Abstract
The endoplasmic reticulum (ER) forms highly organized network structures composed of tubules and cisternae. Many plant species develop additional ER-derived structures, most of which are specific for certain groups of species. In particular, a rod-shaped structure designated as the ER body is produced by plants of the Brassicales order, which includes Arabidopsis thaliana. Genetic analyses and characterization of A. thaliana mutants possessing a disorganized ER morphology or lacking ER bodies have provided insights into the highly organized mechanisms responsible for the formation of these unique ER structures. The accumulation of proteins specific for the ER body within the ER plays an important role in the formation of ER bodies. However, a mutant that exhibits morphological defects of both the ER and ER bodies has not been identified. This suggests that plants in the Brassicales order have evolved novel mechanisms for the development of this unique organelle, which are distinct from those used to maintain generic ER structures. In A. thaliana, ER bodies are ubiquitous in seedlings and roots, but rare in rosette leaves. Wounding of rosette leaves induces de novo formation of ER bodies, suggesting that these structures are associated with resistance against pathogens and/or herbivores. ER bodies accumulate a large amount of β-glucosidases, which can produce substances that potentially protect against invading pests. Biochemical studies have determined that the enzymatic activities of these β-glucosidases are enhanced during cell collapse. These results suggest that ER bodies are involved in plant immunity, although there is no direct evidence of this. In this review, we provide recent perspectives of ER and ER body formation in A. thaliana, and discuss clues for the functions of ER bodies. We highlight defense strategies against biotic stress that are unique for the Brassicales order, and discuss how ER structures could contribute to these strategies.
Keywords: endoplasmic reticulum, ER body, organelle biogenesis, β-glucosidase, plant defenses, secondary metabolites, glucosinolate
Introduction
The endoplasmic reticulum (ER) forms highly organized network structures composed of ER tubules and ER cisternae. In addition to this well-conserved ER network, different plant species develop unique ER-derived compartments that can be regarded as ER domains. Many of the ER-derived compartments accumulate specific types of proteins, such as the protein bodies (PBs) in maize and rice, which contain prolamin and zein, respectively (Herman and Larkins, 1999), the KDEL-tailed protease-accumulating vesicles (KVs) in mungbean (Toyooka et al., 2000), and the ricinosomes in castor bean that accumulate papain-type proteases (Schmid et al., 2001). These structures are thought to function as repositories of particular proteins until they are required.
In this review, we focus on what is called the ER body, also known as a fusiform body. This structure is an ER domain of unique shape and taxonomic distribution. In 1965, the ER body was first discovered in root epidermal and cortical cells of radish (Bonnett and Newcomb, 1965). It was described as dilated cisternae that had luminal continuity to the ER. During the following 15 years, researchers tried to characterize these structures and determine their functions (Iversen and Flood, 1969; Iversen, 1970a; Cresti et al., 1974; Hoefert, 1975; Endress and Sjolund, 1976; Jørgensen et al., 1977; Behnke and Eschlbeck, 1978; Gailhofer et al., 1979; Jørgensen, 1981). Three independent studies revealed that the dilated cisternae were primarily restricted to species of the order Brassicales, which were known to produce thioglucosides named glucosinolates (Iversen, 1970a; Behnke and Eschlbeck, 1978; Jørgensen, 1981). Given that glucosinolates required a specific enzyme called myrosinase to become active, this correlation implied that the dilated cisternae could act as a myrosinase repository. Indeed, activity-labeled transmission electron microscopy revealed that the dilated cisternae contained potential myrosinase activity toward sinigrin (Iversen, 1970b), although this experimental system was subsequently questioned (Behnke and Eschlbeck, 1978). The question as to whether the dilated cisternae were involved in glucosinolate metabolism was left unsolved for decades, possibly due to a lack of appropriate molecular tools. In 1998, after several analyses using the model plant A. thaliana, it was suggested that the rod-shaped structures labeled with ER-localized green fluorescent protein (GFP) were equivalent to the dilated cisternae (Gunning, 1998). Hayashi et al. (2001) showed that these rod-shaped structures resembled the dilated cisternae described in past literature, and designated them as ER bodies. Matsushima et al. (2004) isolated the nai1 mutant that lacked ER bodies (“nai” is a Japanese word for “absence”), which was the very first identification of genetic material in ER body research. Since then, genetic and biochemical studies have provided insights into the functions, importance, and biogenesis of this unique organelle, which will be summarized in this review.
Although most of ER-derived compartments form spherical structures, ER bodies have a rod shape. The constitutive presence of ER bodies in A. thaliana is strictly limited to roots in adult plants; they are absent in most cells in rosette leaves. Both wounding and jasmonic acid treatment induce de novo formation of ER bodies in rosette leaves. In combination with their unique shape, one can presume a tightly regulated and unique mechanism for ER body formation, which is discussed in the first section. The tissue specificity and the inducibility also suggest that ER bodies are involved in the response to wounding and some biotic stresses, as discussed in the second section. In the last section, we will discuss how ER bodies are conserved and evolved in the plant kingdom. Current research indicates that ER bodies are specific to the order Brassicales, especially to the families Brassicaceae, Capparaceae, and Cleomaceae. It is known that Brassicales plants have unique defense strategies against biotic stresses, which may lead to an interesting evolutionary story that includes ER bodies.
Molecular mechanisms underlying ER body formation
ER body is a subdomain of ER that has specific organization mechanism
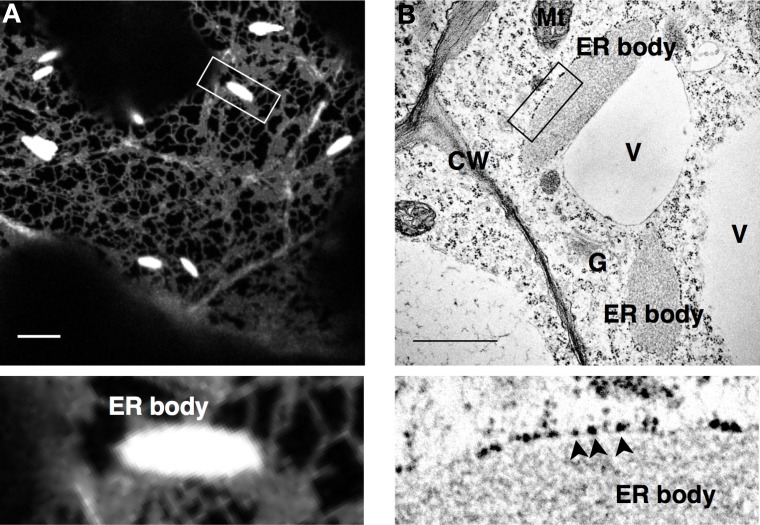
The ER body in A. thaliana was first observed in a transgenic line (GFP-h) expressing ER-targeted GFP. In the cotyledons of GFP-h, bright rod-shaped structures of ER bodies were observed in addition to the generic ER network (Figure 1A; Ridge et al., 1999; Hawes et al., 2001; Hayashi et al., 2001). Electron microscopy analysis revealed that the ER body is covered by a single membrane surrounded by ribosomes, which is a characteristic of the ER (Figure 1B). The ER body is observed as a structure that is connected with ER tubules and/or ER cisternae in electron micrographs (Gunning, 1998; Hayashi et al., 2001). These results indicate that the ER body is continuous to the whole ER network; therefore, it is suggested to be a subdomain of the ER that has specific functions.
Figure 1.
ER bodies in Arabidopsis thaliana. A confocal micrograph (A) and an electron micrograph (B) of cotyledon and root epidermal cells, respectively, of A. thaliana. Arrowheads indicate ribosomes on the surface of the ER body membranes. ER-localized GFP (SP-GFP-HDEL) labels ER bodies as well as the typical ER network, and electron microscopy identifies ribosomes at the cytosolic surface of ER bodies, both of which indicate the luminal continuity between ER and ER bodies. Enlarged images of the squared regions are shown below. CW, cell wall; V, vacuole; Mt, mitochondrion; G, Gold body; Bars, 10 μm (A) and 1 μm (B).
Plants develop several ER-derived structures that are thought to specifically function for protein storage in specific organs or during specific life stages. These include PBs and precursor-accumulating (PAC) vesicles in maturating seeds, KVs and ricinosomes in germinating seeds, and ER bodies in roots and seedlings. PBs accumulate seed-storage proteins in monocot plants such as rice (Oryza sativa) and maize (Zea mays) (Herman and Larkins, 1999). PAC vesicles accumulate precursors of seed-storage proteins in dicot plants such as pumpkin (Cucubita maxima) and castor bean (Ricinus communis), which undergo bulk transport to the protein-storage vacuole (Hara-Nishimura et al., 1998). KVs and ricinosomes accumulate papain-type proteases for the degradation of seed-storage materials in the cotyledon of mungbean (Vigna mungo) or endosperm of castor bean, respectively (Toyooka et al., 2000; Schmid et al., 2001). These vesicles are spherical with diameters of 0.5–1.0 μm and are surrounded by ribosomes, indicating that they are derived from the ER. Compared to these ER-derived vesicles, the ER body is longer and larger (~10 μm long and ~1 μm wide), and accumulates different kinds of proteins, namely the β-glucosidases. Therefore, the ER body is presumably different both in function and in biogenesis mechanisms from other ER-derived structures.
β-glucosidase is the major component of ER body
The major protein component of ER body in A. thaliana is identified as a β-glucosidase called PYK10/BGLU23 (Matsushima et al., 2003). This β-glucosidase was identified by analysis of the nai1 mutant in which ER bodies were absent. PYK10 is actively recruited from the ER network to ER bodies (Matsushima et al., 2003), in contrast to that for generic ER luminal proteins such as SP-GFP-HDEL. The level of PYK10 protein is high enough to be detected as one of the most abundant proteins in A. thaliana roots (Matsushima et al., 2003). Electron microscopy analysis revealed a relatively high electron density in the ER body lumen, suggesting that the ER body contains a large amount of proteins (Gunning, 1998; Hayashi et al., 2001). Consistent with this, ER bodies are enriched in a relatively heavier fraction after a centrifugation-based fractionation of subcellular compartments (Matsushima et al., 2003), which confirms a high protein density within ER bodies. These studies of A. thaliana suggest that the ER body is a vessel for β-glucosidases to separate them from other proteins and substrates. Indeed, when cells are disrupted, PYK10 forms a huge protein complex that contains proteins originating from various subcellular compartments (Nagano et al., 2005, 2008). The A. thaliana genome encodes 47 β-glucosidases within 10 subfamilies (Xu et al., 2004), most of which (40 out of 47) possess signal peptides at their amino-termini, indicating localization in the endomembrane system or secretion to the apoplast (Table 1). All members of the subfamily that includes PYK10 and additional 7 β-glucosidases (BGLU18−25) also contain ER retention signals ([K/H/R][D/E]EL) at their carboxy-termini. Based on their sequence similarity, it can be assumed that these 8 β-glucosidases localize in ER bodies. Consistent with this, the subfamily member BGLU18 accumulates in ER bodies that are induced by wounding in rosette leaves (Ogasawara et al., 2009).
Table 1.
A summary of 47 β-glucosidases encoded in the A. thaliana genome.
| Subfamily§ | Alias | Locus | Signal peptide† | ER retention | Proton donor‡ | Proton acceptor‡ | Aglycone binding‡ | Expression¶ | Notes |
|---|---|---|---|---|---|---|---|---|---|
| 1 | BGLU1 | At1g45191 | + | − | E | E | G | − | − |
| BGLU2 | At5g16580 | − | − | E | E | G | − | ||
| BGLU3 | At4g22100 | + | − | E | E | G | Maturing seeds (late) | − | |
| BGLU4 | At1g60090 | + | − | E | E | G | Very low in all tissues | − | |
| BGLU5 | At1g60260 | + | − | E | E | G | − | − | |
| BGLU6 | At1g60270 | + | − | E | E | G | Aerial tissues except for reproductive tissues | − | |
| BGLU7 | At3g62740 | + | − | E | E | G | Seedlings and roots | − | |
| BGLU8 | At3g62750 | + | − | E | E | A | − | − | |
| BGLU9 | At4g27820 | + | − | E | E | G | Ubiquitous | − | |
| BGLU10 | At4g27830 | + | − | E | E | G | Ubiquitous | − | |
| BGLU11 | At1g02850 | + | − | E | E | G | Reproductive tissues and roots | − | |
| 2 | BGLU12 | At5g42260 | + | − | E | E | Q | − | − |
| BGLU13 | At5g44640 | + | − | E | E | Q | − | − | |
| BGLU14 | At2g25630 | + | − | E | − | Q | Pollen | − | |
| BGLU15 | At2g44450 | + | − | E | E | Q | Maturing seeds (early), flowers, and roots | − | |
| BGLU16 | At3g60130 | + | − | E | E | E | Ubiquitous | − | |
| BGLU17 | At2g44480 | − | − | E | E | L | Maturing seeds (late) and roots | − | |
| 3 | BGLU18 | At1g52400 | + | + | E | E | A | Very high; aerial tissues | Induced by wounding; involved in ABA signaling |
| BGLU19 | At3g21370 | + | + | E | E | S | Maturing seeds (late) | − | |
| BGLU20 | At1g75940 | + | + | E | E | A | Flowers | − | |
| BGLU21 | At1g66270 | + | + | E | E | A | − | − | |
| BGLU22 | At1g66280 | + | + | E | E | A | − | − | |
| BGLU23/PYK10 | At3g09260 | + | + | E | E | A | Seedlings and roots | ER body component | |
| BGLU24 | At5g28510 | + | + | E | E | A | Very low in all tissues | − | |
| BGLU25/GLUC | At3g03640 | + | + | E | G | G | Ubiquitous | − | |
| 4 | BGLU26/PEN2 | At2g44490 | − | − | E | E | A | Ubiquitous, except for maturing seeds | Atypical myrosinase; required for resistance against powdery mildew |
| BGLU27 | At3g60120 | − | − | E | E | G | Very low in all tissues | − | |
| 5 | BGLU28 | At2g44460 | + | − | E | E | A | Weakly in reproductive tissues and roots | − |
| BGLU29 | At2g44470 | + | − | E | E | A | Maturing seeds | − | |
| BGLU30/DIN2/SRG2 | At3g60140 | + | − | E | E | A | Maturing seeds and roots | − | |
| BGLU31 | At5g24540 | + | − | E | E | S | Very low in all tissues | ||
| BGLU32 | At5g24550 | + | − | E | E | S | Very low in all tissues | ||
| 6 | BGLU33 | At2g32860 | + | − | E | E | A | Ubiquitous in vegetative tissues | Involved in ABA signaling |
| 7 | BGLU34/TGG4 | At1g47600 | + | − | Q | E | K | − | Myrosinase in roots; resistance against insects? |
| BGLU35/TGG5 | At1g51470 | + | − | Q | E | K | − | Myrosinase in roots; resistance against insects? | |
| BGLU36/TGG6 | At1g51490 | − | − | Q | P | K | Pollen | Myrosinase; pseudogene | |
| BGLU37/TGG2 | At5g25980 | + | − | Q | E | R | − | Myrosinase in leaves; resistance against insects? | |
| BGLU38/TGG1 | At5g26000 | + | − | Q | E | R | − | Myrosinase in leaves; resistance against insects? | |
| BGLU39/TGG3 | At5g48375 | + | − | Q | E | R | − | Myrosinase; pseudogene | |
| 8 | BGLU40 | At1g26560 | + | − | E | E | Q | Ubiquitous except for roots | − |
| BGLU41 | At5g54570 | + | − | E | E | Q | Reproductive tissues | − | |
| BGLU42 | At5g36890 | − | − | E | E | N | Ubiquitous except for maturing seeds | − | |
| 9 | BGLU43 | At3g18070 | + | − | E | E | L | Very low in all tissues | − |
| BGLU44 | At3g18080 | + | − | E | E | L | Ubiquitous | − | |
| 10 | BGLU45 | At1g61810 | + | − | E | E | L | Stems and maturing seeds | − |
| BGLU46 | At1g61820 | + | − | E | E | L | Maturing seeds and roots | − | |
| BGLU47 | At4g21760 | − | − | E | E | L | Leaves | − |
Xu et al., 2004.
SignalP (http://www.cbs.dtu.dk/services/SignalP/).
Burmeister et al., 1997.
ATTED-II (http://atted.jp/).
NAI2 and PYK10: ER body components required for ER body formation
The protein factors involved in ER body formation in A. thaliana have been identified by analysis of the two mutants nai2 and long ER body (leb). The recessive nai2 mutant lacks ER bodies in seedlings and roots (Yamada et al., 2008). NAI2 accumulates in ER bodies but not in the ER network, indicating that NAI2 is an ER body component that determines ER body formation in A. thaliana. In the absence of NAI2, PYK10 is diffused throughout the ER network, and the protein levels are lower compared to those for wild type (WT). These results show that NAI2 enables the high accumulation and storage of PYK10, presumably by mediating production of the ER body. Based on sequence similarity, NAI2 homologs are only observed in Brassicaceae plants that produce ER bodies, further suggesting that NAI2 has a specific function in the generation of this organelle.
In leb mutant seedlings, there are fewer ER bodies and they have an elongated shape compared to those in WT seedlings (Nagano et al., 2009). A mutation in the PYK10 gene that causes an amino acid substitution is responsible for the leb phenotype, revealing the importance of PYK10 localization in the ER body for ER body formation. PYK10 forms an oligomer linked by a disulfide bond (Nagano et al., 2005), presumably via a cysteine residue that is substituted in the leb mutant, because the mutation resulted in an altered oligomeric structure and reduced accumulation of PYK10 protein (Nagano et al., 2009). The pyk10 single knockout mutant exhibited milder phenotype than that of the leb mutant, indicating that the leb mutation affects other ER body components that contribute to proper ER body organization. BGLU21 and BGLU22 are the two closest homologs of PYK10, which might contribute to ER body formation. A double knockout mutant pyk10 bglu21 showed similar phenotype to the leb mutant, indicating that BGLU21 is involved in ER body formation. The PYK10 mutation affects the nature of BGLU21 protein such as oligomeric states and/or protein conformation and induces the formation of aberrant ER bodies. These data suggest that PYK10, BGLU21, and perhaps BGLU22 are redundantly important for proper organization of this organelle in A. thaliana seedlings. The redundancy between BGLU21 and BGLU22 is also suggested by the fact that these two proteins are more similar to each other than to the closest homolog in Arabidopsis lyrata.
ER body membrane contains specific proteins
The membrane of ER bodies may have an important role in mediating the function and/or formation of ER bodies. Two integral membrane proteins, designated as MEMBRANE OF ER BODY (MEB) 1 and MEB2, have been identified to accumulate specifically at ER body membranes in A. thaliana (Yamada et al., 2013). These proteins were identified by coexpression analysis based on a public microarray database (ATTED-II; http://atted.jp) and transcriptomic analysis using the nai1 mutant. MEB1 and MEB2 are homologous to each other with multiple membrane-spanning regions. They have weak similarity to Ccc1p and VIT1, an iron/manganese transporter in S. cerevisiae and A. thaliana, respectively (Li et al., 2001; Kim et al., 2006). MEB1 and MEB2 appear to possess metal transporter activity (discussed below), although their physiological role for plant fitness is still unknown (Yamada et al., 2013). MEB1 and MEB2 form a protein complex with NAI2, and are diffused throughout the ER network in the nai2 mutant, suggesting the NAI2-dependent recruitment of MEB1 and MEB2 into ER body membranes. ER bodies in the seedlings of the meb1 meb2 double mutant exhibited a comparable number and shape to those in WT seedlings, suggesting that these proteins are not necessary for ER body formation. However, these results indicate that the ER body membrane has a specific composition of proteins that differs from that of the ER network, and suggest that NAI2 is responsible for the organization of these ER body membrane proteins. Because NAI2 alone regulates ER body formation, the NAI2-dependent specification of the membrane by gathering specific proteins may have a crucial role for the biogenesis of ER bodies.
NAI1: the transcription factor for ER body formation
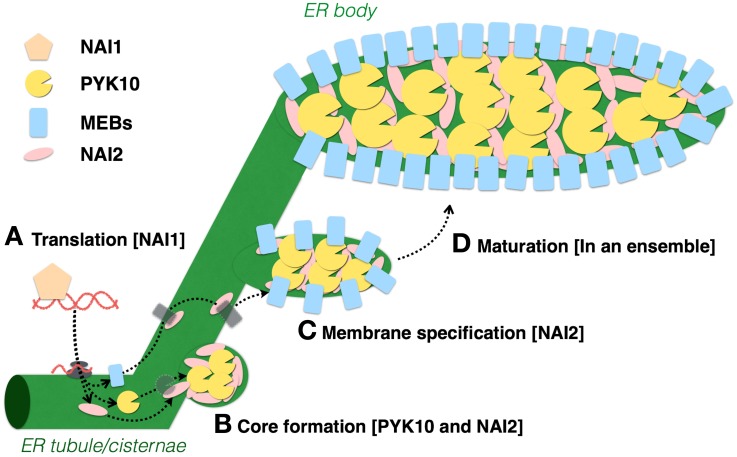
The basic helix-loop-helix- (bHLH) type transcription factor NAI1 (also known as AtbHLH20) solely regulates ER body formation, because disruption of this gene in A. thaliana completely disrupts ER body formation in seedlings and roots (Matsushima et al., 2004). A transcriptomic analysis of the nai1 mutant revealed that ER body proteins, such as those encoded by PYK10, NAI2, MEB1, and MEB2, are expressed in a NAI1-dependent manner. This indicates that NAI1 is the master regulator for ER body formation, and it regulates the expression of these genes. NAI1 also regulates the expression of JACALIN-RELATED LECTIN genes (JAL22, JAL23, JAL31, and PYK10 BINDING PROTEIN 1 (PBP1)/(JAL30), GDSL LIPASE-LIKE PROTEIN genes (GLL23 and GLL25), and MD2-RELATED LIPID RECOGNITION PROTEIN 3 (ML3) (Nagano et al., 2005, 2008; Hakenjos et al., 2013). JAL proteins lack signal peptides and are assumed to localize in the cytosol (Nagano et al., 2005), whereas GLL25 and ML3 have signal peptides and accumulate in vacuoles (Nakano et al., 2012; Hakenjos et al., 2013). JAL and GLL proteins form a large protein complex with PYK10 when cells are collapsed or disrupted (Nagano et al., 2005, 2008; Ahn et al., 2010), suggesting their functional link to PYK10 and ER bodies. Current research indicates that there are no morphological disorders of ER bodies when these proteins are depleted, indicating their lack of importance in ER body formation. Figure 2 shows a proposed model of how the proteins are coordinated during ER body formation. Because a defect of either NAI2 or PYK10 causes disorganized ER body formation, these proteins are important for shaping ER bodies. It is possible that these proteins interact with each other to condense materials for ER bodies, followed by the NAI2-dependent recruitment of MEB1 and MEB2 to the pre-ER body membrane to form the ER bodies.
Figure 2.
Models of ER body formation in A. thaliana seedlings. (A) NAI1 is expressed in epidermal cells to regulate ER body formation by inducing the expression of PYK10, NAI2, MEB1, and MEB2. (B) PYK10 and NAI2 may physically interact to form ER body structures from the ER. (C,D) NAI2 forms a complex with MEB1and MEB2, then recruits them to the ER body to form the ER body-specific membrane.
Relationship of ER network formation and ER body formation
The ER body is derived from the ER network, and some ER proteins also localize in ER bodies, and thus it is possible that the organization of ER and ER bodies is coregulated by similar molecular mechanisms. Understanding how ER networks are formed and maintained provides insights into ER body organization. The molecular basis underlying ER morphology has been intensely studied for more than a decade using a variety of eukaryotic cells including mammals, yeasts, and plants such as A. thaliana and Nicotiana benthamiana (reviewed in Chen et al., 2013). It has been shown that ER network structures are determined by a series of membrane proteins [i.e., tubule-associated proteins such as reticulon family proteins (RTNs), Atlastin/ROOT HAIR DEFECTIVE 3 (RHD3) GTPases, and cisternae-associated proteins including CLIMP-63] (Nziengui et al., 2007; Sparkes et al., 2010; Tolley et al., 2010; Chen et al., 2011; Lee et al., 2011, 2013; Stefano et al., 2011; Zhang et al., 2013). Most studies of RTNs and Atlastin/RHD3 have been carried out using animal cells or N. benthamiana, which does not generate ER bodies. Therefore, it remains unclear how the plants developing ER bodies utilize these conserved mechanisms to generate these unique structures. Because reticulons and other similar proteins prefer localizing to the high curvature of ER membranes to retain a tubular structure, it is possible that these proteins are excluded from the ER bodies that have lower membrane curvature compared to that of the tubules. Alternatively, they might interact with ER body-specific proteins such as MEB1 and MEB2 on the ER body membranes, and contribute to its unique curvatures. In this context, the localization of RTNs and RHD3 in A. thaliana is of great interest.
In A. thaliana, a series of mutants designated as endoplasmic reticulum morphology (ermo) mutants have been isolated in a forward-genetic screen to identify the factors responsible for maintaining ER morphology (Nakano et al., 2009, 2012). In the ermo1 and ermo2 mutants, the cells develop a number of ER-derived spherical bodies, ~1 μm in diameter, in addition to the typical ER network, whereas ermo3 cells develop huge aberrant aggregate structures derived from the ER network and ER bodies. ERMO1 and ERMO2 encode GNOM-LIKE 1 (GNL1) and SEC24a, respectively, which are involved in protein transport between ER and Golgi bodies. The unaffected morphology of ER bodies suggested that both GNL1 and SEC24a were dispensable for the proper organization of ER bodies (Faso et al., 2009; Nakano et al., 2009). ERMO3 encodes a GDSL lipase-like protein also known as MODIFIED VACUOLE PHENOTYPE1 (MVP1) or GOLGI DEFECTIVE36 (GOLD36), which localizes in vacuoles (Agee et al., 2010; Marti et al., 2010; Nakano et al., 2012). The ER aggregations in ermo3 are absent in cells that do not accumulate ER bodies, and are suppressed in whole tissues by the introduction of the nai1 mutation, suggesting a strong relationship between ERMO3 and ER bodies. The ER body structure is not significantly affected in ermo3 cells (Marti et al., 2010; Nakano et al., 2012), and ERMO3 does not interact with NAI2 (Nakano et al., 2012), suggesting that the contribution of ERMO3 to ER body formation is not highly significant. ERMO3 was required for proper protein transport between ER and Golgi bodies, and ERMO3 formed a protein complex with PYK10, JAL, and MATH domain-containing proteins, which are regulated by NAI1 (Nakano et al., 2012). These results suggest that PYK10, JAL, and MATH domain-containing proteins form a large protein aggregate on ER, then alter the ER morphology to inhibit protein secretion in the absence of ERMO3. ERMO3 may be responsible for solving this aggregate in ER, because the expression of ERMO3 in ER rescues the phenotype in the ermo3 mutant (Nakano et al., 2012). This tissue-specific requirement of ERMO3, in addition to the ubiquitous expression of ERMO3, clearly shows that each cell type requires its own regulatory systems to maintain their subcellular organization.
Physiological functions of ER bodies: putative association with innate immunity
Tissue specificity and wounding inducibility indicate the importance of ER body in plant defense
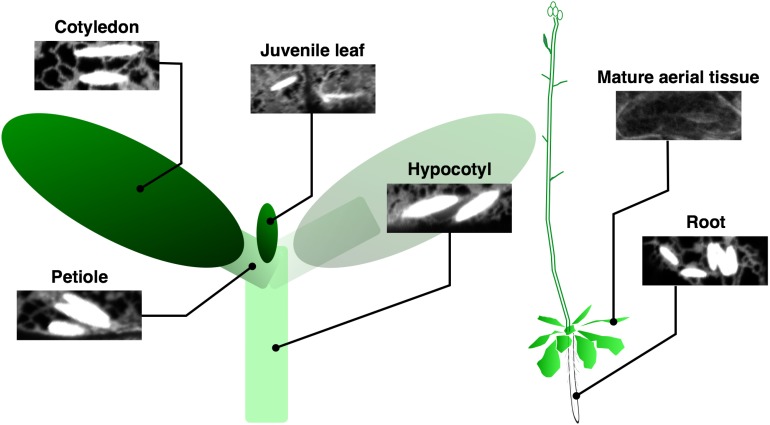
Constitutive ER bodies in A. thaliana are enriched in seedlings and roots of mature plants (Matsushima et al., 2002). In cotyledons, ER bodies develop only in epidermal cells but not in mesophyll cells. Similarly, no ER bodies are detected in the root vascular cylinder, in contrast to their presence in root epidermal, cortical, and endodermal layers (Figure 3). Shoot tissues including rosette and cauline leaves have fewer ER bodies compared to those of the underground tissues. They are absent in most of the rosette leaf cells, except for some epidermal cells along the primary and secondary veins or the edge of the leaves (Nakano et al., 2012). These specificities among plant tissues and cell types suggest that ER bodies are enriched at the interface between plants and surrounding organisms to protect plants from pathogens/herbivores that may enter or feed from the veins or leaf edge. This is supported by the fact that leaf wounding triggers local and systemic de novo formation of ER bodies in a jasmonic acid (JA)-dependent manner (Matsushima et al., 2002; Ogasawara et al., 2009). Atypical, elongated ER bodies are produced after wounding in the nai1 mutant, suggesting that another factor in addition to NAI1 is involved in this response (Matsushima et al., 2003). Consistent with this, the induction of BGLU18, a major component of wound-triggered ER bodies in rosette leaves, is independent of NAI1 (Ogasawara et al., 2009). The observed high abundance of ER bodies in roots may contribute to the interaction between plants and surrounding (potential) pathogens and herbivores that inhabit soil (Yamada et al., 2011). For example, the soil environment contains 106–109 bacteria per gram, which is much greater than in the atmosphere (101–105 per cubic meter) (Bulgarelli et al., 2013). Many other organisms such as insects, worms, nematodes, and fungi also live in the soil and seek opportunities to exploit plant roots.
Figure 3.
Tissue localization of ER bodies. ER bodies are present in the epidermis of whole tissues in young seedlings, but are absent in aerial organs of mature plants. Root epidermal, cortical, and endodermal cells continue to develop ER bodies. Epidermal cells of a transgenic A. thaliana expressing ER-localized GFP from several tissues are shown.
β-glucosidase activity of PYK10 may contribute to the plant immune response
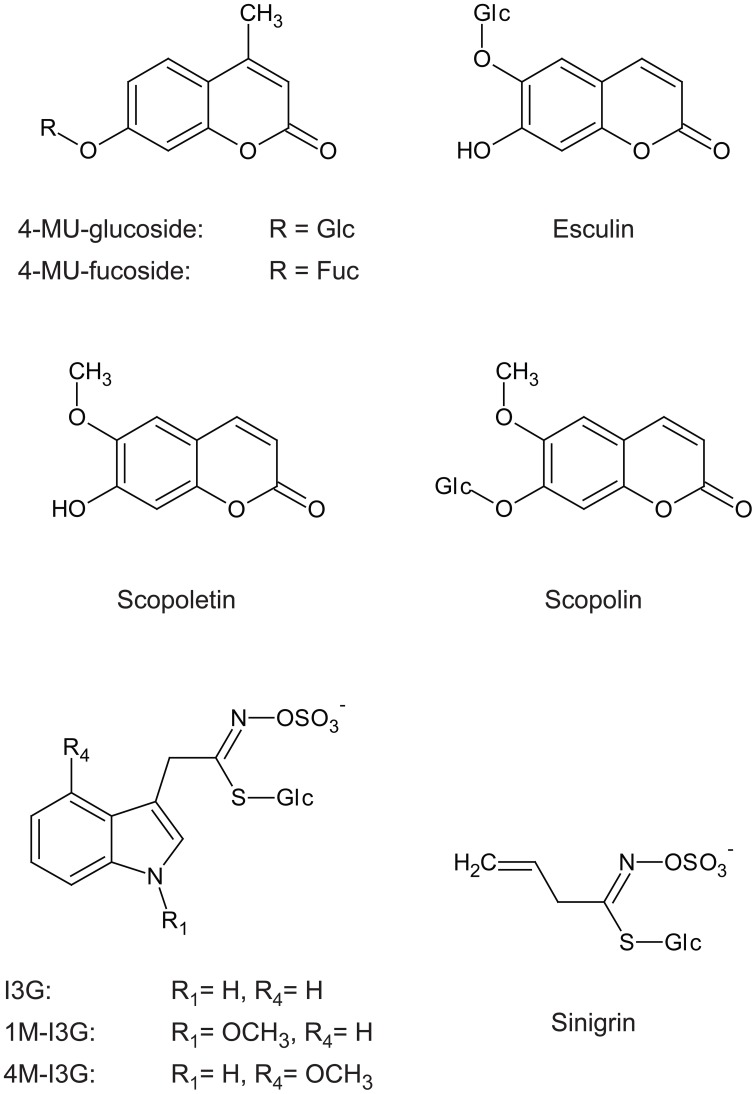
The high abundance of PYK10 and other β-glucosidases in ER bodies implies that the enzymatic activities of these proteins play an important role in the function of this organelle. The reported in vitro substrates of PYK10 include 4-methylumbelliferyl (4MU)-glucoside, 4MU-fucoside (Matsushima et al., 2004), scopolin, and esculin (Ahn et al., 2010). Recombinant PYK10 protein expressed in insect cells hydrolysed with the highest efficiency scopolin (Ahn et al., 2010), a coumarin widely occurring in the plant kingdom, including A. thaliana (Bednarek et al., 2005; Shimizu et al., 2005; Kai et al., 2006; Bayoumi et al., 2008). Scopolin is a β-O-glucoside of scopoletin, which is regarded as a phytoalexin. It was shown that scopoletin and scopolin could inhibit germ tube growth of Sclerotinia sclerotiorum (Prats et al., 2006). Scopoletin was also found to possess toxic activity against Fusarium oxysporum, Fusarium solani, Rhizopus stolonifer, and Lasiodiplodia theobromae (Peterson et al., 2003). Growth inhibition of F. oxysporum was higher with scopoletin than with scopolin, suggesting that a β-glucosidase could have an important role in this activity.
Concerning the biological activity of scopoletin, it is possible that ER bodies are involved in plant defense by hydrolysing scopolin to this aglycone. As discussed above, JAL proteins, GLL proteins, and MATH domain-containing proteins formed a large protein complex with PYK10 after cell disruption (Nagano et al., 2005, 2008; Ahn et al., 2010). These proteins accumulate constitutively in separate cellular organelles: PYK10 in ER body, GLLs in vacuole, JALs in cytosol, and MATHs at plasma membrane (Nagano et al., 2005; Oelmuller et al., 2005; Marti et al., 2010; Nakano et al., 2012). The β-glucosidase activity of PYK10 is higher after complex formation, suggesting that JALs and GLLs serve as activators of PYK10 (Nagano et al., 2005, 2008). Stimulation of β-glucosidase activity can be solely carried out by PBP1, as revealed by an in vitro enzymatic assay (Ahn et al., 2010). This fact, combined with wound inducibility, suggests that these proteins are assembled with PYK10 when cells are damaged by feeding insects or pathogen infection, and that they produce substances that potentially target these invaders.
PYK10 may have myrosinase activity toward indole glucosinolates
β-Glucosidases in the order Brassicales include a unique class of enzymes named myrosinases or β-thioglucoside glucohydrolases (TGGs), which are involved in the defense against insects, fungi, and bacteria (reviewed in Hopkins et al., 2009). Myrosinases are responsible for hydrolysing glucosinolates, thioglucosides that are specific to the order Brassicales (Halkier and Gershenzon, 2006). For a long time, myrosinases were considered to contain a unique amino acid signature that enables in silico prediction of their identity based on nucleotide sequence data (Burmeister et al., 1997). One of the unique features of this signature is a conserved basic residue (lysine or arginine) at the substrate pocket, which can form electrostatic interactions with the negatively charged sulfate group of glucosinolates (see Figure 5). It was found that the glutamate residue that serves as a proton donor in O-glucosidases is substituted by glutamine in myrosinases, resulting in the strict reduction of O-glucosidase activity (Burmeister et al., 1997). In A. thaliana, six genes (TGG1−6) including two pseudogenes [TGG3, (Zhang et al., 2002); TGG6, (Andersson et al., 2009)] encode myrosinases. TGG1 and TGG2 are expressed primarily in leaves (Xue et al., 1995), whereas TGG4 and TGG5 are expressed primarily in roots (Barth and Jander, 2006; Andersson et al., 2009; Zhou et al., 2012). These four myrosinase have conserved lysine/arginine and glutamine residues at the substrate pocket, and have activity against glucosinolates. By contrast, the other β-glucosidases lacking these specific amino acids were thought to be β-glucosidases hydrolysing O-glucosides but not thioglucosides (Rask et al., 2000). However, PEN2, a β-glucosidase lacking these key amino acids, was recently shown to be an atypical myrosinase hydrolysing indol-3-ylmethyl glucosinolate (I3G) and 4-methoxy-I3G (4M-I3G) (Bednarek et al., 2009). PEN2 has a major role in A. thaliana immunity via its myrosinase activity (reviewed in Bednarek, 2012) against various microbes including fungal pathogens such as Blumeria graminis and Plectosphaerella cucumerina (Sanchez-Vallet et al., 2010), Magnaporthe oryzae (Maeda et al., 2009), Leptosphaeria maculans (Elliott et al., 2008), Colletotrichum species (Hiruma et al., 2010), oomycetes [e.g., Phytophthora brassicae (Schlaeppi et al., 2010) and Pythium irregularum (Adie et al., 2007)], and a growth-promoting endophytic fungus (Piriformospora indica; Jacobs et al., 2011).
Figure 5.
Indole glucosinolate structure includes a side chain similar to that of scopolin and esculin, but not similar to that of sinigrin. I3G, indol-3-ylmethylglucosinolate; 1M-I3G, 1-methoxyindol-3-ylmethylglucosino late; 4M-I3G, 4-methoxyindol-3-ylmethylglucosinolate; 4-MU-glucoside, 4-methylumbelliferyl-β-D-glucoside; 4-MU-fucoside, 4-methylumbelliferyl-β-D-fucoside.
PEN2 does not belong to the subfamily of PYK10, but both proteins have high sequence similarity (Figure 4), which suggests that PYK10 also has atypical myrosinase activity, whereas recombinant PYK10 was unable to hydrolyse sinigrin (Ahn et al., 2010). Sinigrin, which represents aliphatic glucosinolates, consists of the glucosinolate core structure with a short aliphatic side chain that differs significantly in its structure from the indolyl group present in I3G and 4M-I3G (indole glucosinolates) (Figure 5). It cannot be excluded that PYK10 may hydrolyse indole glucosinolates in addition to its inactivity toward aliphatic glucosinolates. The reported substrates for PYK10 have aglycones of coumarinyl moieties, suggesting that the substrate pocket of PYK10 is suitable for condensed-ring moieties. This also supports the possible activity of PYK10 to hydrolyse indole glucosinolates, which have condensed-ring moieties as aglycones (Figure 5). According to Brown et al. (2003), indole glucosinolates are the most abundant glucosinolates in A. thaliana roots (at least of Col-0 accession), where ER bodies and PYK10 are also highly abundant. Coexpression analysis based on a public microarray database (ATTED-II; http://atted.jp) revealed a strong correlation between PYK10 and CYP81F4 encoding an enzyme involved in the modification of I3G into 1-methoxy-I3G (Pfalz et al., 2011), further suggesting a functional link between PYK10 and indole glucosinolates.
Figure 4.
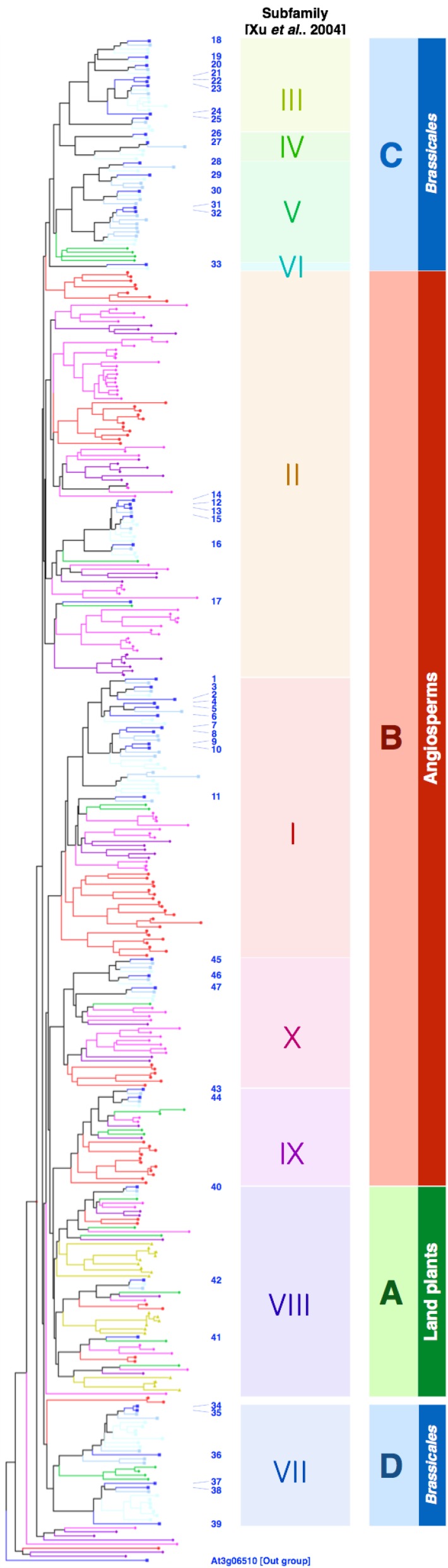
Both classical and atypical myrosinases form Brassicales-specific clades. The full-length protein sequences of β-glucosidases in A. thaliana (blue squares), Capusella rubella (light-blue squares), Thellungiela salsuginea (cyan squares), Carica papaya (green diamonds), Vitis vinifera (dark purple diamonds), Glycine max (magenta diamonds), Oryza sativa (red circles), and Physcomitrella patens (dark yellow triangles) were retrieved from the Phytozome database (http://www.phytozome.net/) and aligned by ClustalW (http://clustalw.ddbj.nig.ac.jp/index.php). BGLU numbers of the β-glucosidases in A. thaliana are indicated by blue letters. At3g06510 was used as an out group. Subfamilies proposed by Xu et al. (2004) are indicated accordingly.
PYK10 in mutualistic relationship with Piriformospora indica and defense against parasitic nematodes
It was reported that transgenic A. thaliana, which produced a novel type of glucosinolates, showed distinct bacterial and fungal communities in its rhizosphere compared to those of WT plants (Bressan et al., 2009). This result strongly suggests the involvement of glucosinolate in pathogenic and mutualistic plant microbe interactions in the soil environment. One of the most extensively studied root-colonizing microorganisms is an endophytic fungus P. indica that has a broad range of host species and a capacity to promote host-plant growth (Varma et al., 1999). Similar to the pen2 mutant, pyk10 and nai1 mutant plants failed to suppress P. indica growth at a preferable level, resulting in fungal overgrowth that stimulated undesirable immune responses and loss of the beneficial interaction (Sherameti et al., 2008; Jacobs et al., 2011). In addition, depletion of two cytochrome P450 enzymes (CYP79B2 and CYP79B3) that are responsible for conversion of tryptophan into indole-3-acetaldoxime (IAOx), the first step of the biosynthesis of indole glucosinolates, also resulted in overgrowth of P. indica (Nongbri et al., 2012). By contrast, the phytoalexin camalexin, which is derived from IAOx and is absent in the cyp79B2 cyp79B3 mutant, was dispensable for this suppression, suggesting that the proper control of this mutualistic fungus may involve indole glucosinolate metabolism. These results suggest that PYK10, PEN2, and indole glucosinolates have important roles in the establishment of the beneficial interaction with P. indica.
PYK10 was originally isolated as a root-specific gene with high expression level that became additionally elevated in nematode-infected tissue (Nitz et al., 2001). Because the degradation products of glucosinolates exhibited inhibitory activity against both a cyst nematode (Globodera rostochiensis; Buskov et al., 2002) and a root-knot nematode (Meloidogyne incognita; Lazzeri et al., 2004), it is possible that PYK10 protects roots from these parasites by its potential myrosinase activity. Cyst nematodes degrade the cell wall and infect intracellularly, whereas root-knot nematodes infect intercellularly (reviewed in Mitchum et al., 2012). In both cases, the cells surrounding infectious nematodes are disrupted and possibly collapse, which can passively trigger hydrolysis of glucosinolates. Alternatively, the plant may recognize the presence of these pests via nematode-associated molecular patterns or effectors injected into plant cells, and actively exert defensive glucosinolate metabolism.
Although both pen2 and pyk10 mutants showed greater P. indica colonization (Sherameti et al., 2008; Jacobs et al., 2011), there was no direct evidence that the control of fungal growth during this interaction was due to the myrosinase activity of both enzymes toward indole glucosinolates. Similarly, in addition to the inhibitory activity of glucosinolates against parasitic nematodes, it is not known if myrosinases are involved in these interactions. In addition to PEN2 and PYK10, A. thaliana roots express two classical myrosinases (TGG4 and TGG5), and TGG4 was shown to metabolize indole glucosinolates when overexpressed in planta (Bednarek et al., 2009). Future work will determine which of these four enzymes, and possibly other atypical myrosinases, are the key players during the interaction with endophytic fungi and parasitic nematodes.
Enhanced secretion of proteins localized in ER bodies (ESPER) is a novel immune response against pathogens
Recently, Watanabe et al. (2013b) reported a novel immune response designated as ESPER. They showed that two plant pathogenesis-related (PR) proteins, defensin (PDF1.2) and PR1, which contain antimicrobial activity, accumulated in ER bodies at a steady-state level and were secreted to the apoplast in response to the non-adapted pathogenic fungi Colletotrichum gloeosporioides. This suggests that ER bodies can serve as storage sites for various PR proteins until pathogenic attack, although these proteins have been reported to localize primarily in the vacuole or apoplast (Keefe et al., 1990; Dixon et al., 1991; Neuhaus et al., 1991; Van Loon et al., 2006; Sels et al., 2008). However, ER bodies can be labeled with GFP fused to proteins in the secretory pathway, including both soluble and membrane proteins, which finally accumulate in other organelles (Teh and Moore, 2007), suggesting the non-selective entry of these proteins into ER bodies. The localization of PDF1.2 and other PR proteins was analyzed using GFP-fused protein constructs driven by the artificial cauliflower mosaic virus (CaMV) 35S promoter (Watanabe et al., 2013b). Therefore, it is possible that the observed protein accumulation in ER bodies could have been an artefact due to gene overexpression. Nevertheless, the drastic change in localization of these GFP-fused PR proteins under fungal attack indicates that the membrane trafficking system in plant cells can dynamically change in response to the surrounding environment. Although the ER body itself is not involved in ESPER, as suggested by the presence of ER bodies even after ESPER occurred, this, in turn, suggests that localization of PYK10 and other ER body components may also dynamically respond to abiotic/biotic conditions to become active.
Potential ER body function in metal tolerance or uptake
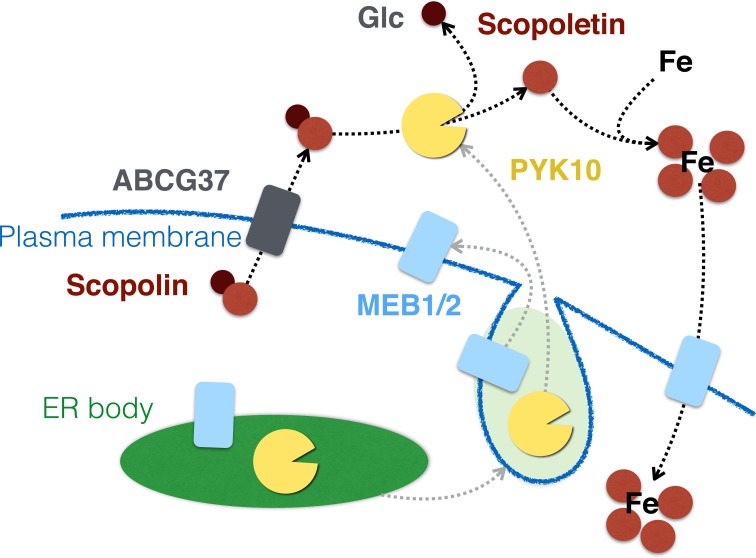
Recently, it was reported that iron starvation stimulated an ATP-binding cassette transporter ABCG37 (PDR9)-dependent secretion of scopoletin (Fourcroy et al., 2014). Roots of the pdr9 mutants that lacked functional ABCG37 failed to secrete scopoletin and showed abnormal accumulation of its derivatives, including scopolin, inside the tissue. These mutants also exhibited decreased tolerance to iron deficiency, suggesting that secretion of scopoletin helped plants to take up iron. This is further supported by the results that some of the coumarin derivatives have the potential to chelate iron and serve as phytosiderophores (Mladenka et al., 2010). Therefore, scopoletin, produced by the action of PYK10 on scopolin, may contribute to iron uptake in A. thaliana roots. Notably, MEB1 and MEB2, the ER body-specific membrane proteins, were recently suggested to be iron transporters that can endow S. cerevisiae with iron tolerance when expressed heterologously (Yamada et al., 2013). These results suggest that ER bodies have a role in metal-stress response apart from plant microbe interactions. One possibility is that under iron-limiting conditions, ER bodies fuse with plasma membrane to release PYK10 into the apoplast, which consequently hydrolyses scopolin that is secreted to the apoplast via ABCG37 (Figure 6). Liberated scopoletin may enhance the availability of iron, which can be taken up via MEB putative transporters that are transferred on the plasma membrane. Mugineic acid, a phytosiderophore in barley, accumulates in ER-derived vesicles peripheral to the plasma membrane of root epidermal cells, and can be secreted to the soil to improve the iron availability (Negishi et al., 2002). To date, there is no report elucidating how ER bodies behave in iron-depleted conditions. In addition, it should be noted that the experiments reported by Fourcroy et al. (2014) were performed with roots of plants grown on agar plates and exposed to light. The accumulation of phenylpropanoids (the group of metabolites that includes scopolin and scopoletin) is strongly upregulated in A. thaliana roots artificially exposed to light, as compared to roots grown in soil (Hemm et al., 2004), suggesting that in soil the scopoletin-dependent iron-uptake mechanism may have reduced significance. Therefore, it is of great interest to test ER body behavior, together with scopoletin production, in A. thaliana roots grown in iron-depleted soil, which may validate the function of ER bodies in metal tolerance or uptake.
Figure 6.
PYK10 and ER bodies may have a role in iron uptake via hydrolysing scopolin. ER bodies may fuse with the plasma membrane under iron-deficient conditions, resulting in the relocation of MEB1 and MEB2 to the plasma membrane and secretion of PYK10 to the apoplast. Scopolin, secreted in an ABCG37-dependent manner, is then converted to scopoletin by PYK10, which in turn helps the cells to take up iron via the chelating activity of scopoletin. This uptake may be carried out by putative MEB transporters.
Stress translation via phytohormone activation
BGLU18/AtBG1 and BGLU33/AtBG2, which belong to the same clade with PYK10 and PEN2 (see Figure 4, discussed below), were reported to be involved in abscisic acid (ABA) signaling and drought resistance by hydrolysing ABA-O-glucoside, which is an inactive pool of ABA (Lee et al., 2006; Han et al., 2012; Xu et al., 2012; Watanabe et al., 2013a). BGLU18 is a member of the PYK10 subfamily. According to the ATTED-II database, BGLU18 is expressed primarily in aerial organs such as leaves and stems, but is limited in roots, which negatively correlates with the presence of constitutive ER bodies. However, BGLU18 is a component of ER bodies that are induced by wounding (Ogasawara et al., 2009). BGLU18 is co-expressed with some homologs of genes related to constitutive ER bodies including TSA1, the closest homolog of NAI2, JAL genes (JAL23 and JAL35), and a GLL gene, strongly suggesting that the mechanisms of BGLU18 accumulation and activation could be similar to those reported for PYK10.
Presuming that ABA is activated through sugar release from ABA-glucoside in drought-stressed leaves, it could be speculated that BGLU18 is engaged in ABA-activation in a manner independent from both constitutive root ER bodies and wound-inducible leaf ER bodies. Because the dynamics of ER bodies under drought stress are still unclear, it is possible that these organelles are induced under water-deficit conditions to mediate the conversion of ABA-O-glucoside to ABA, in response to the signal transduction of drought stresses. According to such a scenario, BGLU18 expression is induced by wounding and by drought stress (ATTED-II), suggesting its dual functions in stress pathways. BGLU19, another member of the PYK10 subfamily, was suggested to be involved in cytokinin activation based on the highest sequence similarity with the β-glucosidase of Brassica napus that hydrolyses zeatin-O-glucoside (Falk et al., 1995; Xu et al., 2004). These results suggest that some β-glucosidases from this clade may have potential roles in phytohormone activation. The accumulation of various glucosylated phytohormones as precursors (Gachon et al., 2005; Poppenberger et al., 2005; Lee et al., 2006) suggests the involvement of glucosidases in the activation of these compounds, where the β-glucosidases may participate. Elucidating the involvement and behavior of ER bodies in phytohormone-mediated stress responses is required to further understand the functions of this organelle.
Evolution of ER bodies and β-glucosidases in the order Brassicales
ER bodies are formed in plants belonging to the Brassicales order
ER bodies were frequently observed by electron microscope using root tissues of plants belonging to the order Brassicales (Bonnett and Newcomb, 1965; Iversen and Flood, 1969; Iversen, 1970a; Cresti et al., 1974; Hoefert, 1975; Endress and Sjolund, 1976; Jørgensen et al., 1977; Behnke and Eschlbeck, 1978; Gailhofer et al., 1979; Jørgensen, 1981). This order includes Brassicaceae (mustard and cabbage family, including A. thaliana), Capparaceae (caper family), Cleomaceae (cleome family), Resedaceae (mignonette family) and Tovariaceae. The plants belonging to Brassicaceae, Cleomaceae, and Capparaceae are closely related to each other (Hall et al., 2004), and develop ER bodies or ER body-like structures in stems or roots. The ER bodies in these species occasionally contain internal filamentous structures (Hoefert, 1975; Jørgensen et al., 1977; Behnke and Eschlbeck, 1978; Gailhofer et al., 1979), suggesting their well-ordered formation. To date, no ER bodies have been observed in the Resedaceae (Iversen, 1970a). The Tovariaceae also do not develop ER bodies; however, filamentous aggregations of electron-dense materials similar to that of ER body were observed in the vacuoles of vascular-bundle cells (Behnke and Eschlbeck, 1978). Considering that Tovariaceae separated from Brassicaceae approximately 40−50 million years ago (Martin-Bravo et al., 2009), these filamentous structures might be the evolutionary intermediates of ER bodies. Collectively, ER bodies and related structures develop exclusively in a taxonomically limited group, raising the question of how this organelle evolved in these families.
The plant order Brassicales includes agriculturally important crops such as Brassica rapa and the model species A. thaliana, and plants in this order have been studied intensely. Many Brassicales plants are characterized with some unique features that evolved in this order, including the production of glucosinolates and other sulfur-containing secondary metabolites (Mithen et al., 2010; Bednarek et al., 2011), and the inability to accept arbuscular mycorrhiza as symbionts (reviewed in Delaux et al., 2013). Based on the public genome sequences, most of the proteins that localize in ER bodies and those that participate in the PYK10 protein complex belong to clades that occur in the family Brassicaceae. This suggests that A. thaliana, and most likely other species in Brassicales, have innovated a set of specific genes for developing these unique organelles.
β-glucosidases of the PYK10 subfamily evolved in the Brassicales order
The A. thaliana genome encodes 47 β-glucosidases, which are grouped into ten subfamilies (Xu et al., 2004). Figure 4 shows a neighbor-joining phylogenic tree generated for the A. thaliana β-glucosidases and related proteins retrieved from the genome sequences of Capsela rubella, Tellungiella salsuginea (formerly T. halophila), Carica papaya, Vitis vinifera, Glycine max, Oryza sativa, and Physcomitrella patens. The clade A includes BGLU40, 41, and 42, which corresponds to subfamily 8 (shaded with green); it also includes the β-glucosidases from P. patens, suggesting that these clades are ancestral among land plants. Clade B (shaded with red) corresponds to subfamilies 1, 2, 9, and 10; it does not include enzymes from P. patens, but does include enzymes from all other species, suggesting that this clade is specific to the angiosperms. By contrast, clades C and D correspond to the subfamilies 3–6, 7, and appear to be specific to Brassicales. Clade D consists of myrosinases, which are known to be specific for this order. The β-glucosidases from C. papaya were excluded from the subfamilies 3–5 that contain PYK10 and PEN2, showing the specificity of these three subfamilies to Brassicaceae, and possibly other species between Brassicaceae and Caricaceae. We identified a different lysine/arginine residue from the one conserved in classical myrosinases, which is located at the surface of the substrate pocket and completely conserved in all members of clade C (unpublished data). This positively charged residue might form electrostatic interactions with glucosinolates and explain the myrosinase activity of PEN2. Furthermore, it suggests that other β-glucosidases grouped into the clade are atypical myrosinases. As the glutamate proton donor residue is conserved within clade C, these enzymes appear to be intermediate forms between typical β-O-glucosidases and classical myrosinases. Twenty-two β-glucosidases in A. thaliana belong to clades C and D (hereafter called EE[K/R]-type and QE[K/R]-type, respectively), which is approximately half of the total β-glucosidases. Although myrosinases have been proposed to be special β-glucosidases, our phylogenetic analyses suggest that myrosinase activity could be a rather common feature of the β-glucosidases in Brassicales. The wide diversity within these β-glucosidases suggests that they have substrate specificity toward specific glucosides that are different in each plant tissue or under distinct conditions. This differential specificity would fit with glucosinolates that are highly diversified within Brassicales (reviewed in Agerbirk and Olsen, 2012).
It is unknown whether QE[K/R]-type β-glucosidases (classical myrosinases) evolved from EE[K/R]-type β-glucosidases (atypical myrosinases), or if they emerged independently. Two myrosinases that were recently isolated from Carica papaya possess the QE signature; however, they do not have any basic [K/R] residues that may mediate electrostatic interactions with glucosinolates. Consequently, both enzymes have a lower affinity for sinigrin compared to that of the classical myrosinases in A. thaliana (Nong et al., 2010). C. papaya is located almost at the edge of the Brassicales phylogenic tree (Hall et al., 2004), and shared a common ancestor with A. thaliana approximately 72 million years ago (Ming et al., 2008), suggesting that myrosinases evolved from these ancient forms that lacked the glucosinolate-binding affinity. Because glucosinolates have been identified in C. papaya (Tang, 1973), evolution of this basic residue [K/R] might have been achieved after the innovation of glucosinolates. The specific positions of these conserved basic residues differ between classical and atypical myrosinases; thus, it is likely that these two groups emerged independently. According to the public genome database, the C. papaya genome does not encode a protein bearing significant similarity to NAI2 from A. thaliana, whereas other species in Brassicaceae do, suggesting that ER bodies have evolved specifically in plants belonging to the Brassicaceae, possibly due to the evolution of NAI2. The evolutionary process of producing ER bodies and atypical myrosinases will be elucidated by analysing species in between C. papaya and Brassicaceae.
Plants of the Brassicales order develop distinct glucosidase-glucoside defense systems
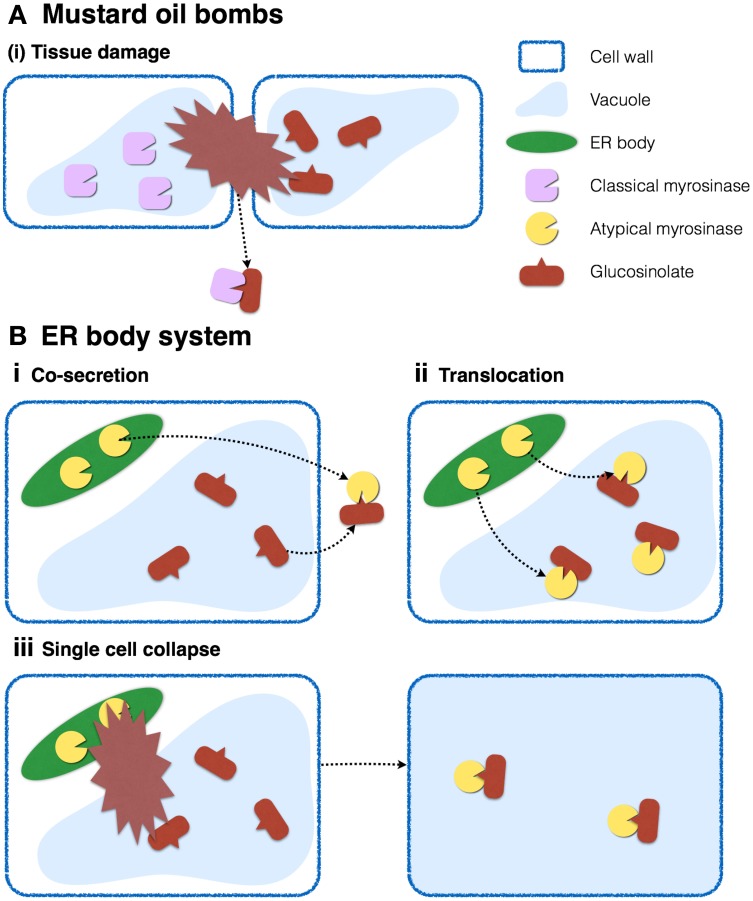
To avoid constitutive hydrolysis of glucosinolates, which are proposed to accumulate in vacuoles, myrosinases have to be separated from their substrates into distinct cellular or subcellular compartments (Figure 7). In the case of classical myrosinases, the mustard-oil bomb system was proposed, in which the enzymes and substrates accumulate in distinct cells called myrosine cells and S cells, respectively, which allows suppression of the enzymatic reaction until the tissue is crushed. This intercellular partitioning does not require specific subcellular compartments for storage, and myrosinases accumulate in the vacuoles of myrosine cells possibly because they have the largest volume.
Figure 7.
Two distinct myrosinase-glucosinolate systems occurring in Brassicales. (A) In leaves, myrosinases and glucosinolates accumulate in vacuoles of distinct cells. Tissue collapse accompanied by destruction of multiple cells results in the mixing of enzymes and substrates, and the production of toxic products against herbivores. (B) In roots, myrosinases and glucosinolates accumulate in ER bodies and vacuoles, respectively, in the same cells. Enzymatic reactions may be activated by (i) cosecretion of enzymes and substrates, (ii) translocation of enzymes into vacuoles, or (iii) destruction of a single cell.
Table 1 shows that many β-glucosidases contain signal peptides, but some of them lost this motif. Differential localization might indicate that β-glucosidase activities are required for various cellular functions including those of endomembrane systems, apoplast, and cytosol. The evolutionary events of losing signal peptides appear to occur independently, suggesting that each of these events could reflect the acquisition of a novel molecular function. For example, PEN2 is known to localize at the periphery of peroxisomal membranes, which enables it to accumulate at the penetration sites in response to Blumeria graminis f. sp. hordei (Bgh) challenge (Lipka et al., 2005). This clearly shows the importance of protein localization for protein function. The mechanism of glucosinolate activation by PEN2 is totally different from that of classical myrosinases, because Bgh challenge does not inflict cell damage or cell disruption.
It is possible that Brassicales plants evolved ER bodies to store atypical myrosinases and separate them from their substrates on the subcellular level. A recent study addressing the cell-type specific metabolome in A. thaliana roots revealed the cellular distribution of glucosinolates (Moussaieff et al., 2013). According to this study, indole glucosinolates accumulate to relatively higher levels in columella and cortex, whereas lower levels accumulate in epidermis, endodermis, and stele, indicating that PYK10 and indole glucosinolates occur concurrently in the same cells in the cortex of A. thaliana roots, but in different organelles. ER bodies contain PYK10, whereas vacuoles presumably store indole glucosinolates, suggesting that plants in the Brassicales order evolved two distinct myrosinase-glucosinolate systems, one of which is the classical mustard-oil bomb system described above. The system proposed here considers that ER bodies separate both components into distinct organelles in a single cell (Figure 7B). This subcellular compartmentalization, which does not require tissue collapse to trigger the response, could be highly effective against fungal/bacterial pathogens that usually start the infection process by invading single plant cells. This system could be triggered by translocation of enzymes and/or substrates in living cells, such as the case of PEN2 (Lipka et al., 2005; Bednarek et al., 2009). It is also possible that chemical or protein factors such as pH or chaperones regulate the activity of ER body-accumulating β-glucosidases.
According to the literature, glucosinolates and myrosine cells are widely distributed throughout the Brassicales (Jørgensen, 1981). By contrast, ER bodies showed higher taxonomic limitation primarily in Brassicaceae, Cleomaceae, and Cappraceae (Iversen, 1970a; Behnke and Eschlbeck, 1978; Jørgensen, 1981), suggesting that ER bodies are a more recent innovation among these families. Due to the lack of genetic/molecular tools in species belonging to other Brassicales families than Brassicaceae, the evolution of specific substrates, enzymes, and organelles remains unclear. Recent improvements in sequencing technologies have facilitated rapid progress in genome sequences for non-model plants, including many Brassicaceae species such as T. salsuginea (Wu et al., 2012; Yang et al., 2013), B. rapa (Wang et al., 2011), C. rubella (Slotte et al., 2013), and A. lyrata (Hu et al., 2011). Molecular and genetic tools are becoming available that will enable the inclusion of many other species in future research.
Concluding remarks and future perspectives
We have shown how ER bodies are unique in their shape, biogenesis mechanism, functions, and evolution. The study of ER bodies provides novel insights into plant innate immunity systems, stress tolerance, and the subcellular organization of eukaryotic cells. Cell biological studies for understanding eukaryotic organelle organization are sometimes too focused on single cell types, including cultured cells. However, cultured cells are very different from endogenous cells in living individuals. For example, tobacco BY-2 cells are one of the most widely utilized plant cultured cell lines, but they have ER structures that are slightly different to those in A. thaliana, lacking ER bodies, and tubules and cisternae that appear different (Nakano et al., 2009). It is necessary to keep in mind what cell types are used for the experiments, and to clearly distinguish what is unique to the cell type and what is common to all eukaryotic cells, or at least to the species in use.
The ER body is a microdomain of the ER. Numerous studies have focused on how microdomains in the plasma membrane are organized and contribute to cell functions (reviewed in Lingwood and Simons, 2010). These membrane microdomains also play important roles in plant cells (reviewed in Malinsky et al., 2013). However, because these studies focused primarily on microdomains in plasma membranes, little is known about the organization of microdomains in organelle membranes including the ER. The organization of lipids and proteins should play an important role in the maintenance of organelle morphology and function. ER bodies may be a good model to understand microdomain organization in ER. The mechanisms underlying ER body formation will provide valuable insights into the general mechanisms of microdomain organization for organelle membranes. Combined with an understanding of physiological functions, ER bodies can be an interesting model case to connect subcellular microdomains to the overall fitness of individual plants.
Yet, we still have some unanswered key questions. The most important piece of the puzzle that is missing is the genetic evidence for ER body function in stress responses. As discussed in the second section, most results achieved in this decade suggest a role of ER bodies in interaction with microbes and in abiotic-stress responses via activation of phytohormones and/or phytosiderophores. However, there is still no direct evidence for the physiological functions of ER bodies. Identification of the native substrate(s) could help us in validating the function(s). Phenotypic analysis of knockout lines deficient either in ER-bodies, PYK10 or substrates of this β-glucosidase, as well as respective multiple mutants subjected to various conditions will be required toward this end. To confirm the involvement of PYK10 and its substrate in the response to a certain environmental stress it will be necessary to show that the mutants depleted in either the enzyme or the substrate show a similar defect at least in regard to that particular response. More importantly, the multiple mutant lacking both the enzyme and the substrate should exhibit comparable, but not additive or synergistic phenotype compared to the mutants lacking only the substrates or the enzymes. Performing such genetic analysis, particularly when both enzymes and substrates are involved in multiple pathways, will provide insights into which combination of enzymes and substrates is registered to the pathway of interest. What is also completely missing is the behavior of ER bodies in response to these stresses; thus understanding the dynamics of ER bodies and PYK10 is crucial. The proper response to stress requires activation of molecules that are involved in the resistance to stress, e.g., toxic compounds should be produced only when plants are subjected to undesirable visitors, because these compounds may also be toxic to the plant. This responsive activation may include dynamic translocation and/or secretion of enzymes, where the ER body β-glucosidases can become activated (Figure 7).
The advantage or disadvantage of developing specific storage organelles is not fully understood. Many plant species evolved specific organelles for protein storage (reviewed in Hara-Nishimura and Matsushima, 2003). The production of special compartments or compounds does require valuable resources, as shown for glucosinolates (Bekaert et al., 2012; Züst et al., 2012), which suggests that the possession of ER bodies should confer a beneficial advantage for plant fitness. To specifically address the significance of accumulating PYK10 in ER bodies, a population genetics approach using generations of nai1, pyk10, and nai2 mutants under appropriate conditions will be required. Revealing the evolutionary processes during the achievement of ER bodies will also provide insight into this issue. Toward this end, a genome sequence of a species in between Brassicaceae and Caricaceae will be a great help.
Conflict of interest statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
References
- Adie B. A. T., Pérez-Pérez J., Pérez-Pérez M. M., Godoy M., Sánchez-Serrano J.-J., Schmelz E. A., et al. (2007). ABA is an essential signal for plant resistance to pathogens affecting JA biosynthesis and the activation of defenses in Arabidopsis. Plant Cell 19, 1665–1681 10.1105/tpc.106.048041 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agee A. E., Surpin M., Sohn E. J., Girke T., Rosado A., Kram B. W., et al. (2010). MODIFIED VACUOLE PHENOTYPE1 is an Arabidopsis myrosinase-associated protein involved in endomembrane protein trafficking. Plant Physiol. 152, 120–132 10.1104/pp.109.145078 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agerbirk N., Olsen C. E. (2012). Glucosinolate structures in evolution. Phytochemistry 77, 16–45 10.1016/j.phytochem.2012.02.005 [DOI] [PubMed] [Google Scholar]
- Ahn Y. O., Shimizu B.-I., Sakata K., Gantulga D., Zhou C., Zhou Z., et al. (2010). Scopolin-hydrolyzing beta-glucosidases in roots of Arabidopsis. Plant Cell Physiol. 51, 132–143 10.1093/pcp/pcp174 [DOI] [PubMed] [Google Scholar]
- Andersson D., Chakrabarty R., Bejai S., Zhang J., Rask L., Meijer J. (2009). Myrosinases from root and leaves of Arabidopsis thaliana have different catalytic properties. Phytochemistry 70, 1345–1354 10.1016/j.phytochem.2009.07.036 [DOI] [PubMed] [Google Scholar]
- Barth C., Jander G. (2006). Arabidopsis myrosinases TGG1 and TGG2 have redundant function in glucosinolate breakdown and insect defense. Plant J. 46, 549–562 10.1111/j.1365-313X.2006.02716.x [DOI] [PubMed] [Google Scholar]
- Bayoumi S. A. L., Rowan M. G., Blagbrough I. S., Beeching J. R. (2008). Biosynthesis of scopoletin and scopolin in cassava roots during post-harvest physiological deterioration: the E-Z-isomerisation stage. Phytochemistry 69, 2928–2936 10.1016/j.phytochem.2008.09.023 [DOI] [PubMed] [Google Scholar]
- Bednarek P. (2012). Chemical warfare or modulators of defence responses - the function of secondary metabolites in plant immunity. Curr. Opin. Plant Biol. 15, 407–414 10.1016/j.pbi.2012.03.002 [DOI] [PubMed] [Google Scholar]
- Bednarek P., Piœlewska-Bednarek M., Svatoš A., Schneider B., Doubsky J., Mansurova M., et al. (2009). A glucosinolate metabolism pathway in living plant cells mediates broad-spectrum antifungal defense. Science 323, 101–106 10.1126/science.1163732 [DOI] [PubMed] [Google Scholar]
- Bednarek P., Piœlewska-Bednarek M., Ver Loren Van Themaat E., Maddula R. K., Svatoš A., Schulze-Lefert P. (2011). Conservation and clade-specific diversification of pathogen-inducible tryptophan and indole glucosinolate metabolism in Arabidopsis thaliana relatives. New Phytol. 192, 713–726 10.1111/j.1469-8137.2011.03824.x [DOI] [PubMed] [Google Scholar]
- Bednarek P., Schneider B., Svatoš A., Oldham N. J., Hahlbrock K. (2005). Structural complexity, differential response to infection, and tissue specificity of indolic and phenylpropanoid secondary metabolism in Arabidopsis roots. Plant Physiol. 138, 1058–1070 10.1104/pp.104.057794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Behnke H., Eschlbeck G. (1978). Dilated cisternae in capparales - an attempt towards the characterization of a specific endoplasmic reticulum. Protoplasma 97, 351–363 10.1007/BF01276292 [DOI] [Google Scholar]
- Bekaert M., Edger P. P., Hudson C. M., Pires J. C., Conant G. C. (2012). Metabolic and evolutionary costs of herbivory defense: systems biology of glucosinolate synthesis. New Phytol. 196, 596–605 10.1111/j.1469-8137.2012.04302.x [DOI] [PubMed] [Google Scholar]
- Bonnett H. T., Newcomb E. H. (1965). Polyribosomes and cisternal accumulations in root cells of radish. J. Cell Biol. 27, 423–432 10.1083/jcb.27.2.423 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bressan M., Roncato M.-A., Bellvert F., Comte G., Haichar F. Z., Achouak W., et al. (2009). Exogenous glucosinolate produced by Arabidopsis thaliana has an impact on microbes in the rhizosphere and plant roots. ISME J. 3, 1243–1257 10.1038/ismej.2009.68 [DOI] [PubMed] [Google Scholar]
- Brown P. D., Tokuhisa J. G., Reichelt M., Gershenzon J. (2003). Variation of glucosinolate accumulation among different organs and developmental stages of Arabidopsis thaliana. Phytochemistry 62, 471–481 10.1016/S0031-9422(02)00549-6 [DOI] [PubMed] [Google Scholar]
- Bulgarelli D., Schlaeppi K., Spaepen S., Ver Loren Van Themaat E., Schulze-Lefert P. (2013). Structure and functions of the bacterial microbiota of plants. Annu. Rev. Plant Biol. 64, 807–838 10.1146/annurev-arplant-050312-120106 [DOI] [PubMed] [Google Scholar]
- Burmeister W., Cottaz S., Driguez H., Iori R., Palmieri S., Henrissat B. (1997). The crystal structures of Sinapis alba myrosinase and a covalent glycosyl-enzyme intermediate provide insights into the substrate recognition and active-site machinery of an S-glycosidase. Structure 5, 663–676 10.1016/S0969-2126(97)00221-9 [DOI] [PubMed] [Google Scholar]
- Buskov S., Serra B., Rosa E., Sørensen H., Sørensen J. C. (2002). Effects of intact glucosinolates and products produced from glucosinolates in myrosinase-catalyzed hydrolysis on the potato cyst nematode (Globodera rostochiensis Cv. Woll). J. Agricult. Food Chem. 50, 690–695 10.1021/jf010470s [DOI] [PubMed] [Google Scholar]
- Chen J., Stefano G., Brandizzi F., Zheng H. (2011). Arabidopsis RHD3 mediates the generation of the tubular ER network and is required for Golgi distribution and motility in plant cells. J. Cell Sci. 124, 2241–2252 10.1242/jcs.084624 [DOI] [PubMed] [Google Scholar]
- Chen S., Novick P., Ferro-Novick S. (2013). ER structure and function. Curr. Opin. Cell Biol. 25, 428–433 10.1016/j.ceb.2013.02.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cresti M., Pacini E., Simoncioli C. (1974). Uncommon paracrystalline structures formed in the endoplasmic reticulum of the integumentary cells of Diplotaxis erucoides ovules. J. Ultrastruct. Res. 49, 219–223 10.1016/S0022-5320(74)80033-X [DOI] [PubMed] [Google Scholar]
- Delaux P.-M., Séjalon-Delmas N., Bécard G., Ané J.-M. (2013). Evolution of the plant–microbe symbiotic ‘toolkit’. Trends Plant Sci. 18, 298–304 10.1016/j.tplants.2013.01.008 [DOI] [PubMed] [Google Scholar]
- Dixon D. C., Cutt J. R., Klessig D. F. (1991). Differential targeting of the tobacco PR-1pathogenesis-related proteins to the extracellular space and vacuoles of crystal idioblasts. EMBO J. 10, 1317–1324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elliott C. E., Harjono, Howlett B. J. (2008). Mutation of a gene in the fungus Leptosphaeria maculans allows increased frequency of penetration of stomatal apertures of Arabidopsis thaliana. Mol. Plant 1, 471–481 10.1093/mp/ssn014 [DOI] [PubMed] [Google Scholar]
- Endress A. G., Sjolund R. D. (1976). Ultrastructural cytology of callus cultures of Streptanthus tortuosus as affected by temperature. Am. J. Bot. 63, 1213–1224 10.2307/2441738 [DOI] [Google Scholar]
- Falk A., Ek B., Rask L. (1995). Characterization of a new myrosinase in Brassica napus. Plant Mol. Biol. 27, 863–874 10.1007/BF00037015 [DOI] [PubMed] [Google Scholar]
- Faso C., Chen Y. N., Tamura K., Held M., Zemelis S., Marti L., et al. (2009). A missense mutation in the Arabidopsis COPII coat protein Sec24A induces the formation of clusters of the endoplasmic reticulum and Golgi apparatus. Plant Cell 21, 3655–3671 10.1105/tpc.109.068262 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fourcroy P., Sisó-Terraza P., Sudre D., Savirón M., Reyt G., Gaymard F., et al. (2014). Involvement of the ABCG37 transporter in secretion of scopoletin and derivatives by Arabidopsis roots in response to iron deficiency. New Phytol. 201, 155–167 10.1111/nph.12471 [DOI] [PubMed] [Google Scholar]
- Gachon C. M., Langlois-Meurinne M., Saindrenan P. (2005). Plant secondary metabolism glycosyltransferases: the emerging functional analysis. Trends Plant Sci. 10, 542–549 10.1016/j.tplants.2005.09.007 [DOI] [PubMed] [Google Scholar]
- Gailhofer M., Thaler I., Rücker W. (1979). Dilated ER in callus cells and in cells from Armoracia plants cultivated in vitro. Protoplasma 98, 263–274 10.1007/BF01281443 [DOI] [Google Scholar]
- Gunning B. E. S. (1998). The identity of mystery organelles in Arabidopsis plants expressing GFP. Trends Plant Sci. 3, 417–417 10.1016/S1360-1385(98)01336-3 [DOI] [PubMed] [Google Scholar]
- Hakenjos J. P., Bejai S., Ranftl Q., Behringer C., Vlot A. C., Absmanner B., et al. (2013). ML3 is a NEDD8- and ubiquitin-modified protein. Plant Physiol. 163, 135–149 10.1104/pp.113.221341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Halkier B. A., Gershenzon J. (2006). Biology and biochemistry of glucosinolates. Annu. Rev. Plant Biol. 57, 303–333 10.1146/annurev.arplant.57.032905.105228 [DOI] [PubMed] [Google Scholar]
- Hall J. C., Iltis H. H., Sytsma K. J. (2004). Molecular phylogenetics of core brassicales, placement of orphan genera emblingia, forchhammeria, tirania, and character evolution. Syst. Bot. 29, 654–669 10.1600/0363644041744491 [DOI] [Google Scholar]
- Han Y.-J., Cho K.-C., Hwang O.-J., Choi Y.-S., Shin A.-Y., Hwang I., et al. (2012). Overexpression of an Arabidopsis β-glucosidase gene enhances drought resistance with dwarf phenotype in creeping bentgrass. Plant Cell Rep. 31, 1677–1686 10.1007/s00299-012-1280-6 [DOI] [PubMed] [Google Scholar]
- Hara-Nishimura I., Matsushima R. (2003). A wound-inducible organelle derived from endoplasmic reticulum: a plant strategy against environmental stresses? Curr. Opin. Plant Biol. 6, 583–588 10.1016/j.pbi.2003.09.015 [DOI] [PubMed] [Google Scholar]
- Hara-Nishimura I., Shimada T., Hatano K., Takeuchi Y., Nishimura M. (1998). Transport of storage proteins to protein storage vacuoles is mediated by large precursor-accumulating vesicles. Plant Cell 10, 825-836 10.1105/tpc.10.5.825 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hawes C., Saint-Jore C., Martin B., Zheng H. Q. (2001). ER confirmed as the location of mystery organelles in Arabidopsis plants expressing GFP! Trends Plant Sci. 6, 245–246 10.1016/S1360-1385(01)01980-X [DOI] [PubMed] [Google Scholar]
- Hayashi Y., Yamada K., Shimada T., Matsushima R., Nishizawa N. K., Nishimura M., et al. (2001). A proteinase-storing body that prepares for cell death or stresses in the epidermal cells of Arabidopsis. Plant Cell Physiol. 42, 894–899 10.1093/pcp/pce144 [DOI] [PubMed] [Google Scholar]
- Hemm M. R., Rider S. D., Ogas J., Murry D. J., Chapple C. (2004). Light induces phenylpropanoid metabolism in Arabidopsis roots. Plant J. 38, 765–778 10.1111/j.1365-313X.2004.02089.x [DOI] [PubMed] [Google Scholar]
- Herman E., Larkins B. (1999). Protein storage bodies and vacuoles. Plant Cell 11, 601–614 10.1105/tpc.11.4.601 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hiruma K., Onozawa-Komori M., Takahashi F., Asakura M., Bednarek P., Okuno T., et al. (2010). Entry mode-dependent function of an indole glucosinolate pathway in Arabidopsis for nonhost resistance against anthracnose pathogens. Plant Cell 22, 2429–2443 10.1105/tpc.110.074344 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoefert L. L. (1975). Tubules in dilated cisternae of endoplasmic reticulum of Thlaspi arvense (Cruciferae). Am. J. Bot. 62, 756–760 10.2307/2442066 [DOI] [Google Scholar]
- Hopkins R. J., Van Dam N. M., Van Loon J. J. A. (2009). Role of glucosinolates in insect-plant relationships and multitrophic interactions. Annu. Rev. Entomol. 54, 57–83 10.1146/annurev.ento.54.110807.090623 [DOI] [PubMed] [Google Scholar]
- Hu T. T., Pattyn P., Bakker E. G., Cao J., Cheng J.-F., Clark R. M., et al. (2011). The Arabidopsis lyrata genome sequence and the basis of rapid genome size change. Nat. Genet. 43, 476–481 10.1038/ng.807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iversen T. (1970a). The morphology, occurrence, and distribution of dilated cisternae of the endoplasmic reticulum in tissues of plants of the cruciferae. Protoplasma 71, 467–477 10.1007/BF01279689 [DOI] [Google Scholar]
- Iversen T. H. (1970b). Cytochemical localization of myrosinase (β-thioglucosidase) in root tips of Sinapis alba. Protoplasma 71, 451–466 10.1007/BF01279688 [DOI] [Google Scholar]
- Iversen T.-H., Flood P. R. (1969). Rod-shaped accumulations in cisternae of the endoplasmic reticulum in root cells of Lepidium sativum seedlings. Planta 86, 295–298 10.1007/BF00386462 [DOI] [PubMed] [Google Scholar]
- Jacobs S., Zechmann B., Molitor A., Trujillo M., Petutschnig E., Lipka V., et al. (2011). Broad-spectrum suppression of innate immunity is required for colonization of Arabidopsis roots by the fungus Piriformospora indica. Plant Physiol. 156, 726–740 10.1104/pp.111.176446 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jørgensen L. B. (1981). Myrosin cells and dilated cisternae of the endoplasmic reticulum in the order Capparales. Nordic J. Bot. 1, 433–445 10.1111/j.1756-1051.1981.tb00709.x [DOI] [Google Scholar]
- Jørgensen L. B., Behnke H. D., Mabry T. J. (1977). Protein-accumulating cells and dilated cisternae of the endoplasmic reticulum in three glucosinolate-containing genera: Armoracia, Capparis, Drypetes. Planta 137, 215–224 10.1007/BF00388153 [DOI] [PubMed] [Google Scholar]
- Kai K., Shimizu B.-I., Mizutani M., Watanabe K., Sakata K. (2006). Accumulation of coumarins in Arabidopsis thaliana. Phytochemistry 67, 379–386 10.1016/j.phytochem.2005.11.006 [DOI] [PubMed] [Google Scholar]
- Keefe D., Hinz U., Meins F. J. (1990). The effect of ethylene on the cell-typespecific and intracellular localization of b-1,3-glucanase and chitinase in tobacco leaves. Planta 182, 43–51 [DOI] [PubMed] [Google Scholar]
- Kim S. A., Punshon T., Lanzirotti A., Li L., Alonso J. M., Ecker J. R., et al. (2006). Localization of iron in arabidopsis seed requires the vacuolar membrane transporter VIT1. Science 314, 1295–1298 10.1126/science.1132563 [DOI] [PubMed] [Google Scholar]
- Lazzeri L., Curto G., Leoni O., Dallavalle E. (2004). Effects of glucosinolates and their enzymatic hydrolysis products via myrosinase on the root-knot nematode Meloidogyne incognita (Kofoid et White) Chitw. J. Agricult. Food Chem. 52, 6703–6707 10.1021/jf030776u [DOI] [PubMed] [Google Scholar]
- Lee H. Y., Bowen C. H., Popescu G. V., Kang H.-G., Kato N., Ma S., et al. (2011). Arabidopsis RTNLB1 and RTNLB2 reticulon-like proteins regulate intracellular trafficking and activity of the FLS2 immune receptor. Plant Cell 23, 3374–3391 10.1105/tpc.111.089656 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee H., Sparkes I., Gattolin S., Dzimitrowicz N., Roberts L. M., Hawes C., et al. (2013). An Arabidopsis reticulon and the atlastin homologue RHD3-like2 act together in shaping the tubular endoplasmic reticulum. New Phytol. 197, 481–489 10.1111/nph.12038 [DOI] [PubMed] [Google Scholar]
- Lee K. H., Piao H. L., Kim H.-Y., Choi S. M., Jiang F., Hartung W., et al. (2006). Activation of glucosidase via stress-induced polymerization rapidly increases active pools of abscisic acid. Cell 126, 1109–1120 10.1016/j.cell.2006.07.034 [DOI] [PubMed] [Google Scholar]
- Li L., Chen O. S., McVey Ward D., Kaplan J. (2001). CCC1 is a transporter that mediates vacuolar iron storage in yeast. J. Biol. Chem. 276, 29515–29519 10.1074/jbc.M103944200 [DOI] [PubMed] [Google Scholar]
- Lingwood D., Simons K. (2010). Lipid rafts as a membrane-organizing principle. Science 327, 46–50 10.1126/science.1174621 [DOI] [PubMed] [Google Scholar]
- Lipka V., Dittgen J., Bednarek P., Bhat R., Wiermer M., Stein M., et al. (2005). Pre- and postinvasion defenses both contribute to nonhost resistance in Arabidopsis. Science 310, 1180–1183 10.1126/science.1119409 [DOI] [PubMed] [Google Scholar]
- Maeda K., Houjyou Y., Komatsu T., Hori H., Kodaira T., Ishikawa A. (2009). AGB1 and PMR5 contribute to PEN2-mediated preinvasion resistance to Magnaporthe oryzae in Arabidopsis thaliana. Mol. Plant-Microbe Interact. 22, 1331–1340 10.1094/MPMI-22-11-1331 [DOI] [PubMed] [Google Scholar]
- Malinsky J., Opekarová M., Grossmann G., Tanner W. (2013). Membrane microdomains, rafts, and detergent-resistant membranes in plants and fungi. Annu. Rev. Plant Biol. 64, 501–529 10.1146/annurev-arplant-050312-120103 [DOI] [PubMed] [Google Scholar]
- Marti L., Stefano G., Tamura K., Hawes C., Renna L., Held M. A., et al. (2010). A missense mutation in the vacuolar protein GOLD36 causes organizational defects in the ER and aberrant protein trafficking in the plant secretory pathway. Plant J. 63, 901–913 10.1111/j.1365-313X.2010.04296.x [DOI] [PubMed] [Google Scholar]
- Martin-Bravo S., Vargas P., Luceno M. (2009). Is Oligomeris (Resedaceae) indigenous to North America? Molecular evidence for a natural colonization from the Old World. Am. J. Bot. 96, 507–518 10.3732/ajb.0800216 [DOI] [PubMed] [Google Scholar]
- Matsushima R., Fukao Y., Nishimura M., Hara-Nishimura I. (2004). NAI1 gene encodes a basic-helix-loop-helix-type putative transcription factor that regulates the formation of an endoplasmic reticulum-derived structure, the ER body. Plant Cell 16, 1536–1549 10.1105/tpc.021154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima R., Hayashi Y., Kondo M., Shimada T., Nishimura M., Hara-Nishimura I. (2002). An endoplasmic reticulum-derived structure that is induced under stress conditions in Arabidopsis. Plant Physiol. 130, 1807–1814 10.1104/pp.009464 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsushima R., Kondo M., Nishimura M., Hara-Nishimura I. (2003). A novel ER-derived compartment, the ER body, selectively accumulates a beta-glucosidase with an ER-retention signal in Arabidopsis. Plant J. 33, 493–502 10.1046/j.1365-313X.2003.01636.x [DOI] [PubMed] [Google Scholar]
- Ming R., Hou S., Feng Y., Yu Q., Dionne-Laporte A., Saw J. H., et al. (2008). The draft genome of the transgenic tropical fruit tree papaya (Carica papaya Linnaeus). Nature 452, 991–996 10.1038/nature06856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchum M. G., Wang X., Wang J., Davis E. L. (2012). Role of nematode peptides and other small molecules in plant parasitism. Annu. Rev. Phytopathol. 50, 175–195 10.1146/annurev-phyto-081211-173008 [DOI] [PubMed] [Google Scholar]
- Mithen R., Bennett R., Marquez J. (2010). Glucosinolate biochemical diversity and innovation in the Brassicales. Phytochemistry 71, 2074–2086 10.1016/j.phytochem.2010.09.017 [DOI] [PubMed] [Google Scholar]
- Mladenka P., Macáková K., Zatloukalová L., Reháková Z., Singh B. K., Prasad A. K., et al. (2010). In vitro interactions of coumarins with iron. Biochimie 92, 1108–1114 10.1016/j.biochi.2010.03.025 [DOI] [PubMed] [Google Scholar]
- Moussaieff A., Rogachev I., Brodsky L., Malitsky S., Toal T. W., Belcher H., et al. (2013). High-resolution metabolic mapping of cell types in plant roots. Proc. Natl. Acad. Sci. U.S.A. 110, E1232–E1241 10.1073/pnas.1302019110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nagano A. J., Fukao Y., Fujiwara M., Nishimura M., Hara-Nishimura I. (2008). Antagonistic jacalin-related lectins regulate the size of ER body-type β-glucosidase complexes in Arabidopsis thaliana. Plant Cell Physiol. 49, 969–980 10.1093/pcp/pcn075 [DOI] [PubMed] [Google Scholar]
- Nagano A. J., Maekawa A., Nakano R. T., Miyahara M., Higaki T., Kutsuna N., et al. (2009). Quantitative analysis of ER body morphology in an Arabidopsis mutant. Plant Cell Physiol. 50, 2015–2022 10.1093/pcp/pcp157 [DOI] [PubMed] [Google Scholar]
- Nagano A. J., Matsushima R., Hara-Nishimura I. (2005). Activation of an ER-body-localized beta-glucosidase via a cytosolic binding partner in damaged tissues of Arabidopsis thaliana. Plant Cell Physiol. 46, 1140–1148 10.1093/pcp/pci126 [DOI] [PubMed] [Google Scholar]
- Nakano R. T., Matsushima R., Nagano A. J., Fukao Y., Fujiwara M., Kondo M., et al. (2012). ERMO3/MVP1/GOLD36 is involved in a cell type-specific mechanism for maintaining er morphology in Arabidopsis thaliana. PLoS ONE 7:e49103 10.1371/journal.pone.0049103 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakano R. T., Matsushima R., Ueda H., Tamura K., Shimada T., Li L., et al. (2009). GNOM-LIKE1/ERMO1 and SEC24a/ERMO2 are required for maintenance of endoplasmic reticulum morphology in Arabidopsis thaliana. Plant Cell 21, 3672–3685 10.1105/tpc.109.068270 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Negishi T., Nakanishi H., Yazaki J., Kishimoto N., Fujii F., Shimbo K., et al. (2002). cDNA microarray analysis of gene expression during Fe-deficiency stress in barley suggests that polar transport of vesicles is implicated in phytosiderophore secretion in Fe-deficient barley roots. Plant J. 30, 83–94 10.1046/j.1365-313X.2002.01270.x [DOI] [PubMed] [Google Scholar]
- Neuhaus J. M., Sticher L., Meins F. J., Boller T. (1991). A short C-terminal sequence is necessary and sufficient for the targeting of chitinases to the plant vacuole. Proc. Natl. Acad. Sci. U.S.A. 88, 10362–10366 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitz I., Berkefeld H., Puzio P. S., Grundler F. M. W. (2001). Pyk10, a seedling and root specific gene and promoter from Arabidopsis thaliana. Plant Sci. 161, 337–346 10.1016/S0168-9452(01)00412-5 [DOI] [PubMed] [Google Scholar]
- Nong H., Zhang J.-M., Li D.-Q., Wang M., Sun X.-P., Zhu Y. J., et al. (2010). Characterization of a novel β-thioglucosidase CpTGG1 in Carica papaya and its substrate-dependent and ascorbic acid-independent O-β-glucosidase activity. J. Integr. Plant Biol. 52, 879–890 10.1111/j.1744-7909.2010.00988.x [DOI] [PubMed] [Google Scholar]
- Nongbri P. L., Johnson J. M., Sherameti I., Glawischnig E., Halkier B. A., Oelmüller R. (2012). Indole-3-acetaldoxime-derived compounds restrict root colonization in the beneficial interaction between Arabidopsis roots and the endophyte Piriformospora indica. Mol. Plant-Microbe Interact. 25, 1186–1197 10.1094/MPMI-03-12-0071-R [DOI] [PubMed] [Google Scholar]
- Nziengui H., Bouhidel K., Pillon D., Der C., Marty F., Schoefs B. (2007). Reticulon-like proteins in Arabidopsis thaliana: structural organization and ER localization. FEBS Lett. 581, 3356–3362 10.1016/j.febslet.2007.06.032 [DOI] [PubMed] [Google Scholar]
- Oelmuller R., Peskan-Berghofer T., Shahollari B., Trebicka A., Sherameti I., Varma A. (2005). MATH domain proteins represent a novel protein family in Arabidopsis thaliana, and at least one member is modified in roots during the course of a plant-microbe interaction. Physiol. Plant. 124, 152–166 10.1111/j.1399-3054.2005.00505.x [DOI] [Google Scholar]
- Ogasawara K., Yamada K., Christeller J. T., Kondo M., Hatsugai N., Hara-Nishimura I., et al. (2009). Constitutive and inducible ER bodies of Arabidopsis thaliana accumulate distinct beta-glucosidases. Plant Cell Physiol. 50, 480–488 10.1093/pcp/pcp007 [DOI] [PubMed] [Google Scholar]
- Peterson J. K., Harrison H. F., Jackson D. M., Snook M. E. (2003). Biological activities and contents of scopolin and scopoletin in sweetpotato clones. Hortscience 38, 1129–1133 [Google Scholar]
- Pfalz M., Mikkelsen M. D., Bednarek P., Olsen C. E., Halkier B. A., Kroymann J. (2011). Metabolic engineering in Nicotiana benthamiana reveals key enzyme functions in Arabidopsis indole glucosinolate modification. Plant Cell 23, 716–729 10.1105/tpc.110.081711 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poppenberger B., Fujioka S., Soeno K., George G. L., Vaistij F. E., Hiranuma S., et al. (2005). The UGT73C5 of Arabidopsis thaliana glucosylates brassinosteroids. Proc. Natl. Acad. Sci. U.S.A. 102, 15253–15238 10.1073/pnas.0504279102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prats E., Bazzalo M. E., León A., Jorrín J. V. (2006). Fungitoxic effect of scopolin and related coumarins on Sclerotinia sclerotiorum. A way to overcome sunflower head rot. Euphytica 147, 451–460 10.1007/s10681-005-9045-8 [DOI] [Google Scholar]
- Rask L., Andréasson E., Ekbom B., Eriksson S., Pontoppidan B., Meijer J. (2000). Myrosinase: gene family evolution and herbivore defense in Brassicaceae. Plant Mol. Biol. 42, 93–113 10.1023/A:1006380021658 [DOI] [PubMed] [Google Scholar]
- Ridge R. W., Uozumi Y., Plazinski J., Hurley U. A., Williamson R. E. (1999). Developmental transitions and dynamics of the cortical ER of Arabidopsis cells seen with green fluorescent protein. Plant Cell Physiol. 40, 1253–1261 10.1093/oxfordjournals.pcp.a029513 [DOI] [PubMed] [Google Scholar]
- Sanchez-Vallet A., Ramos B., Bednarek P., López G., Piślewska-Bednarek M., Schulze-Lefert P., et al. (2010). Tryptophan-derived secondary metabolites in Arabidopsis thaliana confer non-host resistance to necrotrophic Plectosphaerella cucumerina fungi. Plant J. 63, 115–127 10.1111/j.1365-313X.2010.04224.x [DOI] [PubMed] [Google Scholar]
- Schlaeppi K., Abou-Mansour E., Buchala A., Mauch F. (2010). Disease resistance of Arabidopsis to Phytophthora brassicae is established by the sequential action of indole glucosinolates and camalexin. Plant J. 62, 840–851 10.1111/j.1365-313X.2010.04197.x [DOI] [PubMed] [Google Scholar]
- Schmid M., Simpson D. J., Sarioglu H., Lottspeich F., Gietl C. (2001). The ricinosomes of senescing plant tissue bud from the endoplasmic reticulum. Proc. Natl. Acad. Sci. U.S.A. 98, 5353–5358 10.1073/pnas.061038298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sels J., Mathys J., De Coninck B. M., Cammue B. P., De Bolle M. F. (2008). Plant pathogenesis-related (PR) proteins: a focus on PR peptides. Plant Physiol. Biochem. 46, 941–950 10.1016/j.plaphy.2008.06.011 [DOI] [PubMed] [Google Scholar]
- Sherameti I., Venus Y., Drzewiecki C., Tripathi S., Dan V. M., Nitz I., et al. (2008). PYK10, a beta-glucosidase located in the endoplasmatic reticulum, is crucial for the beneficial interaction between Arabidopsis thaliana and the endophytic fungus Piriformospora indica. Plant J. 54, 428–439 10.1111/j.1365-313X.2008.03424.x [DOI] [PubMed] [Google Scholar]
- Shimizu B.-I., Miyagawa H., Ueno T., Sakata K., Watanabe K., Ogawa K. (2005). Morning glory systemically accumulates scopoletin and scopolin after interaction with Fusarium oxysporum. Z. Naturforsch. C 60, 83–90 Available online at: http://www.znaturforsch.com/ac/v60c/c60c.htm; http://www.znaturforsch.com/ac/v60c/s60c0083.pdf [DOI] [PubMed] [Google Scholar]
- Slotte T., Hazzouri K. M., Ågren J. A., Koenig D., Maumus F., Guo Y.-L., et al. (2013). The Capsella rubella genome and the genomic consequences of rapid mating system evolution. Nat. Genet. 45, 831–835 10.1038/ng.2669 [DOI] [PubMed] [Google Scholar]
- Sparkes I., Tolley N., Aller I., Svozil J., Osterrieder A., Botchway S., et al. (2010). Five Arabidopsis reticulon isoforms share endoplasmic reticulum location, topology, and membrane-shaping properties. Plant Cell 22, 1333–1343 10.1105/tpc.110.074385 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stefano G., Renna L., Moss T., McNew J. A., Brandizzi F. (2011). In Arabidopsis, the spatial and dynamic organization of the endoplasmic reticulum and Golgi apparatus is influenced by the integrity of the C-terminal domain of RHD3, a non-essential GTPase. Plant J. 69, 957–966 10.1111/j.1365-313X.2011.04846.x [DOI] [PubMed] [Google Scholar]
- Tang C.-S. (1973). Localization of benzyl glucosinolate and thioglucosidase in Carica papaya fruit. Phytochemistry 12, 769–773 10.1016/0031-9422(73)80676-4 [DOI] [Google Scholar]
- Teh O.-K., Moore I. (2007). An ARF-GEF acting at the Golgi and in selective endocytosis in polarized plant cells. Nature 448, 493–496 10.1038/nature06023 [DOI] [PubMed] [Google Scholar]
- Tolley N., Sparkes I., Craddock C. P., Eastmond P. J., Runions J., Hawes C., et al. (2010). Transmembrane domain length is responsible for the ability of a plant reticulon to shape endoplasmic reticulum tubules in vivo. Plant J. 64, 411–418 10.1111/j.1365-313X.2010.04337.x [DOI] [PubMed] [Google Scholar]
- Toyooka K., Okamoto T., Minamikawa T. (2000). Mass transport of proform of a KDEL-tailed cysteine proteinase (SH-EP) to protein storage vacuoles by endoplasmic reticulum-derived vesicle is involved in protein mobilization in germinating seeds. J. Cell Biol. 148, 453–464 10.1083/jcb.148.3.453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Loon L. C., Rep M., Pieterse C. M. (2006). Significance of inducible defenserelated proteins in infected plants. Annu. Rev. Phytopathol. 44, 135–162 [DOI] [PubMed] [Google Scholar]
- Varma A., Savita V., Sudha, Sahay N., Butehorn B., Franken P. (1999). Piriformospora indica, a cultivable plant-growth-promoting root endophyte. Appl. Environ. Microbiol. 65, 2741–2744 10.1007/s40003-012-0019-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X., Wang H., Wang J., Sun R., Wu J., Liu S., et al. (2011). The genome of the mesopolyploid crop species Brassica rapa. Nat. Genet. 43, 1035–1039 10.1038/ng.919 [DOI] [PubMed] [Google Scholar]
- Watanabe S., Matsumoto M., Hakomori Y., Takagi H., Shimada H., Sakamoto A. (2013a). The purine metabolite allantoin enhances abiotic stress tolerance through synergistic activation of abscisic acid metabolism. Plant Cell Environ. [Epub ahead of print]. 10.1111/pce.12218 [DOI] [PubMed] [Google Scholar]
- Watanabe S., Shimada T. L., Hiruma K., Takano Y. (2013b). Pathogen infection trial increases the secretion of proteins localized in the endoplasmic reticulum body of Arabidopsis. Plant Physiol. 163, 659–664 10.1104/pp.113.217364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H.-J., Zhang Z., Wang J.-Y., Oh D.-H., Dassanayake M., Liu B., et al. (2012). Insights into salt tolerance from the genome of Thellungiella salsuginea. Proc. Natl. Acad. Sci. U.S.A. 109, 12219–12224 10.1073/pnas.1209954109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu Z., Escamilla-Treviño L., Zeng L., Lalgondar M., Bevan D., Winkel B., et al. (2004). Functional genomic analysis of Arabidopsis thaliana glycoside hydrolase family 1. Plant Mol. Biol. 55, 343–367 10.1007/s11103-004-0790-1 [DOI] [PubMed] [Google Scholar]
- Xu Z.-Y., Lee K. H., Dong T., Jeong J. C., Jin J. B., Kanno Y., et al. (2012). A vacuolar β-glucosidase homolog that possesses glucose-conjugated abscisic acid hydrolyzing activity plays an important role in osmotic stress responses in Arabidopsis. Plant Cell 24, 2184–2199 10.1105/tpc.112.095935 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue J., Jørgensen M., Pihlgren U., Rask L. (1995). The myrosinase gene family in Arabidopsis thaliana: gene organization, expression and evolution. Plant Mol. Biol. 27, 911–922 10.1007/BF00037019 [DOI] [PubMed] [Google Scholar]
- Yamada K., Hara-Nishimura I., Nishimura M. (2011). Unique defense strategy by the endoplasmic reticulum body in plants. Plant Cell Physiol. 52, 2039–2049 10.1093/pcp/pcr156 [DOI] [PubMed] [Google Scholar]
- Yamada K., Nagano A. J., Nishina M., Hara-Nishimura I., Nishimura M. (2008). NAI2 is an endoplasmic reticulum body component that enables ER body formation in Arabidopsis thaliana. Plant Cell 20, 2529–2540 10.1105/tpc.108.059345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamada K., Nagano A. J., Nishina M., Hara-Nishimura I., Nishimura M. (2013). Identification of two novel endoplasmic reticulum body-specific integral membrane proteins. Plant Physiol. 161, 108–120 10.1104/pp.112.207654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang R., Jarvis D. E., Chen H., Beilstein M. A., Grimwood J., Jenkins J., et al. (2013). The reference genome of the halophytic plant Eutrema salsugineum. Front. Plant Sci. 4:46 10.3389/fpls.2013.00046 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Pontoppidan B., Xue J., Rask L., Meijer J. (2002). The third myrosinase gene TGG3 in Arabidopsis thaliana is a pseudogene specifically expressed in stamen and petal. Physiol. Plant. 115, 25–34 10.1034/j.1399-3054.2002.1150103.x [DOI] [PubMed] [Google Scholar]
- Zhang M., Wu F., Shi J., Zhu Y., Zhu Z., Gong Q., et al. (2013). ROOT HAIR DEFECTIVE3 family of dynamin-like GTPases mediates homotypic endoplasmic reticulum fusion and is essential for Arabidopsis development. Plant Physiol. 163, 713–720 10.1104/pp.113.224501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou C., Tokuhisa J., Bevan D., Esen A. (2012). Properties of β-thioglucoside hydrolases (TGG1 and TGG2) from leaves of Arabidopsis thaliana. Plant Sci. 191–192, 82–92 10.1016/j.plantsci.2012.02.004 [DOI] [PubMed] [Google Scholar]
- Züst T., Heichinger C., Grossniklaus U., Harrington R., Kliebenstein D. J., Turnbull L. A. (2012). Natural enemies drive geographic variation in plant defenses. Science 338, 116–119 10.1126/science.1226397 [DOI] [PubMed] [Google Scholar]