Abstract
Melioidosis is a severe infection caused by the flagellated bacterium Burkholderia pseudomallei. The nonsense polymorphism TLR51174C>T is associated with improved outcome in Thais with melioidosis. We hypothesized that other TLR5 variants may modulate the host response and determine outcome in melioidosis. We genotyped 12 TLR5 variants selected de novo from the HapMap database and examined the association of each with cytokines induced by flagellin stimulation of whole blood from healthy Thai subjects. We found a blunted cytokine response for three related markers that were in linkage disequilibrium with a non-synonymous variant, TLR51846T>C. Carriers of TLR51846T>C had broadly impaired cytokine responses induced by flagellin. TLR51846T>C was associated with protection against death in melioidosis patients (OR 0.62, 95% CI: 0.42-0.93, p=0.021). We observed no impairment in TLR51846C-dependent NF-κB activation, however, suggesting an alternative mechanism for the effect. We found that TLR51846T>C was in strong linkage disequilibrium with TLR51174C>T. Many of the blunted cytokine responses observed and the association of TLR51846T>C with survival in melioidosis patients may be attributable to TLR51174C>T, but we could not exclude an independent effect of TLR51846T>C. These data identify novel associations for TLR51846T>C, enhance our understanding of TLR5 genetic architecture in Thais, and provide future directions of study.
Keywords: melioidosis, TLR5, innate immune response, flagellin, genetic variation
Introduction
A primary host defence mechanism against invading pathogens is the innate immune response which is initiated upon the recognition of pathogen-associated molecular patterns (PAMPs) by pathogen sensors such as toll-like receptors (TLRs).1 These trans-membrane-receptors use leucine-rich repeat extracellular domains to distinguish the highly conserved structure of pathogens from self-components and mediate the signal via a cytoplasmic receptor homology domain.2
At least ten types of TLRs are present with different density on various cell types in blood and tissue such as macrophage, monocyte, dendritic cells, neutrophils and epithelial cells.1, 3 Each TLR recognizes a different group of PAMPs including lipolysaccharide (LPS) and flagellin from Gram-negative bacteria, lipoteichoic acid from Gram-positive bacteria, bacterial DNA, viral RNA and envelope glycoproteins. The activation of TLRs leads to a massive production of a variety of immune mediators, which are important triggers for inflammatory responses and the connection to adaptive immune response to infections.4-6 TLR5 recognizes flagellin and signals in a MyD88-dependent fashion to activate NF-κB.7, 8 This is in contrast to TLR2 or TLR4 which utilize the additional adaptor molecule TIRAP to recruit MyD88. TLR4 may also signal in a MyD88-independent but TRIF/TRAM-dependent fashion. The final result of these different PAMPs and TLR engagement is the activation of downstream signalling pathways, ultimately resulting in the production of several immune mediators.
TLR5 is the only TLR that responds to flagellin (FliC), a main protein of flagella. Human TLR5 is a type I transmembrane protein with a 642-amino acid leucine-rich extracellular domain, an 18-residue transmembrane domain, and a 198-residue cytoplasmic Toll homology signalling domain.9 Several Gram-negative bacteria use flagella to direct movement and facilitate pathogenicity in hosts.10-12 The flagellins from flagellated bacteria share a common structure which binds to TLR5 and precipitates a MyD88- and NF-κB-dependent host response.3
Melioidosis is a tropical disease caused by Burkholderia pseudomallei, a flagellated Gram-negative soil saprophyte. The disease is endemic mostly in Southeast Asia and Northern Australia.13 In northeast Thailand, the overall melioidosis mortality rate is 40%, and reaches 90% in severe infection.14, 15 The importance of the innate immune response in control of infection and pathophysiology of lethal melioidosis is documented in several studies. In septicaemic melioidosis, TLRs – including TLR5 – and associated molecules are highly expressed.16 Melioidosis patients who die have elevated plasma levels of pro-inflammatory cytokines IL-12, IL-18, IL-15 and IFN-γ compared to those who survive.17
We have previously demonstrated that a common nonsense TLR5 polymorphism that truncates the receptor and abolishes flagellin signalling (TLR51174C>T)9 is associated with improved outcome from melioidosis.18 These data demonstrate the importance of flagellin sensing in melioidosis. Others have shown strong evidence for selection of TLR5 polymorphisms. 19 We hypothesized that other TLR5 genetic variants may modulate flagellin signalling and determine outcome from melioidosis independently of TLR51174C>T. To test this hypothesis, we analysed the association of candidate TLR5 polymorphisms identified de novo from Asian HapMap databases with whole blood responses to flagellin in healthy Thai subjects; examined the association of a tagged coding variant with death from melioidosis; and assayed innate immune activation by the coding variant in transfected cells.
Results
Blood cytokine responses to flagellin are highly correlated
We stimulated blood from 300 healthy Thai subjects with Salmonella typhimurium flagellin and measured eight cytokine and chemokine levels 6 h later using a multiplex bead assay (Chantratita et al, submitted for publication).18 The median age of subjects was 33 years, IQR 24-41. One hundred and twenty seven (42.3%) of the 300 subjects were female. Given non-normal distributions of the cytokine data, we performed log transformations before analyses, resulting in normally distributed residuals. We next examined the data to determine whether any of the eight mediators were highly correlated. We found that IL-6 was highly correlated (pairwise correlation coefficient (r)>0.73) with all mediators (IL-8, TNF-α, IL-10, MCP-1, G-CSF, and IL-1β) except IL-1ra (Table 1). As a result, we restricted our subsequent analyses to IL-6 and IL-1ra.
Table 1. Correlation between blood cytokines induced by flagellin.
| Inter-cytokine correlation1 | ||||||||
|---|---|---|---|---|---|---|---|---|
|
| ||||||||
| IL-6 | IL-8 | TNF-α | IL-10 | MCP-1 | IL-1ra | G-CSF | IL-1β | |
|
|
||||||||
| IL-6 | 1.00 | |||||||
| IL-8 | 0.87 | 1.00 | ||||||
| TNF-α | 0.94 | 0.88 | 1.00 | |||||
| IL-10 | 0.86 | 0.75 | 0.79 | 1.00 | ||||
| MCP-1 | 0.73 | 0.65 | 0.63 | 0.70 | 1.00 | |||
| IL-1ra | 0.56 | 0.41 | 0.51 | 0.55 | 0.58 | 1.00 | ||
| G-CSF | 0.87 | 0.82 | 0.85 | 0.82 | 0.61 | 0.46 | 1.00 | |
| IL-1β | 0.83 | 0.87 | 0.84 | 0.71 | 0.66 | 0.50 | 0.75 | 1.00 |
Pairwise correlation coefficient (r) between log-transformed cytokines induced by stimulation of whole blood from 300 healthy subjects with 500 ng/mL flagellin.
TLR5 variants are associated with flagellin-induced cytokine responses in blood
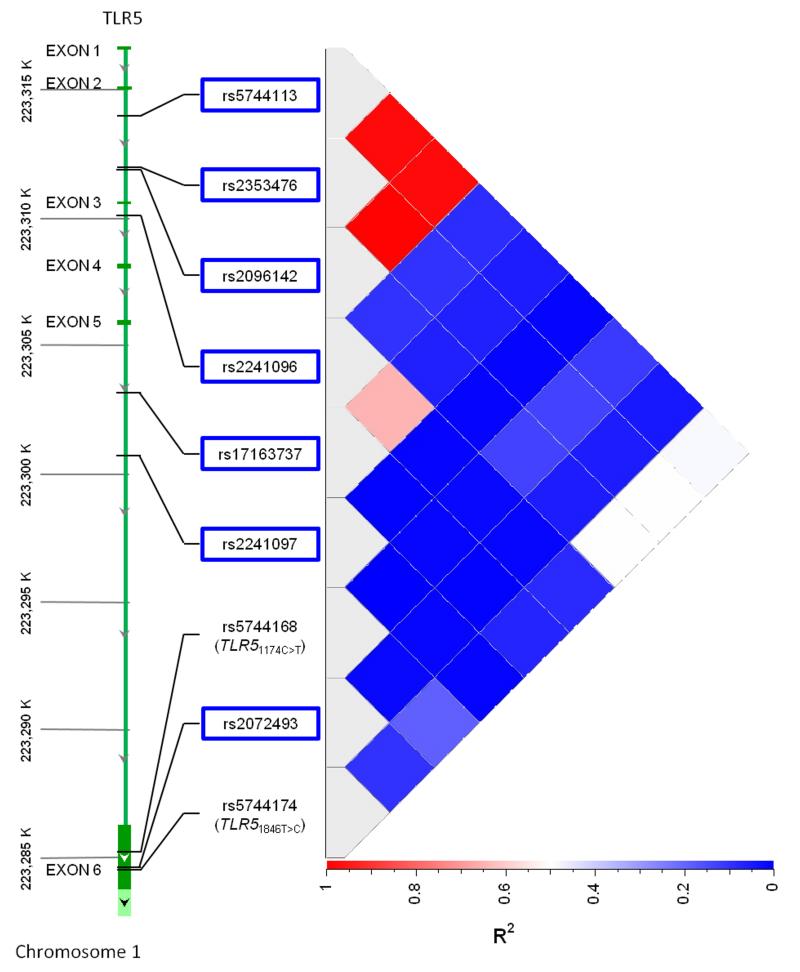
We then tested whether genetic variants in TLR5 were associated with IL-6 or IL-1ra induced by flagellin stimulation of blood. We identified 12 candidate single nucleotide polymorphisms (SNPs) in TLR5 selected as described in the methods (rs2072493, rs5744173, rs2096142, rs5744175, rs2241096, rs5744176, rs2241097, rs2353476, rs5744113, rs5744166, rs17163737, rs5744167). We genotyped these variants in the 300 subjects. No assays failed but there was no variation in five variants so these were not considered further. The call rates for the seven remaining variants (rs2072493, rs2096142, rs2241096, rs2241097, rs2353476, rs5744113, rs17163737) were 100% and no variant failed a test for Hardy Weinberg equilibrium. We found that three variants, rs2096142, rs2353476, and rs5744113 were significantly associated with both IL-6 and IL-1ra in additive models (Table 2). We did not adjust for multiple comparisons given the possibility that these variants were not independent. On further investigation, rs2096142, rs2353476, and rs5744113 were in very high linkage disequilibrium with each other (r2 = 0.98-1.0) (Figure 1). We then searched the HapMap Han Chinese in Beijing (HCB) population using the Genome Variation Server (www.gvs.gs.washington.edu) to identify whether the haplotype containing these variants also contained a non-synonymous coding variant that might be functional. We found that the linkage disequilibrium (LD) among the three variants was also very high in this population (r2=0.93-1.0). The only coding variant we identified in LD with the three variants was rs5744174 (r2=0.63-0.68). This non-synonymous coding variant exchanges T for C at position 1846, corresponding to an amino acid change from phenylalanine to leucine at position 616 in the extracellular domain. Recently published evidence suggests that this variant has been subjected to significant selective pressure in Yorubans.19 The genomic evolutionary rate profiling (GERP) score, indicating variability across mammals, is −11.2.20
Table 2. Association of TLR5 variants with blood cytokine response to flagellin.
| Variant | Minor allele frequency |
IL-6 | Cytokine | IL-1ra | |||
|---|---|---|---|---|---|---|---|
|
P values1 |
|||||||
| Dominant | Recessive | Additive | Dominant | Recessive | Additive | ||
| rs2072493 | 0.26 | 0.202 | 0.72 | 0.246 | 0.468 | 0.957 | 0.579 |
| rs2096142 | 0.23 | 0.027 | 0.039 | 0.010 | 0.261 | 0.002 | 0.032 |
| rs2241096 | 0.25 | 0.127 | 0.139 | 0.070 | 0.043 | 0.128 | 0.027 |
| rs2241097 | 0.04 | 0.324 | 0.259 | 0.245 | 0.051 | 0.745 | 0.058 |
| rs2353476 | 0.23 | 0.027 | 0.039 | 0.010 | 0.261 | 0.002 | 0.032 |
| rs5744113 | 0.23 | 0.023 | 0.058 | 0.011 | 0.253 | 0.005 | 0.042 |
| rs17163737 | 0.21 | 0.017 | 0.619 | 0.051 | 0.050 | 0.509 | 0.052 |
P values determined by linear regression of association of each variant with log transformed cytokine level induced by stimulation of whole blood from 300 healthy subjects with 500 ng/mL flagellin.
Figure 1. TLR5 variant linkage disequilibrium map in healthy Thai subjects.
r2 values for seven screening TLR5 variants outlined by blue boxes, TLR51846T>C (rs5744174) and TLR51174C>T (rs5744168) in 300 healthy subjects.
TLR51846T>C is associated with flagellin-induced cytokine responses in blood
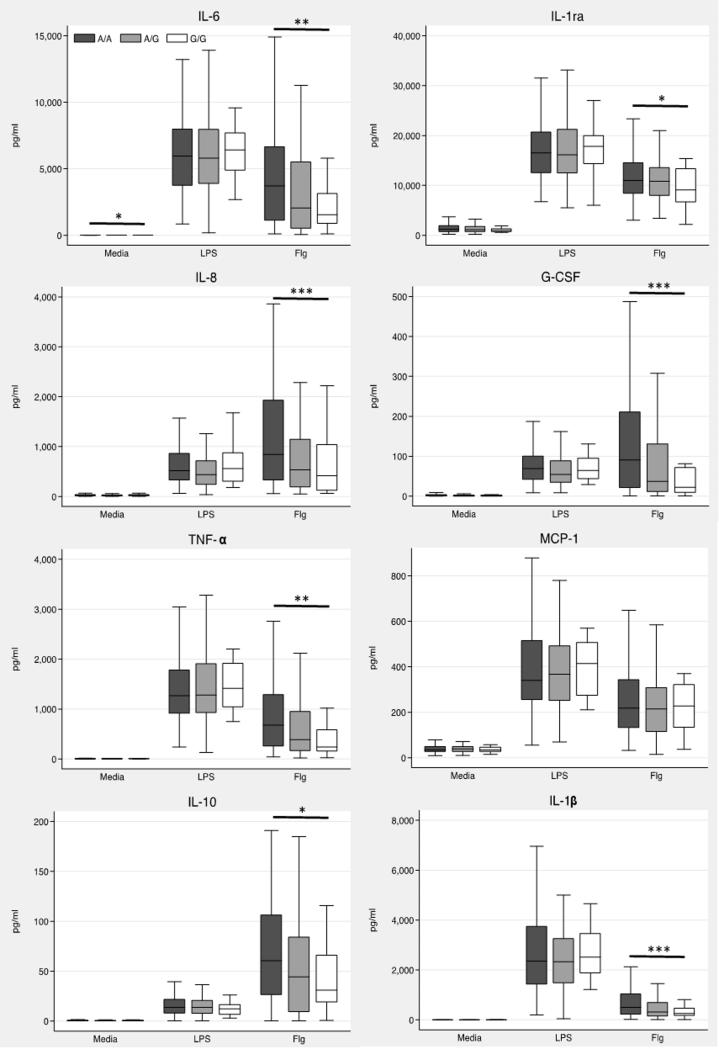
We then genotyped TLR51846T>C in the 300 Thai subjects (call rate 100%) and found that the LD with our initial three variants was modest (r2 ~0.5) (Figure 1). The minor allele frequency of TLR51846T>C was 0.22 and the variant was in Hardy Weinberg equilibrium (p=0.76). We tested whether TLR51846T>C was associated with cytokine responses to flagellin using an additive genetic model. We found that the variant was highly associated with lower levels of all mediators except MCP-1 (Table 3 and Figure 2). Monocyte counts, age, and sex are associated with cytokine responses to various bacterial ligands (Chantratita et al, unpublished observations). Inclusion of these variables in the model had only modest effects on the point estimates (Supplemental Table 1).
Table 3. Association of TLR51846T>C with blood cytokine release induced by flagellin.
| Cytokine | Coefficient1 | 95% CI | P |
|---|---|---|---|
| IL-6 | −0.168 | −0.280 – −0.056 | 0.003 |
| IL-8 | −0.180 | −0.277 – −0.084 | 2.8 × 10−4 |
| TNF-α | −0.153 | −0.252 – −0.054 | 0.003 |
| IL-10 | −0.151 | −0.273 – −0.029 | 0.015 |
| MCP-1 | −0.048 | −0.114 – 0.018 | 0.155 |
| IL-1ra | −0.038 | −0.072 – −0.004 | 0.028 |
| G-CSF | −0.253 | −0.400 – −0.107 | 7.7 × 10−4 |
| IL-1β | −0.169 | −0.263 – −0.076 | 4.1 × 10−4 |
Linear regression coefficient for TLR51846T>C on log transformed cytokine levels induced by stimulation of whole blood from 300 healthy subjects with 500 ng/mL flagellin.
Figure 2. Cytokine responses induced by flagellin are lower in carriers of the TLR51846T>C.
Whole blood from 300 healthy subjects was incubated with media alone, flagellin 500 ng/mL or E. coli LPS 10 ng/mL for 6 h at 37 °C. Plasma cytokines were assayed by multiplex bead assay. Boxes show the median and interquartile range; whiskers show upper and lower adjacent values. Outside values are not shown for clarity. *, p≤0.05; **, p≤0.01; ***, p≤0.001 by linear regression for an additive genetic model.
TLR51846T>C is associated with survival from melioidosis
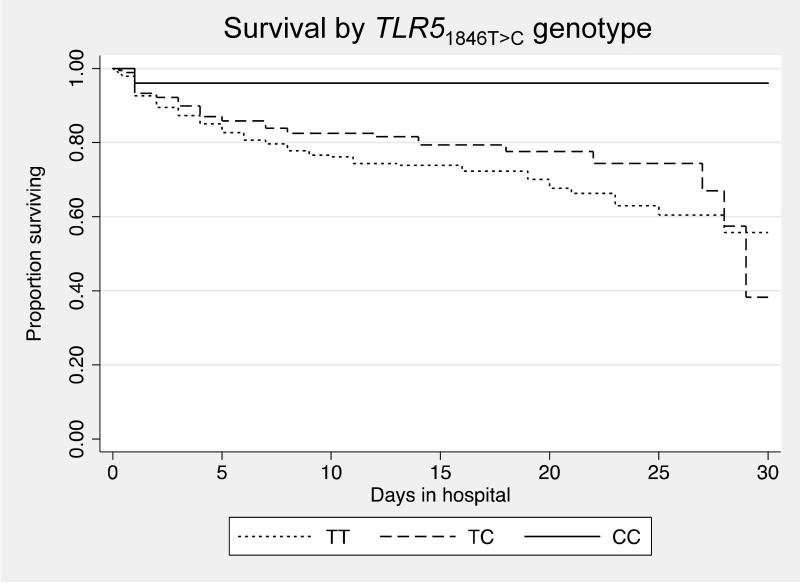
To determine whether TLR51846T>C was associated with clinical outcome from flagellated infection, we genotyped the variant in a cohort of Thai subjects with melioidosis. Melioidosis is infection caused by the Gram-negative and flagellated soil saprophyte B. pseudomallei. Mortality is very high in northeast Thailand. This cohort has been previously described.18, 21 Of 600 individuals with melioidosis, the median age was 49, IQR 39-60, and 274 (45.7%) were female. One hundred and forty three (23.8%) died during their hospitalization or were discharged home in extremis for palliative care. DNA was available for genotyping in 588 subjects for whom outcome at hospital discharge was known. The variant was genotyped in 580 individuals (call rate 98.6%) and did not deviate from Hardy Weinberg equilibrium in survivors to hospital discharge (p=0.89). We found that TLR51846T>C was significantly associated with protection against in-hospital death in an unadjusted analysis (Table 4). We then tested whether the variant was associated with any pre-existing condition (diabetes, chronic liver disease, renal insufficiency, and steroid use) to identify potential confounders to consider in the analysis but found no associations. In the adjusted model we included factors that were independently associated with death: age, diabetes, renal insufficiency, pneumonia, bacteraemia, and urinary tract infection. This adjusted model confirmed a significantly protective effect of TLR51846T>C against death from melioidosis (odds ratio (OR) 0.62, 95% confidence interval (CI): 0.42-0.93, p=0.021). Kaplan-Meier curves demonstrating in-hospital survival for carriers of each genotype were significantly different (p=0.029) (Fig. 3).
Table 4. Unadjusted association of TLR51846T>C with death in subjects with melioidosis.
| Death | |||||
|---|---|---|---|---|---|
| Genotype | No | Yes | OR | 95% CI | P 1 |
| TT | 263 (73.5%) | 95 (26.5%) | |||
| TC | 156 (79.6%) | 40 (20.4%) | 0.64 | 0.44-0.91 | 0.014 |
| CC | 24 (92.3%) | 2 (7.7%) | |||
Additive model P value calculated by logistic regression.
Figure 3. Survival from melioidosis is enhanced for carriers of TLR51846T>C.
Kaplan–Meier in-hospital survival curves are plotted for melioidosis subjects, grouped by genotype. Curves are significantly different by the test for trend (p = 0.029).
TLR51174C>T underlies some of the effect of TLR51846T>C on death in melioidosis
We have previously shown a protective association of TLR51174C>T with death in melioidosis 18. TLR51174C>T is not listed in the HapMap database but we hypothesized that TLR51174C>T might be in LD with TLR51846T>C and potentially explain the association of TLR51846T>C with outcome from melioidosis. We first tested whether TLR51846T>C and TLR51174C>T were in LD in the 300 healthy subjects. We found that r2=0.19 (Figure 1) but D′=1.0 and the association between genotypes displayed in a contingency table was significant (p=4 × 10−15) indicating that the two variants were in LD. No subjects with the major genotype TLR51846TT carried a TLR51174T allele but 25% of the TLR51846TC subjects and 44% of the TLR51846CC subjects did (Table 5). We next tested the association of TLR51846T>C with death in melioidosis in the 500 hospitalized subjects who did not carry the rare variant of TLR51174C>T and found that the effect was no longer significant (adjusted OR 0.87, 95% CI 0.56-1.33, p=0.51). However, the power to reject the null hypothesis of no association between death and TLR51846T>C in this subset that independent of TLR51174C>T is ≤50% when the true OR is ≥0.62. A haplotype analysis of both variants on death confirmed a non-significant effect of TLR51846C on death in the absence of TLR51174T and showed a significant protective association of both TLR51174T and TLR51846C with death (Table 6). The frequency of the haplotype containing TLR51846C but not TLR51174T was too low to test the association with death. Thus, we concluded that, for these two non-synonymous TLR5 variants in LD, there is strong evidence for an effect on death of both variants together while the effect of TLR51846T>C alone is indeterminate.
Table 5. Relationship between TLR51846T>C and TLR51174C>T.
| Variant | TLR5 1174C>T | |||
|---|---|---|---|---|
| Genotype1 | CC | CT | TT | |
| TLR5 1846T>C | TT | 183 (100%) | 0 (0.0%) | 0 (0.0%) |
| TC | 77 (76.2%) | 24 (24.8%) | 0 (0.0%) | |
| CC | 9 (56.3%) | 7 (43.8%) | 0 (0.0%) |
Genotypes determined in 300 healthy subjects.
Table 6. Association of TLR5 haplotype with death in melioidosis patients.
| Haplotype1 |
TLR51174C>T _TLR51846T>C |
|||
|---|---|---|---|---|
| 00 | 01 | 10 | 11 | |
| Frequency | 0.77 | 0.15 | 0.001 | 0.08 |
| OR (95% CI) | 0.85 (0.57-1.25) |
- | 0.25 (0.11-0.57) |
|
| P value | 0.41 | - | 0.001 | |
0 indicates common allele and 1 indicates rare allele at each of the two TLR5 loci. The major haplotype (00) is set as the reference; the 10 haplotype is not analyzed due to its low frequency. Associations are determined by logistic regression, adjusting for age, diabetes, renal insufficiency, pneumonia, bacteraemia, and urinary tract infection.
The association of TLR51846T>C on flagellin-induced cytokine release is partly attributable to TLR51174C>T
We have previously shown that TLR51174C>T significantly reduces all eight cytokine responses induced by stimulation of blood by flagellin.18 This work extended original studies by Hawn et al who also demonstrated the lack of flagellin-induced NF-κB activation by this variant, independent of TLR51846T>C.9 To investigate whether TLR51846T>C was associated with reduced cytokine release in whole blood independent of TLR51174C>T we repeated our analysis of the effect of TLR51846T>C on cytokine release in response to flagellin excluding carriers of the TLR51174C>T variant allele. In comparison to all subjects, we found that the effect of TLR51846T>C on lowered cytokine response in this subset was attenuated for all cytokines (excluding MCP-1). The relative reduction in effect of TLR51846T>C ranged from 43% for IL-6 to 16% for IL-1ra. The point estimates were uniformly negative and the p value for a joint test of all eight differences, accounting for correlation among the cytokines, was 0.08. The associations between TLR51846T>C and reduced cytokine response remained significant individually only for G-CSF and IL-1β (Table 7). These data suggest that the association of TLR51846T>C with impaired cytokine release may be partly attributable to TLR51174C>T.
Table 7. Association of TLR51846T>C with blood cytokine release induced by flagellin independent of TLR51174C>T.
| Cytokine | Coefficient1 | 95% CI | P |
|---|---|---|---|
| IL-6 | −0.095 | −0.226 – 0.035 | 0.152 |
| IL-8 | −0.110 | −0.223 – 0.003 | 0.057 |
| TNF-α | −0.080 | −0.196 – 0.036 | 0.174 |
| IL-10 | −0.095 | −0.237 – 0.047 | 0.188 |
| MCP-1 | −0.022 | −0.100 – 0.056 | 0.575 |
| IL-1ra | −0.032 | −0.072 – 0.007 | 0.115 |
| G-CSF | −0.205 | −0.381 – −0.030 | 0.022 |
| IL-1β | −0.120 | −0.229 – −0.010 | 0.032 |
Linear regression coefficient for TLR51846T>C on log transformed cytokine levels induced by stimulation of whole blood from 269 healthy TLR51174CC subjects with 500 ng/mL flagellin.
TLR51846T>C is not dysfunctional in NF-κB signalling
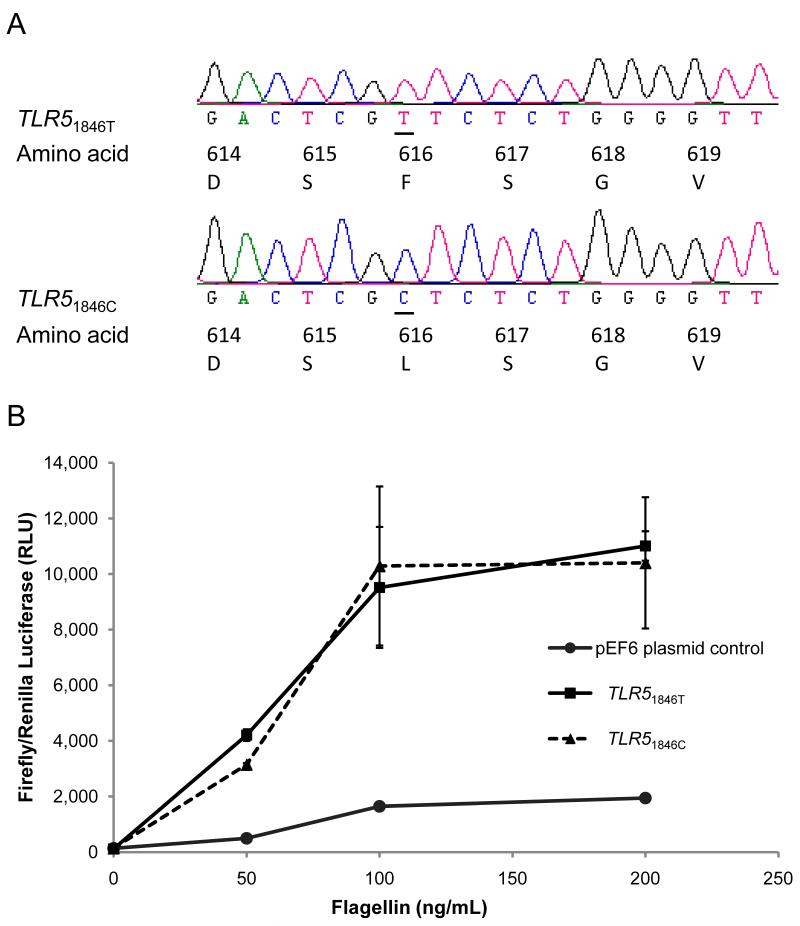
To further investigate whether TLR51846T>C was independently associated with altered flagellin signalling we performed site directed mutagenesis on a previously described TLR5 construct9 to construct a mutant TLR5 plasmid corresponding to TLR51846T>C. Sequence verification confirmed the base change at position 1846 and the absence of any other known non-synonymous variation in the full TLR5 coding sequence in the plasmid (Figure 4A). We transfected HEK 293 cells with TLR51846T and TLR51846C and assayed NF-κB activation by luciferase reporter assay following stimulation with 50, 100 and 200 ng/mL flagellin. The cells transfected with TLR51846C showed no difference in NF-κB activation compared to TLR51846T-transfected cells (Figure 4B). These data indicated that in this construct TLR51846T>C does not impair flagellin-driven innate immune activation via NF-κB.
Figure 4. TLR51846T>C does not impair flagellin-induced NF-κB activation.
(A) Sequence chromatogram indicating a single T to C substitution in TLR5 at 1846 in the mutant plasmid (lower picture) compared to the wild type plasmid (upper picture). Alteration of corresponding amino acid from phenylalanine to leucine at position 616 is shown. (B) HEK 293 cells were transfected with 40 ng of pEF6 plasmid control, TLR51846T or TLR51846C. Cells were stimulated with 50,100, or 200 ng/mL of purified flagellin from S. typhimurium and NF-κB activity was quantified by dual luciferase assay.
Differential linkage disequilibrium of variants with TLR51846T>C across populations
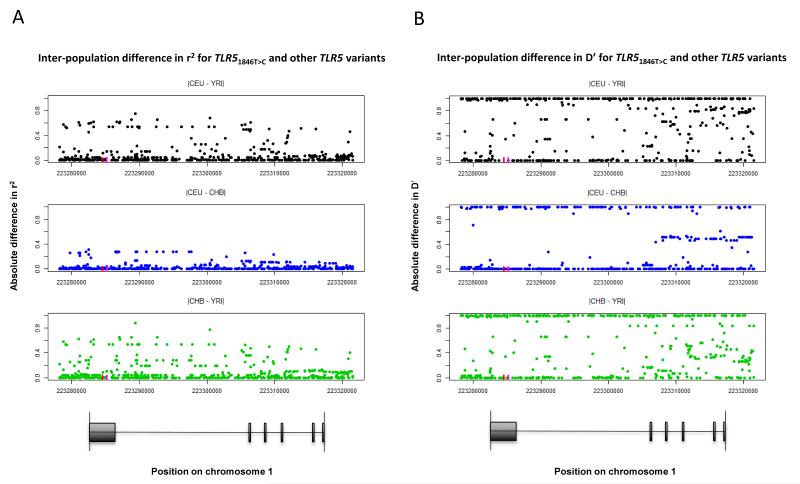
In light of the evidence in favour of selection for TLR51846T>C in Yorubans19, but our inability to conclusively determine a clinical effect of TLR51846T>C independent of TLR51174C>T in Thais, we hypothesized that there may be significant population-level differences in LD structure in TLR5. We examined the distribution of variants in LD with TLR51846T>C in three populations (Yorubans, Utah residents with ancestry from northern and western Europe, and Han Chinese) sequenced by the 1000 Genomes Project.22 The degree of LD between TLR51846T>C and TLR51174C>T was comparable in the three populations (r2: 0.26, 0.12, 0.16; D′: 1.0, 1.0, 1.0, respectively) raising the possibility that positive selection for TLR51846T>C in Yorubans may be related to TLR51174C>T. However, there were marked inter-population differences in the degree of LD between other variants and TLR51846T>C (Figure 5). Overall, the Yoruban population differed the most from the other two populations. Moreover, the LD structure of the entire TLR5 gene differed greatly across the three populations (Supplemental Figures 1 and 2). Thus, additional variation in TLR5 may confound differential evolutionary and clinical associations.
Figure 5. Population differences in TLR51846T>C LD structure.
(A) r2 and (B) D′ was calculated for TLR51846T>C and each of 733 polymorphic sites in the TLR5 gene region from Phase 1 1000 Genomes data for Utah residents with ancestry from northern and western Europe (CEU), Yoruban (YRI) and Han Chinese (CHB) populations. The pairwise difference in these LD measures for each population pair is shown for the 233 variants for which at least one difference was non-zero. The red line indicates the position of TLR51846T>C and the magenta line indicates the position of TLR51174C>T on chromosome 1. A map of TLR5 is shown for reference.
Discussion
The results of our study identify a tagged TLR5 non-synonymous coding variant, TLR51846T>C, that is associated with reduced cytokine responses to flagellin in human blood and further show that it is associated with protection against death in melioidosis. In vitro studies demonstrate that the variant does not alter NF-κB activation induced by flagellin, however, and the cytokine and clinical effects observed are in large part driven by previously described hypofunctional variant that is in LD with TLR51846T>C, TLR51174C>T. Collectively, these data identify new biological and clinical associations for TLR51846T>C, advance our understanding of genetic variation in TLR5 in Thais, and confirm the relevance of TLR5 variation in determining outcome in melioidosis.
Although it is known that TLRs are important in regulating innate immune responses3, only a few reports have demonstrated the clinical significance of TLR5 polymorphisms in bacterial infections. Studies demonstrating an association have mostly been restricted to a functional nonsense polymorphism TLR51174C>T.9, 18 Accordingly, we searched for additional genetic variation in TLR5 that might have functional effects and clinical relevance in melioidosis. Knowledge of innate immune genetic variation in Thais is limited, necessitating that we choose candidate variants based on publically available data from other populations. Our data therefore provide the first published data on frequency and LD patterns of TLR5 genetic variation in Thais.
Our main finding in this work is that TLR51846T>C (F616L) is associated with broadly reduced cytokine responses to flagellin and protection against death in melioidosis. Other investigators have reported associations of TLR51846T>C with immune responses and clinical disease. The variant is associated with earlier spontaneous Hepatitis B virus e antigen (HBeAg) seroconversion (disappearance of serum HBeAg and the presence of anti- HBeAg for >6 months) and higher levels of IFN-γ production in Taiwanese patients with chronic Hepatitis B infection.23 TLR51846T>C is also associated with gastric cancer and interacts with Helicobacter pylori infection in a cohort of Chinese subjects.24 The variant is also associated with higher measles-specific IFN-γ responses.25 The associations with viral infections raise the question of a role for this variant that is unrelated to flagellin sensing.
A key question that arises from our data is whether TLR51846T>C has a biological or clinical effect that is independent of the nonsense polymorphism with which it is in LD, TLR51174C>T. Notably, we did not observe any impairment in NF-κB activation upon stimulation of cells expressing TLR51846C alone. This observation confirms the results of other investigators.9, 26 In contrast, a more recent study reports a TLR51846C-dependent decrement in NF-κB activation.19 Although in the extracellular domain, amino acid 616 is outside the known flagellin binding sites27, 28 and variation at this site is predicted to be benign by Polyphen2.29 A potential explanation for observed differences between investigators is the source of the TLR5 plasmids studied in these experiments. We show marked differences in LD structure in TLR5 across populations and potentially confounding effects of additional TLR5 variation in plasmids from subjects of different ancestries. In addition, a plasmid generated by mutagenesis of a single locus may yield different functional effects than DNA derived from a carrier of the desired genotype (and accompanying haplotype). We largely attribute our associations of TLR51846T>C with blunted whole blood cytokine responses and improved clinical outcome to TLR51174C>T. However, some of the effects of TLR51846T>C on G-CSF and IL-1β release in whole blood persisted despite excluding carriers of TLR51174C>T from our analysis. Our study of the association of TLR51846T>C with death from melioidosis in the absence of TLR51174C>T was underpowered and therefore inconclusive. Moreover, in contrast to their observations for TLR51846T>C, Wu et al did not find that TLR51174C>T was associated with age of HBeAg seroconversion.23 Thus, we cannot exclude the possibility that TLR51846T>C has an immunomodulatory effect independent of TLR51174C>T. Further work to elucidate the functional effects of TLR5 variants in different populations is needed.
Our study has several limitations. We cannot exclude that there may be additional interactive effects of unmeasured TLR5 variation that could alter our results. We analysed a relatively restricted number of TLR5 variants in our subjects but we anticipate analysing this region more intensively in the future. Although population stratification may confound any genetic analysis, we have previously shown that we did not detect significant evidence of this in our Sappasithiprasong Hospital cohort.18
In conclusion, we demonstrate novel biological and clinical associations for TLR51846T>C, present new information about TLR5 genetic structure in Thais, and underscore the importance of understanding the role of TLR5 in modulating outcome in melioidosis. Further work is needed to dissect the relative biological contributions of differential TLR5 variation in diverse populations.
Materials and methods
Human subjects
Immuno-assay studies: Three-hundred healthy Thai subjects donating blood at the blood donation centre at Sappasithiprasong Hospital in 2010 were recruited for participation as previously described.18 Subjects were included if their stated age was between 18 and 60 years, and did not report any history of immunodeficiency or inflammatory conditions, chronic diseases, pregnancy in the past six months, anti-inflammatory medication use in the past week, antibiotic use in the past month, vaccination in the past six months, heavy exercise or alcohol consumption in the past 24 h, or smoking in the past month. Those who met enrollment criteria gave written informed consent to participate and provided a post-donation blood sample.
Melioidosis cohort: The study cohort has been previously described.18 Clinical data and blood were obtained from 588 patients with melioidosis admitted to Sappasithiprasong Hospital, Ubon Ratchathani, Thailand from 1999 to 2005. A study team screening patients with clinical signs of infection cultured blood, urine and other relevant samples (for example, abscess aspirates) for B. pseudomallei. Inclusion into the melioidosis cohort required a positive culture for B. pseudomallei from a sample collected by the study team or independently by hospital clinicians. For the analyses of genotype with death from melioidosis, deaths were defined as those individuals who died during their hospitalization or were discharged home in extremis for palliative care. Consent for enrollment into clinical studies of melioidosis was obtained from subjects or their representatives at the time of recruitment.
The Ethical Review Committee for Research in Human Subjects, Ministry of Public Health, Thailand; the Ethical Review Committee for Research in Human Subjects, Sappasithiprasong Hospital, Ubon Ratchathani, Thailand; the Ethics Committee of the Faculty of Tropical Medicine, Mahidol University, Bangkok, Thailand; and the University of Washington Human Subjects Division Institutional Review Board approved the studies.
Immuno-assays
Three hundred and eighty microliters of fresh whole blood in citrate mixed 1:1 with RPMI media was added to pre-prepared plates containing 20 μL of various stimulants. For this study, the stimulants analysed were S. typhimurium flagellin at a final concentration of 500 ng/mL and Escherichia coli LPS 10 ng/mL (List Biologicals). Plates were incubated at 37 °C on a shaking incubator under 5% CO2 for 6 h before being spun down and plasma removed and frozen at −80 °C. IL-6, IL-8, TNF-α, IL-10, MCP-1, IL-1ra, G-CSF, and IL-1β were later assayed in duplicate on a multiplex bead system (Luminex, Austin, TX) using reagents from R&D Systems (Minneapolis, MN). If either (or both) duplicate(s) was less than the lower limit of detection of the assay, it was assigned the lowest detectable value of any of the 300 subjects’ duplicate raw results for that cytokine induced by the same stimulant, a form of data imputation. The amended duplicate values were then averaged to provide a final cytokine concentration. A complete blood count with differential was performed in the hospital clinical laboratory for each subject at the time of phlebotomy.
Genotyping
DNA was extracted from whole blood of healthy donors using the QIAamp DNA Blood Midi Kit (Qiagen, Hilden, Germany) and from whole blood of patients with melioidosis using Nucleon BACC3 kits (GE Healthcare, Buckinghamshire, UK). SNP selection of non-synonymous coding variants was performed by searching the HapMap project database (http://hapmap.ncbi.nlm.nih.gov) and based on functional prediction using a FastSNP analysis (http://fastsnp.ibms.sinica.edu.tw).30 Tag SNPs were selected from the Han Chinese in Beijing and Japanese in Tokyo populations in the HapMap database for variants with a minor allele frequency at least 2% using the HapMap tag-SNP picker option. TLR51174C>T (rs5744168) was not listed in the HapMap database for these populations. Genotyping was performed using Fluidigm SNPtype assays on a Biomark microfluidics real time PCR system or using ABI TaqMan assays on an ABI Prism 7900 (Carlsbad, CA).
Cloning and site-directed mutagenesis
A T to C mutation in TLR5 at position 1846 was constructed with a QuikChange II site-directed mutagenesis kit (Stratagene, CA). The coding region of TLR5 was amplified from 7.5 ng plasmid template TLR5-RNF-V5-pEF6 which contained full-length human TLR5 coding region cloned into the pEF6/V5-His TOPO expression vector (Invitrogen, CA) 9 in a 25 μL reaction volume. TLR5-RNF-V5-pEF6 was a gift from Thomas R. Hawn, University of Washington, Seattle, WA. Mutagenic primers were designed from TLR5 mRNA sequence (Gen Bank accession no. AB060695.1) using software from http://labtools.stratagene.com/QC. The forward primer was modified to replace T with C at the position 1846 corresponding to rs5744174. The sequences of primers from 5′ to 3′ were as follows: F-GTGTACCCTGACTCGCTCTCTGGGGTTTCCC and R-GGGAAACCCCAGAGAGCGAGTCAGGGTACAC. The PCR reaction mixture was performed with PfuUltra high-fedelity (HF) DNA polymerase according to the protocol described by manufacturer and the conditions were as follows: initial denaturation at 95 °C for 1min; 12 cycles of denaturation at 95 °C for 1 min, annealing at 60 °C for 1 min, and extension at 68 °C for 15 min; and the reaction was cooled to 4 °C for 1 min. The parental methylated and hemimethylated DNA remaining in amplification product was digested with 1 μL of 10 U/μL Dpn I restriction enzyme at 37 °C for 1 h. The mutant TLR5 plasmid (containing 5,840 bp plasmid and 2,574 bp TLR5 insert) was transformed into E. coli DH5α. The clones with mutant constructs were selected from plates containing 100 μg/mL ampicillin. Colonies containing the desired plasmids were analysed by PCR using T7 promoter forward and BGH reverse primers flanking the TLR5 mutant alleles from 5′ to 3′ as follows: F- TAATACGACTCACTATAGGG and R- TAGAAGGCACAGTCGAGG. The plasmids were extracted from E. coli using a QIAprep spin miniprep kit (Qiagen) and DNA sequence was verified by DNA sequencing (Macrogen, Korea). The sequence of the mutant clone was analysed and aligned with the sequence of TLR5-RNF-V5-pEF6 wild type using Seqman software (Laser gene) version 8 (DNASTAR Inc., WI, USA) and MEGA version 5.31 The plasmid from the selected clone was extracted for HEK 293 cell transfection experiment using Endo free plasmid purification (Qiagen).
Transfections
HEK 293 cells (CRL-1573, ATCC) were transfected and stimulated as described previously with some modifications.9, 18 HEK 293 cells were cultured in complete Dulbecco’s Modification of Eagle’s Medium (DMEM) without sodium pyruvate and were supplemented with L-glutamine, 4.5 g/L glucose (Mediatech Cellgro, VA) and 10% fetal bovine serum (FBS) (Hyclone, Illinois). Transfection was performed with 5 × 104 cells/well HEK 293 cells in a 96-well plate with 100 μL transfection reagent containing 0.5 μL Polyfect reagent (Qiagen, Hilden, Germany), 10 ng pNiFty-Luc firefly luciferase reporter (Invivogen, CA), 1 ng pRL-TK Renilla luciferase control reporter (Promega, WI) and 40 ng of plasmid (TLR51846T wild type or TLR51846C variant or pEF6 control). Before transfection, the plasmids were prepared in DMEM without FBS and mixed with Polyfect reagent by vortexing for 10 s, incubated at room temperature for 10 min and the volume adjusted to 100 μL with complete DMEM medium.
Transfected HEK 293 cells were stimulated with 100 μL of 100, 200 and 400 ng/mL of ultra purified flagellin from S. typhimurium (Invivogen) to obtain final concentration 50, 100 and 200 ng/mL. The plate was centrifuged at 200 ×g at room temperature for 3 min and incubated at 37 °C with 5% CO2 for 24 h. The cells were washed 2 times with 100 μL Dulbecco’s Phosphate-Buffered Saline (DPBS) without Ca2+ or Mg2+ (Mediatech Cellgro, VA), lysed with 40 μL passive lysis buffer with shaking at room temperature for 15 min. Twenty microliters of lysate was measured for luminescence using the Dual-Lucifearase Reporter Assay (Promega) on a GloMax Multi Detection System (Promega). The experiment was performed in triplicate.
Statistics
As immune-assay cytokine concentrations were often non-normally distributed, they were log10 transformed to obtain normally distributed residuals for estimation of pairwise correlations and for estimation of genotype association with cytokine levels. Associations between genotype as a predictor of cytokine concentration were performed using linear regression, coding the genotype to reflect dominant, recessive, or additive genetic models. This is equivalent to testing for differences in mean cytokine levels between genotype groups (and allows adjustment for confounding). Where indicated, this model was adjusted for age, sex, and monocyte count. Associations between genotype or haplotype and death were tested using logistic regression, assuming an additive genetic model. This model was adjusted for age, diabetes, renal insufficiency, pneumonia, bacteraemia, and urinary tract infection. Survival analyses were performed with a test for trend across genotypes. Statistics were performed with R (version 2.15.2; GNU project [http://www.r-project.org]) or with Stata 11.2 (College Station, TX), incorporating the functions pwld for test for linkage disequilibrium, genhw to test for deviation from Hardy-Weinberg equilibrium using the exact test, and haplologit for haplotype analyses. Golden Helix SNP & Variation Suite software (Bozeman, MT) was used to calculate linkage disequilibrium and generate maps. A two-sided p value ≤0.05 was considered significant.
Supplementary Material
Supplemental Figure 1. TLR5 LD Map (r2) LD map (r2) of TLR5 variation in Yoruban (YRI), Utah residents with ancestry from northern and western Europe(CEU), and Han Chinese (CHB) populations using Phase 1 1000 Genomes Project data.
Supplemental Figure 2. TLR5 LD Map (D′)LD map (D′) of TLR5 variation in Yoruban (YRI), Utah residents with ancestry from northern and western Europe (CEU), and Han Chinese (CHB) populations using Phase 1 1000 Genomes Project data.
Supplemental Table 1. Adjusted association of TLR51846T>C with blood cytokine release induced by flagellin
Acknowledgements
The authors acknowledge the support of the staff and patients at Sappasithiprasong Hospital and Mahidol-Oxford Tropical Medicine Research Unit; DNA extraction by Premjit Amornchai, Aunchalee Thanwisai, and Malinee Oyuchua; assistance generating immuno-assays from Mark Wurfel; and generation of LD maps by Johanna Robertson.
This work was supported by the Wellcome Trust [087769/Z/08/Z to N.C.]; the NIHR Cambridge Biomedical Research Centre [to S.J.P.]; National Institutes of Health awards [K08HL094759 and R01HL113382 to T.E.W.] and by the Doris Duke Charitable Foundation [to T.E.W.]. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Footnotes
Conflict of Interest The authors declare there is no conflict of interest.
References
- 1.Takeda K, Kaisho T, Akira S. Toll-like receptors. Annu Rev Immunol. 2003;21:335–76. doi: 10.1146/annurev.immunol.21.120601.141126. [DOI] [PubMed] [Google Scholar]
- 2.O’Neill LA, Bowie AG. The family of five: TIR-domain-containing adaptors in Toll-like receptor signalling. Nat Rev Immunol. 2007;7(5):353–64. doi: 10.1038/nri2079. [DOI] [PubMed] [Google Scholar]
- 3.Mogensen TH. Pathogen recognition and inflammatory signaling in innate immune defenses. Clin Microbiol Rev. 2009;22(2):240–73. doi: 10.1128/CMR.00046-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Liu L, Botos I, Wang Y, Leonard JN, Shiloach J, Segal DM, et al. Structural basis of toll-like receptor 3 signaling with double-stranded RNA. Science. 2008;320(5874):379–81. doi: 10.1126/science.1155406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Jin MS, Kim SE, Heo JY, Lee ME, Kim HM, Paik SG, et al. Crystal structure of the TLR1-TLR2 heterodimer induced by binding of a tri-acylated lipopeptide. Cell. 2007;130(6):1071–82. doi: 10.1016/j.cell.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 6.Park BS, Song DH, Kim HM, Choi BS, Lee H, Lee JO. The structural basis of lipopolysaccharide recognition by the TLR4-MD-2 complex. Nature. 2009;458(7242):1191–5. doi: 10.1038/nature07830. [DOI] [PubMed] [Google Scholar]
- 7.Zhao Y, Yang J, Shi J, Gong YN, Lu Q, Xu H, et al. The NLRC4 inflammasome receptors for bacterial flagellin and type III secretion apparatus. Nature. 2011;477(7366):596–600. doi: 10.1038/nature10510. [DOI] [PubMed] [Google Scholar]
- 8.Yoon SI, Kurnasov O, Natarajan V, Hong M, Gudkov AV, Osterman AL, et al. Structural basis of TLR5-flagellin recognition and signaling. Science. 2012;335(6070):859–64. doi: 10.1126/science.1215584. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hawn TR, Verbon A, Lettinga KD, Zhao LP, Li SS, Laws RJ, et al. A common dominant TLR5 stop codon polymorphism abolishes flagellin signaling and is associated with susceptibility to legionnaires’ disease. J Exp Med. 2003;198(10):1563–72. doi: 10.1084/jem.20031220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Hayashi F, Smith KD, Ozinsky A, Hawn TR, Yi EC, Goodlett DR, et al. The innate immune response to bacterial flagellin is mediated by Toll-like receptor 5. Nature. 2001;410(6832):1099–103. doi: 10.1038/35074106. [DOI] [PubMed] [Google Scholar]
- 11.Andersen-Nissen E, Smith KD, Strobe KL, Barrett SL, Cookson BT, Logan SM, et al. Evasion of Toll-like receptor 5 by flagellated bacteria. Proc Natl Acad Sci U S A. 2005;102(26):9247–52. doi: 10.1073/pnas.0502040102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Smith KD, Andersen-Nissen E, Hayashi F, Strobe K, Bergman MA, Barrett SL, et al. Toll-like receptor 5 recognizes a conserved site on flagellin required for protofilament formation and bacterial motility. Nat Immunol. 2003;4(12):1247–53. doi: 10.1038/ni1011. [DOI] [PubMed] [Google Scholar]
- 13.Wiersinga WJ, Currie BJ, Peacock SJ. Melioidosis. N Engl J Med. 2012;367(11):1035–44. doi: 10.1056/NEJMra1204699. [DOI] [PubMed] [Google Scholar]
- 14.Wiersinga WJ, van der Poll T, White NJ, Day NP, Peacock SJ. Melioidosis: insights into the pathogenicity of Burkholderia pseudomallei. Nat Rev Microbiol. 2006;4(4):272–82. doi: 10.1038/nrmicro1385. [DOI] [PubMed] [Google Scholar]
- 15.Limmathurotsakul D, Peacock SJ. Melioidosis: a clinical overview. Br Med Bull. 2011;99:125–39. doi: 10.1093/bmb/ldr007. [DOI] [PubMed] [Google Scholar]
- 16.Wiersinga WJ, Wieland CW, Dessing MC, Chantratita N, Cheng AC, Limmathurotsakul D, et al. Toll-like receptor 2 impairs host defense in gram-negative sepsis caused by Burkholderia pseudomallei (Melioidosis) PLoS Med. 2007;4(7):e248. doi: 10.1371/journal.pmed.0040248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Friedland JS, Suputtamongkol Y, Remick DG, Chaowagul W, Strieter RM, Kunkel SL, et al. Prolonged elevation of interleukin-8 and interleukin-6 concentrations in plasma and of leukocyte interleukin-8 mRNA levels during septicemic and localized Pseudomonas pseudomallei infection. Infect Immun. 1992;60(6):2402–8. doi: 10.1128/iai.60.6.2402-2408.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.West TE, Chantratita N, Chierakul W, Limmathurotsakul D, Wuthiekanun V, Myers ND, et al. Impaired TLR5 Functionality Is Associated with Survival in Melioidosis. J Immunol. 2013;190(7):3373–9. doi: 10.4049/jimmunol.1202974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grossman SR, Andersen KG, Shlyakhter I, Tabrizi S, Winnicki S, Yen A, et al. Identifying recent adaptations in large-scale genomic data. Cell. 2013;152(4):703–13. doi: 10.1016/j.cell.2013.01.035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Cooper GM, Stone EA, Asimenos G, Green ED, Batzoglou S, Sidow A. Distribution and intensity of constraint in mammalian genomic sequence. Genome Res. 2005;15(7):901–13. doi: 10.1101/gr.3577405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.West TE, Chierakul W, Chantratita N, Limmathurotsakul D, Wuthiekanun V, Emond MJ, et al. Toll-like receptor 4 region genetic variants are associated with susceptibility to melioidosis. Genes Immun. 2012;13(1):38–46. doi: 10.1038/gene.2011.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, et al. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491(7422):56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wu JF, Chen CH, Ni YH, Lin YT, Chen HL, Hsu HY, et al. Toll-like receptor and hepatitis B virus clearance in chronic infected patients: a long-term prospective cohort study in Taiwan. J Infect Dis. 2012;206(5):662–8. doi: 10.1093/infdis/jis420. [DOI] [PubMed] [Google Scholar]
- 24.Zeng HM, Pan KF, Zhang Y, Zhang L, Ma JL, Zhou T, et al. Genetic variants of toll-like receptor 2 and 5, helicobacter pylori infection, and risk of gastric cancer and its precursors in a chinese population. Cancer Epidemiol Biomarkers Prev. 2011;20(12):2594–602. doi: 10.1158/1055-9965.EPI-11-0702. [DOI] [PubMed] [Google Scholar]
- 25.Dhiman N, Ovsyannikova IG, Vierkant RA, Ryan JE, Pankratz VS, Jacobson RM, et al. Associations between SNPs in toll-like receptors and related intracellular signaling molecules and immune responses to measles vaccine: preliminary results. Vaccine. 2008;26(14):1731–6. doi: 10.1016/j.vaccine.2008.01.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Merx S, Zimmer W, Neumaier M, Ahmad-Nejad P. Characterization and functional investigation of single nucleotide polymorphisms (SNPs) in the human TLR5 gene. Hum Mutat. 2006;27(3):293. doi: 10.1002/humu.9409. [DOI] [PubMed] [Google Scholar]
- 27.Mizel SB, West AP, Hantgan RR. Identification of a sequence in human toll-like receptor 5 required for the binding of Gram-negative flagellin. J Biol Chem. 2003;278(26):23624–9. doi: 10.1074/jbc.M303481200. [DOI] [PubMed] [Google Scholar]
- 28.Andersen-Nissen E, Smith KD, Bonneau R, Strong RK, Aderem A. A conserved surface on Toll-like receptor 5 recognizes bacterial flagellin. J Exp Med. 2007;204(2):393–403. doi: 10.1084/jem.20061400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Adzhubei IA, Schmidt S, Peshkin L, Ramensky VE, Gerasimova A, Bork P, et al. A method and server for predicting damaging missense mutations. Nat Methods. 2010;7(4):248–9. doi: 10.1038/nmeth0410-248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yuan HY, Chiou JJ, Tseng WH, Liu CH, Liu CK, Lin YJ, et al. FASTSNP: an always up-to-date and extendable service for SNP function analysis and prioritization. Nucleic Acids Res. 2006;34(Web Server issue):W635–41. doi: 10.1093/nar/gkl236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tamura K, Peterson D, Peterson N, Stecher G, Nei M, Kumar S. MEGA5: molecular evolutionary genetics analysis using maximum likelihood, evolutionary distance, and maximum parsimony methods. Mol Biol Evol. 2011;28(10):2731–9. doi: 10.1093/molbev/msr121. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplemental Figure 1. TLR5 LD Map (r2) LD map (r2) of TLR5 variation in Yoruban (YRI), Utah residents with ancestry from northern and western Europe(CEU), and Han Chinese (CHB) populations using Phase 1 1000 Genomes Project data.
Supplemental Figure 2. TLR5 LD Map (D′)LD map (D′) of TLR5 variation in Yoruban (YRI), Utah residents with ancestry from northern and western Europe (CEU), and Han Chinese (CHB) populations using Phase 1 1000 Genomes Project data.
Supplemental Table 1. Adjusted association of TLR51846T>C with blood cytokine release induced by flagellin