Abstract
Nuclear factor-E2-related factor 2 (Nrf2) is a regulator of lipid metabolism as well as various cytoprotective enzymes and may be involved in the pathogenesis of non-alcoholic fatty liver disease. Although, bile acids affect lipid metabolism, the role of Nrf2 in bile acid metabolism remains unclear. In this study, it was tested how Nrf2 modulates lipid and bile acid homeostasis in liver in response to changes of cholesterol absorption under high-fat diet using Nrf2-null mice. Eight-week-old male wild-type and Nrf2-null mice (n = 6/group) were divided into three groups fed the following diets: 1) control diet containing 4% soybean oil and 16% lard, 2) control diet plus ezetimibe, 3) control diet plus cholesterol. Blood and livers were removed after 4 weeks feeding. High cholesterol diet increased hepatic expression of liver X receptor α target genes related to fatty acid metabolism (FAS, ACC1, SREBP-1c, SCD-1c and CD36), cholesterol transport (Abcg5/abcg8) and bile acid synthesis (Cyp7a1) in wild type mice. However, these genes were not induced in Nrf2-null mice. These findings suggest that Nrf2 has a relation to liver X receptor α and controls the regulation of gene expressions related to lipid and bile acid metabolism.
Keywords: nuclear factor-E2-related factor 2, non-alcoholic fatty liver disease, cholesterol metabolism, bile acid metabolism, liver X receptor α
Introduction
High fat diet is a well-known contributor to the pathogenesis of non-alcoholic fatty liver disease (NAFLD). NAFLD represents the hepatic manifestation of the metabolic syndrome, characterized by insulin resistance, dyslipidemia, and hypertension.(1,2) The term NAFLD contains a broad spectrum of liver diseases ranging from simple steatosis to non-alcoholic steatohepatitis (NASH).(3,4) Oxidative stress has been suggested to be important for the progression from simple steatosis to NASH and several studies have investigated the efficacy of antioxidative supplements against progression of NASH.(5,6)
Recent studies showed that dietary cholesterol is important for the progression of hepatic steatosis.(7–10) Ezetimibe is an inhibitor of the cholesterol uptake transporter, Niemann-Pick C1-like 1 (NPC1L1), locates in the apical membrane of enterocytes.(11,12) Several studies showed that ezetimibe attenuates liver steatosis in experimental models.(13,14) These findings suggest that the management of cholesterol intake is one of the therapeutic targets for NAFLD. In addition to the cholesterol intake and biosynthesis, catabolism to bile acids is also critical for maintaining cholesterol homeostasis. Administration of natural (colic acid) or synthetic (GW4064) ligand for the nuclear bile acid receptor farnesoid X receptor (FXR) is known to prevent a high-fat diet-induced hepatic steatosis in mice.(15,16)
The Nuclear factor-E2-related factor 2 (Nrf2), which is a cellular sensor for oxidative and electrophilic stress, induces the various cytoprotective enzymes.(17) Recent studies showed that Nrf2 deletion affected lipid metabolism. Nrf2-null mice showed the dysregulation of β-oxidation and the increase in hepatic triglycerides.(18,19) In Nrf2-null mice, the methionine- and choline-deficient diet caused rapid onset and progression of nutritional steatohepatits.(20) Furthermore, Nrf2 activator prevented a high-fat diet induced hepatic lipid accumulation in wild type mice but not in Nrf2-null mice.(21) Nrf2 may play a role in lipid metabolism and may be involved in the pathogenesis of NAFLD. Nrf2 also regulates hepatic bile acid metabolism. Sulforaphane, an Nrf2 activator induced the hepatic FXR mRNA expression in wild-type mice but not in Nrf2-null mice.(22)
The purpose of this study is to evaluate the role of Nrf2 on bile acid as well as lipid metabolism in high fat diet induced NAFLD model. In this study, we investigated the effect of cholesterol absorption on lipid and bile acid metabolism in Nrf2-null mice and wild type mice. To change the cholesterol absorption, mice were fed a diet containing high cholesterol or ezetimibe. In human and rat, NPC1L1 is expressed both in intestine and liver. Therefore, ezetimibe may affect hepatic lipid metabolism. On the other hand, NPC1L1 is predominantly expressed in the intestine in mice and is hardly expressed in the liver.(11) Ezetimibe only inhibits intestinal cholesterol absorption and does not directly affect hepatic lipid metabolism in mice. Here, we reported that the induction of the gene expression related to lipid and bile acid metabolism by cholesterol influx was at least partially controlled by Nrf2.
Materials and Methods
Materials and chemicals
Milk casein, corn starch, α-corn starch, mineral AIN-93 mixture and vitamin AIN-93 VX mixture were purchased from CLEA Japan (Osaka, Japan). Soybean oil and lard were purchased from Oriental Yeast (Tokyo, Japan) and Yamakei (Osaka, Japan), respectively. Ezetimibe was kindly provided by Schering-Plough Co., Ltd. (Osaka, Japan). All other chemicals were purchased from Wako Pure Chem. Ind., Ltd. (Osaka, Japan), unless noted.
Animals and diets
A colony of wild-type and Nrf2-null mice, generated originally by Itoh et al.(23), were backcrossed with C57BL/6 mice for ten generations. All mice were housed in the same animal care facility controlling for temperature (22 ± 2°C), humidity (55 ± 5%), and light (lights on; 07:00–19:00). Eight-week-old male wild-type and Nrf2-null mice (n = 6/group) were divided into three groups fed the following diets: 1) the control diet group (C); containing 4% soybean oil,16% lard, 14% milk casein, 0.18% L-cystine, 3.5% AIN-93G mineral mixture, 1% AIN-93VX vitamine mixture, 5% cellulose powder, 30.57% corn starch, 15.5% α-corn starch, 10% sucrose, 0.25% choline bitartrate for 4 weeks; 2) the ezetimibe group (EZ); a control diet containing ezetimibe (0.3 mg/day) for 4 weeks; 3) the cholesterol diet group (CH); containing 4% soybean oil, 16% lard and 1% cholesterol, 14% milk casein, 0.18% L-cystine, 3.5% AIN-93G mineral mixture, 1% AIN-93VX vitamine mixture, 5% cellulose powder, 29.57% corn starch, 15.5% α-corn starch, 10% sucrose, 0.25% choline bitartrate, 1% cholesterol, for 4 weeks. After 4 weeks, food was removed 2 h prior to collecting tissues and blood samples were collected by heart puncture after anesthesia with pentobarbital (50 mg/kg, intra-peritoneally), and livers and duodenum were harvested and stored at –80°C until use. Studies were approved by Kinki University Faculty of Medicine Animal Care and Use Committee.
Biochemical and histological analysis
Serum biochemical markers were quantified using biochemistry autoanalyzer Labospect 008 (Hitach High-Technologies Corporation, Ibaragi, Japan). Liver lipid content was extracted according to the method of Folch et al.(24) Hepatic cholesterol and triglyceride concentrations were determined by standard enzymatic-colorimetric assays using commercially available kits (Wako Pure Chem. Ind.). Hepatic bile acid concentrations were quantified.(25) In brief, 100 mg of liver was homogenized in 900 µl of 1:1 t-butylalcohol/double distilled water (v/v) and incubated overnight at room temperature. The homogenate was centrifuged at 5,000 rpm for 10 min. Hepatic bile acid levels were measured in the supernatant by enzymatic-colorimetric assays using bile acid assay kits (Wako Pure Chem. Ind.). Liver tissues were fixed in 10% paraformaldehyde, embedded in paraffin and stained with hematoxylin-eosin.
RNA isolation and real-time polymerase chain reaction
Total RNA was isolated using TRIzol reagent (Life Technologies, Tokyo, Japan) according to the manufacturer’s protocol. The concentration of total RNA in each sample was quantified spectrophotometrically at 260 nm. The mRNA expression of lipid and bile acid metabolism-related genes and 18s rRNA were quantified by SYBR real-time polymerase chain reaction (PCR) (SYBR Premix Ex Taq; Takara bio Inc., Shiga, Japan). Primers were used from previous reports.(18,26–34) The amplification reactions were carried out in an ABI Prism 7900 HT sequence detection system (Applied Biosystems, Foster, CA). The amount of mRNA was calculated using the comparative CT method which determines the amount of target normalized to an endogenous reference. Each gene was normalized to 18s rRNA.
Statistical analysis
Statistical analysis was performed using the software package, SYSTAT, ver. 11 (Systat Inc., Evanston, IL). Comparisons among the groups were analyzed by analysis of variance, followed by a Tukey’s multiple range test. All results are expressed as means ± SEM. Significance was set at p<0.05.
Results
Serum and hepatic concentrations of lipids and bile acids in wild-type and Nrf2-null mice
Lipid and bile acid profiles in wild-type and Nrf2-null mice are shown in Table 1. Serum cholesterol was higher in wild-type mice compared to Nrf2-null mice feeding the control diet. Ezetimibe decreased serum cholesterol and HDL-cholesterol in wild-type mice. Serum triglyceride and bile acids were not changed in each group after feeding the respective diet. Hepatic triglycerides tended to be higher in Nrf2-null control mice compared to wild-type control mice. Ezetimibe decreased hepatic triglyceride both in wild-type mice and Nrf2-null mice. Hepatic cholesterol concentration was not changed by ezetimibe administration and was significantly increased by high cholesterol diet both in wild-type mice and Nrf2-null mice. There were no significant differences in hepatic bile acid concentrations between genotypes of each group.
Table 1.
Serum and hepatic concentrations of lipids and bile acids in wild-type and Nrf2-null mice
| Wild-type mice | Nrf2-null mice | |||||
|---|---|---|---|---|---|---|
| Control | EZ | CH | Control | EZ | CH | |
| Serum concentration | ||||||
| Triglycerides (mg/dl) | 27 ± 2 | 19 ± 3 | 15 ± 2 | 19 ± 3 | 23 ± 2 | 13 ± 1 |
| Cholesterol (mg/dl) | 144 ± 12 | 101 ± 7a | 122 ± 13 | 102 ± 14c | 101 ± 6 | 107 ± 16 |
| HDL-cholesterol (mg/dl) | 79 ± 5 | 58 ± 3a | 72 ± 8 | 55 ± 10 | 49 ± 11 | 51 ± 8 |
| Bile acids (µmol/l) | 4.6 ± 0.7 | 8.3 ± 3.9 | 4.3 ± 0.5 | 4.0 ± 1.2 | 5.3 ± 1.2 | 7.6 ± 1.4 |
| Hepatic concentration | ||||||
| Triglycerides (mg/g liver) | 22 ± 4 | 9 ± 1a | 66 ± 6a | 45 ± 11 | 16 ± 2a | 68 ± 10 |
| Cholesterol (mg/g liver) | 2.5 ± 0.2 | 2.5 ± 0.1 | 11.5 ± 2.0b | 3.3 ± 0.2 | 2.6 ± 0.3 | 10.6 ± 0.6b |
| Bile acids (µmol/g liver) | 0.051 ± 0.008 | 0.092 ± 0.042 | 0.048 ± 0.006 | 0.044 ± 0.014 | 0.058 ± 0.013 | 0.084 ± 0.015 |
ap<0.05 significant difference from mice fed with control diet. bp<0.01 significant difference from mice fed with control diet. cp<0.05 significant difference between genotypes.
Histological findings of liver in wild-type and Nrf2-null mice
Histological findings of liver in wild-type and Nrf2-null mice are shown in Fig. 1. Consistent with hepatic triglyceride level, lipid droplets in hepatocytes tended to increase in Nrf2-null control mice compared to wild-type control mice. Ezetimibe decreased hepatic steatosis both in wild-type mice and Nrf2-null mice. Hepatic steatosis level was increased by cholesterol feeding both in wild-type mice and Nrf2-null mice.
Fig. 1.
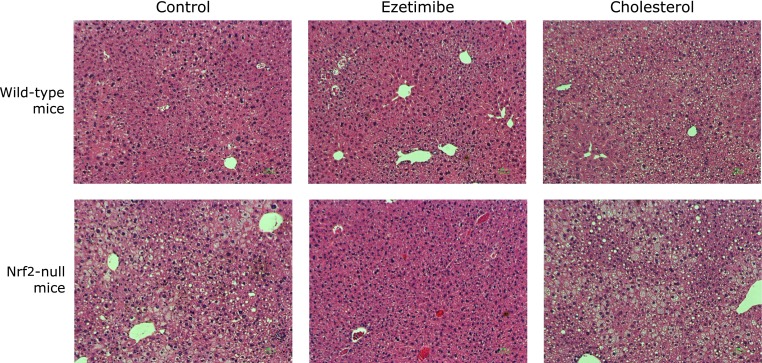
Hematoxylin and eosin-stained liver sections from wild-type and Nrf2-null mice fed with three experimental diets.
Hepatic expression of Nrf2 target genes and lipid metabolism-related genes in wild-type and Nrf2-null mice
The changes in Nrf2 target genes and lipid metabolism-related genes expression are summarized in Table 2. In regard to Nrf2 target genes, hepatic expression of glutamate-cysteine ligase, catalytic subunit (Gclc) and heme oxygenase-1 (Ho-1) were measured. Hepatic gene expression of Gclc was lower in Nrf2-null mice with cholesterol diet compared to the CH group in wild-type mice. Hepatic expression of Ho-1 was unchanged among the three groups of both mice. With respect to the hepatic fatty acid synthesis, the expression of fatty acid synthase (Fas) and acetyl coenzyme-A carboxylase-1 (Acc1) were unchanged among the three groups of both mice. Hepatic gene expression of sterol regulatory element-binding protein (Srebp)-1c, stearoyl-CoA desaturase(Scd1)and CD36, an fatty acid transporter, were higher in the CH group compared to the EZ group in wild-type mice, and the expression of these mRNA were unchanged among the three groups of Nrf2-null mice. In regard to the fatty acid oxidation, hepatic gene expression of peroxisome proliferator-activated receptors (Ppar) α, carnitine palmitoyltransferase (Cpt) 1, and acyl-coenzyme A oxidase (Acox) 1 were higher in the CH group than the EZ group in wild-type mice, and the mRNA expressions of these genes were unchanged among the three groups of Nrf2-null mice. The expression of Srebp-2 was lower in the CH group than the EZ group in Nrf2-null mice. However, there were no differences in the expression of this gene among the three groups in wild-type mice. The mRNA expression of 3-hydroxy-3-methylglutaryl coenzyme A-reductase (Hmg-CoA-R) was unchanged among the three groups of both genotypes.
Table 2.
Hepatic expression of Nrf2 target genes and lipid metabolism-related genes in wild-type and Nrf2-null mice
| Wild-type mice | Nrf2-null mice | |||||
|---|---|---|---|---|---|---|
| Control | EZ | CH | Control | EZ | CH | |
| Gclc | 1.00 ± 0.22 | 0.49 ± 0.34 | 1.01 ± 0.13 | 0.50 ± 0.20 | 0.51 ± 0.16 | 0.34 ± 0.05d |
| Ho-1 | 1.00 ± 0.38 | 0.43 ± 0.15 | 1.11 ± 0.23 | 0.73 ± 0.10 | 0.56 ± 0.11 | 0.51 ± 0.11 |
| Pparα | 1.00 ± 0.22 | 0.61 ± 0.13 | 1.69 ± 0.19c | 1.31 ± 0.16 | 1.16 ± 0.16 | 0.89 ± 0.11 |
| Acox | 1.00 ± 0.27 | 0.52 ± 0.10 | 1.78 ± 0.32c | 1.05 ± 0.19 | 0.80 ± 0.15 | 0.78 ± 0.13 |
| Cpt1 | 1.00 ± 0.27 | 0.50 ± 0.10 | 1.37 ± 0.40b | 1.12 ± 0.18 | 0.97 ± 0.15 | 0.78 ± 0.13 |
| Srebp-1c | 1.00 ± 0.42 | 0.26 ± 0.06 | 2.03 ± 0.40c | 0.72 ± 0.16 | 0.58 ± 0.11 | 1.28 ± 0.18 |
| Fas | 1.00 ± 0.40 | 0.54 ± 0.11 | 0.72 ± 0.17 | 0.87 ± 0.20 | 0.90 ± 0.08 | 0.40 ± 0.05 |
| Acc1 | 1.00 ± 0.24 | 0.55 ± 0.12 | 1.09 ± 0.14 | 1.08 ± 0.20 | 0.83 ± 0.11 | 0.58 ± 0.06 |
| CD36 | 1.00 ± 0.31 | 0.34 ± 0.09 | 1.57 ± 0.33c | 0.84 ± 0.21 | 0.37 ± 0.07 | 0.78 ± 0.16 |
| SCD1 | 1.00 ± 0.29 | 0.36 ± 0.05 | 1.84 ± 0.29b | 1.52 ± 0.43 | 0.76 ± 0.14 | 1.24 ± 0.31 |
| Srebp-2 | 1.00 ± 0.19 | 0.79 ± 0.19 | 0.69 ± 0.12 | 0.95 ± 0.14 | 1.21 ± 0.13 | 0.47 ± 0.04b |
| Hmg-CoA-R | 1.00 ± 0.42 | 1.60 ± 0.57 | 0.86 ± 0.20 | 1.56 ± 0.38 | 1.70 ± 0.13 | 0.58 ± 0.07 |
| LXRα | 1.00 ± 0.17 | 0.57 ± 0.12 | 1.18 ± 0.15b | 1.08 ± 0.15 | 0.87 ± 0.13 | 0.71 ± 0.11 |
| Abcg5 | 1.00 ± 0.34 | 0.47± 0.07 | 4.39 ± 1.11a,c | 0.77 ± 0.07 | 0.63 ± 0.04 | 1.70 ± 0.19 |
| Abcg8 | 1.00 ± 0.32 | 0.69 ± 0.12 | 4.52 ± 0.95a,c | 0.87 ± 0.11 | 1.00 ± 0.07 | 1.78 ± 0.26 |
ap<0.01 significant difference from mice fed with control diet. bp<0.05 significant difference from mice fed with ezetimibe diet. cp<0.01 significant difference from mice fed with ezetimibe diet. dp<0.05 significant difference between genotypes.
The mRNA expression of liver X receptor α (LXRα) was induced by feeding the cholesterol diet in wild-type mice, however, it was unchanged among the three groups of Nrf2-null mice. The mRNA expression of ATP-binding cassette sub-family G member 5 (Abcg5) and ATP-binding cassette sub-family G member 8 (Abcg8), the target genes of Lxrα, was induced by cholesterol feeding in wild-type mice. On the other hand, the expression of Abcg5 and Abcg8 were only slightly induced by cholesterol feeding in Nrf2-null mice.
Hepatic expression of bile acid metabolism-related genes in wild-type and Nrf2-null mice
The changes in bile acids metabolism-related gene expression are summarized in Table 3. In wild-type mice, hepatic gene expression of Fxr and small heterodimer partner (Shp) were higher in the CH group compared to the EZ group. In Nrf2-null mice, there were no differences in the expression of Fxr and Shp among the three groups. In regard to the bile acid synthesis, hepatic gene expression of cholesterol 7 α-hydroxylase (Cyp7a1), sterol 12-α-hydroxylase (Cyp8b1) and 25-hydroxycholesterol 7-α-hydroxylase (Cyp7b1) of wild-type mice were higher in the CH group compared to the EZ group and in Nrf2-null mice there were no differences in the mRNA expression of these genes among the three groups. Hepatic mRNA expression of sterol 27-hydroxylase (Cyp27a1) was unchanged in both genotypes of each group. With respect to hepatic bile acid transporters, the expression of Na+-taurocholate cotransporting polypeptide (Ntcp), organic anion-transporting polypeptide (Oatp)1a1, Oatp1a4 and bile salt export pump (Bsep) were unchanged among the three groups of both genotypes. Hepatic multidrug resistance-associated protein (Mrp) 2 and Mrp3 gene expression of wild-type mice were higher in the CH group compared to the EZ group, though, in Nrf2-null mice there were no differences in the expressions of these mRNA among the three groups.
Table 3.
Hepatic expression of bile acid metabolism-related genes in wild-type and Nrf2-null mice
| Wild-type mice | Nrf2-null mice | |||||
|---|---|---|---|---|---|---|
| Control | EZ | CH | Control | EZ | CH | |
| Fxr | 1.00 ± 0.20 | 0.43 ± 0.07 | 1.39 ± 0.24c | 0.81 ± 0.06 | 1.11 ± 0.16 | 0.52 ± 0.04 |
| Shp | 1.00 ± 0.25 | 0.67 ± 0.27 | 1.95 ± 0.40c | 0.81 ± 0.26 | 1.34 ± 0.22 | 0.91 ± 0.24 |
| Cyp7a1 | 1.00 ± 0.30 | 0.77 ± 0.22 | 5.01 ± 1.06b,c | 3.21 ± 1.52 | 2.01 ± 0.43 | 1.90 ± 0.29 |
| Cyp8b1 | 1.00 ± 0.24 | 0.66 ± 0.17 | 1.71 ± 0.32c | 1.08 ± 0.18 | 1.26 ± 0.41 | 0.59 ± 0.13 |
| Cyp27a1 | 1.00 ± 0.25 | 0.60 ± 0.13 | 1.26 ± 0.18 | 0.80 ± 0.11 | 0.79 ± 0.07 | 0.61 ± 0.09 |
| Cyp7b1 | 1.00 ± 0.29 | 0.78 ± 0.12 | 1.85 ± 0.27a,d | 0.86 ± 0.12 | 1.17 ± 0.18 | 0.58 ± 0.08 |
| Ntcp | 1.00 ± 0.27 | 0.65 ± 0.11 | 1.34 ± 0.24 | 1.33 ± 0.16 | 1.22 ± 0.20 | 0.66 ± 0.14 |
| Oatp1a1 | 1.00 ± 0.36 | 0.66 ± 0.14 | 1.20 ± 0.16 | 1.02 ± 0.23 | 0.76 ± 0.14 | 0.82 ± 0.18 |
| Oatp1a4 | 1.00 ± 0.16 | 0.32 ± 0.07 | 1.39 ± 0.50 | 0.98 ± 0.16 | 0.97 ± 0.23 | 0.74 ± 0.14 |
| Bsep | 1.00 ± 0.20 | 0.58 ± 0.14 | 1.08 ± 0.16 | 0.91 ± 0.16 | 0.74 ± 0.09 | 0.59 ± 0.07 |
| Mrp2 | 1.00 ± 0.19 | 0.50 ± 0.06 | 1.66 ± 0.24a,d | 1.00 ± 0.17 | 0.94 ± 0.07 | 0.75 ± 0.07 |
| Mrp3 | 1.00 ± 0.25 | 0.57 ± 0.12 | 1.80 ± 0.43d | 0.21 ± 0.03e | 0.16 ± 0.03e | 0.23 ± 0.04e |
| Baat | 1.00 ± 0.34 | 0.47 ± 0.09 | 0.99 ± 0.17 | 0.71 ± 0.17 | 0.53 ± 0.12 | 0.60 ± 0.11 |
ap<0.05 significant difference from mice fed with control diet. bp<0.01 significant difference from mice fed with control diet. cp<0.05 significant difference from mice fed with ezetimibe diet. dp<0.01 significant difference from mice fed with ezetimibe diet. ep<0.05 significant difference between genotypes.
Discussion
In this study, we investigated the roles of Nrf2 on cholesterol and bile acid metabolism under high fat diet by using Nrf2-null mice. Our results showed that Nrf2 is necessary for the gene regulation related to cholesterol and bile acid metabolism. SREBP2 and LXRα are the regulators for hepatic cholesterol metabolisam.(35,36) High cholesterol diet is known to induce the expression of Abcg5 and Abcg8, which are target of LXRα.(37) In this experiment, the expression level of Abcg5 and Abcg8 were not increased by cholesterol feeding in Nrf2-null mice. This finding suggests that Nrf2 may be required for LXRα activation. Administration of LXRα agonists also induced hepatic mRNA expression of fatty acid metabolism related genes, such as FAS, ACC1, SREBP-1c, SCD-1c and CD36.(38–40) In accordance with this phenomenon, high cholesterol diet induced these genes in wild type mice. Though, in Nrf2-null mice these genes were not induced by high cholesterol diet. These findings indicate that Nrf2 has roles in regulation of the expression of LXRα related genes and controls fatty acid metabolism as well as cholesterol metabolism.
Although the defects in the alteration of LXRα related gene expression were observed in Nrf2-null mice, high cholesterol feeding increased hepatic triglyceride level as well as hepatic cholesterol levels in both genotypes. Some factor other than LXRα may control hepatic lipid concentrations. With respect to cholesterol synthesis, the regulation of HMG-CoA-R is mediated by SREBP cleavage-activating protein (SCAP)-SREBP2 pathway.(35) As expected, in this experiment, mRNA expression of SREBP2 and HMG-CoA-R were decreased by high cholesterol diet in both genotypes. This finding suggests that Nrf2 may not be involved in cholesterol synthesis regulated by SCAP-SREBP2 pathway.
Regarding bile acid synthesis, FXR plays a central role in the regulation of Cyp7a1, a rate-limiting enzyme of bile acid synthesis. Several recent studies have shown that Nrf2 controls the gene expression level related to bile acid synthesis.(41,42) Kay HY et al.(19) demonstrated that Nrf2 has some roles in FXR activation. In mice but not in human, high cholesterol diet induces bile acid synthesis because Cyp7a1 is a LXRα target gene.(43) Similar to their study, we also observed the induction of hepatic Cyp7a1mRNA by high cholesterol feeding in wild-type mice. However, high cholesterol diet did not affect the expression of Cyp7a1 in Nrf2-null mice. This phenomenon suggests that Nrf2 is required for the regulation of classic pathway of bile acid synthesis via LXRα in mice.
With respect to bile acid transporters, it has been reported that Nrf2 regulates MRP2 and BSEP in human hepatocytes,(44,45) which excrete bile acids into bile. Recent study demonstrated that human MRP2 is induced by LXRα in HepG2 cells.(46) However, it has never reported that rodent Mrp2 was induced by LXRα. In this study, we demonstrated that Mrp2 was decreased in the EZ group and was increased in the CH group in wild-type mice, suggesting that rodent Mrp2 may be possibly induced by LXRα.
In conclusion, it was tested how Nrf2 modulates lipid and bile acid homeostasis in liver in response to cholesterol supplementation under the high fat diet. Nrf2 deletion dysregulates the expression of hepatic LXRα target genes which are related to lipid and bile acid metabolism.
Acknowledgments
The authors would like to thank Yoko Saito and Mariko Shibayama for technical assistance. This work was partly supported by a Grant-in-Aid for Scientific Research (No.24501010) from the Japan Society for the Promotion of Science.
Conflict of Interest
No potential conflicts of interest were disclosed.
References
- 1.Schreuder TC, Verwer BJ, van Nieuwkerk CM, Mulder CJ. Nonalcoholic fatty liver disease: an overview of current insights in pathogenesis, diagnosis and treatment. World J Gastroenterol. 2008;14:2474–2486. doi: 10.3748/wjg.14.2474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Angulo P. Nonalcoholic fatty liver disease. N Engl J Med. 2002;346:1221–1231. doi: 10.1056/NEJMra011775. [DOI] [PubMed] [Google Scholar]
- 3.Krawczyk M, Bonfrate L, Portincasa P. Nonalcoholic fatty liver disease. Best Pract Res Clin Gastroenterol. 2010;24:695–708. doi: 10.1016/j.bpg.2010.08.005. [DOI] [PubMed] [Google Scholar]
- 4.Smith BW, Adams LA. Non-alcoholic fatty liver disease. Crit Rev Clin Lab Sci. 2011;48:97–113. doi: 10.3109/10408363.2011.596521. [DOI] [PubMed] [Google Scholar]
- 5.Yachi R, Muto C, Ohtaka N, et al. Effects of tocotrienol on tumor necrosis factor-α/d-galactosamine-induced steatohepatitis in rats. J Clin Biochem Nutr. 2013;52:146–153. doi: 10.3164/jcbn.12-101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pak W, Takayama F, Mine M, et al. Anti-oxidative and anti-inflammatory effects of spirulina on rat model of non-alcoholic steatohepatitis. J Clin Biochem Nutr. 2012;51:227–234. doi: 10.3164/jcbn.12-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Matsuzawa N, Takamura T, Kurita S, et al. Lipid-induced oxidative stress causes steatohepatitis in mice fed an atherogenic diet. Hepatology. 2007;46:1392–1403. doi: 10.1002/hep.21874. [DOI] [PubMed] [Google Scholar]
- 8.Musso G, Gambino R, De Michieli F. Dietary habits and their relations to insulin resistance and postprandial lipemia in nonalcoholic steatohepatitis. Hepatology. 2003;37:909–916. doi: 10.1053/jhep.2003.50132. [DOI] [PubMed] [Google Scholar]
- 9.Yasutake K, Nakamuta M, Shima Y, et al. Nutritional investigation of non-obese patients with non-alcoholic fatty liver disease: the significance of dietary cholesterol. Scand J Gastroenterol. 2009;44:471–477. doi: 10.1080/00365520802588133. [DOI] [PubMed] [Google Scholar]
- 10.Okada K, Warabi E, Sugimoto H, et al. Deletion of Nrf2 leads to rapid progression of steatohepatitis in mice fed atherogenic plus high-fat diet. J Gastroenterol. 2013;48:620–632. doi: 10.1007/s00535-012-0659-z. [DOI] [PubMed] [Google Scholar]
- 11.Altmann SW, Davis HR, Jr, Zhu L, et al. Niemann-Pick C1 Like 1 protein is critical for intestinal cholesterol absorption. Science. 2004;303:1201–1204. doi: 10.1126/science.1093131. [DOI] [PubMed] [Google Scholar]
- 12.Garcia-Calvo M, Lisnock JM, Bull HG, et al. The target of ezetimibe is Niemann–Pick C1-Like 1 (NPC1L1) Proc Natl Acad Sci U S A. 2005;102:8132–8137. doi: 10.1073/pnas.0500269102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zheng S, Hoos L, Cook J, et al. Ezetimibe improves high fat and cholesterol diet-induced non-alcoholic fatty liver disease in mice. Eur J Pharmacol. 2008;584:118–124. doi: 10.1016/j.ejphar.2008.01.045. [DOI] [PubMed] [Google Scholar]
- 14.Muraoka T, Aoki K, Iwasaki T, et al. Ezetimibe decreases SREBP-1c expression in liver and reverses hepatic insulin resistance in mice fed a high-fat diet. Metabolism. 2011;60:617–628. doi: 10.1016/j.metabol.2010.06.008. [DOI] [PubMed] [Google Scholar]
- 15.Watanabe M, Houten SM, Wang L, et al. Bile acids lower triglyceride levels via a pathway involving FXR, SHP, and SREBP-1c. J Clin Invest. 2004;113:1408–1418. doi: 10.1172/JCI21025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Ma Y, Huang Y, Yan L, Gao M, Liu D. Synthetic FXR agonist GW4064 prevents diet-induced hepatic steatosis and insulin resistance. Pharm Res. 2013;30:1447–1457. doi: 10.1007/s11095-013-0986-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Itoh K, Tong KI, Yamamoto M. Molecular mechanism activating Nrf2-Keap1 pathway in regulation of adaptive response to electrophiles. Free Radic Biol Med. 2004;36:1208–1213. doi: 10.1016/j.freeradbiomed.2004.02.075. [DOI] [PubMed] [Google Scholar]
- 18.Tanaka Y, Ikeda T, Yamamoto K, Ogawa H, Kamisako T. Dysregulated expression of fatty acid oxidation enzymes and iron-regulatory genes in livers of Nrf2-null mice. J Gastroenterol Hepatol. 2012;27:1711–1717. doi: 10.1111/j.1440-1746.2012.07180.x. [DOI] [PubMed] [Google Scholar]
- 19.Tanaka Y, Aleksunes LM, Yeager RL, et al. NF-E2-related factor 2 inhibits lipid accumulation and oxidative stress in mice fed a high-fat diet. J Pharmacol Exp Ther. 2008;325:655–664. doi: 10.1124/jpet.107.135822. [DOI] [PubMed] [Google Scholar]
- 20.Sugimoto H, Okada K, Shoda J, et al. Deletion of nuclear factor-E2-related factor-2 leads to rapid onset and progression of nutritional steatohepatitis in mice. Am J Physiol Gastrointest Liver Physiol. 2010;298:G283–G294. doi: 10.1152/ajpgi.00296.2009. [DOI] [PubMed] [Google Scholar]
- 21.Shin S, Wakabayashi J, Yates MS, et al. Role of Nrf2 in prevention of high-fat diet-induced obesity by synthetic triterpenoid CDDO-imidazolide. Eur J Pharmacol. 2009;620:138–144. doi: 10.1016/j.ejphar.2009.08.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Kay HY, Kim WD, Hwang SJ, et al. Nrf2 inhibits LXRα-dependent hepatic lipogenesis by competing with FXR for acetylase binding. Antioxid Redox Signal. 2011;15:2135–2146. doi: 10.1089/ars.2010.3834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Itoh K, Chiba T, Takahashi S, et al. An Nrf2/small Maf heterodimer mediates the induction of phase II detoxifying enzyme genes through antioxidant response elements. Biochem Biophys Res Commun. 1997;236:313–322. doi: 10.1006/bbrc.1997.6943. [DOI] [PubMed] [Google Scholar]
- 24.Folch J, Lee M, Sloane Stanley GH. A simple method for the isolation and purification of total lipids from animal tissues. J Biol Chem. 1957;226:497–509. [PubMed] [Google Scholar]
- 25.Bohan A, Chen WS, Denson LA, Held MA, Boyer JL. Tumor necrosis factor alpha-dependent up-regulation of Lrh-1 and Mrp3(Abcc3) reduces liver injury in obstructive cholestasis. J Biol Chem. 2003;278:36688–36698. doi: 10.1074/jbc.M304011200. [DOI] [PubMed] [Google Scholar]
- 26.Lin X, Chen Z, Yue P, et al. A targeted apoB38.9 mutation in mice is associated with reduced hepatic cholesterol synthesis and enhanced lipid peroxidation. Am J Physiol Gastrointest Liver Physiol. 2006;290:G1170–G1176. doi: 10.1152/ajpgi.00402.2005. [DOI] [PubMed] [Google Scholar]
- 27.Ballatori N, Fang F, Christian WV, Li N, Hammond CL. Ostalpha-Ostbeta is required for bile acid and conjugated steroid disposition in the intestine, kidney, and liver. Am J Physiol Gastrointest Liver Physiol. 2008;295:G179–G186. doi: 10.1152/ajpgi.90319.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kim I, Ahn SH, Inagaki T, et al. Differential regulation of bile acid homeostasis by the farnesoid X receptor in liver and intestine. J Lipid Res. 2007;48:2664–2672. doi: 10.1194/jlr.M700330-JLR200. [DOI] [PubMed] [Google Scholar]
- 29.Oiwa A, Kakizawa T, Miyamoto T, et al. Synergistic regulation of the mouse orphan nuclear receptor SHP gene promoter by CLOCK-BMAL1 and LRH-1. Biochem Biophys Res Commun. 2007;353:895–901. doi: 10.1016/j.bbrc.2006.12.131. [DOI] [PubMed] [Google Scholar]
- 30.Inoue Y, Yu AM, Yim SH, et al. Regulation of bile acid biosynthesis by hepatocyte nuclear factor 4alpha. J Lipid Res. 2006;47:215–227. doi: 10.1194/jlr.M500430-JLR200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wiwi CA, Gupte M, Waxman DJ. Sexually dimorphic P450 gene expression in liver-specific hepatocyte nuclear factor 4alpha-deficient mice. Mol Endocrinol. 2004;18:1975–1987. doi: 10.1210/me.2004-0129. [DOI] [PubMed] [Google Scholar]
- 32.Miyata M, Tozawa A, Otsuka H, et al. Role of farnesoid X receptor in the enhancement of canalicular bile acid output and excretion of unconjugated bile acids: a mechanism for protection against cholic acid-induced liver toxicity. J Pharmacol Exp Ther. 2005;312:759–766. doi: 10.1124/jpet.104.076158. [DOI] [PubMed] [Google Scholar]
- 33.Teng S, Piquette-Miller M. The involvement of the pregnane X receptor in hepatic gene regulation during inflammation in mice. J Pharmacol Exp Ther. 2005;312:841–848. doi: 10.1124/jpet.104.076141. [DOI] [PubMed] [Google Scholar]
- 34.Matsushima S, Maeda K, Hayashi H, et al. Involvement of multiple efflux transporters in hepatic disposition of fexofenadine. Mol Pharmacol. 2008;73:1474–1483. doi: 10.1124/mol.107.041459. [DOI] [PubMed] [Google Scholar]
- 35.Horton JD, Shimomura I. Sterol regulatory element-binding proteins: activators of cholesterol and fatty acid biosynthesis. Curr Opin Lipidol. 1999;10:143–150. doi: 10.1097/00041433-199904000-00008. [DOI] [PubMed] [Google Scholar]
- 36.Millatt LJ, Bocher V, Fruchart JC, Staels B. Liver X receptors and the control of cholesterol homeostasis: potential therapeutic targets for the treatment of atherosclerosis. Biochim Biophys Acta. 2003;1631:107–118. doi: 10.1016/s1388-1981(02)00366-9. [DOI] [PubMed] [Google Scholar]
- 37.Repa JJ, Berge KE, Pomajzl C, Richardson JA, Hobbs H, Mangelsdorf DJ. Regulation of ATP-binding cassette sterol transporters ABCG5 and ABCG8 by the liver X receptors alpha and beta. J Biol Chem. 2002;277:18793–18800. doi: 10.1074/jbc.M109927200. [DOI] [PubMed] [Google Scholar]
- 38.Repa JJ, Liang G, Ou J, et al. Regulation of mouse sterol regulatory element-binding protein-1c gene (SREBP-1c) by oxysterol receptors, LXRalpha and LXRbeta. Genes Dev. 2000;14:2819–2830. doi: 10.1101/gad.844900. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zhang X, Liu J, Su W, et al. Liver X receptor activation increases hepatic fatty acid desaturation by the induction of SCD1 expression through an LXRα-SREBP1c-dependent mechanism J Diabetes 2013. DOI: 10.1111/1753-0407.12081 [DOI] [PubMed] [Google Scholar]
- 40.Zhou J, Febbraio M, Wada T, et al. Hepatic fatty acid transporter Cd36 is a common target of LXR, PXR, and PPARgamma in promoting steatosis. Gastroenterology. 2008;134:556–567. doi: 10.1053/j.gastro.2007.11.037. [DOI] [PubMed] [Google Scholar]
- 41.Weerachayaphorn J, Mennone A, Soroka CJ, et al. Nuclear factor-E2-related factor 2 is a major determinant of bile acid homeostasis in the liver and intestine. Am J Physiol Gastrointest Liver Physiol. 2012;302:G925–G936. doi: 10.1152/ajpgi.00263.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Huang J, Tabbi-Anneni I, Gunda V, Wang L. Transcription factor Nrf2 regulates SHP and lipogenic gene expression in hepatic lipid metabolism. Am J Physiol Gastrointest Liver Physiol. 2010;299:G1211–G1221. doi: 10.1152/ajpgi.00322.2010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Peet DJ, Turley SD, Ma W, et al. Cholesterol and bile acid metabolism are impaired in mice lacking the nuclear oxysterol receptor LXR alpha. Cell. 1998;93:693–704. doi: 10.1016/s0092-8674(00)81432-4. [DOI] [PubMed] [Google Scholar]
- 44.Vollrath V, Wielandt AM, Iruretagoyena M, Chianale J. Role of Nrf2 in the regulation of the Mrp2 (ABCC2) gene. Biochem J. 2006;395:599–609. doi: 10.1042/BJ20051518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Weerachayaphorn J, Cai SY, Soroka CJ, Boyer JL. Nuclear factor erythroid 2-related factor 2 is a positive regulator of human bile salt export pump expression. Hepatology. 2009;50:1588–1596. doi: 10.1002/hep.23151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Adachi T, Nakagawa H, Hagiya Y, Yasuoka T, Ishikawa T. Transport-metabolism interplay: LXRalpha-mediated induction of human ABC transporter ABCC2 (cMOAT/MRP2) in HepG2 cells. Mol Pharm. 2009;6:1678–1688. doi: 10.1021/mp9001156. [DOI] [PubMed] [Google Scholar]


