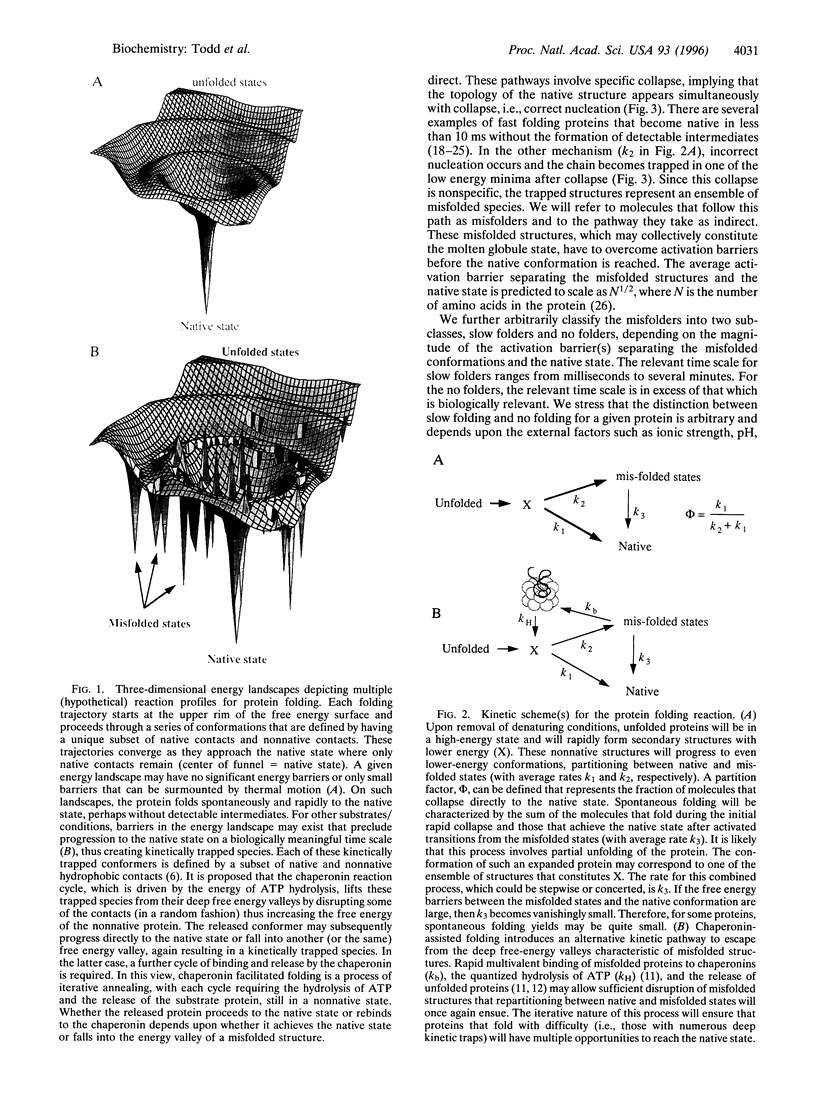
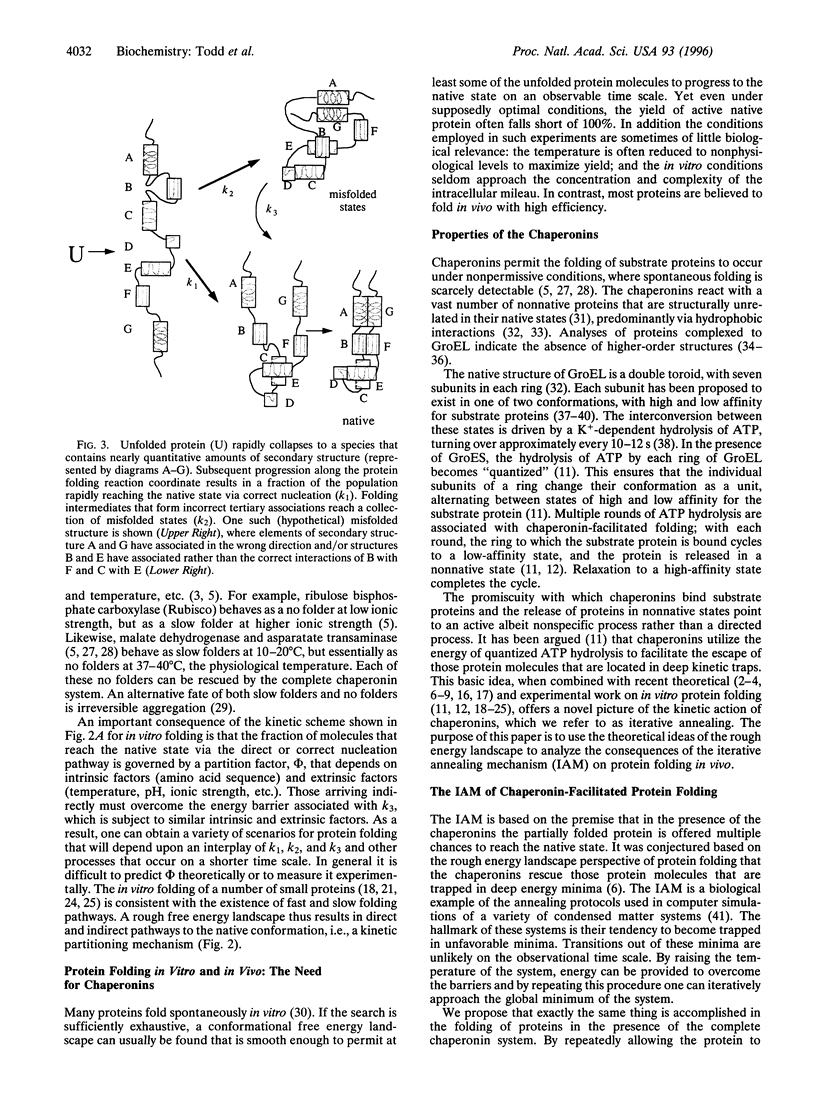
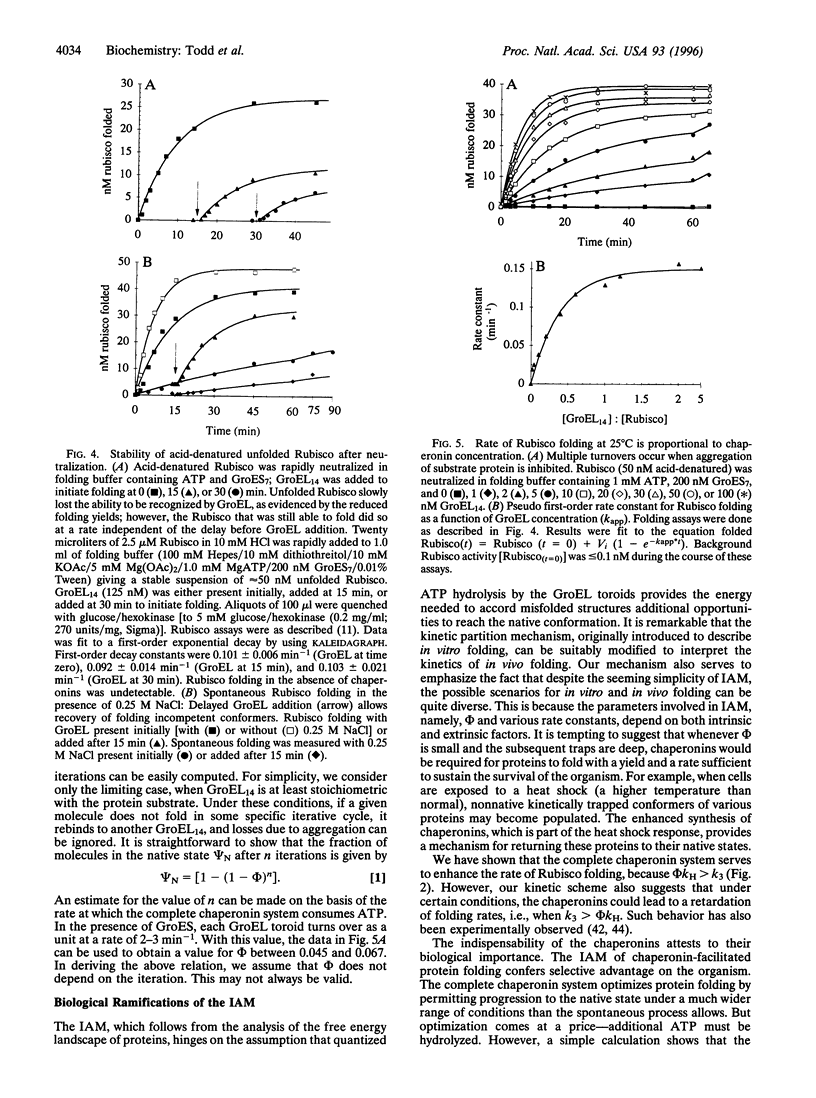
Abstract
We develop a heuristic model for chaperonin-facilitated protein folding, the iterative annealing mechanism, based on theoretical descriptions of "rugged" conformational free energy landscapes for protein folding, and on experimental evidence that (i) folding proceeds by a nucleation mechanism whereby correct and incorrect nucleation lead to fast and slow folding kinetics, respectively, and (ii) chaperonins optimize the rate and yield of protein folding by an active ATP-dependent process. The chaperonins GroEL and GroES catalyze the folding of ribulose bisphosphate carboxylase at a rate proportional to the GroEL concentration. Kinetically trapped folding-incompetent conformers of ribulose bisphosphate carboxylase are converted to the native state in a reaction involving multiple rounds of quantized ATP hydrolysis by GroEL. We propose that chaperonins optimize protein folding by an iterative annealing mechanism; they repeatedly bind kinetically trapped conformers, randomly disrupt their structure, and release them in less folded states, allowing substrate proteins multiple opportunities to find pathways leading to the most thermodynamically stable state. By this mechanism, chaperonins greatly expand the range of environmental conditions in which folding to the native state is possible. We suggest that the development of this device for optimizing protein folding was an early and significant evolutionary event.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Anfinsen C. B. Principles that govern the folding of protein chains. Science. 1973 Jul 20;181(4096):223–230. doi: 10.1126/science.181.4096.223. [DOI] [PubMed] [Google Scholar]
- Bochkareva E. S., Girshovich A. S. ATP induces non-identity of two rings in chaperonin GroEL. J Biol Chem. 1994 Sep 30;269(39):23869–23871. [PubMed] [Google Scholar]
- Braig K., Otwinowski Z., Hegde R., Boisvert D. C., Joachimiak A., Horwich A. L., Sigler P. B. The crystal structure of the bacterial chaperonin GroEL at 2.8 A. Nature. 1994 Oct 13;371(6498):578–586. doi: 10.1038/371578a0. [DOI] [PubMed] [Google Scholar]
- Bryngelson J. D., Onuchic J. N., Socci N. D., Wolynes P. G. Funnels, pathways, and the energy landscape of protein folding: a synthesis. Proteins. 1995 Mar;21(3):167–195. doi: 10.1002/prot.340210302. [DOI] [PubMed] [Google Scholar]
- Chan H. S., Dill K. A. A simple model of chaperonin-mediated protein folding. Proteins. 1996 Mar;24(3):345–351. doi: 10.1002/(SICI)1097-0134(199603)24:3<345::AID-PROT7>3.0.CO;2-F. [DOI] [PubMed] [Google Scholar]
- Dill K. A., Bromberg S., Yue K., Fiebig K. M., Yee D. P., Thomas P. D., Chan H. S. Principles of protein folding--a perspective from simple exact models. Protein Sci. 1995 Apr;4(4):561–602. doi: 10.1002/pro.5560040401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Elöve G. A., Bhuyan A. K., Roder H. Kinetic mechanism of cytochrome c folding: involvement of the heme and its ligands. Biochemistry. 1994 Jun 7;33(22):6925–6935. doi: 10.1021/bi00188a023. [DOI] [PubMed] [Google Scholar]
- Gordon C. L., Sather S. K., Casjens S., King J. Selective in vivo rescue by GroEL/ES of thermolabile folding intermediates to phage P22 structural proteins. J Biol Chem. 1994 Nov 11;269(45):27941–27951. [PubMed] [Google Scholar]
- Gray T. E., Fersht A. R. Refolding of barnase in the presence of GroE. J Mol Biol. 1993 Aug 20;232(4):1197–1207. doi: 10.1006/jmbi.1993.1471. [DOI] [PubMed] [Google Scholar]
- Gulukota K., Wolynes P. G. Statistical mechanics of kinetic proofreading in protein folding in vivo. Proc Natl Acad Sci U S A. 1994 Sep 27;91(20):9292–9296. doi: 10.1073/pnas.91.20.9292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Harrison S. C., Durbin R. Is there a single pathway for the folding of a polypeptide chain? Proc Natl Acad Sci U S A. 1985 Jun;82(12):4028–4030. doi: 10.1073/pnas.82.12.4028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson G. S., Staniforth R. A., Halsall D. J., Atkinson T., Holbrook J. J., Clarke A. R., Burston S. G. Binding and hydrolysis of nucleotides in the chaperonin catalytic cycle: implications for the mechanism of assisted protein folding. Biochemistry. 1993 Mar 16;32(10):2554–2563. doi: 10.1021/bi00061a013. [DOI] [PubMed] [Google Scholar]
- Jackson S. E., Fersht A. R. Folding of chymotrypsin inhibitor 2. 1. Evidence for a two-state transition. Biochemistry. 1991 Oct 29;30(43):10428–10435. doi: 10.1021/bi00107a010. [DOI] [PubMed] [Google Scholar]
- Kiefhaber T., Rudolph R., Kohler H. H., Buchner J. Protein aggregation in vitro and in vivo: a quantitative model of the kinetic competition between folding and aggregation. Biotechnology (N Y) 1991 Sep;9(9):825–829. doi: 10.1038/nbt0991-825. [DOI] [PubMed] [Google Scholar]
- Kirkpatrick S., Gelatt C. D., Jr, Vecchi M. P. Optimization by simulated annealing. Science. 1983 May 13;220(4598):671–680. doi: 10.1126/science.220.4598.671. [DOI] [PubMed] [Google Scholar]
- Kovalenko O., Yifrach O., Horovitz A. Residue lysine-34 in GroES modulates allosteric transitions in GroEL. Biochemistry. 1994 Dec 20;33(50):14974–14978. doi: 10.1021/bi00254a004. [DOI] [PubMed] [Google Scholar]
- Kuszewski J., Clore G. M., Gronenborn A. M. Fast folding of a prototypic polypeptide: the immunoglobulin binding domain of streptococcal protein G. Protein Sci. 1994 Nov;3(11):1945–1952. doi: 10.1002/pro.5560031106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Z., Schwartz F. P., Eisenstein E. The hydrophobic nature of GroEL-substrate binding. J Biol Chem. 1995 Jan 20;270(3):1011–1014. doi: 10.1074/jbc.270.3.1011. [DOI] [PubMed] [Google Scholar]
- Lorimer G. H. A quantitative assessment of the role of the chaperonin proteins in protein folding in vivo. FASEB J. 1996 Jan;10(1):5–9. doi: 10.1096/fasebj.10.1.8566548. [DOI] [PubMed] [Google Scholar]
- Mattingly J. R., Jr, Iriarte A., Martinez-Carrion M. Homologous proteins with different affinities for groEL. The refolding of the aspartate aminotransferase isozymes at varying temperatures. J Biol Chem. 1995 Jan 20;270(3):1138–1148. doi: 10.1074/jbc.270.3.1138. [DOI] [PubMed] [Google Scholar]
- Okazaki A., Ikura T., Nikaido K., Kuwajima K. The chaperonin GroEL does not recognize apo-alpha-lactalbumin in the molten globule state. Nat Struct Biol. 1994 Jul;1(7):439–446. doi: 10.1038/nsb0794-439. [DOI] [PubMed] [Google Scholar]
- Peralta D., Hartman D. J., Hoogenraad N. J., Høj P. B. Generation of a stable folding intermediate which can be rescued by the chaperonins GroEL and GroES. FEBS Lett. 1994 Feb 14;339(1-2):45–49. doi: 10.1016/0014-5793(94)80381-1. [DOI] [PubMed] [Google Scholar]
- Radford S. E., Dobson C. M., Evans P. A. The folding of hen lysozyme involves partially structured intermediates and multiple pathways. Nature. 1992 Jul 23;358(6384):302–307. doi: 10.1038/358302a0. [DOI] [PubMed] [Google Scholar]
- Ranson N. A., Dunster N. J., Burston S. G., Clarke A. R. Chaperonins can catalyse the reversal of early aggregation steps when a protein misfolds. J Mol Biol. 1995 Jul 28;250(5):581–586. doi: 10.1006/jmbi.1995.0399. [DOI] [PubMed] [Google Scholar]
- Robinson C. V., Gross M., Eyles S. J., Ewbank J. J., Mayhew M., Hartl F. U., Dobson C. M., Radford S. E. Conformation of GroEL-bound alpha-lactalbumin probed by mass spectrometry. Nature. 1994 Dec 15;372(6507):646–651. doi: 10.1038/372646a0. [DOI] [PubMed] [Google Scholar]
- Schindler T., Herrler M., Marahiel M. A., Schmid F. X. Extremely rapid protein folding in the absence of intermediates. Nat Struct Biol. 1995 Aug;2(8):663–673. doi: 10.1038/nsb0895-663. [DOI] [PubMed] [Google Scholar]
- Schmidt M., Buchner J., Todd M. J., Lorimer G. H., Viitanen P. V. On the role of groES in the chaperonin-assisted folding reaction. Three case studies. J Biol Chem. 1994 Apr 8;269(14):10304–10311. [PubMed] [Google Scholar]
- Sfatos C. D., Gutin A. M., Abkevich V. I., Shakhnovich E. I. Simulations of chaperone-assisted folding. Biochemistry. 1996 Jan 9;35(1):334–339. doi: 10.1021/bi952033a. [DOI] [PubMed] [Google Scholar]
- Sosnick T. R., Mayne L., Hiller R., Englander S. W. The barriers in protein folding. Nat Struct Biol. 1994 Mar;1(3):149–156. doi: 10.1038/nsb0394-149. [DOI] [PubMed] [Google Scholar]
- Todd M. J., Viitanen P. V., Lorimer G. H. Dynamics of the chaperonin ATPase cycle: implications for facilitated protein folding. Science. 1994 Jul 29;265(5172):659–666. doi: 10.1126/science.7913555. [DOI] [PubMed] [Google Scholar]
- Todd M. J., Viitanen P. V., Lorimer G. H. Hydrolysis of adenosine 5'-triphosphate by Escherichia coli GroEL: effects of GroES and potassium ion. Biochemistry. 1993 Aug 24;32(33):8560–8567. doi: 10.1021/bi00084a024. [DOI] [PubMed] [Google Scholar]
- Udgaonkar J. B., Baldwin R. L. Early folding intermediate of ribonuclease A. Proc Natl Acad Sci U S A. 1990 Nov;87(21):8197–8201. doi: 10.1073/pnas.87.21.8197. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Dyk T. K., Gatenby A. A., LaRossa R. A. Demonstration by genetic suppression of interaction of GroE products with many proteins. Nature. 1989 Nov 23;342(6248):451–453. doi: 10.1038/342451a0. [DOI] [PubMed] [Google Scholar]
- Viitanen P. V., Donaldson G. K., Lorimer G. H., Lubben T. H., Gatenby A. A. Complex interactions between the chaperonin 60 molecular chaperone and dihydrofolate reductase. Biochemistry. 1991 Oct 8;30(40):9716–9723. doi: 10.1021/bi00104a021. [DOI] [PubMed] [Google Scholar]
- Viitanen P. V., Gatenby A. A., Lorimer G. H. Purified chaperonin 60 (groEL) interacts with the nonnative states of a multitude of Escherichia coli proteins. Protein Sci. 1992 Mar;1(3):363–369. doi: 10.1002/pro.5560010308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weissman J. S., Kashi Y., Fenton W. A., Horwich A. L. GroEL-mediated protein folding proceeds by multiple rounds of binding and release of nonnative forms. Cell. 1994 Aug 26;78(4):693–702. doi: 10.1016/0092-8674(94)90533-9. [DOI] [PubMed] [Google Scholar]
- Wetlaufer D. B. Nucleation, rapid folding, and globular intrachain regions in proteins. Proc Natl Acad Sci U S A. 1973 Mar;70(3):697–701. doi: 10.1073/pnas.70.3.697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolynes P. G., Onuchic J. N., Thirumalai D. Navigating the folding routes. Science. 1995 Mar 17;267(5204):1619–1620. doi: 10.1126/science.7886447. [DOI] [PubMed] [Google Scholar]
- Zahn R., Spitzfaden C., Ottiger M., Wüthrich K., Plückthun A. Destabilization of the complete protein secondary structure on binding to the chaperone GroEL. Nature. 1994 Mar 17;368(6468):261–265. doi: 10.1038/368261a0. [DOI] [PubMed] [Google Scholar]



