Abstract
Ubiquitylation is a posttranslational protein modification that modulates various cellular processes of key significance, including protein degradation and DNA damage repair. In animals subjected to transient cerebral ischemia, ubiquitin-conjugated proteins accumulate in Triton-insoluble aggregates. Although this process is widely considered to modulate the fate of postischemic neurons, few attempts have been made to characterize the ubiquitin-modified proteome in these aggregates. We performed proteomics analyses to identify ubiquitylated proteins in postischemic aggregates. Mice were subjected to 10 minutes of forebrain ischemia and 4 hours of reperfusion. The hippocampi were dissected, aggregates were isolated, and trypsin-digested after spiking with GG-BSA as internal standard. K-ɛ-GG-containing peptides were immunoprecipitated and analyzed by label-free quantitative liquid chromatography tandem mass spectrometry (LC-MS/MS) analysis. We identified 1,664 peptides to 520 proteins containing at least one K-ɛ-GG. Sixty-six proteins were highly ubiquitylated, with 10 or more K-ɛ-GG peptides. Based on selection criteria of greater than fivefold increase and P<0.001, 763 peptides to 272 proteins were highly enriched in postischemic aggregates. These included proteins involved in important neuronal functions and signaling pathways that are impaired after ischemia. Results of this study could serve as an important platform to uncover the mechanisms linking insoluble ubiquitin aggregates to the functions of postischemic neurons.
Keywords: cerebral ischemia, protein aggregates, proteomics analysis, ubiquitin conjugation
Introduction
Transient cerebral ischemia activates various posttranslational protein modifications. These include ubiquitylation, small ubiquitin-like modifier conjugation, and ISGylation, all of which modify lysine residues in target proteins.1, 2, 3, 4 Transient cerebral ischemia triggers accumulation of ubiquitylated proteins that form Triton X-100-insoluble aggregates.3 Aggregates of ubiquitylated proteins start to appear after ischemia during early reperfusion when cells are still morphologically intact.5 Several groups have reported postischemic accumulation of ubiquitin-conjugated proteins in Triton X-100-insoluble aggregates,2, 3, 5, 6, 7, 8, 9, 10 but the significance of this process for the fate and functions of postischemic neurons has not yet been uncovered. No attempts have been made to perform proteomics analysis to characterize the ubiquitin-modified proteome in postischemic Triton X-100-insoluble aggregates.
Ubiquitin proteomic analysis is hampered by low levels of ubiquitylated proteins. Without efficient and specific enrichment before LC-MS/MS analysis, co-immunoprecipitation (IP) of proteins that are not ubiquitylated together with ubiquitylated proteins could be a major problem. To overcome problems associated with conventional ubiquitin proteomics analyses, a new strategy has recently been established that enables highly specific enrichment of ubiquitylated peptides derived from trypsin-digested ubiquitylated proteins.11, 12, 13 This approach takes advantage of antibodies that bind specifically to the di-glycine remnant (K-ɛ-GG) derived from the two carboxy-terminal glycine residues of ubiquitin that are covalently conjugated to the ɛ-amino groups of lysine residues in target proteins after trypsin digestion. The mass shift of 114.04 kDa caused by the di-glycine remnant enables identification of ubiquitylated proteins and also precise localization of ubiquitylation sites of target proteins. This new approach is, therefore, highly specific because it allows ubiquitylated proteins to be distinguished from unspecific contaminants based on MS/MS sequence confirmation. In the present study, we used this approach for a comprehensive analysis of the ubiquitin-modified proteome in Triton X-100-insoluble protein aggregates in postischemic brain samples. This is, to the best of our knowledge, the first study to use K-ɛ-GG peptide IP to characterize the ubiquitin-modified proteome in a pathologic state and to use an internal standard to correct for recovery variability between samples.
Materials and Methods
Animal Experiments
The following study was approved by the Duke University Animal Care and Use Committee (Durham, NC, USA) and complies with the Guide for the Care and Use of Animals published by the National Institutes of Health. Male C57Bl/6J mice (Jackson Laboratories, Bar Harbor, ME, USA) 8- to 10-weeks-old and weighing 20 to 25 g were fasted overnight with free access to water.1 Mice were anesthetized with 5% isoflurane. The trachea was intubated with a 20-gauge intravenous catheter (Insyte-W, Becton-Dickenson, Sandy, UT, USA), and the isoflurane concentration was reduced to 1.8% of the inspired air. The rectal temperature was servocontrolled at 37°C by surface cooling or heating during ischemia and for 30 minutes after onset of reperfusion. The right internal jugular vein was cannulated to withdraw blood.
Forebrain ischemia was induced by bilateral common carotid artery occlusion and blood withdrawal to reduce mean arterial blood pressure to 30 mm Hg. After 10 minutes of ischemia, carotid arteries were deoccluded, withdrawn blood was reinfused, and catheters were removed. The wound was infiltrated with bupivacaine and closed. After 4 hours of reperfusion, mice were reanesthetized with 5% isoflurane and decapitated. The brains were quickly removed, and hippocampi were excised, immediately frozen, and stored at −80°C. Sham-operated control mice underwent the same procedures as experimental animals, except that the carotid arteries were not occluded and blood was not withdrawn. For the present study, animals were randomized and all mice exposed to sham or ischemia surgery for used for analysis. In earlier studies, using the same animal model, we found that ∼40% of CA1 hippocampal neurons were damaged.
Isolation of Triton X-100-Insoluble Protein Aggregates
Hippocampi from 6 to 8 animals were pooled, and Triton X-100-insoluble protein aggregates were isolated using three consecutive steps. Combined hippocampi were first weighed while still frozen and then homogenized with 800 μL buffer #1/100 mg tissue at 4°C using a Dounce homogenizer (Thermo Fisher Scientific, Pittsburgh, PA, USA) and 20 gentle strokes. Buffer #1 was composed of 15 mmol/L Tris-HCl pH 7.6, 250 mmol/L sucrose, 1 mmol/L MgCl2, 2.5 mmol/L EDTA, 1 mmol/L EGTA, 1 mmol/L Na3VO4, 20 mmol/L N-ethylmaleimide (NEM), 1 mmol/L dithiothreitol, 1 mmol/L PMSF, and 1% protease inhibitor cocktail (Sigma, St Louis, MO, USA). Homogenates (T1) were centrifuged for 10 minutes at 4°C to yield pellets and supernatant fractions (S1). Pellets were homogenized by sonication using buffer #1 supplemented with 2% Triton X-100 and 150 mmol/L KCl (buffer #2). Homogenates (T2) were then centrifuged to yield supernatants (S2) and pellets. For western blot analysis, pellets, representing Triton X-100-insoluble protein aggregates, were solubilized by sonication in buffer #2 supplemented with 8 mol/L urea (T3). For ubiquitin proteomics analysis, pellets were processed as described below.
Western Blot Analysis
For western blot analysis, homogenates and supernatant fractions were mixed 1:2 with 2 × Laemmli sample buffer and heated to 99°C for 10 minutes. Immunoblotting was performed using 4% to 15% SDS-PAGE gels (Bio-Rad, Hercules, CA, USA). After electrophoresis, proteins were transferred to polyvinylidene difluoride membranes (Bio-Rad). Membranes were blocked with 0.1% TBST and 5% skim milk powder, and then incubated with the primary antibody overnight at 4°C, followed by incubation with the secondary antibody for 1 hour at room temperature. Protein bands were visualized using the ECL western blotting detection reagents (GE Healthcare, Pittsburgh, PA, USA). The following primary and secondary antibodies were used: mouse anti-ubiquitin, rabbit anti-ubiquitin K48 linkage, rabbit anti-Nedd8, rabbit anti-ISG15, rabbit anti-CaMKIIα, rabbit anti-eIF2α (Cell Signaling Technology, Danvers, MA, USA), goat anti-rabbit or anti-mouse horseradish peroxidase conjugates (Santa Cruz Biotechnology, Santa Cruz, CA, USA).
Immunofluorescence Staining
Immunofluorescence staining was performed as described previously.14 In short, after transcardial perfusion, brains were harvested and embedded in paraffin. After deparaffinization, sections were incubated with rabbit anti-hnRNP A/0 (1:400; Cell Signaling Technology), mouse anti-ubiquitin (1:500; Millipore, Billerica, MA, USA), or mouse anti-CaMKII (1:500; Abcam, Cambridge, MA, USA) at 4°C overnight. Sections were then incubated with Alexa Fluor 594-conjugated goat anti-rabbit IgG and Alexa Fluor 488-conjugated goat anti-mouse IgG (1:500; Invitrogen) at 20°C for 1 hour. Images were captured on a Leica SP5 confocal microscope (Leica Microsystems, Mannheim, Germany).
Proteomics Analysis
Generation of K-ɛ-GG-modified bovine serum albumin internal standard
Boc-Gly-Gly-OSu was prepared in the Duke Small Molecule Synthesis Facility by carbodiimide-mediated coupling of N-hydroxysuccinimide and Boc-Gly-Gly-OH (Indofine Chemical Company, Hillsborough, NJ, USA). Diglycyl-modified bovine serum albumin was then generated by incubating BSA (Calbiochem) at 2 mg/mL in phosphate-buffered saline with 10 mM Boc-Gly-Gly-OSu for 1 hour at room temperature with gentle mixing. Boc was removed by acidification to 1% TFA and heating to 60°C for 1 hour. Buffer exchange into 50 mmol/L ammonium bicarbonate was performed using Amicon 4 10-kDa MWCO filters (Millipore), and concentration of GG-BSA was determined by mini-Bradford assay (Bio-Rad). Diglycyl-modified bovine serum albumin stock was stored at 1.4 μg/μL at −80 °C until use. The GG-BSA was independently characterized by tryptic digestion and LC-MS/MS and verified to contain K-ɛ-GG modification on at least 12 lysine residues (data not shown).
Protein isolation and trypsin digestion
Ubiquitin proteomics analysis was performed on Triton X-100-insoluble protein aggregates isolated from hippocampi from sham and postischemic animals. Hippocampi from 6 to 8 animals were pooled and processed together to yield 1.1 mg of total protein in the insoluble aggregates/sample, which was considered one biologic replicate. To account for biologic variability in the response to ischemia three biologic replicates were processed per group. Triton X-100-insoluble pellets were resuspended in 1.2 mL urea (8 mol/L) supplemented with ammonium bicarbonate (50 mmol/L, AmBic). Samples were then diluted with 50 mmol/L AmBic to 1.8 mol/L urea. As an internal standard to control for quantitative recovery, 30 ng GG-BSA was added to each sample. Cysteine residues were reduced with 10 mmol/L dithiothreitol for 25 minutes at 32°C, and samples were alkylated at 20 mmol/L iodoacetamide for 20 minutes at room temperature in the dark. TPCK trypsin (USB Corporation, Santa Clara, CA, USA) was then added to each sample (25:1 substrate:enzyme ratio), and digestion was allowed to proceed at 32°C overnight. Samples were acidified to 0.5% v/v TFA, and desalted using 500 mg SEP-PAK C18 cartridges (Waters Corporation, Milford, MA, USA), per manufacturer's protocol. Peptide-containing eluates (2.25 mL, 50:49.9:0.1v/v/v MeCN/H2O/TFA) were diluted with 3 mL water, frozen at −80°C, and lyophilized overnight to yield peptide samples ready for IP.
K-ɛ-GG peptide immunoprecipitation
K-ɛ-GG peptides were immunoprecipitated using the UbiScan K-ɛ-GG peptide enrichment kit (Cell Signaling Technologies), with minor modifications to manufacturer's protocol. To improve IP specificity and render the resin reusable,15 the antibody was first covalently cross-linked to the beads using 20 mmol/L dimethylpimelimidate (Pierce Chemical, Rockford, IL, USA) in 0.2 mol/L sodium borate buffer, pH 9. Cross-linking was quenched in 0.2 mol/L ethanolamine, and the resin was then washed 2 × with IAP buffer (Cell Signaling Technology) before use. Digested peptide 1.1 mg for each sample, spiked with GG-BSA internal standard, was resuspended in 1.4 mL IAP buffer and transferred to UbiScan enrichment beads containing ∼100 μg cross-linked antibody. Immunoprecipitation was performed overnight at 4°C. After centrifugation, beads were washed with 1 mL 0.25% NP40 in IAP buffer, followed by 1 mL IAP buffer and 2 times with water. Elution was performed with two 60-mL aliquots of 0.15% TFA in water for ∼10 minutes. Combined eluates were desalted using p10 pipet tips self-packed with C18 Empore disks (3M Corporation, St Paul, MN, USA), and resulting peptides were dried by lyophilization. Samples were then resuspended in 12 μL of 1/2/97 v/v/v TFA/MeCN/H2O containing 10 fmol/μL ADH1_YEAST MassPrep Standard (Waters) and transferred to autosampler vials.
Liquid chromatography tandem mass spectrometry analysis, database searching, and peptide quantitation
Label-free quantitative LC-MS/MS was performed in duplicate for each sample (5 μL per injection), using a nanoAcquity UPLC system (Waters) coupled to an LTQ-Orbitrap XL tandem mass spectrometer (Thermo Scientific, West Palm Beach, FL, USA) via a nanoelectrospray ionization source. Briefly, the sample was first trapped on a Symmetry C18 20 mm × 180 μm trapping column (5 μL/minute at 99.9/0.1v/v water/acetonitrile), after which the analytical separation was performed with a 1.7 μm Acquity BEH130 C18 75 μm × 250 mm column (Waters Corporation) 90-minute gradient of 5% to 40% acetonitrile with 0.1% formic acid at a flow rate of 400 nL/minute and a column temperature of 55°C. Data collection on the LTQ-Orbitrap XL was performed with MS1 analysis in the Orbitrap mass analyzer at 60,000 resolution and AGC target of 1e6 ions. Tandem mass spectrometry data was collected in the LTQ mass analyzer for the top five most intense precursor ions per MS1 scan, with AGC target of 5e4 ions and normalized collision energy of 35. Dynamic inclusion was enabled with a repeat count of 2 and 120 seconds exclusion window.
The order of data collection was interwoven between conditions to minimize temporal bias, alternating sample analysis between biologic conditions with the technical replicates of each sample run back to back. After the 12 analyses, data were imported into Rosetta Elucidator v3.3 (Rosetta Biosoftware, Seattle, WA, USA), and all LC-MS/MS runs were aligned based on the accurate mass and retention time of detected ions using PeakTeller algorithm (Elucidator). The relative peptide abundance was calculated based on area-under-the-curve of aligned features across all runs. The overall data set had 50,569 quantified isotope (peptide) groups. In addition, 77,539 MS/MS spectra were acquired for peptide sequencing by database searching. These MS/MS data were searched against a custom NCBI RefSeq database with mus musculus taxonomy, which also contained a reversed-sequence ‘decoy' database for false positive rate determination as well as several proteins used as surrogate standards (ADH1_YEAST, ALBU_BOVIN, PYGM_RABBIT, CASA1_BOVIN, and ENO1_YEAST).
Database searching was performed in an automated fashion from the Elucidator software package using Mascot search engine v2.2, assigning a precursor ion tolerance of 5 p.p.m. and product ion tolerance of 0.8 kDa. Searching allowed variable modification of N and Q (deamidation, +1 kDa), M (oxidation, +16 kDa), and K (diglycyl, +114 kDa). After aggregating all search results, assignment of peptide sequences to the quantitative MS signal was performed for peptides with an ion score greater than 24, to obtain a 1% false discovery rate at the peptide level according to the ratio of decoy database identifications to forward identifications. For quantification, the data were first curated to contain only K-ɛ-GG-modified peptides with appropriate chromatographic peak shape (peak time score >0.8). To enable statistical filtering, the fold-change between treatment groups was calculated as a ratio of the average intensity between ischemia and sham conditions, so that positive fold-change indicated upregulated in ischemia, and negative indicated downregulated in ischemia versus sham. Additionally, the data were log2-transformed and two statistical tests were performed, a two-sample t-test and an error-weighted analysis of variance.
Bioinformatics analysis
Hierarchical clustering analysis was performed within Rosett Elucidator (Rosetta Biosoftware) using z-score-normalized peptide intensity values as input into the 2-D agglomerative clustering method, which used cosine correlation similarity measures. Gene Ontology terms for proteins isolated via K-ɛ-GG-modified peptide pulldown were annotated with the db2db tool in BioDBnet (http://www.biobnet.abcc.ncifcrf.gov/), using the protein GI numbers as input. PANTHER Analysis (http://www.pantherdb.org) and Ingenuity Pathway analysis (http://www.analysis.ingenuity.com/) were used to assist with biologic contextualization of the ubiquitylated proteins as a function of ischemia (fivefold upregulated, and P<0.001 ischemia versus sham). Motif-X analysis (http://www.motif-x.med.harvard.edu) was performed to extract statistically significant motifs surrounding ubiquitylated lysine sites identified by LC-MS/MS analysis, using as input the entire list of modified peptides identified or the statistically significant data set.
Results
Results from several experimental studies suggest that transient cerebral ischemia triggers accumulation of ubiquitylated proteins in Triton X-100-insoluble aggregates. This aggregation is believed to trap proteins required for physiologic cell functions. In the present study, we have used a model of transient global cerebral ischemia and focused ubiquitin proteomics analysis on the hippocampus, a brain region particularly affected in this model. We also used 4 hours of reperfusion to be able to compare our results with findings from an earlier study.16
First, we verified that transient cerebral ischemia activated ubiquitin conjugation in our model. Hippocampi were dissected from sham and postischemic brains, and Triton-insoluble proteins were isolated as detailed in Materials and Methods. Western blots of whole-cell homogenates showed a marked postischemic activation of global ubiquitin conjugation (Supplementary Figure S1, ubiquitin). After differential centrifugation to isolate Triton X-100-insoluble protein aggregates, a significant fraction of ubiquitylated proteins was still present in postischemia samples but markedly less in sham samples (Supplementary Figure S1, ubiquitin T3). When we stripped membranes and re-probed with an antibody against polyubiquitin with K48 linkage, the pattern was quite similar to the ubiquitin pattern. We used eIF2α and β-actin antibodies as loading controls. The soluble protein eIF2α was found only in T1 and S1 samples, as expected. However, β-actin was still present in T3 samples, suggesting that structural proteins contributed to the Triton X-100-insoluble fractions both in sham and postischemia samples.
In the present study, we used a recently established strategy for specific enrichment of ubiquitylated peptides. This strategy takes advantage of an antibody that binds specifically to the di-glycine remnant on lysine residues, which is derived from trypsin digestion of ubiquitin-conjugated proteins. This di-glycine remnant is also generated by trypsin digestion of Nedd8- or ISG15-conjugated proteins. To determine whether Nedd8- or ISG15-conjugation could potentially contribute to K-ɛ-GG-modified peptide IP, we performed western blots using sham and ischemia samples and Nedd8- and ISG15-specific antibodies (Supplementary Figure S1). T3 samples representing Triton X-100-insoluble protein aggregates contained only a minor fraction of the Nedd8- and ISG15-conjugated proteins found in total cell homogenates (T1). This suggests that neddylation and ISGylation did not have a major role in the present proteomics analyses performed on T3 samples.
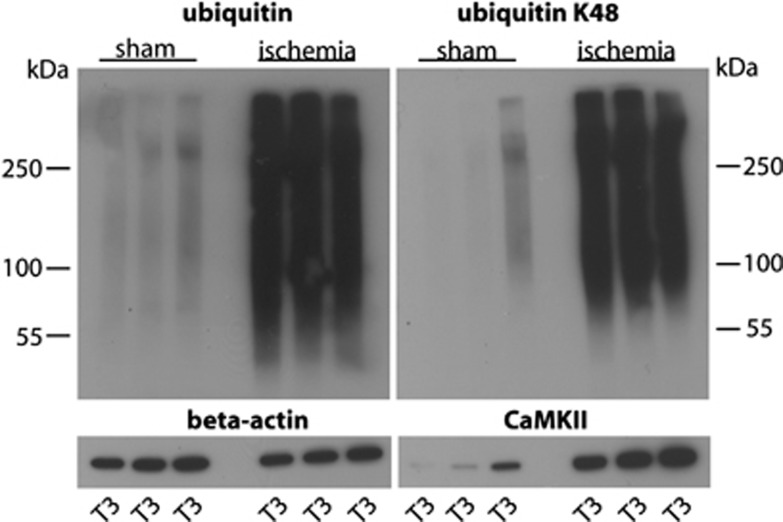
Next, we verified the extent of postischemic accumulation of ubiquitylated proteins in Triton X-100-insoluble protein aggregates by running western blots using all samples prepared from sham and postischemic hippocampi for proteomics analysis. Levels of ubiquitin-conjugated proteins and of K48 linkage polyubiquitin were dramatically increased in postischemic samples (Figure 1). Optical density measurements and statistical analysis revealed that transient ischemia caused a 11.0±0.4% increase in levels of ubiquitin-conjugated proteins in Triton X-100-insoluble aggregates (n=3/group; P<0.001; analysis of variance and Fisher's protected least significant difference test). We also found that in T3 samples representing Triton X-100-insoluble protein aggregates levels of CaMKII were much higher in ischemia than in sham samples (Figure 1), thus confirming results from earlier studies.8, 10, 17, 18
Figure 1.
Transient cerebral ischemia results in accumulation of ubiquitin-conjugated proteins in Triton X-100-insoluble aggregates. Western blot analysis depicts the pattern of ubiquitin conjugation and K48 linkage polyubiquitylation in sham and postischemic T3 samples, representing Triton X-100-insoluble protein aggregates used for proteomics analysis. Note the dramatic increase in ubiquitin and K48 linkage ubiquitin conjugates in postischemic hippocampi. CaMKII was markedly enriched in postischemic T3 samples. Antibody against β-actin was used as loading control.
Proteomics analysis identified a total of 5,483 peptides to 1,320 proteins, including 1,664 peptides to 520 proteins containing at least one K-ɛ-GG-modified lysine. These 1,664 peptides were then scaled to the robust mean intensity of the internal standard (GG-BSA). The accurate mass- and retention time-aligned label-free intensity values for each peptide and each sample, including mass, retention time, best ion score, peptide sequence, and raw peak intensity within each sample are reported in Supplementary Table S1. MS/MS spectra used to make peptide identifications for each sample have been made available as a Scaffold v4 file (http://www.proteomesoftware.com), available for downloading at https://www.discovery.genome.duke.edu/express/resources/3403/3403_MSMS_Sup plement_050813.sf3.
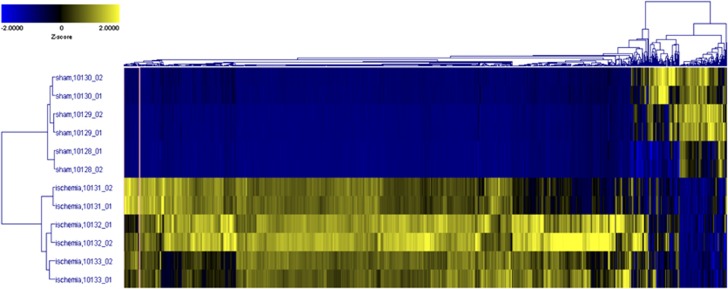
To assess the global pattern of ubiquitylated proteins in Triton X-100-insoluble aggregates and visually compare this pattern with that observed via western blotting, we performed a 2-D Agglomerative Cluster analysis using z-score transformed data from Supplementary Table S1 for all peptides (Figure 2). In this unsupervised 2-D cluster, we saw drastic and consistent differences between sham and ischemia conditions, and the samples within each analytical and biologic group cluster appropriately with the samples expected to be most biologically similar. This suggests that no strong biologic outliers were in the group, and based on this clustering, we observed that most regions of the cluster showed upregulation in the postischemic samples, as expected from the pattern of ubiquitylated proteins in Triton X-100-insoluble aggregates (Figure 1).
Figure 2.
Two-dimensional (2-D) hierarchical clustering across all K-ɛ-GG peptides after intensity normalization to diglycyl-modified bovine serum albumin (GG-BSA) internal standard. Z-Score values are displayed in a 2-D agglomerative cluster model, with cluster similarity along vertical- (sample) and horizontal- (peptide) axes calculated using cosine correlation. Note that the majority of peptides were significantly enriched in the postischemic Triton X-100-insoluble protein aggregates, and are highly correlated.
Next, we defined selection criteria of >fivefold increase and P<0.001 false discovery rate-corrected t-test from Supplementary Table S1 (ischemia versus sham) to characterize, with high confidence, the ubiquitin-conjugated proteome in Triton X-100-insoluble aggregates regulated by transient ischemia. These selection criteria were fulfilled by 763 peptides to 272 proteins (Supplementary Table S2). Thirty-four of these proteins had 10 or more quantified K-ɛ-GG peptides (Table 1). CaMKII subunit alpha isoform 2 was at the top of the list with 73 quantified K-ɛ-GG peptides; 27 out of all 28 lysine residues were ubiquitylated. Among the highly ubiquitylated proteins were the vesicle-fusing ATPase (50 K-ɛ-GG peptides), protein kinase C gamma and beta (31 and 28 K-ɛ-GG peptides, respectively), NMDA receptor subunits epsilon-1 and 2 (15 and 11 K-ɛ-GG peptides, respectively), the neuron-specific calcium-binding protein hippocalcin (11 K-ɛ-GG peptides), the eukaryotic elongation factor 1-alpha 1-like isoform 9 (11 K-ɛ-GG peptides), and the heat shock 70 kDa protein 12A (10 K-ɛ-GG peptides). The ubiquitin protein was identified with 14 K-ɛ-GG peptides (Supplementary Table S2). Ubiquitin K6, K11, K48, and K63 linkages were found with a sixfold, 14-fold, 40-fold, and 11-fold increase in the ischemia versus sham samples, respectively. This indicated that the postischemia Triton X-100-insoluble pellets were highly enriched with polyubiquitin chains, and specifically that K48 appeared to be induced to a higher degree than the other linkages.
Table 1. Proteins with 10 or more K-ɛ-GG-modified peptides.
| GI Number | Protein description | GG(K) peptides | Fold-change |
|---|---|---|---|
| 28916677 | Calcium/calmodulin-dependent protein kinase type II subunit alpha isoform 2 | 73 | 7.3–192 |
| 31543349 | Vesicle-fusing ATPase | 50 | 9.9–189 |
| 226693349 | Calcium/calmodulin-dependent protein kinase type II subunit beta isoform 2 | 42 | 25–101 |
| 6755080 | Protein kinase C gamma type | 31 | 9.9–70 |
| 6679345 | Protein kinase C beta type | 28 | 9.3–114 |
| 51339023 | Disks large-associated protein 1 isoform 1 | 22 | 17–57 |
| 124486887 | Adenylate kinase isoenzyme 5 | 22 | 18–185 |
| 6754012 | Guanine nucleotide-binding protein G(o) subunit alpha isoform A | 20 | 5.2–22 |
| 21704242 | caM kinase-like vesicle-associated protein | 19 | 9.2–1000 |
| 157909820 | Disks large homolog 4 isoform 2 | 19 | 7.1–46 |
| 124486885 | Leucine-rich repeat-containing protein 7 | 17 | 7.3–36 |
| 34740335 | Tubulin alpha-1B chain | 15 | 6.3–25 |
| 117168299 | Glutamate [NMDA] receptor subunit epsilon-2 precursor | 15 | 7.6–25 |
| 225543210 | Disks large-associated protein 2 isoform 1 | 15 | 13–56 |
| 45598372 | Brain acid soluble protein 1 | 14 | 1000 |
| 76443694 | Ubiquitin-40S ribosomal protein S27a precursor | 14 | 8.7–40 |
| 10946574 | Creatine kinase B-type | 13 | 18–224 |
| 21362303 | Synaptosomal-associated protein 47 | 13 | 17–45 |
| 149273202 | Glyceraldehyde-3-phosphate dehydrogenase-like isoform 2 | 13 | 13–58 |
| 309270933 | Ras GTPase-activating protein SynGAP | 13 | 7.2–55 |
| 6755901 | Tubulin alpha-1A chain | 12 | 13–26 |
| 29244248 | Connector enhancer of kinase suppressor of ras 2 | 12 | 8.8–75 |
| 79750129 | Calcium/calmodulin-dependent protein kinase type 1D | 12 | 16–109 |
| 6671509 | Actin, cytoplasmic 1 | 11 | 5.9–11 |
| 7305619 | Ubiquitin carboxyl-terminal hydrolase 5 | 11 | 7.7–123 |
| 41680705 | Glutamate [NMDA] receptor subunit epsilon-1 precursor | 11 | 9.7–37 |
| 70906479 | Calcium/calmodulin-dependent protein kinase type II subunit delta isoform 1 | 11 | 26–98 |
| 83816899 | Diacylglycerol kinase beta | 11 | 14–32 |
| 84370300 | Arf-GAP with GTPase, ANK repeat and PH domain-containing protein 2 | 11 | 24–61 |
| 194354004 | Neuron-specific calcium-binding protein hippocalcin | 11 | 27–52 |
| 407261712 | PREDICTED: elongation factor 1-alpha 1-like isoform 9 | 11 | 6.2–21 |
| 22122643 | Inositol-trisphosphate 3-kinase A | 10 | 22–70 |
| 28461135 | Heat shock 70 kDa protein 12A | 10 | 9.1–101 |
| 116089329 | SRC kinase signaling inhibitor 1 | 10 | 7.9–55 |
List of proteins enriched at least fivefold (P<0.001; ischemia versus sham) and with 10 or more identified K-ɛ-GG peptides. Fold-change: range from the K-ɛ-GG peptide with the smallest to that of the largest fold increase in ischemic versus sham samples.
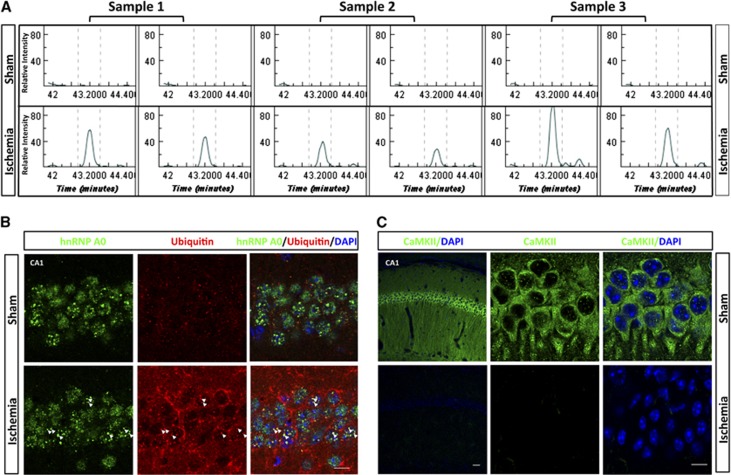
To visually illustrate the differential expression data obtained by LC-MS/MS analysis, Figure 3A shows an example of the raw chromatographic data for a double-ubiquitylated peptide to the ETS-related transcription factor Elf-1. The peptide K.GGAATILK*PGNS[KAANPK]*.D was virtually absent in the LC-MS/MS analyses of sham samples, and clearly abundant in the ischemia samples. In Figures 3B, C confocal microscopic images illustrate the distribution of ubiquitin and 2 of the ubiquitin-conjugated proteins identified in Triton X-100-insoluble aggregates—heterogeneous nuclear ribonucleoprotein A/0 (hnRNPA0; Figure 3B) and CaMKII, the protein that showed the most pronounced postischemic increase in ubiquitylation (Figure 3C). Ubiquitin immunoreactivity was markedly increased after ischemia (Figure 3B), as expected from ubiquitin western blots (Supplementary Figure S1, ubiquitin). No clear colocalization of ubiquitin and hnRNP A/0 was seen in the sham brain. After ischemia, however, many of the hnRNP A/0-positive nuclear dots were also positive for ubiquitin, as indicated by the yellow dots in the merged image (Figure 3B, arrowheads). The sham brain displayed strong CaMKII immunoreactivity; but after ischemia, CaMKII immunoreactivity disappeared almost completely (Figure 3C). This pattern is consistent with earlier observations that hypoxia-ischemia in neonatal rats triggered trapping of CaMKII in Triton X-100-insoluble aggregates and disappearance of CaMKII immunoreactivity.19 This postischemic disappearance of CaMKII immunoreactivity may have been caused by inability of the antibody to bind to the protein because of the high ubiquitylation rate or the trapping in protein aggregates.
Figure 3.
Visual illustration of ubiquitin proteomics results. (A) Raw chromatographic data for the double-ubiquitylated peptide K.GGAATILK*PGNS[KAANPK]*.D (m/z 482.01, charge=4). This peptide to the ETS-related transcription factor Elf-1 is prominent in the ischemic condition, and absence in the sham condition by label-free liquid chromatography tandem mass spectrometry analysis. (B, C) Postischemic colocalization of heterogeneous nuclear ribonucleoprotein A/0 (hnRNPA0) with ubiquitin, and disappearance of CaMKII immunoreactivity in hippocampal CA1 neurons. (B) Confocal microscopic images of sham and postischemic hippocampal CA1 neurons labeled with hnRNP A/0 (green) and ubiquitin (red) together with 4′,6-diamidino-2-phenylindole (DAPI) (blue). Colocalization of ubiquitin with hnRNP A/0 in postischemic neurons is illustrated by color shift to yellow, highlighted by arrowheads (scale bar: 10 μm). (C) Confocal microscopic images of sham and postischemic hippocampal CA1 neurons labeled with CaMKII (green) together with DAPI (blue). Sham neurons displayed strong CaMKII immunoreactivity that disappeared almost completely in postischemic neurons (scale bars, left, low magnification, 50 μm; right, high magnification, 10 μm).
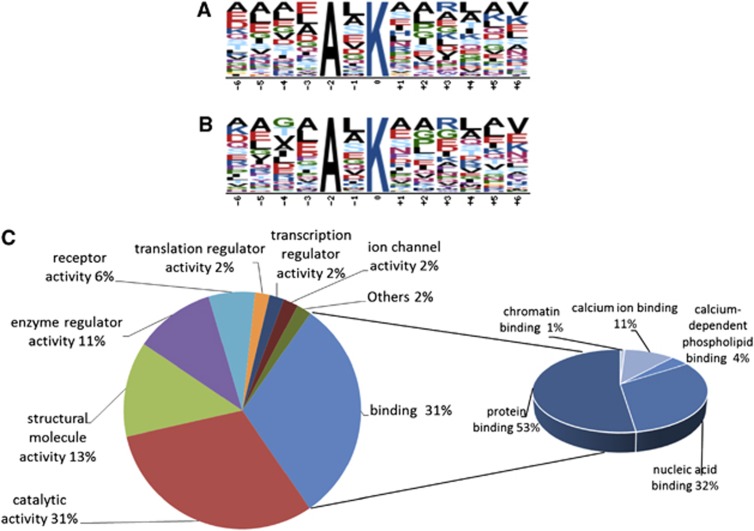
As we were also interested to investigate whether any sequence patterns surrounding the ubiquitylation sites were preferentially targeted in postischemic aggregates, we performed a Motif-X analysis (http://www.motif-x.med.harvard.edu) using either the entire list of modified peptides identified (Supplementary Table S1) or the statistically significant data set (Supplementary Table S2).20, 21 Using these data sets as the input, there was a single AxK motif that was identified in the full data set (n=131, P<1e-6; Figure 4A) and the statistically significant data set (n=67, P<1e-5; Figure 4B). At the time of this publication, it is believed that this sequence motif has not been published as a specific recognition motif for ubiquitin modification. Two recent studies of global ubiquitylation with over 5,000 unique sites in human Jurkat cells, and over 200 sites in the rat brain, did not contain this motif as statistically significant.15, 22 This could indicate that the AxK domain has some significance about increased ubiquitylation in Triton X-100-insoluble aggregates in the brain. However, it must be stated that only ∼10% of the modified peptides identified in this study obey this motif. Thus, the AxK domain is likely to be a contributing factor only toward selective protein ubiquitylation in the insoluble aggregates. Molecular function analysis using the Panther program revealed that 30.7% of 272 proteins in postischemic Triton X-100-insoluble aggregates belonged to a group of binding proteins of which protein binding and nucleic acid binding accounted for 52.5% and 31.7%, respectively (Figure 4C).
Figure 4.
Bioinformatic analysis. Motif-X analysis of the entire ubiquitin-modified proteome of the Triton X-100-insoluble fraction (A) and those ubiquitylated peptides that were highly enriched in the Triton X-100-insoluble fraction of ischemia versus sham hippocampi (B). This analysis shows that the motif AxK is overrepresented in the data set. (C) PANTHER analysis of proteins accumulated in postischemic aggregates. Molecular functions of the 272 ubiquitin-conjugated proteins highly enriched in postischemic Triton X-100-insoluble aggregates.
Ingenuity Pathway Analysis was performed on the 272 proteins that fulfilled our strict selection criteria, to identify the neuronal functions and canonical pathways associated with the data set. Postischemic Triton X-100-insoluble protein aggregates were highly enriched with proteins associated with long-term potentiation (26 of the identified proteins; P=2.5E-15), plasticity of synapse (17 proteins; P=4.9E-13), neurotransmission (28 proteins; P=8.4E-11), and morphology of the nervous system (33 proteins; P=1.6E-6) (Supplementary Table S3). Canonical pathways associated with the data set included nNOS signaling in neurons (12 proteins; P =1.1E-12), gap junction signaling (18 proteins; P=4.8E-12), 14-3-3-mediated signaling (15 proteins; P=6.6E-11), calcium signaling (15 proteins; 2.8E-8), and p70S6K signaling (8 proteins; P=2.4E-4).
Discussion
We report here the first comprehensive analysis of the ubiquitin-modified proteome regulated by transient forebrain ischemia. Proteomics analyses were performed on Triton X-100-insoluble protein aggregates isolated from hippocampi, the brain region most vulnerable to even a short period of vascular occlusion.23 We made slight modifications to a recently developed highly specific ubiquitin proteomics approach.11 To account for inter-sample variability in the efficacy of di-glycine remnant motif IP and the consecutive proteomics analysis, we used a novel approach by adding a chemically distinct (exogenous) internal standard (GG-BSA) to all samples before IP. To account for biologic variability, we analyzed three independent samples/group, and defined stringent selection criteria for statistical significance in the ischemia-induced ubiquitin-modified proteome (fivefold increase in ischemia versus sham samples and P<0.001). We are therefore confident that the list of 763 K-ɛ-GG-modified peptides to 272 proteins that fulfilled these criteria is a reliable representation of the ubiquitin-modified proteome that is regulated by transient forebrain ischemia and accumulated in Triton X-100-insoluble aggregates in postischemic hippocampi.
Lysine is the preferred amino acid modulated by various posttranslational protein modifications, including conjugation by acetate, ubiquitin, small ubiquitin-like modifier, ISG15, and Nedd8. Ubiquitin, ISG15, and Nedd8 all have the identical C-terminal sequence–RGG, and form K-ɛ-GG-modified peptides upon trypsin digestion of conjugated proteins. K-ɛ-GG peptides identified in this study could therefore result from ubiquitin, ISG15, and Nedd8 conjugation. However, in a recent study, Nedd8- and ISG15-conjugated proteins were quantified in the brain, and were found to be less than 2% of ubiquitin-conjugated proteins.22 Furthermore, we performed a western blot analysis to evaluate ischemia-induced changes in Nedd8 and ISG15 conjugation. Levels of Nedd8- and ISG15-modified proteins were markedly lower in Triton X-100-insoluble aggregates than in whole-cell lysates (Supplementary Figure S1). This suggests that Nedd8- and ISG15-conjugated proteins did not significantly contribute, if at all, to the list of ubiquitylated proteins in Triton X-100-insoluble aggregates.
Earlier studies reported postischemic translocation of CaMKII and protein kinase C from the soluble to the particulate (insoluble) fraction.8, 10, 17, 18 We found these proteins to be highly ubiquitylated in postischemic Triton X-100-insoluble aggregates (Table 1). Furthermore, proteins of the translational machinery were reported to be highly ubiquitylated in postischemic aggregates, and it has been proposed that this trapping of components of the translational complex contributes to the irreversible suppression of proteins synthesis.16, 24, 25 In our list of 272 ubiquitylated proteins highly enriched in postischemic protein aggregates, we identified five chaperons (heat shock 70 kDa proteins 12A and 4L, and Dnaj homologs subfamily A member 1, and subfamily B members 2 and 6), three eukaryotic initiation factors (eIF4H, eIF4A-1, and eIF5A-1), and two elongation factors (eEFalpha 1 isoforms 3 and 9), thus supporting earlier observations.
The role of the postischemic accumulation of proteins in Triton X-100-insoluble aggregates for the fate and functions of postischemic neurons has not yet been uncovered. At least two different scenarios can be envisioned. Protein aggregates per se could be lethal to cells, and the trapping of proteins in insoluble aggregates could contribute to the impairment of neuronal functioning triggered by transient cerebral ischemia. No direct evidence has yet been presented to suggest that insoluble aggregates could be lethal to postischemic cells, and conflicting data have been reported regarding the association of aggregate formation and ischemic cell damage. After transient forebrain ischemia, the postischemic increase in levels of ubiquitin-conjugated protein was found to be dependent on ischemic damage.6 However, after transient focal cerebral ischemia, a short period of ischemia that caused no tissue damage resulted in ubiquitin aggregate levels similar to those induced by a longer period of ischemia that caused substantial infarction.5
The mechanisms underlying the postischemic accumulation of ubiquitylated proteins in Triton X-100-insoluble aggregates still need to be uncovered. Two different scenarios can be envisioned. The accumulation could be an active process or a non-specific sequestration owing to an overall postischemic activation of protein ubiquitylation. Indeed, we reported here that transient cerebral ischemia resulted in an increase in protein ubiquitylation, as indicated by the more pronounced smear of bands of ubiquitin-conjugated proteins on western blots of whole tissue homogenates from postischemic samples (Supplementary Figure S1, T1 ischemia versus T1 sham). This overall postischemic increase in levels of ubiquitylated proteins resulted from a specific and dramatic trapping of ubiquitin-conjugated proteins in Triton X-100-insoluble aggregates (Supplementary Figure S1, T3 ischemia versus T3 sham), while changes in the soluble fractions were minor (Supplementary Figure S1 ischemia versus S1 sham). This suggests the postischemic trapping of ubiquitylated proteins in insoluble aggregates to be an active process. Whether proteins were first sequestered in those aggregates and then ubiquitylated or vice versa needs to be elucidated in future studies. The observation that ubiquitylated CaMKII trapped in postischemic aggregates was not accessible any more for the CaMKII antibody (Figure 3C) supports a scenario whereby proteins were ubiquitylated before being sequestered in aggregates.
In the brain, ubiquitin conjugation is a physiologic process, and 1,786 K-ɛ-GG sites on 2,064 peptides to 921 proteins have recently been identified in normal rat brains, including CaMKII subunits alpha, delta, and gamma, and NMDA receptor subunits NR1 and NR2B.22 CaMKII and the NMDA receptor that were found in postischemic insoluble aggregates (Table 1) form a complex that has a role in long-term potentiation.26 Long-term potentiation is one of the major mechanisms underlying learning and memory.27 Activity levels of neurons control the postsynaptic composition and signaling by modulating the composition of the postsynaptic density by reversible activation of ubiquitin conjugation/deconjugation.28 Although the ubiquitin–proteasome system is an integral component of neuronal functioning, it is reasonable to predict that trapping ubiquitin-conjugated proteins in insoluble aggregates may impair neuronal function in the postischemic brain. Indeed, many of the ubiquitylated proteins that were trapped in postischemic Triton X-100-insoluble aggregates have pivotal roles in protein synthesis, and learning and memory processes, both of which are impaired after ischemia.29, 30, 31, 32
A hallmark of transient ischemia-induced neuronal cell death is an irreversible translational arrest.33, 34 This translational arrest is believed to be triggered by ischemia-induced impairment of endoplasmic reticulum functions that activates the unfolded protein response, resulting in shutdown of protein synthesis.35, 36, 37 The initiation factor eIF3 and the chaperons HSP40 and HSC70 were earlier reported to be trapped in postischemic protein aggregates.16 We also found that many initiation and elongation factors and various chaperons were ubiquitylated after ischemia and trapped in Triton X-100-insoluble aggregates. Further, the calcium-binding protein hippocalcin was highly ubiquitylated after ischemia. Hippocalcin protects cells against endoplasmic reticulum stress-induced cell death and functions as a calcium sensor in synaptic plasticity.38, 39 Furthermore, 26 of the proteins that were highly ubiquitylated after ischemia and trapped in Triton X-100-insoluble aggregates have a role in long-term potentiation. It is therefore reasonable to conclude that the postischemic accumulation of ubiquitylated proteins in insoluble aggregates contributes to the pathologic process induced by transient cerebral ischemia and results in impaired endoplasmic reticulum function, suppression of protein synthesis, and impairment of neuronal functions associated with learning and memory.
Our ubiquitin proteomics analyses identified ubiquitin conjugated to ubiquitin at K6, K11, K48, and K63, indicating postischemic formation of polyubiquitin chains with different linkages. The K48 linkage, which targets proteins for degradation at the proteasome, was the most dysregulated with ischemia. Polyubiquitin chain formation with K6, K11, and K63 linkages are involved in various cellular processes, including DNA damage repair, endocytosis, inflammation, and kinase signaling. Of note, K63 linkage polyubiquitylation is involved in the formation of protein inclusions associated with neurodegenerative diseases.40 Whether K6, K11, and K63 linkage polyubiquitylation and trapping of these modified proteins in postischemic aggregates indeed have a role in the fate and functions of postischemic neurons must be verified in future studies.
In conclusion, we presented here the first comprehensive proteomics analysis of the ubiquitin-modified proteome regulated by transient forebrain ischemia. We focused our analysis on the Triton X-100-insoluble fractions isolated from hippocampi of sham and postischemic brains, and we spiked samples, for the first time, with GG-BSA as internal standard before trypsin digestion—a step critical to successful label-free quantitative analysis. The 763 K-ɛ-GG-modified peptides to 272 proteins that fulfilled our selection criteria of fivefold increase and P<0.001 in ischemia versus sham samples is a comprehensive list of the ubiquitin-modified proteome regulated by transient forebrain ischemia and accumulated in Triton X-100-insoluble aggregates. Many of these proteins have important roles in protein synthesis, synaptic plasticity, and learning and memory—processes that are impaired after ischemia. The data presented here could, therefore, serve as an important platform for future studies to better understand the mechanisms underlying ischemia-induced impairment of neuronal functions.
Acknowledgments
The authors thank Pei Miao for her excellent technical support and Kathy Gage for her excellent editorial contribution in the preparation of this manuscript.
Note added to the proof
Raw mass spectrometry data for this study has been made available as part of the Chorus Project, and is downloadable using the following link: https://chorusproject.org/anonymous/download/experiment/333874003018238886.
The authors declare no conflict of interest.
Footnotes
Supplementary Information accompanies the paper on the Journal of Cerebral Blood Flow & Metabolism website (http://www.nature.com/jcbfm)
This study was supported by funds from the Department of Anesthesiology and by NIH R01 grant NS081299 (to WP).
Supplementary Material
References
- Yang W, Sheng H, Warner DS, Paschen W. Transient global cerebral ischemia induces a massive increase in protein sumoylation. J Cereb Blood Flow Metabol. 2008;28:269–279. doi: 10.1038/sj.jcbfm.9600523. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Takada K, Matsuda M. Changes in ubiquitin and ubiquitin-protein conjugates in the Ca1 neurons after transient sublethal ischemia. Mol Chem Neuropathol. 1991;15:75–82. doi: 10.1007/BF03161057. [DOI] [PubMed] [Google Scholar]
- Hu BR, Janelidze S, Ginsberg MD, Busto R, Perez-Pinzon M, Sick TJ, et al. Protein aggregation after focal brain ischemia and reperfusion. J Cereb Blood Flow Metabol. 2001;21:865–875. doi: 10.1097/00004647-200107000-00012. [DOI] [PubMed] [Google Scholar]
- Nakka VP, Lang BT, Lenschow DJ, Zhang DE, Dempsey RJ, Vemuganti R. Increased cerebral protein ISGylation after focal ischemia is neuroprotective. J Cereb Blood Flow Metabol. 2011;31:2375–2384. doi: 10.1038/jcbfm.2011.103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hochrainer K, Jackman K, Anrather J, Iadecola C. Reperfusion rather than ischemia drives the formation of ubiquitin aggregates after middle cerebral artery occlusion. Stroke. 2012;43:2229–2235. doi: 10.1161/STROKEAHA.112.650416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi T, Tanaka J, Kamikubo T, Takada K, Matsuda M. Increase in ubiquitin conjugates dependent on ischemic damage. Brain Res. 1993;620:171–173. doi: 10.1016/0006-8993(93)90288-x. [DOI] [PubMed] [Google Scholar]
- Liu CL, Martone ME, Hu BR. Protein ubiquitination in postsynaptic densities after transient cerebral ischemia. J Cereb Blood Flow Metab. 2004;24:1219–1225. doi: 10.1097/01.WCB.0000136706.77918.21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mengesdorf T, Althausen S, Mies G, Olah L, Paschen W. Phosphorylation state, solubility, and activity of calcium/calmodulin-dependent protein kinase II alpha in transient focal ischemia in mouse brain. Neurochem Res. 2002;27:477–484. doi: 10.1023/a:1019844518704. [DOI] [PubMed] [Google Scholar]
- Hayashi T, Takada K, Matsuda M. Subcellular distribution of ubiquitin-protein conjugates in the hippocampus following transient ischemia. J Neurosci Res. 1992;31:561–564. doi: 10.1002/jnr.490310321. [DOI] [PubMed] [Google Scholar]
- Aronowski J, Grotta JC, Waxham MN. Ischemia-induced translocation of Ca2+/calmodulin-dependent protein kinase II: potential role in neuronal damage. J Neurochem. 1992;58:1743–1753. doi: 10.1111/j.1471-4159.1992.tb10049.x. [DOI] [PubMed] [Google Scholar]
- Xu G, Paige JS, Jaffrey SR. Global analysis of lysine ubiquitination by ubiquitin remnant immunoaffinity profiling. Nat Biotechnol. 2010;28:868–873. doi: 10.1038/nbt.1654. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wagner SA, Beli P, Weinert BT, Nielsen ML, Cox J, Mann M, et al. A proteome-wide, quantitative survey of in vivo ubiquitylation sites reveals widespread regulatory roles Mol Cell Proteomics 201110M111 013284. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim W, Bennett EJ, Huttlin EL, Guo A, Li J, Possemato A, et al. Systematic and quantitative assessment of the ubiquitin-modified proteome. Mol Cell. 2011;44:325–340. doi: 10.1016/j.molcel.2011.08.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Ma Q, Yang W, Mackensen GB, Paschen W. Moderate hypothermia induces marked increase in levels and nuclear accumulation of SUMO2/3-conjugated proteins in neurons. J Neurochem. 2012;123:349–359. doi: 10.1111/j.1471-4159.2012.07916.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Udeshi ND, Mani DR, Eisenhaure T, Mertins P, Jaffe JD, Clauser KR, et al. Methods for quantification of in vivo changes in protein ubiquitination following proteasome and deubiquitinase inhibition. Mol Cell Proteomics. 2012;11:148–159. doi: 10.1074/mcp.M111.016857. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CL, Ge P, Zhang F, Hu BR. Co-translational protein aggregation after transient cerebral ischemia. Neurosci. 2005;134:1273–1284. doi: 10.1016/j.neuroscience.2005.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katsura KI, Kurihara J, Kato H, Katayama Y. Ischemic pre-conditioning affects the subcellular distribution of protein kinase C and calcium/calmodulin-dependent protein kinase II in the gerbil hippocampal CA1 neurons. Neurol Res. 2001;23:751–754. doi: 10.1179/016164101101199126. [DOI] [PubMed] [Google Scholar]
- Wieloch T, Cardell M, Bingren H, Zivin J, Saitoh T. Changes in the activity of protein-kinase-C and the differential subcellular redistribution of its isozymes in the rat striatum during and following transient forebrain ischemia. J Neurochem. 1991;56:1227–1235. doi: 10.1111/j.1471-4159.1991.tb11415.x. [DOI] [PubMed] [Google Scholar]
- Tang K, Liu C, Kuluz J, Hu B. Alterations of CaMKII after hypoxia–ischemia during brain development. J Neurochem. 2004;91:429–437. doi: 10.1111/j.1471-4159.2004.02733.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chou MF, Schwartz D. Biological sequence motif discovery using motif-x. Curr Protoc Bioinformatics. 2011;35:13.15.1–13.15.24. doi: 10.1002/0471250953.bi1315s35. [DOI] [PubMed] [Google Scholar]
- Schwartz D, Gygi SP. An iterative statistical approach to the identification of protein phosphorylation motifs from large-scale data sets. Nat Biotechnol. 2005;23:1391–1398. doi: 10.1038/nbt1146. [DOI] [PubMed] [Google Scholar]
- Na CH, Jones DR, Yang Y, Wang X, Xu Y, Peng J. Synaptic protein ubiquitination in rat brain revealed by antibody-based ubiquitome analysis. J Proteome Res. 2012;11:4722–4732. doi: 10.1021/pr300536k. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirino T. Delayed neuronal death in the gerbil hippocampus following ischemia. Brain Res. 1982;239:57–69. doi: 10.1016/0006-8993(82)90833-2. [DOI] [PubMed] [Google Scholar]
- Zhang F, Liu CL, Hu BR. Irreversible aggregation of protein synthesis machinery after focal brain ischemia. J Neurochem. 2006;98:102–112. doi: 10.1111/j.1471-4159.2006.03838.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeGracia DJ, Hu BR. Irreversible translation arrest in the reperfused brain. J Cereb Blood Flow Metabol. 2007;27:875–893. doi: 10.1038/sj.jcbfm.9600388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanhueza M, Fernandez-Villalobos G, Stein IS, Kasumova G, Zhang P, Bayer KU, et al. Role of the CaMKII/NMDA receptor complex in the maintenance of synaptic strength. J Neurosci. 2011;31:9170–9178. doi: 10.1523/JNEUROSCI.1250-11.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bliss TVP, Collingridge GL. A synaptic model of memory—long-term potentiation in the hippocampus. Nature. 1993;361:31–39. doi: 10.1038/361031a0. [DOI] [PubMed] [Google Scholar]
- Ehlers MD. Activity level controls postsynaptic composition and signaling via the ubiquitin-proteasome system. Nat Neurosci. 2003;6:231–242. doi: 10.1038/nn1013. [DOI] [PubMed] [Google Scholar]
- Paschen W, Proud CG, Mies G. Shut-down of translation, a global neuronal stress response: mechanisms and pathological relevance. Curr Pharm Des. 2007;13:1887–1902. doi: 10.2174/138161207780858401. [DOI] [PubMed] [Google Scholar]
- Kleihues P, Hossmann KA, Pegg AE, Kobayashi K, Zimmermann V. Resuscitation of the monkey brain after one hour complete ischemia. III. Indications of metabolic recovery. Brain Res. 1975;95:61–73. doi: 10.1016/0006-8993(75)90207-3. [DOI] [PubMed] [Google Scholar]
- Murphy TH, Corbett D. Plasticity during stroke recovery: from synapse to behaviour. Nat Rev Neurosci. 2009;10:861–872. doi: 10.1038/nrn2735. [DOI] [PubMed] [Google Scholar]
- Yukie M, Yamaguchi K, Yamashima T. Impairments in recognition memory for object and for location after transient brain ischemia in monkeys. Rev Neurosci. 2006;17:201–214. doi: 10.1515/revneuro.2006.17.1-2.201. [DOI] [PubMed] [Google Scholar]
- Thilmann R, Xie Y, Kleihues P, Kiessling M. Persistent inhibition of protein synthesis precedes delayed neuronal death in postischemic gerbil hippocampus. Acta Neuropathol. 1986;71:88–93. doi: 10.1007/BF00687967. [DOI] [PubMed] [Google Scholar]
- Mengesdorf T, Proud CG, Mies G, Paschen W. Mechanisms underlying suppression of protein synthesis induced by transient focal cerebral ischemia in mouse brain. Exp Neurol. 2002;177:538–546. doi: 10.1006/exnr.2002.8002. [DOI] [PubMed] [Google Scholar]
- Kumar R, Azam S, Sullivan JM, Owen C, Cavener DR, Zhang P, et al. Brain ischemia and reperfusion activates the eukaryotic initiation factor 2alpha kinase, PERK. J Neurochem. 2001;77:1418–1421. doi: 10.1046/j.1471-4159.2001.00387.x. [DOI] [PubMed] [Google Scholar]
- Paschen W, Doutheil J. Disturbance of endoplasmic reticulum functions: a key mechanism underlying cell damage. Acta Neurochir Suppl. 1999;73:1–5. doi: 10.1007/978-3-7091-6391-7_1. [DOI] [PubMed] [Google Scholar]
- Paschen W, Aufenberg C, Hotop S, Mengesdorf T. Transient cerebral ischemia activates processing of xbp1 messenger RNA indicative of endoplasmic reticulum stress. J Cereb Blood Flow Metabol. 2003;23:449–461. doi: 10.1097/01.WCB.0000054216.21675.AC. [DOI] [PubMed] [Google Scholar]
- Palmer CL, Lim W, Hastie PG, Toward M, Korolchuk VI, Burbidge SA, et al. Hippocalcin functions as a calcium sensor in hippocampal LTD. Neuron. 2005;47:487–494. doi: 10.1016/j.neuron.2005.06.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Korhonen L, Hansson I, Kukkonen JP, Brannvall K, Kobayashi M, Takamatsu K, et al. Hippocalcin protects against caspase-12-induced and age-dependent neuronal degeneration. Mol Cell Neurosci. 2005;28:85–95. doi: 10.1016/j.mcn.2004.08.015. [DOI] [PubMed] [Google Scholar]
- Lim KL, Lim GG. K63-linked ubiquitination and neurodegeneration. Neurobiol Dis. 2011;43:9–16. doi: 10.1016/j.nbd.2010.08.001. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.