repaired by the pathways which are normally used in metazoan cells to avoid malignant transformation, but if the repair machinery is defective, genetic aberrations might arise [25]. Indeed, inhibition of the p53 or p16 pathways greatly improves reprogramming efficiency, but at the expense of DNA integrity [26–28].
NOT ALL iPS CELLS HARBOR GENETIC DAMAGE
These findings rise the possibility that cell reprogramming might cause genomic damage and that, as a consequence, all iPSCs harbor genetic defects that may compromise their use for disease modeling or cell therapy. Several studies challenge this view. The karyotype analysis of more than 1700 human ESC and iPSC cultures (40 ESC and 219 iPSC lines) from 97 investigators in 29 laboratories did not reveal major differences between ESCs and iPSCs [14]. The overall incidence of karyotype abnormalities was 12.5% in human iPSCs and 12.9% and hESCs, suggesting that cell reprogramming is not a significant source of karyotype abnormality. On the other hand, there were differences concerning the occurrence of some abnormalities, such as higher incidence of trisomy 12 in hESCs and of trisomy 8 in iPCs, and most strikingly almost 10% of trisomy 17 in hESCs, but none in iPSCs. Interestingly, in this very large data set, recurrent translocations were not found, differently from what is observed in many cancers. This is, however, a retrospective analysis and it is not clear whether the observed differences relate to the methods used to generate these PSC lines or whether they only reflect biases in the in vitro cell expansion protocols.
The idea that subkaryotypic abnormalities are unavoidable during cell reprogramming was also recently questioned. Qinlan et al. used whole-genome paired-end DNA sequencing to compare three iPSC lines that were generated by reprogramming mouse embryonic fibroblasts (MEF) with four lentiviral constructs coding for OCT4, SOX2, CMYC and KLF4 under the control of a doxycycline inducible promoter [29]. These low-passage iPSC lines were previously used to generate viable mice in tetraploid embryo complementation assays, undisputedly demonstrating that complete reprogramming had occurred. Unexpectedly, the authors found only one or two de novo structural genomic variations in each iPSC line. As a matter of fact, the most important source of de novo genomic alteration was the insertion of endogenous retroviral elements from the mouse leukemia virus (MLV) family that most likely originated from the CF-1 MEFs which were used as feeder cells. In another study, Young et al. analyzed by whole genome sequencing iPSC clones that were obtained using an OCT4, SOX2 and KLF4 polycistronic lentiviral vector and their parental fibroblasts. Their findings suggest that the single-nucleotide variations and the rare structural variations (SV) detected in the iPSC lines reflected the parental genomic background rather than the consequences of reprogramming [30]. These findings have been extended in human iPSC generated by using the canonical retroviral method. The authors analyzed the iPSC lines by whole genome sequencing and digital droplet PCR and found that most of the iPSC CNVs reflect somatic mosaicism in the human skin [31]. These observations are in line with the hypothesis that cell reprogramming “captures” pre-existing mutations in the parental cell line during cloning, which is a necessary consequence of reprogramming. These data obtained by whole genome sequencing of mouse iPSC lines were extended to human iPSC lines that were obtained using non-integrating Epstein-Barr nuclear antigen-1 (oriP/EBNA1) episomal vectors. Although more than 1,000 single nucleotide variations (SNVs) were found in each iPSC line, very few were within coding sequences [32]. Among those, 50% were synonymous changes, whereas SNVs that modify the coding sequence were not enriched for known cancer-associated genes. Moreover, no CNV was found in the analyzed iPSC lines.
Overall, the studies suggesting that cell reprogramming may be inherently linked to DNA damage are counterbalanced by other works showing that the mutation status of iPSC lines is similar to that of their parental cells. Nevertheless the reports about the occurrence of genome alterations during cell reprogramming are alarming as they highlight bona fide DNA damage. These contradictory reports could be explained mainly by two reasons: (1) different reprogramming techniques and/or primary cell types could result in dramatic differences in genome integrity; (2) different in vitro cell culture conditions could differentially affect PSC integrity and therefore be an important confounding variable. For instance, comparison of sequencing data of different iPSC lines derived by different groups, as done by Quinlan et al, demonstrated the huge variability in mutation frequency of such cells, strongly suggesting that some reprogramming protocols/culture conditions might more easily result in DNA damage [29]. From the data published to date, it is not possible to conclude whether the source cell type, the nature of the reprogramming vector (for instance with viral integration versus without integration) or the choice of the reprogramming gene combination, are correlated with the incidence of genomic abnormalities. In one report, some of these factors could be evaluated but none of these factors influenced the results [20], but these results are too preliminary to reach any conclusion.
CELL CULTURE-INDUCED GENOMIC ALTERATIONS
It will not be possible to precisely quantify the genetic abnormalities generated by cell reprogramming without knowing the role played by the cell culture conditions in the mutational process. Indeed, a cell culture protocol that favors the appearance of DNA alterations in PSCs will also do so during cell reprogramming and during the first passages which are required to amplify newly derived human iPSCs. In this regard, much can be learned from ESCs. ESCs are karyotypically normal at derivation [33,34] and the occurrence of secondary genetic alterations has been thoroughly investigated. Aneuploid hESC clones can appear during cell culture. Such chromosomal abnormalities are often recurrent and gains of chromosome 12 (most frequently 12p), 17 (particularly 17q), 20 (most frequently 20q) or X have been often detected [35–37]. These abnormalities might bestow a growth advantage and, as a consequence, cells carrying mutations are often selected during cell culture. The biological repercussions comprise increased proliferation capacity and decreased growth factor-dependence as well as modifications of the physiological differentiation capacities of such cells [38]. It must be noted that trisomy of chromosome 12, 17 or X is also detected in germ cell tumors and embryonal carcinoma cells. The same survival and/or cell proliferation pathways that are involved in germ and pluripotent cell tumor development could thus be involved also in the selection of aneuploid hESC clones during cell culture. As reported above, the extensive study of 40 hESC lines in which 1163 karyotypes were analyzed concluded that 12.9% of the hESC cultures displayed chromosomal aberrations [14]. This frequency could still increase if more sensitive methods were used, such as microarrays [19,39,40]. Several factors could influence the incidence of these genetic aberrations, particularly the duration and the type of culture conditions. Enzymatic passaging might favor clonal selection of aneuploid clones, but this hypothesis has not been formally proven [39,41]. We recently observed that karyotypic abnormalities in hESCs can occur rapidly within ten passages and sub-karyotypic abnormalities even faster when using single-cell dissociation passaging (unpublished data). These findings in ESCs indicate that the culture conditions used for cell reprogramming and cell expansion are a critical factor for DNA integrity during iPSC generation. Moreover, as several studies on genomic alterations in iPSCs were based on DNA samples isolated from cells before passage 20, it is not possible to exclude a contribution of the cell culture conditions to the DNA abnormalities that were ascribed to cell reprogramming [22]. Many issues need to be addressed to prevent these deleterious mutational events, particularly: (1) to identify the optimal culture conditions for PSC expansion and (2) to understand the mechanisms that drive the generation of genomic alterations. Therefore, a quantitative assay is required to measure the genomic integrity of PSCs in order to compare the impact of different parameters on the incidence of mutational events.
IMPROVING CELL REPROGRAMMING
Overall, the genome integrity of human iPSCs and ESCs is comparable, although some differences have been reported. We will focus here on the measures that can be taken to diminish DNA damage during cell reprogramming.
To avoid insertional mutagenesis, several non-integrating approaches are currently available (see [3] for a discussion on these techniques). They include transient transfection [42] or the use of adenoviral vectors [43], non-integrating viral vectors such as the oriP/EBNA1 episomal vectors [12], non-integrating lentiviral vectors [44] and temperature-sensitive Sendai virus vectors [45]. Another strategy consists in removing the reprogramming vectors in the selected iPSC clones by using the “piggyBac” [46] or the Cre-LoxP excision system [47] after a transient integration step. Nevertheless, even when the vectors do not usually integrate into the DNA, or are excised after cell reprogramming, their insertion in the genome cannot be totally excluded. Thus, techniques that avoid DNA should preferred, such as the direct delivery of reprogramming cell-permeant proteins [48], or of synthetic modified mRNAs [49], which seem to have higher reprogramming efficacy. Direct protein delivery is less efficient probably because the innate immunity program needs to be activated via induction of the TLR3 pathway for effective cell reprogramming [50]. Indeed, in the classical reprogramming protocols, TLR3 is activated by the viral particles. Interestingly, addition of polyinosinic-polycytidylic acid (Poly I:C) to the culture medium as an alternative non-viral approach to activate TLR3, significantly improves and accelerates cell reprogramming of both mouse and human cells by cell-permeant proteins [50].
The other concern is reprogramming-induced DNA damage. As its causes are still not clearly understood, the solution of this type of damage is still speculative. One direction could be to improve the reprograming transcription factor cocktail. Combinations that may improve cell reprogramming could associate the classical reprogramming factors with genes coding for components of the ATP-dependent BAF chromatin-remodeling complex [51], or the use of the PSC-specific microRNAs miR-302 and miR-372 [52]. To reduce oncogene-induced replication stress, c-MYC should be avoided. Of note, strategies that employ genes coding for viral oncoproteins (SV40) or inhibition of the p53 or p16 tumor suppressor pathways are clearly inacceptable when trying to minimize DNA damage [25]. As somatic-cell nuclear transfer and cell fusion, two other cell reprogramming techniques, are considered more efficient than the iPS technique [53], these approaches might shed light on the molecular mechanisms underlying cell reprogramming and serve as a blue print to improve iPSC generation protocols. Finally, culture conditions are critical for the maintenance of PSC genome integrity. It will therefore be important to focus on how to improve PSC expansion in vitro.
SETTING GUIDELINES TO ASSESS iPS CELLS
Currently, there is no consensus on the extent of DNA damage that can occur during PSC generation/culture, the underlying mechanisms and the methods to avoid it. Therefore, a technique to analyze DNA integrity in PSCs and guidelines to interpret DNA variations are mandatory.
PSC DNA integrity is still mainly assessed by karyotype analysis. Other approaches have been tested to overcome the obvious resolution limitations of the classic karyotyping techniques, for example CGH arrays or SNP microarrays; however, there is no consensus on the method to use to discriminate between the really worrying, possibly carcinogenic mutations and the DNA modifications with no or barely any impact on the biological behavior of PSCs, or the simple polymorphisms. For instance, in the preliminary report on the only published clinical trial based on human PSCs, the DNA integrity of the ESC line was confirmed by karyotyping of 20 G-banded metaphase cells and FISH analysis of chromosomes 12 and 17 in 200 interphase nuclei [54].
As highlighted by Prockop and Keating, the assessment of genomic stability in cultured cells is an old issue that was raised more than 50 years ago for cells in culture [55,56]. Indeed, genomic instability is a general feature of cultured cells and has been observed in many cell types, including adult stem cells [57]. Yet, standards or guidelines about what should be considered as acceptable genome integrity of normal cells in culture are still lacking. As DNA sequencing technologies and their resolution (whole genome maps at single-base resolution) are improving very fast and their price rapidly decreasing [58], it is time to anticipate the near future when the routine analysis of the whole PSC genome will be available at a reasonable price.
PSCs without genome damage are obviously required for tissue modeling or medical applications. Next-generation sequencing technologies may be the solution to demonstrate that PSC lines are free of deleterious mutations [25]. However, a 100% undamaged DNA may not be necessary. The notion that only cells with pristine genetic information should be considered for use is certainly a preconception. Not only because the massive occurrence of polymorphisms in normal cells precludes a strict definition of pristine genetic information, but also because of the cell/tissue DNA information heterogeneity. DNA modifications arise continuously from early embryo development to adulthood and their occurrence increases during aging, leading to various degree of cell mosaicism [59–61]. The causes of these lesions are multiple and include, but are not limited to, random events promoted notably by reactive oxygen species (ROS), retrotransposition, UV (for skin fibroblasts) or senescence. Cell mosaicism can be readily observed in primary fibroblasts used for cell reprogramming when these cells are analyzed by using high-throughput techniques [22, 31]. As this genetic heterogeneity is uncovered by whole genome sequencing, there is an acute need to set up a demarcation line between acceptable and off-limit DNA alterations in the primary parental cells as well as in the freshly reprogrammed PSCs and long-term cultured PSCs. To this aim, the lesions that alter the cellular physiology or that pose a risk of malignant transformation must be identified [19]. As there is no means to assess the functionality of every known gene or to test the tumorigenicity of cultured cells, the only way is to make some assumptions about what kind of abnormalities could affect the physiology or safety of PSCs. Ideally, a PSC line should not display:
Karyotype abnormalities, either in number or structure
Genetic defects that have been reported in cancer cells, such as activating mutations of known oncogenes or inactivation of tumor suppressor genes (the COSMIC catalogue of somatic mutations in cancer database may help)[62]
Mutations that target a coding region and that are predicted to alter the protein sequence by giving rise to a new stop codon, a frameshift or a new splicing event.
Genetic defects that occur recurrently in cultured PSCs. To this aim, the creation of a PSC sequence database should be a key objective of scientists working on human PSCs. These recurrent mutations could be “driving” mutations (as opposed to “passenger” mutations) that confer a growth advantage to the cells. In addition, integration of these data with metabolic and phenotypic information will be instrumental for hierarchizing the abnormalities. Validation of these recurrent genetic abnormalities on large series of samples by independent techniques, for instance by digital droplet PCR for CNV, will be necessary [31]. In addition, measuring the impact of these abnormalities on the expression of the genes present in the vicinity by expression analysis will help in determining their functional consequences on the biology of PSC.
High-resolution, whole-genome sequencing data are ideally needed for the implementation of these guidelines, but they are not routinely available at the moment due to the actual costs of sequencing and the still complex pipeline analysis of such massive amount of data. Meanwhile, karyotyping will still be the reference test together with FISH analysis of chromosome 12, 17 and 20, and/or low-resolution CGH to exclude gross karyotype abnormalities.
CONCLUSION
The genetic integrity of iPSCs has been questioned and invoked as an argument against their medical use. However, these results have been challenged by reports that described the generation of iPSCs with low incidence of DNA sequence variations. Two factors may be central to PSC genetic stability: (i) the culture conditions, which can rapidly affect PSC genomic stability and (ii) the reprogramming protocols. As specific culture conditions are associated with very different rates of genetic variations, it can be anticipated that different starting cell types, reprogramming transcription factors and vectors, reprogramming protocols and early culture/passaging conditions can specifically affect the type/load of genomic damage linked to cell reprogramming. We therefore call for a careful quantification of the impact on genome integrity of all these factors in order to optimize the reprogramming protocols and ultimately provide safe iPSCs, both for research and clinical applications.
Acknowledgments
We thank E. Andermarcher for critical reading of this manuscript. Supported by grants from the Association Française contre les Myopathies (AFM), the Région Languedoc-Roussillon (Chercheur d’Avenir 09-13198 01) and the Agence Nationale de la Recherche (ANR-08-BIOT-012-02).
LIST OF ABBREVIATIONS
- iPSC
induced pluripotent stem cells
- ESC
embryonic stem cells
- PSC
pluripotent stem cells
- SV
structural variants
- SNV
single nucleotide variation
Footnotes
DISCLOSURE OF POTENTIAL CONFLICTS OF INTEREST
The authors indicate no potential conflicts of interest.
References
- 1.Ramirez JM, Bai Q, Dijon-Grinand M, Assou S, Gerbal-Chaloin S, Hamamah S, De Vos J. Human pluripotent stem cells: From biology to cell therapy. World J Stem Cells. 2010;2:24–33. doi: 10.4252/wjsc.v2.i2.24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Landry DW, Zucker HA. Embryonic death and the creation of human embryonic stem cells. J Clin Invest. 2004;114:1184–1186. doi: 10.1172/JCI23065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Yamanaka S. Induced pluripotent stem cells: past, present, and future. Cell Stem Cell. 2012;10:678–684. doi: 10.1016/j.stem.2012.05.005. [DOI] [PubMed] [Google Scholar]
- 4.Kiskinis E, Eggan KC. Progress toward the clinical application of patient-specific pluripotent stem cells. J Clin Invest. 2010;120:51–59. doi: 10.1172/JCI40553. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Gourraud P-A, Gilson L, Girard M, Peschanski M. The Role of HLA Matching in the Development of Multiethnic “Haplobank” of iPSC Lines. Stem Cells. 2011 doi: 10.1002/stem.772. [DOI] [PubMed] [Google Scholar]
- 6.Lapasset L, Milhavet O, Prieur A, Besnard E, Babled A, Aït-Hamou N, Leschik J, Pellestor F, Ramirez JM, De Vos J, Lehmann S, Lemaitre J-M. Rejuvenating senescent and centenarian human cells by reprogramming through the pluripotent state. Genes Dev. 2011;25:2248–2253. doi: 10.1101/gad.173922.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu SM, Hochedlinger K. Harnessing the potential of induced pluripotent stem cells for regenerative medicine. Nat Cell Biol. 2011;13:497–505. doi: 10.1038/ncb0511-497. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Takahashi K, Yamanaka S. Induction of pluripotent stem cells from mouse embryonic and adult fibroblast cultures by defined factors. Cell. 2006;126:663–676. doi: 10.1016/j.cell.2006.07.024. [DOI] [PubMed] [Google Scholar]
- 9.Hacein-Bey-Abina S, Kalle Von C, Schmidt M, McCormack MP, Wulffraat N, Leboulch P, Lim A, Osborne CS, Pawliuk R, Morillon E, Sorensen R, Forster A, Fraser P, Cohen JI, de Saint Basile G, Alexander I, Wintergerst U, Frebourg T, Aurias A, Stoppa-Lyonnet D, Romana S, Radford-Weiss I, Gross F, Valensi F, Delabesse E, Macintyre E, Sigaux F, Soulier J, Leiva LE, Wissler M, Prinz C, Rabbitts TH, Le Deist F, Fischer A, Cavazzana-Calvo M. LMO2-associated clonal T cell proliferation in two patients after gene therapy for SCID-X1. Science. 2003;302:415–419. doi: 10.1126/science.1088547. [DOI] [PubMed] [Google Scholar]
- 10.Okita K, Ichisaka T, Yamanaka S. Generation of germline-competent induced pluripotent stem cells. Nature. 2007;448:313–317. doi: 10.1038/nature05934. [DOI] [PubMed] [Google Scholar]
- 11.Takahashi K, Tanabe K, Ohnuki M, Narita M, Ichisaka T, Tomoda K, Yamanaka S. Induction of pluripotent stem cells from adult human fibroblasts by defined factors. Cell. 2007;131:861–872. doi: 10.1016/j.cell.2007.11.019. [DOI] [PubMed] [Google Scholar]
- 12.Yu J, Hu K, Smuga-Otto K, Tian S, Stewart R, Slukvin II, Thomson JA. Human induced pluripotent stem cells free of vector and transgene sequences. Science. 2009;324:797–801. doi: 10.1126/science.1172482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Mayshar Y, Ben-David U, Lavon N, Biancotti J-C, Yakir B, Clark AT, Plath K, Lowry WE, Benvenisty N. Identification and classification of chromosomal aberrations in human induced pluripotent stem cells. Cell Stem Cell. 2010;7:521–531. doi: 10.1016/j.stem.2010.07.017. [DOI] [PubMed] [Google Scholar]
- 14.Taapken SM, Nisler BS, Newton MA, Leonhard KA, McIntire EM, Montgomery KD, Sampsell-Barron TL. Karotypic abnormalities in human induced pluripotent stem cells and embryonic stem cells. Nat Biotechnol. 2011;29:313–314. doi: 10.1038/nbt.1835. [DOI] [PubMed] [Google Scholar]
- 15.Chen Q, Shi X, Rudolph C, Yu Y, Zhang D, Zhao X, Mai S, Wang G, Schlegelberger B, Shi Q. Recurrent trisomy and Robertsonian translocation of chromosome 14 in murine iPS cell lines. Chromosome Res. 2011;19:857–868. doi: 10.1007/s10577-011-9239-y. [DOI] [PubMed] [Google Scholar]
- 16.Ben-David U, Benvenisty N. High Prevalence of Evolutionarily-conserved and Species-Specific Genomic Aberrations in Mouse Pluripotent Stem Cells. Stem Cells. 2012 doi: 10.1002/stem.1057. [DOI] [PubMed] [Google Scholar]
- 17.Martins-Taylor K, Xu R-H. Determinants of pluripotency: from avian, rodents, to primates. J Cell Biochem. 2010;109:16–25. doi: 10.1002/jcb.22402. [DOI] [PubMed] [Google Scholar]
- 18.Pasi CEzAD, Negrini S, Friedli M, Fragola G, Lombardo A, Van Houwe G, Naldini L, Casola S, Testa G, Trono D, Pelicci PG, Halazonetis TD. Genomic instability in induced stem cells. Cell Death Differ. 2011;18:745–753. doi: 10.1038/cdd.2011.9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Laurent LC, Ulitsky I, Slavin I, Tran H, Schork A, Morey R, Lynch C, Harness JV, Lee S, Barrero MJ, Ku S, Martynova M, Semechkin R, Galat V, Gottesfeld J, Izpisua Belmonte JC, Murry C, Keirstead HS, Park H-S, Schmidt U, Laslett AL, Muller F-J, Nievergelt CM, Shamir R, Loring JF. Dynamic changes in the copy number of pluripotency and cell proliferation genes in human ESCs and iPSCs during reprogramming and time in culture. Cell Stem Cell. 2011;8:106–118. doi: 10.1016/j.stem.2010.12.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Hussein SMI, Batada NN, Vuoristo S, Ching RW, Autio R, Närvä E, Ng S, Sourour M, Hämäläinen R, Olsson C, Lundin K, Mikkola M, Trokovic R, Peitz M, Brüstle O, Bazett-Jones DP, Alitalo K, Lahesmaa R, Nagy A, Otonkoski T. Copy number variation and selection during reprogramming to pluripotency. Nature. 2011;471:58–62. doi: 10.1038/nature09871. [DOI] [PubMed] [Google Scholar]
- 21.Ji J, Ng SH, Sharma V, Neculai D, Hussein SMI, Sam M, Trinh Q, Church GM, McPherson JD, Nagy A, Batada NN. Elevated Coding Mutation Rate During the Reprogramming of Human Somatic Cells into Induced Pluripotent Stem Cells. Stem Cells. 2011 doi: 10.1002/stem.1011. [DOI] [PubMed] [Google Scholar]
- 22.Gore A, Li Z, Fung H-L, Young JE, Agarwal S, Antosiewicz-Bourget J, Canto I, Giorgetti A, Israel MA, Kiskinis E, Lee J-H, Loh Y-H, Manos PD, Montserrat N, Panopoulos AD, Ruiz S, Wilbert ML, Yu J, Kirkness EF, Izpisua Belmonte JC, Rossi DJ, Thomson JA, Eggan KC, Daley GQ, Goldstein LSB, Zhang K. Somatic coding mutations in human induced pluripotent stem cells. Nature. 2011;471:63–67. doi: 10.1038/nature09805. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blasco MA, Serrano M, Fernandez-Capetillo O. Genomic instability in iPS: time for a break. EMBO J. 2011;30:991–993. doi: 10.1038/emboj.2011.50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Pera MF. Stem cells: The dark side of induced pluripotency. Nature. 2011;471:46–47. doi: 10.1038/471046a. [DOI] [PubMed] [Google Scholar]
- 25.Krizhanovsky V, Lowe SW. Stem cells: The promises and perils of p53. Nature. 2009;460:1085–1086. doi: 10.1038/4601085a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hong H, Takahashi K, Ichisaka T, Aoi T, Kanagawa O, Nakagawa M, Okita K, Yamanaka S. Suppression of induced pluripotent stem cell generation by the p53-p21 pathway. Nature. 2009;460:1132–1135. doi: 10.1038/nature08235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Marión RM, Strati K, Li H, Murga M, Blanco R, Ortega S, Fernandez-Capetillo O, Serrano M, Blasco MA. A p53-mediated DNA damage response limits reprogramming to ensure iPS cell genomic integrity. Nature. 2009;460:1149–1153. doi: 10.1038/nature08287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li H, Collado M, Villasante A, Strati K, Ortega S, Cañamero M, Blasco MA, Serrano M. The Ink4/Arf locus is a barrier for iPS cell reprogramming. Nature. 2009;460:1136–1139. doi: 10.1038/nature08290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Quinlan AR, Boland MJ, Leibowitz ML, Shumilina S, Pehrson SM, Baldwin KK, Hall IM. Genome Sequencing of Mouse Induced Pluripotent Stem Cells Reveals Retroelement Stability and Infrequent DNA Rearrangement during Reprogramming. Cell Stem Cell. 2011;9:366–373. doi: 10.1016/j.stem.2011.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Young MA, Larson DE, Sun C-W, George DR, Ding L, Miller CA, Lin L, Pawlik KM, Chen K, Fan X, Schmidt H, Kalicki-Veizer J, Cook LL, Swift GW, Demeter RT, Wendl MC, Sands MS, Mardis ER, Wilson RK, Townes TM, Ley TJ. Background mutations in parental cells account for most of the genetic heterogeneity of induced pluripotent stem cells. Cell Stem Cell. 2012;10:570–582. doi: 10.1016/j.stem.2012.03.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Abyzov A, Mariani J, Palejev D, Zhang Y, Haney MS, Tomasini L, Ferrandino AF, Rosenberg Belmaker LA, Szekely A, Wilson M, Kocabas A, Calixto NE, Grigorenko EL, Huttner A, Chawarska K, Weissman S, Urban AE, Gerstein M, Vaccarino FM. Somatic copy number mosaicism in human skin revealed by induced pluripotent stem cells. Nature. 2012 doi: 10.1038/nature11629. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cheng L, Hansen NF, Zhao L, Du Y, Zou C, Donovan FX, Chou B-K, Zhou G, Li S, Dowey SN, Ye Z, Chandrasekharappa SC, Yang H, Mullikin JC, Liu PP NISC Comparative Sequencing Program. Low incidence of DNA sequence variation in human induced pluripotent stem cells generated by nonintegrating plasmid expression. Cell Stem Cell. 2012;10:337–344. doi: 10.1016/j.stem.2012.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Evans MJ, Kaufman MH. Establishment in culture of pluripotential cells from mouse embryos. Nature. 1981;292:154–156. doi: 10.1038/292154a0. [DOI] [PubMed] [Google Scholar]
- 34.Thomson JA, Itskovitz-Eldor J, Shapiro SS, Waknitz MA, Swiergiel JJ, Marshall VS, Jones JM. Embryonic stem cell lines derived from human blastocysts. Science. 1998;282:1145–1147. doi: 10.1126/science.282.5391.1145. [DOI] [PubMed] [Google Scholar]
- 35.Draper JS, Smith K, Gokhale P, Moore HD, Maltby E, Johnson J, Meisner L, Zwaka TP, Thomson JA, Andrews PW. Recurrent gain of chromosomes 17q and 12 in cultured human embryonic stem cells. Nat Biotechnol. 2003;22:53–54. doi: 10.1038/nbt922. [DOI] [PubMed] [Google Scholar]
- 36.The International Stem Cell Initiative. Amps KJ, Andrews PW, Anyfantis G, Armstrong L, Avery S, Baharvand H, Baker J, Baker D, Munoz MB, Beil S, Benvenisty N, Ben-Yosef D, Biancotti J-C, Bosman A, Brena RM, Brison D, Caisander G, Camarasa MV, Chen J, Chiao E, Choi YM, Choo ABH, Collins D, Colman A, Crook JM, Daley GQ, Dalton A, De Sousa PA, Denning C, Downie J, Dvorak P, Montgomery KD, Feki A, Ford A, Fox V, Fraga AM, Frumkin T, Ge L, Gokhale PJ, Golan-Lev T, Gourabi H, Gropp M, Guangxiu L, Hampl A, Harron K, Healy L, Herath W, Holm F, Hovatta O, Hyllner J, Inamdar MS, Irwanto AK, Ishii T, Jaconi M, Jin Y, Kimber S, Kiselev S, Knowles BB, Kopper O, Kukharenko V, Kuliev A, Lagarkova MA, Laird PW, Lako M, Laslett AL, Lavon N, Lee DR, Lee JE, Li C, Lim LS, Ludwig TE, Ma Y, Maltby E, Mateizel I, Mayshar Y, Mileikovsky M, Minger SL, Miyazaki T, Moon SY, Moore H, Mummery C, Nagy A, Nakatsuji N, Narwani K, Oh SKW, Oh SK, Olson C, Otonkoski T, Pan F, Park I-H, Pells S, Pera MF, Pereira LV, Qi O, Raj GS, Reubinoff BE, Robins A, Robson P, Rossant J, Salekdeh GH, Schulz TC, Sermon K, Mohamed JS, Shen H, Sherrer E, Sidhu K, Sivarajah S, Skottman H, Spits C, Stacey GN, Strehl R, Strelchenko N, Suemori H, Sun B, Suuronen R, Takahashi K, Tuuri T, Venu P, Verlinsky Y, Oostwaard DW-V, Weisenberger DJ, Wu Y, Yamanaka S, Young L, Zhou Q. Screening ethnically diverse human embryonic stem cells identifies a chromosome 20 minimal amplicon conferring growth advantage. Nat Biotechnol. 2011 doi: 10.1038/nbt.2051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Lefort N, Feyeux M, Bas C, Féraud O, Bennaceur-Griscelli A, Tachdjian G, Peschanski M, Perrier LAL. Human embryonic stem cells reveal recurrent genomic instability at 20q11.21. Nat Biotechnol. 2008;26:1364–1366. doi: 10.1038/nbt.1509. [DOI] [PubMed] [Google Scholar]
- 38.Ben-David U, Benvenisty N. The tumorigenicity of human embryonic and induced pluripotent stem cells. Nat Rev Cancer. 2011;11:268–277. doi: 10.1038/nrc3034. [DOI] [PubMed] [Google Scholar]
- 39.Maitra A, Arking DE, Shivapurkar N, Ikeda M, Stastny V, Kassauei K, Sui G, Cutler DJ, Liu Y, Brimble SN, Noaksson K, Hyllner J, Schulz TC, Zeng X, Freed WJ, Crook J, Abraham S, Colman A, Sartipy P, Matsui S-I, Carpenter MK, Gazdar AF, Rao M, Chakravarti A. Genomic alterations in cultured human embryonic stem cells. Nature genetics. 2005;37:1099–1103. doi: 10.1038/ng1631. [DOI] [PubMed] [Google Scholar]
- 40.Närvä E, Autio R, Rahkonen N, Kong L, Harrison N, Kitsberg D, Borghese L, Itskovitz-Eldor J, Rasool O, Dvorak P, Hovatta O, Otonkoski T, Tuuri T, Cui W, Brüstle O, Baker D, Maltby E, Moore HD, Benvenisty N, Andrews PW, Yli-Harja O, Lahesmaa R. High-resolution DNA analysis of human embryonic stem cell lines reveals culture-induced copy number changes and loss of heterozygosity. Nat Biotechnol. 2010;28:371–377. doi: 10.1038/nbt.1615. [DOI] [PubMed] [Google Scholar]
- 41.Mitalipova MM, Rao RR, Hoyer DM, Johnson JA, Meisner LF, Jones KL, Dalton S, Stice SL. Preserving the genetic integrity of human embryonic stem cells. Nat Biotechnol. 2005;23:19–20. doi: 10.1038/nbt0105-19. [DOI] [PubMed] [Google Scholar]
- 42.Okita K, Nakagawa M, Hyenjong H, Ichisaka T, Yamanaka S. Generation of mouse induced pluripotent stem cells without viral vectors. Science. 2008;322:949–953. doi: 10.1126/science.1164270. [DOI] [PubMed] [Google Scholar]
- 43.Stadtfeld M, Nagaya M, Utikal J, Weir G, Hochedlinger K. Induced Pluripotent Stem Cells Generated Without Viral Integration. Science. 2008 doi: 10.1126/science.1162494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Mali P, Ye Z, Hommond H, Yu X, Lin J, Chen G, Zou J, Cheng L. Improved Efficiency and Pace of Generating Induced Pluripotent Stem Cells from Human Adult and Fetal Fibroblasts. Stem Cells. 2008 doi: 10.1634/stemcells.2008-0346. [DOI] [PubMed] [Google Scholar]
- 45.Ban H, Nishishita N, Fusaki N, Tabata T, Saeki K, Shikamura M, Takada N, Inoue M, Hasegawa M, Kawamata S, Nishikawa S-I. Efficient generation of transgene-free human induced pluripotent stem cells (iPSCs) by temperature-sensitive Sendai virus vectors. Proc Natl Acad Sci USA. 2011 doi: 10.1073/pnas.1103509108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Woltjen K, Michael I, Mohseni P, Desai R, Mileikovsky M, Hämäläinen R, Cowling R, Wang W, Liu P, Gertsenstein M, Kaji K, Sung H, Nagy A. piggyBac transposition reprograms fibroblasts to induced pluripotent stem cells. Nature. 2009 doi: 10.1038/nature07863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Soldner F, Hockemeyer D, Beard C, Gao Q, Bell GW, Cook EG, Hargus G, Blak A, Cooper O, Mitalipova MM, Isacson O, Jaenisch R. Parkinson’s disease patient-derived induced pluripotent stem cells free of viral reprogramming factors. Cell. 2009;136:964–977. doi: 10.1016/j.cell.2009.02.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kim D, Kim C-H, Moon J-I, Chung Y-G, Chang M-Y, Han B-S, Ko S, Yang E, Cha KY, Lanza RP, Kim K-S. Generation of human induced pluripotent stem cells by direct delivery of reprogramming proteins. Cell Stem Cell. 2009;4:472–476. doi: 10.1016/j.stem.2009.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Warren L, Manos PD, Ahfeldt T, Loh Y-H, Li H, Lau F, Ebina W, Mandal PK, Smith ZD, Meissner A, Daley GQ, Brack AS, Collins JJ, Cowan CA, Schlaeger TM, Rossi DJ. Highly Efficient Reprogramming to Pluripotency and Directed Differentiation of Human Cells with Synthetic Modified mRNA. Cell Stem Cell. 2010 doi: 10.1016/j.stem.2010.08.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Lee J, Sayed N, Hunter A, Au KF, Wong WH, Mocarski ES, Reijo Pera RA, Yakubov E, Cooke JP. Activation of innate immunity is required for efficient nuclear reprogramming. Cell. 2012;151:547–558. doi: 10.1016/j.cell.2012.09.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Singhal N, Graumann J, Wu G, Araúzo-Bravo MJ, Han DW, Greber B, Gentile L, Mann M, Schöler HR. Chromatin-Remodeling Components of the BAF Complex Facilitate Reprogramming. Cell. 2010;141:943–955. doi: 10.1016/j.cell.2010.04.037. [DOI] [PubMed] [Google Scholar]
- 52.Subramanyam D, Lamouille S, Judson RL, Liu JY, Bucay N, Derynck R, Blelloch RH. Multiple targets of miR-302 and miR-372 promote reprogramming of human fibroblasts to induced pluripotent stem cells. Nat Biotechnol. 2011 doi: 10.1038/nbt.1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jullien J, Pasque V, Halley-Stott RP, Miyamoto K, Gurdon JB. Mechanisms of nuclear reprogramming by eggs and oocytes: a deterministic process? Vol. 12. Nature Publishing Group; 2011. pp. 453–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Schwartz SD, Hubschman J-P, Heilwell G, Franco-Cardenas V, Pan CK, Ostrick RM, Mickunas E, Gay R, Klimanskaya I, Lanza RP. Embryonic stem cell trials for macular degeneration: a preliminary report. Lancet. 2012 doi: 10.1016/S0140-6736(12)60028-2. [DOI] [PubMed] [Google Scholar]
- 55.Prockop DJ, Keating A. Re-Learning the Lessons of Genomic Stability of Human Cells During Expansion in Culture: Implications for Clinical Research. Stem Cells. 2012 doi: 10.1002/stem.1103. [DOI] [PubMed] [Google Scholar]
- 56.Hayflick L, Perkins F, Stevenson RE. Human Diploid Cell Strains. Science. 1964;143:976. doi: 10.1126/science.143.3609.976. [DOI] [PubMed] [Google Scholar]
- 57.Ben-David U, Mayshar Y, Benvenisty N. Large-scale analysis reveals acquisition of lineage-specific chromosomal aberrations in human adult stem cells. Cell Stem Cell. 2011;9:97–102. doi: 10.1016/j.stem.2011.06.013. [DOI] [PubMed] [Google Scholar]
- 58.Eisenstein M. The battle for sequencing supremacy. Vol. 30. Nature Publishing Group; 2012. pp. 1023–1026. [DOI] [PubMed] [Google Scholar]
- 59.Kirkwood TBL. Understanding the odd science of aging. Cell. 2005;120:437–447. doi: 10.1016/j.cell.2005.01.027. [DOI] [PubMed] [Google Scholar]
- 60.Piotrowski A, Bruder CEG, Andersson R, Diaz de Ståhl T, Menzel U, Sandgren J, Poplawski A, Tell von D, Crasto C, Bogdan A, Bartoszewski R, Bebok Z, Krzyzanowski M, Jankowski Z, Partridge EC, Komorowski J, Dumanski JP. Somatic mosaicism for copy number variation in differentiated human tissues. Hum Mutat. 2008;29:1118–1124. doi: 10.1002/humu.20815. [DOI] [PubMed] [Google Scholar]
- 61.Youssoufian H, Pyeritz RE. Mechanisms and consequences of somatic mosaicism in humans. Nat Rev Genet. 2002;3:748–758. doi: 10.1038/nrg906. [DOI] [PubMed] [Google Scholar]
- 62.Forbes SA, Bindal N, Bamford S, Cole C, Kok CY, Beare D, Jia M, Shepherd R, Leung K, Menzies A, Teague JW, Campbell PJ, Stratton MR, Futreal PA. COSMIC: mining complete cancer genomes in the Catalogue of Somatic Mutations in Cancer. Nucleic Acids Res. 2011;39:D945–50. doi: 10.1093/nar/gkq929. [DOI] [PMC free article] [PubMed] [Google Scholar]


