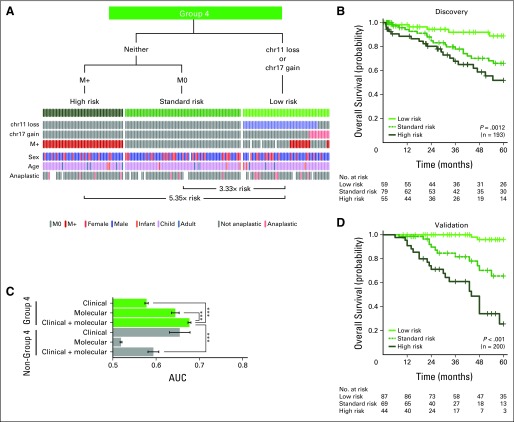
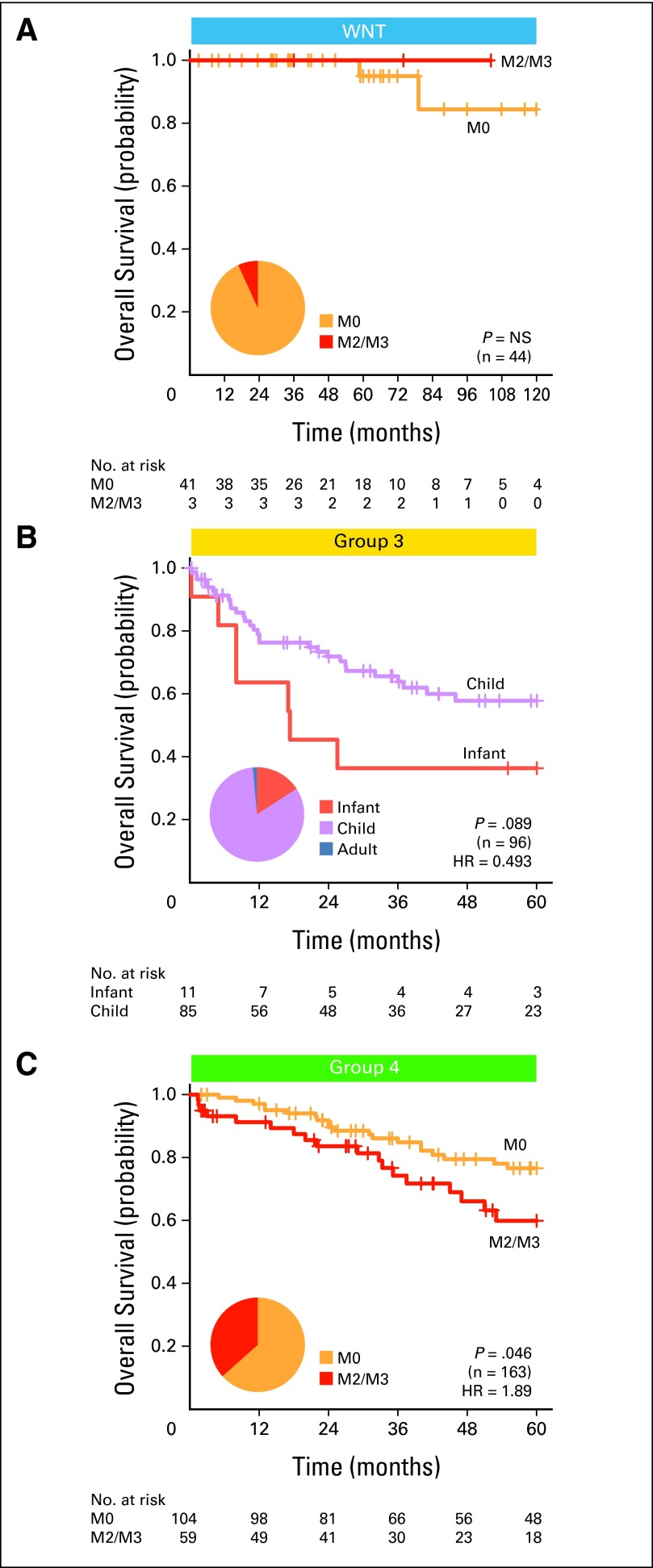
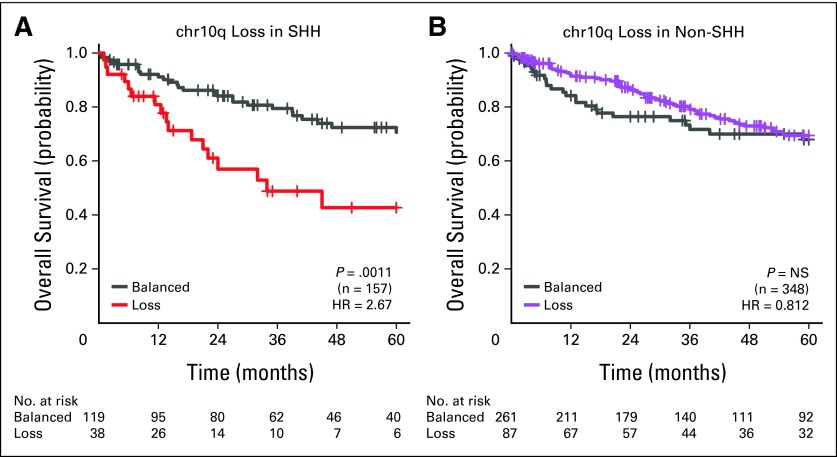
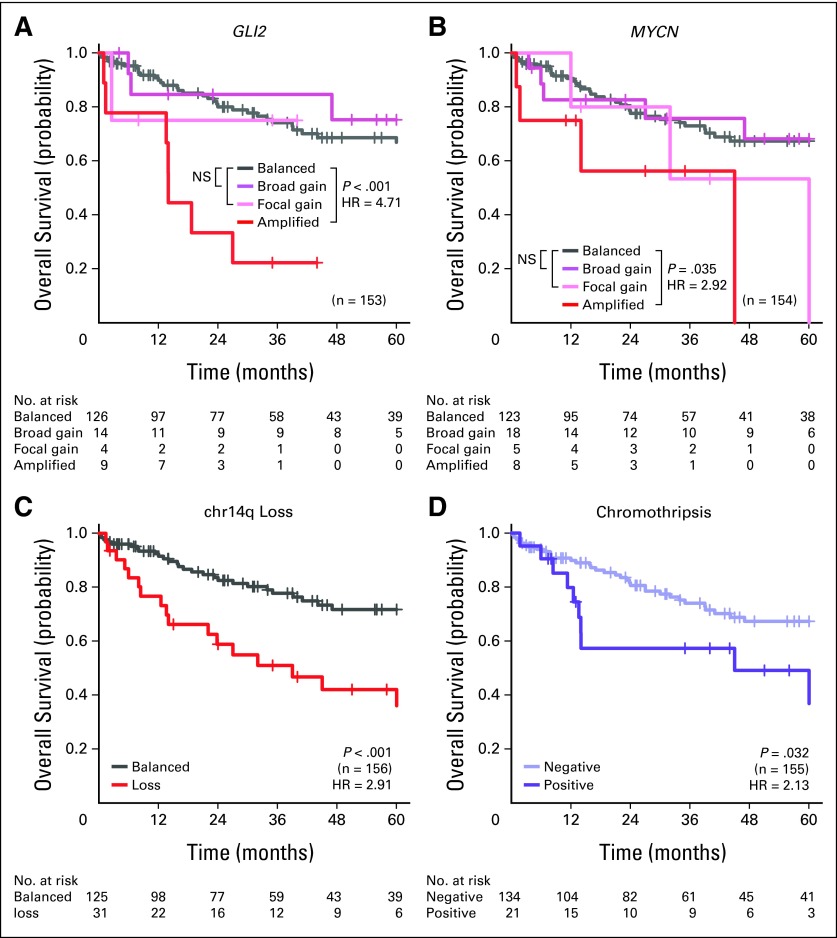
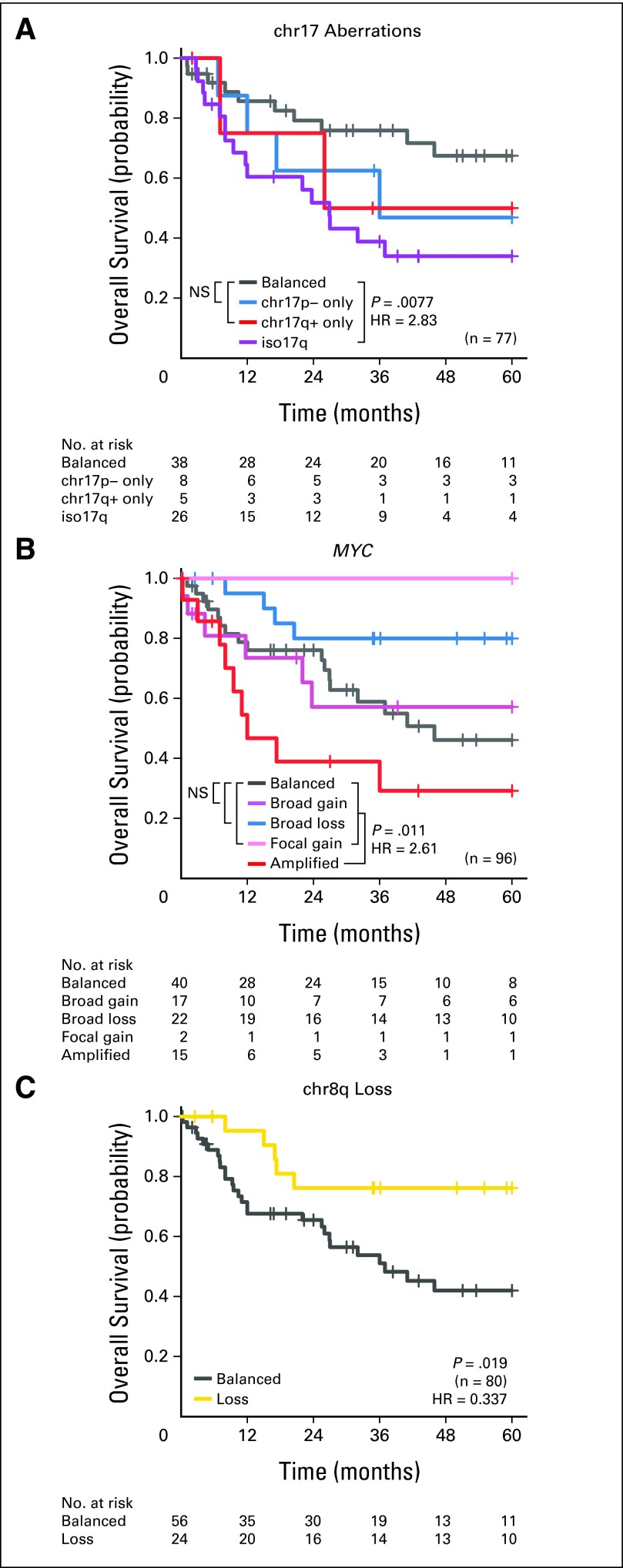
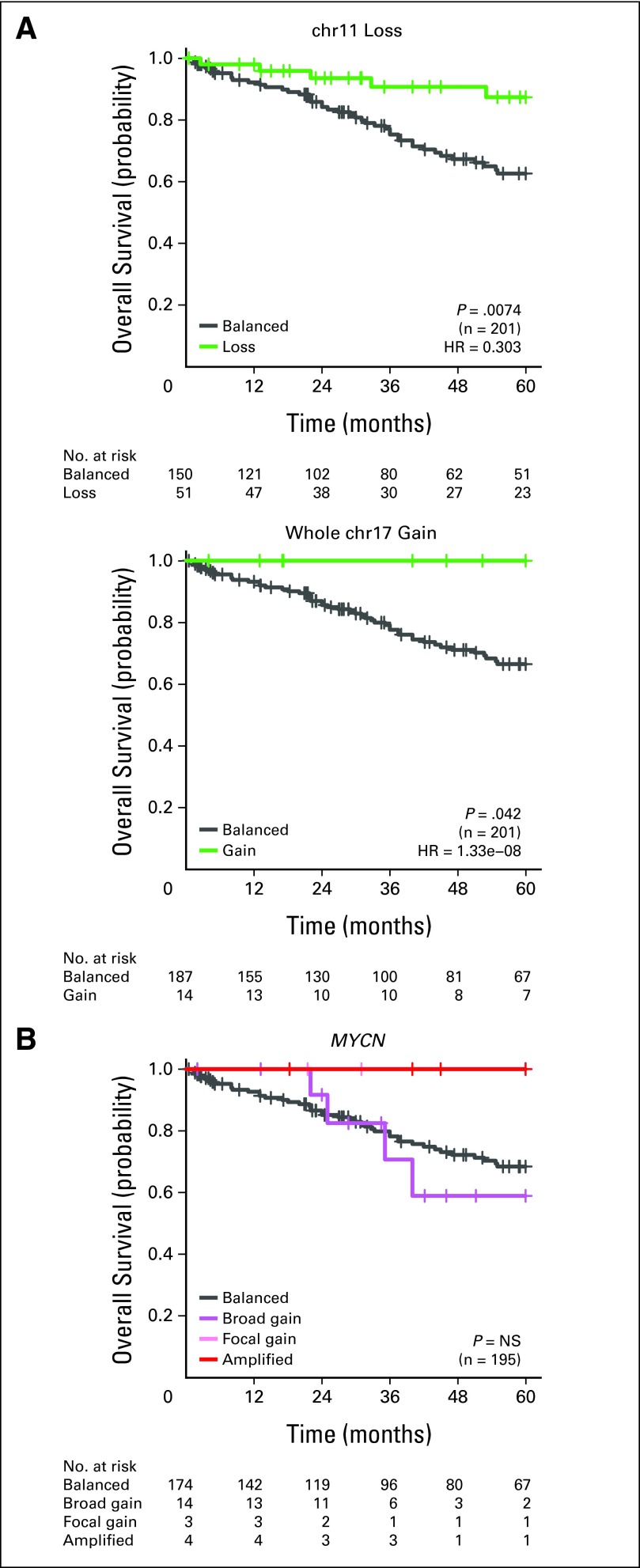
David JH Shih
David JH Shih
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Paul A Northcott
Paul A Northcott
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Marc Remke
Marc Remke
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Andrey Korshunov
Andrey Korshunov
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Vijay Ramaswamy
Vijay Ramaswamy
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Marcel Kool
Marcel Kool
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Betty Luu
Betty Luu
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Yuan Yao
Yuan Yao
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Xin Wang
Xin Wang
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Adrian M Dubuc
Adrian M Dubuc
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Livia Garzia
Livia Garzia
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
John Peacock
John Peacock
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Stephen C Mack
Stephen C Mack
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Xiaochong Wu
Xiaochong Wu
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Adi Rolider
Adi Rolider
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
A Sorana Morrissy
A Sorana Morrissy
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Florence MG Cavalli
Florence MG Cavalli
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
David TW Jones
David TW Jones
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Karel Zitterbart
Karel Zitterbart
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Claudia C Faria
Claudia C Faria
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Ulrich Schüller
Ulrich Schüller
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Leos Kren
Leos Kren
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Toshihiro Kumabe
Toshihiro Kumabe
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Teiji Tominaga
Teiji Tominaga
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Young Shin Ra
Young Shin Ra
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Miklós Garami
Miklós Garami
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Peter Hauser
Peter Hauser
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Jennifer A Chan
Jennifer A Chan
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Shenandoah Robinson
Shenandoah Robinson
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
László Bognár
László Bognár
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Almos Klekner
Almos Klekner
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Ali G Saad
Ali G Saad
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Linda M Liau
Linda M Liau
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Steffen Albrecht
Steffen Albrecht
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Adam Fontebasso
Adam Fontebasso
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Giuseppe Cinalli
Giuseppe Cinalli
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Pasqualino De Antonellis
Pasqualino De Antonellis
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Massimo Zollo
Massimo Zollo
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Michael K Cooper
Michael K Cooper
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Reid C Thompson
Reid C Thompson
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Simon Bailey
Simon Bailey
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Janet C Lindsey
Janet C Lindsey
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Concezio Di Rocco
Concezio Di Rocco
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Luca Massimi
Luca Massimi
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Erna MC Michiels
Erna MC Michiels
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Stephen W Scherer
Stephen W Scherer
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Joanna J Phillips
Joanna J Phillips
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Nalin Gupta
Nalin Gupta
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Xing Fan
Xing Fan
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Karin M Muraszko
Karin M Muraszko
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Rajeev Vibhakar
Rajeev Vibhakar
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Charles G Eberhart
Charles G Eberhart
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Maryam Fouladi
Maryam Fouladi
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Boleslaw Lach
Boleslaw Lach
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Shin Jung
Shin Jung
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Robert J Wechsler-Reya
Robert J Wechsler-Reya
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Michelle Fèvre-Montange
Michelle Fèvre-Montange
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Anne Jouvet
Anne Jouvet
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Nada Jabado
Nada Jabado
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Ian F Pollack
Ian F Pollack
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
William A Weiss
William A Weiss
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Ji-Yeoun Lee
Ji-Yeoun Lee
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Byung-Kyu Cho
Byung-Kyu Cho
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Seung-Ki Kim
Seung-Ki Kim
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Kyu-Chang Wang
Kyu-Chang Wang
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Jeffrey R Leonard
Jeffrey R Leonard
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Joshua B Rubin
Joshua B Rubin
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Carmen de Torres
Carmen de Torres
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Cinzia Lavarino
Cinzia Lavarino
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Jaume Mora
Jaume Mora
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Yoon-Jae Cho
Yoon-Jae Cho
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Uri Tabori
Uri Tabori
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
James M Olson
James M Olson
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Amar Gajjar
Amar Gajjar
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Roger J Packer
Roger J Packer
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Stefan Rutkowski
Stefan Rutkowski
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Scott L Pomeroy
Scott L Pomeroy
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Pim J French
Pim J French
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Nanne K Kloosterhof
Nanne K Kloosterhof
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Johan M Kros
Johan M Kros
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Erwin G Van Meir
Erwin G Van Meir
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Steven C Clifford
Steven C Clifford
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Franck Bourdeaut
Franck Bourdeaut
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Olivier Delattre
Olivier Delattre
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
François F Doz
François F Doz
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Cynthia E Hawkins
Cynthia E Hawkins
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
David Malkin
David Malkin
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Wieslawa A Grajkowska
Wieslawa A Grajkowska
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Marta Perek-Polnik
Marta Perek-Polnik
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Eric Bouffet
Eric Bouffet
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
James T Rutka
James T Rutka
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Stefan M Pfister
Stefan M Pfister
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,
Michael D Taylor
Michael D Taylor
1David J.H. Shih, Marc Remke, Vijay Ramaswamy, Betty Luu, Yuan Yao, Xin Wang, Adrian M. Dubuc, Livia Garzia, John Peacock, Stephen C. Mack, Xiaochong Wu, Adi Rolider, A. Sorana Morrissy, Florence M.G. Cavalli, Claudia C. Faria, Stephen W. Scherer, Uri Tabori, Cynthia E. Hawkins, David Malkin, Eric Bouffet, James T. Rutka, and Michael D. Taylor, Hospital for Sick Children; David J.H. Shih, Marc Remke, Vijay Ramaswamy, Yuan Yao, Xin Wang, Adrian M. Dubuc, John Peacock, Stephen C. Mack, and Michael D. Taylor, University of Toronto, Toronto; Boleslaw Lach, McMaster University, Hamilton, Ontario; Jennifer A. Chan, University of Calgary, Calgary, Alberta; Steffen Albrecht, Adam Fontebasso, and Nada Jabado, McGill University, Montreal, Quebec, Canada; Paul A. Northcott, Andrey Korshunov, Marcel Kool, David T.W. Jones, and Stefan M. Pfister, German Cancer Research Center; Stefan M. Pfister, University Hospital Heidelberg, Heidelberg; Ulrich Schüller, Ludwig-Maximilians-University, Munich; Stefan Rutkowski, University Medical Center Hamburg-Eppendorf, Hamburg, Germany; Karel Zitterbart, Masaryk University School of Medicine; Karel Zitterbart and Leos Kren, University Hospital Brno, Brno, Czech Republic; Toshihiro Kumabe and Teiji Tominaga, Tohoku University Graduate School of Medicine, Sendai, Japan; Young Shin Ra, University of Ulsan, Asan Medical Center; Ji-Yeoun Lee, Byung-Kyu Cho, Seung-Ki Kim, and Kyu-Chang Wang, Seoul National University Children's Hospital, Seoul; Shin Jung, Chonnam National University Research Institute of Medical Sciences, Chonnam National University Hwasun Hospital and Medical School, Chonnam, South Korea; Peter Hauser and Miklós Garami, Semmelweis University, Budapest; László Bognár and Almos Klekner, University of Debrecen, Medical and Health Science Centre, Debrecen, Hungary; Shenandoah Robinson, Boston Children's Hospital; Scott L. Pomeroy, Harvard Medical School, Boston, MA; Ali G. Saad, University of Arkansas for Medical Sciences, Little Rock, AR; Linda M. Liau, David Geffen School of Medicine, University of California Los Angeles, Los Angeles; Joanna J. Phillips, Nalin Gupta, and William A. Weiss, University of California San Francisco, San Francisco; Robert J. Wechsler-Reya, Sanford-Burnham Medical Research Institute, La Jolla; Yoon-Jae Cho, Stanford University School of Medicine, Stanford, CA; Giuseppe Cinalli, Ospedale Santobono-Pausilipon; Pasqualino De Antonellis and Massimo Zollo, University of Naples; Massimo Zollo, Ceinge Biotecnologie Avanzate, Naples; Concezio Di Rocco and Luca Massimi, Catholic University Medical School, Rome, Italy; Michael K. Cooper and Reid C. Thompson, Vanderbilt Medical Center, Nashville; Amar Gajjar, St Jude Children's Research Hospital, Memphis, TN; Simon Bailey, Janet C. Lindsey, and Steven C. Clifford, Newcastle University, Northern Institute for Cancer Research, Newcastle upon Tyne, United Kingdom; Erna M.C. Michiels, Pim J. French, Nanne K. Kloosterhof, and Johan M. Kros, Erasmus Medical Center, Rotterdam, the Netherlands; Xing Fan and Karin M. Muraszko, University of Michigan Medical School, Ann Arbor, MI; Rajeev Vibhakar, University of Colorado Denver, Aurora, CO; Charles G. Eberhart, Johns Hopkins University School of Medicine, Baltimore, MD; Maryam Fouladi, University of Cincinnati, Cincinnati Children's Hospital Medical Center, Cincinnati, OH; Michelle Fèvre-Montange and Anne Jouvet, Université de Lyon, Lyon; Franck Bourdeaut, Olivier Delattre, and François F. Doz, Institut Curie and Institut National de la Santé et de la Recherche Médicale U830; François F. Doz, University Paris Descartes, Paris, France; Ian F. Pollack, University of Pittsburgh School of Medicine, Pittsburgh, PA; Jeffrey R. Leonard and Joshua B. Rubin, Washington University School of Medicine, St Louis Children's Hospital, St Louis, MO; Carmen de Torres, Cinzia Lavarino, and Jaume Mora, Hospital Sant Joan de Déu, Barcelona, Spain; James M. Olson, Fred Hutchinson Cancer Research Center, Seattle, WA; Roger J. Packer, Children's National Medical Center, Washington, DC; Erwin G. Van Meir, Emory University School of Medicine, Winship Cancer Institute, Atlanta, GA; Wieslawa A. Grajkowska and Marta Perek-Polnik, Children's Memorial Health Institute, Warsaw, Poland; and Claudia C. Faria, Hospital de Santa Maria, Lisbon, Portugal.
1,✉