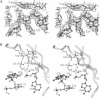
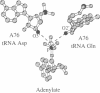
Abstract
The crystal structures of the various complexes formed by yeast aspartyl-tRNA synthetase (AspRS) and its substrates provide snapshots of the active site corresponding to different steps of the aminoacylation reaction. Native crystals of the binary complex tRNA-AspRS were soaked in solutions containing the two other substrates, ATP (or its analog AMPPcP) and aspartic acid. When all substrates are present in the crystal, this leads to the formation of the aspartyl-adenylate and/or the aspartyl-tRNA. A class II-specific pathway for the aminoacylation reaction is proposed which explains the known functional differences between the two classes while preserving a common framework. Extended signature sequences characteristic of class II aaRS (motifs 2 and 3) constitute the basic functional unit. The ATP molecule adopts a bent conformation, stabilized by the invariant Arg531 of motif 3 and a magnesium ion coordinated to the pyrophosphate group and to two class-invariant acidic residues. The aspartic acid substrate is positioned by a class II invariant acidic residue, Asp342, interacting with the amino group and by amino acids conserved in the aspartyl synthetase family. The amino acids in contact with the substrates have been probed by site-directed mutagenesis for their functional implication.
Full text
PDF


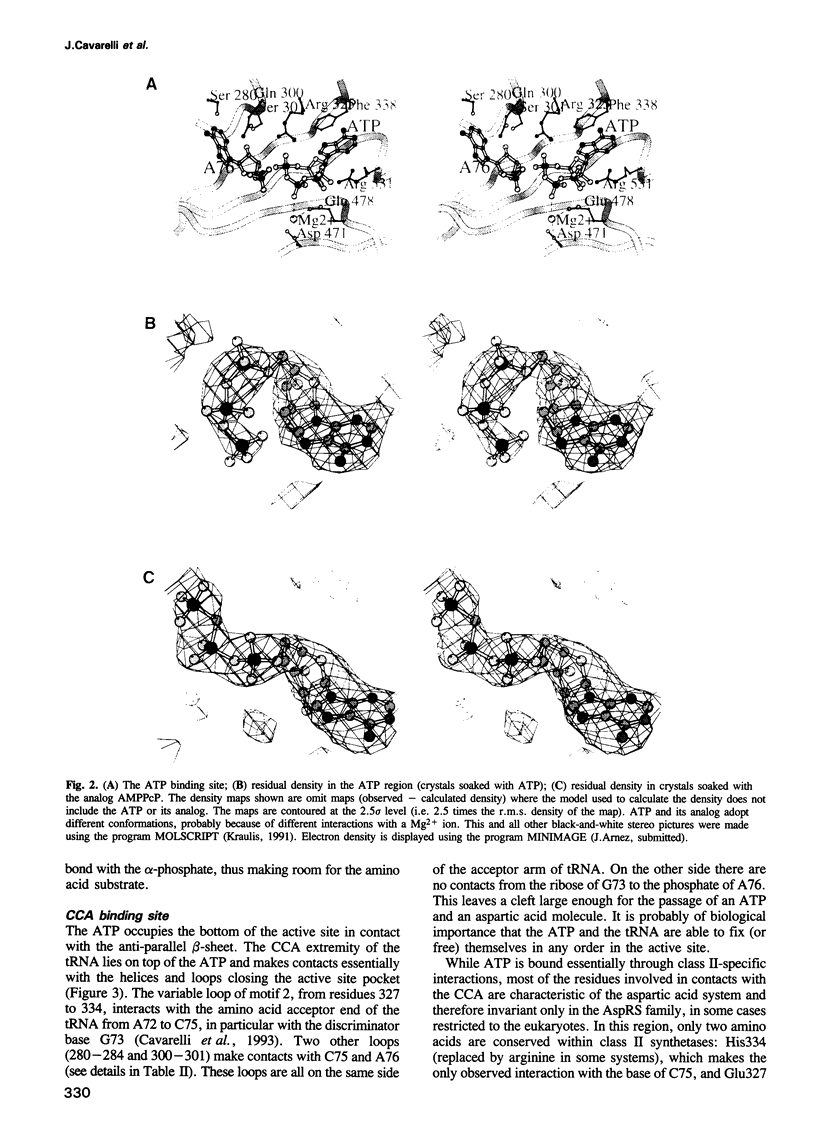



Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Bossemeyer D., Engh R. A., Kinzel V., Ponstingl H., Huber R. Phosphotransferase and substrate binding mechanism of the cAMP-dependent protein kinase catalytic subunit from porcine heart as deduced from the 2.0 A structure of the complex with Mn2+ adenylyl imidodiphosphate and inhibitor peptide PKI(5-24). EMBO J. 1993 Mar;12(3):849–859. doi: 10.1002/j.1460-2075.1993.tb05725.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brick P., Bhat T. N., Blow D. M. Structure of tyrosyl-tRNA synthetase refined at 2.3 A resolution. Interaction of the enzyme with the tyrosyl adenylate intermediate. J Mol Biol. 1989 Jul 5;208(1):83–98. doi: 10.1016/0022-2836(89)90090-9. [DOI] [PubMed] [Google Scholar]
- Brunie S., Zelwer C., Risler J. L. Crystallographic study at 2.5 A resolution of the interaction of methionyl-tRNA synthetase from Escherichia coli with ATP. J Mol Biol. 1990 Nov 20;216(2):411–424. doi: 10.1016/S0022-2836(05)80331-6. [DOI] [PubMed] [Google Scholar]
- Brünger A. T. Crystallographic refinement by simulated annealing. Application to a 2.8 A resolution structure of aspartate aminotransferase. J Mol Biol. 1988 Oct 5;203(3):803–816. doi: 10.1016/0022-2836(88)90211-2. [DOI] [PubMed] [Google Scholar]
- Burbaum J. J., Starzyk R. M., Schimmel P. Understanding structural relationships in proteins of unsolved three-dimensional structure. Proteins. 1990;7(2):99–111. doi: 10.1002/prot.340070202. [DOI] [PubMed] [Google Scholar]
- Cavarelli J., Rees B., Ruff M., Thierry J. C., Moras D. Yeast tRNA(Asp) recognition by its cognate class II aminoacyl-tRNA synthetase. Nature. 1993 Mar 11;362(6416):181–184. doi: 10.1038/362181a0. [DOI] [PubMed] [Google Scholar]
- Courtney M., Buchwalder A., Tessier L. H., Jaye M., Benavente A., Balland A., Kohli V., Lathe R., Tolstoshev P., Lecocq J. P. High-level production of biologically active human alpha 1-antitrypsin in Escherichia coli. Proc Natl Acad Sci U S A. 1984 Feb;81(3):669–673. doi: 10.1073/pnas.81.3.669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cusack S., Berthet-Colominas C., Härtlein M., Nassar N., Leberman R. A second class of synthetase structure revealed by X-ray analysis of Escherichia coli seryl-tRNA synthetase at 2.5 A. Nature. 1990 Sep 20;347(6290):249–255. doi: 10.1038/347249a0. [DOI] [PubMed] [Google Scholar]
- Delarue M., Moras D. The aminoacyl-tRNA synthetase family: modules at work. Bioessays. 1993 Oct;15(10):675–687. doi: 10.1002/bies.950151007. [DOI] [PubMed] [Google Scholar]
- Eriani G., Delarue M., Poch O., Gangloff J., Moras D. Partition of tRNA synthetases into two classes based on mutually exclusive sets of sequence motifs. Nature. 1990 Sep 13;347(6289):203–206. doi: 10.1038/347203a0. [DOI] [PubMed] [Google Scholar]
- Eriani G., Prevost G., Kern D., Vincendon P., Dirheimer G., Gangloff J. Cytoplasmic aspartyl-tRNA synthetase from Saccharomyces cerevisiae. Study of its functional organisation by deletion analysis. Eur J Biochem. 1991 Sep 1;200(2):337–343. doi: 10.1111/j.1432-1033.1991.tb16190.x. [DOI] [PubMed] [Google Scholar]
- Fersht A. R. Dissection of the structure and activity of the tyrosyl-tRNA synthetase by site-directed mutagenesis. Biochemistry. 1987 Dec 15;26(25):8031–8037. doi: 10.1021/bi00399a001. [DOI] [PubMed] [Google Scholar]
- Fersht A. R., Knill-Jones J. W., Bedouelle H., Winter G. Reconstruction by site-directed mutagenesis of the transition state for the activation of tyrosine by the tyrosyl-tRNA synthetase: a mobile loop envelopes the transition state in an induced-fit mechanism. Biochemistry. 1988 Mar 8;27(5):1581–1587. doi: 10.1021/bi00405a028. [DOI] [PubMed] [Google Scholar]
- Hountondji C., Dessen P., Blanquet S. Sequence similarities among the family of aminoacyl-tRNA synthetases. Biochimie. 1986 Sep;68(9):1071–1078. doi: 10.1016/s0300-9084(86)80181-x. [DOI] [PubMed] [Google Scholar]
- Jeltsch A., Alves J., Wolfes H., Maass G., Pingoud A. Substrate-assisted catalysis in the cleavage of DNA by the EcoRI and EcoRV restriction enzymes. Proc Natl Acad Sci U S A. 1993 Sep 15;90(18):8499–8503. doi: 10.1073/pnas.90.18.8499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones T. A., Thirup S. Using known substructures in protein model building and crystallography. EMBO J. 1986 Apr;5(4):819–822. doi: 10.1002/j.1460-2075.1986.tb04287.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kunkel T. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Proc Natl Acad Sci U S A. 1985 Jan;82(2):488–492. doi: 10.1073/pnas.82.2.488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perona J. J., Rould M. A., Steitz T. A. Structural basis for transfer RNA aminoacylation by Escherichia coli glutaminyl-tRNA synthetase. Biochemistry. 1993 Aug 31;32(34):8758–8771. doi: 10.1021/bi00085a006. [DOI] [PubMed] [Google Scholar]
- Rossmann M. G., Moras D., Olsen K. W. Chemical and biological evolution of nucleotide-binding protein. Nature. 1974 Jul 19;250(463):194–199. doi: 10.1038/250194a0. [DOI] [PubMed] [Google Scholar]
- Rould M. A., Perona J. J., Steitz T. A. Structural basis of anticodon loop recognition by glutaminyl-tRNA synthetase. Nature. 1991 Jul 18;352(6332):213–218. doi: 10.1038/352213a0. [DOI] [PubMed] [Google Scholar]
- Rould M. A., Perona J. J., Söll D., Steitz T. A. Structure of E. coli glutaminyl-tRNA synthetase complexed with tRNA(Gln) and ATP at 2.8 A resolution. Science. 1989 Dec 1;246(4934):1135–1142. doi: 10.1126/science.2479982. [DOI] [PubMed] [Google Scholar]
- Ruff M., Cavarelli J., Mikol V., Lorber B., Mitschler A., Giege R., Thierry J. C., Moras D. A high resolution diffracting crystal form of the complex between yeast tRNAAsp and aspartyl-tRNA synthetase. J Mol Biol. 1988 May 5;201(1):235–236. doi: 10.1016/0022-2836(88)90450-0. [DOI] [PubMed] [Google Scholar]
- Ruff M., Krishnaswamy S., Boeglin M., Poterszman A., Mitschler A., Podjarny A., Rees B., Thierry J. C., Moras D. Class II aminoacyl transfer RNA synthetases: crystal structure of yeast aspartyl-tRNA synthetase complexed with tRNA(Asp). Science. 1991 Jun 21;252(5013):1682–1689. doi: 10.1126/science.2047877. [DOI] [PubMed] [Google Scholar]
- Schimmel P. R., Söll D. Aminoacyl-tRNA synthetases: general features and recognition of transfer RNAs. Annu Rev Biochem. 1979;48:601–648. doi: 10.1146/annurev.bi.48.070179.003125. [DOI] [PubMed] [Google Scholar]
- Schimmel P. Aminoacyl tRNA synthetases: general scheme of structure-function relationships in the polypeptides and recognition of transfer RNAs. Annu Rev Biochem. 1987;56:125–158. doi: 10.1146/annurev.bi.56.070187.001013. [DOI] [PubMed] [Google Scholar]
- Söll D. The accuracy of aminoacylation--ensuring the fidelity of the genetic code. Experientia. 1990 Dec 1;46(11-12):1089–1096. doi: 10.1007/BF01936918. [DOI] [PubMed] [Google Scholar]
- Yarus M., Berg P. Recognition of tRNA by aminoacyl tRNA synthetases. J Mol Biol. 1967 Sep 28;28(3):479–490. doi: 10.1016/s0022-2836(67)80098-6. [DOI] [PubMed] [Google Scholar]
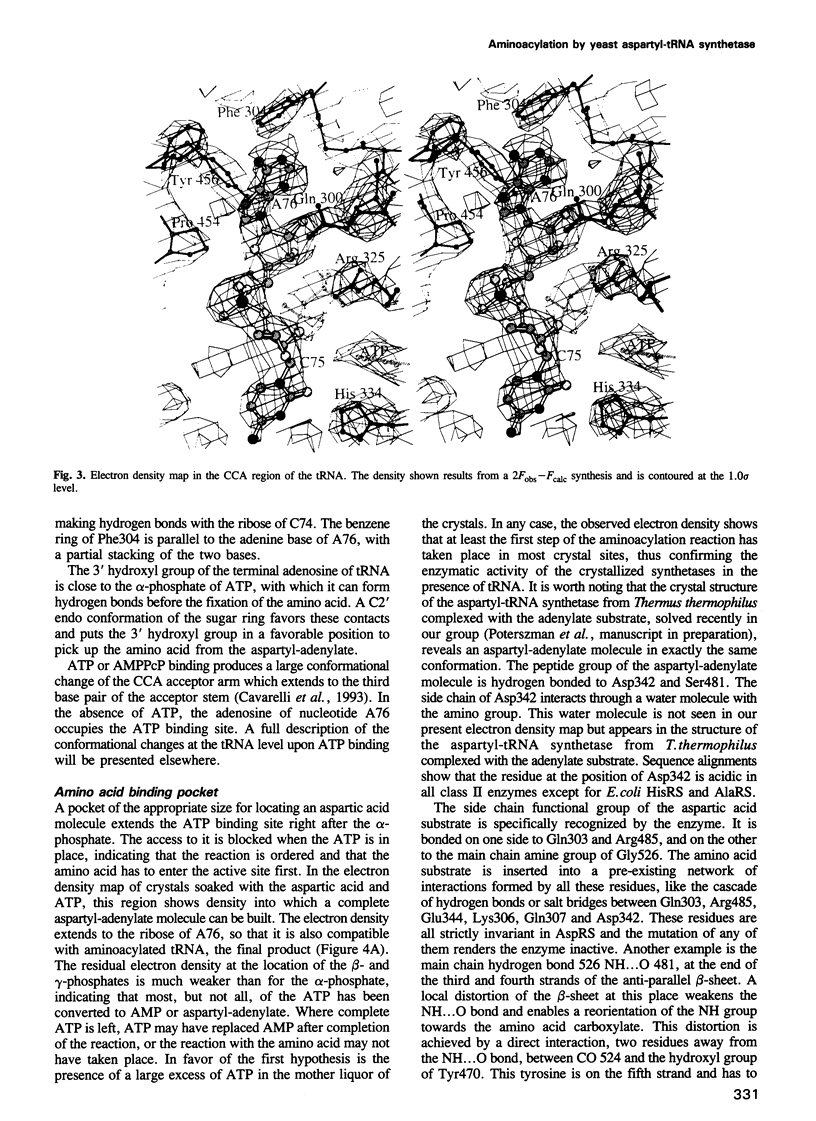
- de Prat Gay G., Duckworth H. W., Fersht A. R. Modification of the amino acid specificity of tyrosyl-tRNA synthetase by protein engineering. FEBS Lett. 1993 Mar 1;318(2):167–171. doi: 10.1016/0014-5793(93)80014-l. [DOI] [PubMed] [Google Scholar]