Abstract
Phosphate is critical in multiple biological processes (phosphorylation reactions, ATP production, and DNA structure and synthesis). It remains unclear how individual cells initially sense changes in phosphate availability and the cellular consequences of these changes. PiT1 (or SLC20A1) is a constitutively expressed, high-affinity sodium-dependent phosphate import protein. In vitro data suggest that PiT1 serves a direct role in mediating cellular proliferation; its role in vivo is unclear. We have discovered that mice lacking PiT1 develop a profound underproduction anemia characterized by mild macrocytosis, dyserythropoiesis, increased apoptosis, and a near complete block in terminal erythroid differentiation. In addition, the animals are severely B cell lymphopenic because of a defect in pro–B cell development and mildly neutropenic. The phenotype is intrinsic to the hematopoietic system, is associated with a defect in cell cycle progression, and occurs in the absence of changes in serum phosphate or calcium concentrations and independently of a change in cellular phosphate uptake. The severity of the anemia and block in terminal erythroid differentiation and B cell lympho-penia are striking and suggest that PiT1 serves a fundamental and nonredundant role in murine terminal erythroid differentiation and B cell development. Intriguingly, as the anemia mimics the ineffective erythropoiesis in some low-grade human myelodysplastic syndromes, this murine model might also provide pathologic insight into these disorders.
Inorganic phosphorous, in the form of phosphate (Pi), is required in multiple biological processes, ranging from energy homeostasis, intracellular signaling and maintenance of membrane integrity, and nucleic acid synthesis to bone mineralization. In humans, approximately 85% of total body phosphorous is in bone, 14% is intracellular, and only 1% is in the extracellular fluid [1]. A variety of mechanisms, primarily mediated by the effects of parathyroid hormone and the vitamin D endocrine system in bone, parathyroid, kidney, and intestine have evolved to increase the efficiency of phosphate retention in states of phosphate deficiency and conversely to decrease retention in states of phosphate excess [2]. Although our understanding of the mechanisms of systemic phosphate homeostasis has advanced [3], it remains unclear how individual cells maintain phosphate homeostasis and whether the same mechanism exists and cellular consequences of dyshomeostasis occur within all cell types.
In mammals, sodium-dependent Pi import proteins mediate the movement of Pi into cells. There are currently three identified cotransporter families, which differ in their Pi affinity, tissue distribution, and physiologic regulation and function (termed type I, II, and III and assigned to the solute carrier series SLC17, SLC34, and SLC20, respectively). The role of type I cotransporters remains to be fully established. Type II cotransporters function to maintain whole body phosphate homeostasis; members include NPT2a and NPT2c, which regulate renal phosphate absorption [4], and NPT2b, which regulates intestinal Pi absorption [5]. Type III sodium-dependent cotransporters include PiT1 (SLC20A1) and PiT2 (SLC20A2), which were initially identified as the human retroviral receptor for Gibbon Ape Leukemia Virus (GALV) [6] and the receptor (Ram-1) for amphotropic murine retrovirus [7], respectively, and subsequently as sodium-dependent phosphate importers [8]. PiT1 also functions as the retroviral receptor for feline leukemia virus subgroup B (FeLV-B) [9]. In humans, PiT1 and PiT2 are ubiquitously expressed [8], share approximately 60% amino acid sequence homology with one another and no significant sequence homology with the type I or type II cotransporters, and have the highest substrate affinity among the three families [10].
In vitro studies suggest that PiT1 has a role in cellular proliferation [11,12]; however, studies of its in vivo role were only recently initiated with the development of two null mouse models. Constitutive deletion of exon five results in embryonic lethality at embryonic day 12.5 because of severely hypoplastic liver development and resultant anemia [13]; similarly, mice with constitutive deletion of exons three and four die in mid-gestation and display pale livers suggesting anemia [14]. The embryos display no clear independent defect in vascular or bone development. Because the fetal liver at mid-gestation is the site of definitive erythropoiesis [15], we suspected that PiT1 is required for red blood cell development. In this study, we report that adult mice lacking PiT1 develop a profound underproduction anemia characterized by mild macrocytosis, dyserythropoiesis, increased apoptosis, and a near complete block in terminal erythroid differentiation. In addition, the animals are severely B cell lymphopenic due to a defect in pro–B cell development and mildly neutropenic.
Methods
Generation of inducible PiT1 mutant mice
Animals were housed in a specific pathogen-free facility at the University of Washington (Seattle, WA). The Institutional Animal Care and Use Committee approved all studies. To induce MxCre expression and delete exons 3 and 4 of Pit1, 5–7-day-old neonatal pups were treated with 50 μg of polyinosinic:polycytidylic acid (poly[I]poly[C]; Amersham/GE Life Sciences, Piscataway, NJ, USA) intraperitoneally (IP) every other day for three doses.
Genotyping of transgenic mice
Genomic DNA was isolated from tail snips and directly amplified by polymerase chain reaction (PCR) for genotyping using the REDExtract-N-Amp Tissue PCR Kit (Sigma Aldrich, Saint Louis, MO, USA).
Blood collection and analyses
Mice were bled retroorbitally into microtainer tubes treated with ethylenediaminetetraacetic acid (Becton Dickinson, Franklin Lakes, NJ). Reticulocyte counts and serum chemistries were obtained at Phoenix Central Laboratory (Everett, WA, USA) on an ADVIA120 and a Siemens ADVIA1650, respectively. Complete blood counts were obtained on a Hemavet HV950FS analyzer (Drew Scientific, Oxford, CT, USA). Erythropoietin was measured using Mouse/Rat EPO Immunoassay Kit (R&D Systems, Minne-apolis, MN, USA) according to manufacturer’s protocol. Bone marrow cytospins were generated using a Thermo Shandon Cyto-spin cytocentrifuge and stained with Hema 3 stain (Fisher, Pitts-burg, PA, USA).
Murine transplantation studies
Bone marrow cells harvested from Pit1flox/flox;MxCre or MxCre mice were transplanted into 6–8-week-old Pep3b lethally irradiated (11 cGy) recipients (2 × 106 cells/animal). After stable engraftment (5–6 weeks after transplant), mice were treated with 125 μg of poly(I)poly(C) IP every other day for three doses and sacrificed 7–9 weeks later for analyses.
Quantitative real-time reverse transcriptase PCR
Mice were sacrificed under anesthesia (2, 2, 2-tribromoethanol) by cervical dislocation to reduce the possibility of hypoxic regulation of gene expression. Total RNA was isolated from murine tissues using the AllPrep DNA/RNA Mini Kit (Qiagen Sciences, Mary-land, USA). B cells (for RNA studies) were isolated from fresh bone marrow by first staining cells with a biotin-labeled anti-B220 antibody, followed by incubation with a streptavidin-coupled bead (Dynabead M280), and subsequent magnetic separation (using a Dynamag-15; Invitrogen Life Technologies, Grand Island, NY, USA). RNA integrity was verified by the 260:230 ratio using the Eppendorf BioPhotometer Plus. cDNA was synthesized using the iScript cDNA Synthesis kit (Bio-Rad Life Science Research, Hercules, CA, USA) according to the manufacturer’s directions. Levels of messenger RNA (mRNA) were determined by quantitative real-time reverse transcriptase PCR with SsoFast EvaGreen Supermix (Bio-Rad) reagents and a Bio-Rad CFX96 qPCR system. All PCR reactions were performed in triplicate. Quantifi-cation of gene expression was calculated using the Pfaffl method and normalized to a housekeeping gene (18S, β-actin, or GAPDH). All primers are available on request.
Percent deletion of floxed-allele by quantitative real-time PCR
Genomic quantitative PCR was performed to discern the percent deletion of the floxed-allele using primers recognizing both Pit1 wild type and floxed alleles and, in a separate PCR reaction, primers recognizing the Pit1 deleted allele. Primers are available on request. All PCR reactions were performed in triplicate. Absolute quantification of percent deletion was calculated using a standard curve constructed from a diluted standard DNA template encoding the wild type allele and the deleted allele.
Flow cytometry and cell sorting
Single-cell suspensions were prepared from freshly isolated bone marrow or spleen using Iscove’s modified Dulbecco’s medium containing 2% fetal calf serum. Cells were immuno-stained with antibodies to Ter119, CD44, CD71, B220, Gr1, and Mac1 [16,17]. For hematopoietic progenitor studies, cells were immunostained and analyzed as described previously [18]. For B and T cell studies in the peripheral blood, cells were immunostained with antibodies to CD4, CD8, CD19, and B220, and red blood cells were lysed with hemolytic buffer before FACS analysis. For B cell progenitor studies, marrow cells were immunostained and analyzed as described previously [19,20]. Apoptosis was measured using an FITC Annexin V Apoptosis Detection Kit (BD Biosciences, San Jose, CA, USA) according to the manufacturer’s protocol. Cell staining was quantified with a FACSCanto flow cytometer (BD Biosciences). Flow cytometry data were analyzed with FlowJo software (Tree Star, Ashland, OR, USA). Cell sorting was performed on a FACSAria cell sorter (BD Biosciences).
Hematopoietic colony assays
Granulocyte-macrophage colony-forming unit assays were performed as described previously [21]. To detect burst-forming units-erythroid and colony-forming units-erythroid colonies, 4 × 105 and 2 × 105 cells/plate, respectively, were plated from Pit1-deleted or control mouse marrows in duplicate in semisolid medium (MethocultM3434; StemCell Technologies, Vancouver, BC, Canada) according to the manufacturer’s protocol.
Cell cycle analyses
Bromodeoxyuridine (BrdU; 2 mg/mouse) was injected intraperito-neally into mice, and marrow was harvested 30 and 90 minutes later. Cell cycle profiles of discreet populations of terminally differentiating erythroid cells (CD44, CD71, Ter119 methods in [16]) were obtained by the analyses of erythroid enriched progenitors (B220−, Gr1−, CD11b−, CD3−, CD4−, CD8−, CD5−). BrdU incorporation was detected using a BrdU flow kit (BD Biosciences) according to manufacturer’s directions. DNA content was assessed by 7-aminoactinomycin-D (7-AAD), or 4′6′-diamidino-2-phenylindole (DAPI) staining after membrane permeabilization with 0.1% saponin.
Phosphate uptake studies
Phosphate uptake studies were performed as described previously [22]. Uptake values were normalized to cellular protein content.
Western blot analysis studies
CD71+Glycophorin A− erythroid precursors were generated by culturing human peripheral blood CD34+ cells in Stemspan SFEM (StemCell Technologies, Vancouver, BC, Canada) containing 10% charcoal/dextran-treated fetal bovine serum (FBS; Atlanta Biologicals, Lawrenceville, GA, USA), rhSCF (50 ng/ml, PeproTech, Rocky Hills, NJ, USA), rhIL-3 (10 ng/ mL; PeproTech), rhIL-6 (20 ng/mL; PeproTech), rhIGF-I (40 ng/mL; PeproTech), rhEpo (2 U/mL; University of Washington Drug Services), 1% bovine serum albumin (Lampire Biotech, Lampire Biological Laboratories, Pipersville, PA, USA), 1 μmol/L dexamethasone (Sigma), holo-human transferrin (200 μg/mL; ProSpect-Tany Technogene Ltd., East Brunswick, NJ, USA), rh-insulin (10 μg/mL; University of Washington Drug Services), 2 mmol/L glutamine (Gibco), 1 μmol/L β-mercaptoe-thanol (Sigma), 0.5% Pen-Strep (Gibco), and 0.5% Fungizone (Gibco). K562 cells were cultured in RPMI medium (Gibco, Life Technologies, Auckland, NZ) containing 10% FBS (Sigma). 293T cells were culture in Dulbecco’s modified Eagle medium (Gibco) containing 10% FBS (Sigma). All cells were lysed in radioimmunoprecipitation assay (RIPA) buffer. Fresh mouse brain was lysed in RIPA buffer. One million cells per sample or greater were loaded on a 4%–20% gel (Bio-Rad), and a rabbit polyclonal antibody generated with AA408 421(CGDSGDKPLRRNNSY) of human/mouse SLC20A1 (in-house antibody) was used to detect the protein by Western blot analysis. Band intensity was quantified using ChemiDoc XRS+ with Image Lab software (Bio-Rad) and normalized to β-actin.
Results
Generation of mice lacking PiT1
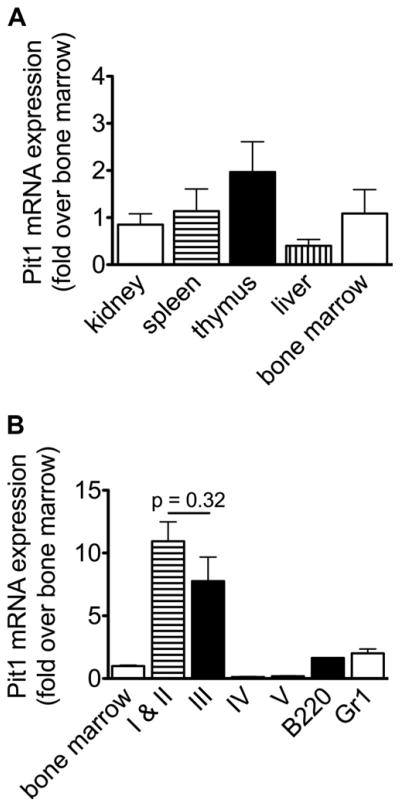
To study the role of PiT1 in murine hematopoiesis, we first measured Pit1 mRNA in adult wild type murine tissues. We confirmed that Pit1 is ubiquitously expressed, and its bone marrow expression is high in proerythroblasts and baso-philic erythroblasts; it declines in more terminally differentiated erythroid cells and is present in Gr1+ myeloid and B220+ B cells (Fig. 1). In our hands, no currently available anti-mouse PiT1 protein antibody reliably detects PiT1 in primary murine cells; we confirmed that PiT1 (SLC20A1) is expressed in primary human erythroid precursors (Supplementary Figure E1, online only, available at www.exphem.org). We then obtained mice expressing a Pit1 allele with loxP sites flanking exons 3 and 4, which allows for the conditional deletion of these exons by Cre recombi-nase, resulting in expression of a nonfunctional protein [14], and we bred an inducible mutant line that expresses Cre recombinase under the interferon-inducible Mx1 promoter [23] (Pit1flox/flox;MxCre). Mice that are heterozygous and homozygous for the conditional allele are viable, healthy, fertile, and grossly indistinguishable from wild type littermates. Treatment of neonatal experimental mice with poly(I)poly(C) to induce MxCre expression results in greater than 95% deletion of the floxed-allele in hematopoi-etic tissues (whole bone marrow, spleen, and flow cytomet-rically sorted marrow erythroid precursors, Gr1+ and B220+ cells, and peripheral blood B220+ cells). Deletion is also robust in the liver but marginal in the thymus, circulating peripheral blood CD4+ and CD8+ T cells, and kidney (Supplementary Figures E2A and E2B, online only, available at www.exphem.org). As expected, mRNA expression correlates with the percent deletion of the floxed allele and is nearly absent in Pit1-deleted hematopoietic tissues compared with controls (Supplementary Figure E2C, online only, available at www.exphem.org).
Figure 1.
Pit1 mRNA expression in wild type murine tissues. Tissues were harvested from 11-week-old C57BL/6 wild type mice. (A) Expression in kidney, spleen, thymus, liver, and whole bone marrow relative to expression in whole bone marrow. (B) Expression in flow cytometrically sorted bone marrow populations relative to whole bone marrow. Populations I–V include proerythroblasts/basophilic erythroblasts (I and II), poly-chromatophilic erythroblasts (III), orthochromatophilic erythroblasts and other maturing erythroid precursors (IV), and reticulocytes and mature red blood cells (V)16 (gates are shown in Fig. 3B). For each sample, mRNA levels were measured by reverse transcriptase quantitative real-time PCR. Sample PCR reactions were performed in triplicate and normalized to 18S expression. Bone marrow and sorted marrow populations: n = 2 samples, each sample contains pooled marrow from 2 male or 2 female mice; tissues: n = 4 mice. All data represent mean ± SEM, two-tailed Student t test.
Mice lacking PiT1 have multilineage hematopoietic cell defects
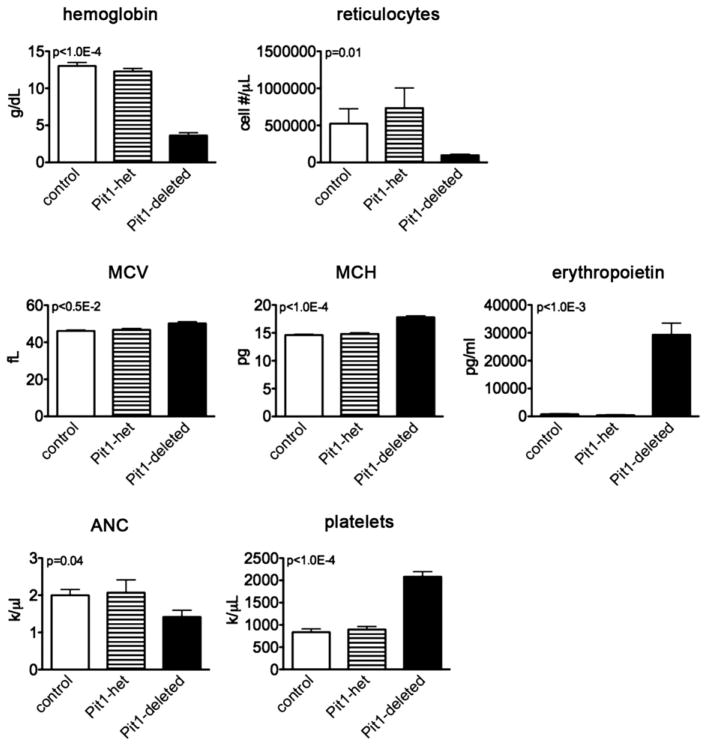
Within 4 weeks of Pit1 deletion, the mice are runted with pale paws. On necropsy, their spleens (a site of stress erythropoi-esis in mice) are massively enlarged due to a dramatic expansion of erythroid precursors in the red pulp space (Spleen weight/Body weight, 48.4 ± 2.5 × 10−3; n =10 Pit1-deleted vs. 4.7 ± 0.6 × 10−3; n = 8 control mice; p < 1.0E−4, mean ± SEM, two-tailed Student t test; Supplementary Figure E3, online only, available at www.exphem.org). Peripheral blood analyses demonstrate a severe anemia characterized by a low absolute reticulocyte count, mild elevation in the mean corpuscular volume and mean corpuscular hemoglobin, and elevated erythropoietin levels. In addition, the animals are mildly neutropenic and have thrombocytosis (Fig. 2 and Supplementary Table E1, online only, available at www.exphem.org). Peripheral blood smears, apart from macrocytosis, were unremarkable compared with controls (data not shown). The MxCre trans-gene by itself had no effect on blood counts.
Figure 2.
PiT1-deleted mice develop a macrocytic anemia, mild neutropenia, and thrombocytosis. Hematologic parameters: control mice, n = 7; Pit1-heterozygously deleted mice (Pit1-het), n = 7; Pit1-deleted mice, n = 11; platelets, n = 10. Reticulocyte count measurements: control mice, n = 7; Pit1-het, n = 6; Pit1-deleted mice, n = 13. Erythropoietin levels: control mice, n = 5; Pit1-het, n = 2; Pit1-deleted mice, n = 8. Data represent mean ± SEM, two-tailed Student t test. MCH = mean cell hemoglobin; MCV = mean corpuscular volume; ANC = absolute neutrophil count.
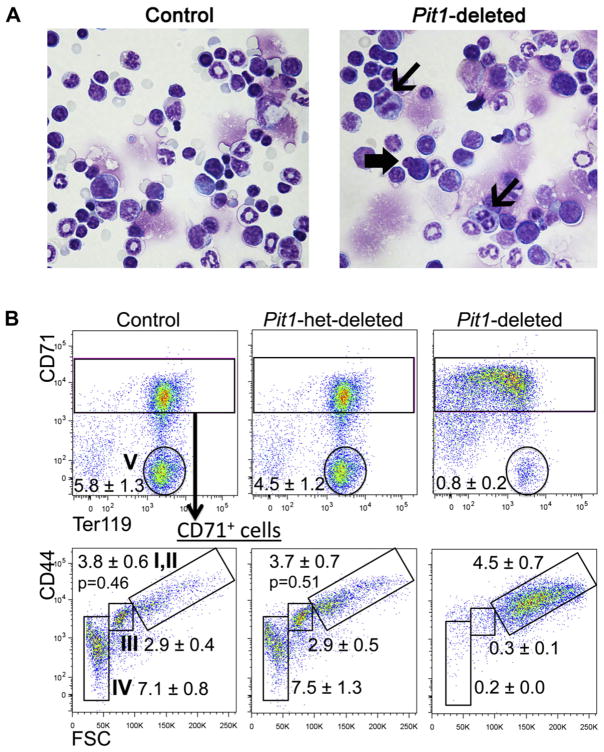
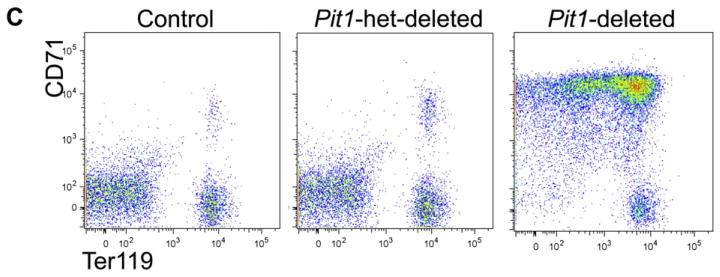
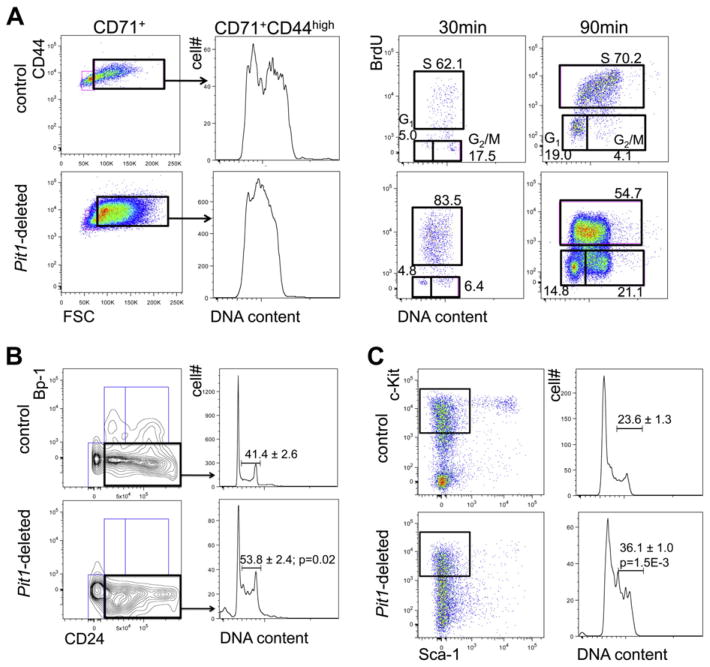
To explore the ontogeny of the anemia, we studied the bone marrow and spleen. Deleted mice have reduced numbers of total marrow mononuclear cells (16.1 × 106 cells per two femurs [±3.3; n = 4 Pit1-deleted mice] vs.47.0 × 106 ± 5.5 [n = 8 control mice]; p < 0.001). Bone marrow cytospins show erythroid maturation arrest at the proerythroblast/basophilic erythroblast stage and dys-erythropoiesis in the rare maturing erythroid forms, characterized by nuclear blebbing and occasional abnormal mitotic figures (Fig. 3A and Supplementary Figure E4, online only, available at www.exphem.org). Flow cytometric analyses demonstrate a relative and marked expansion of CD71highCD44highTer119+FSChigh cells (proerythroblasts to basophilic erythroblasts) [16] in deleted mice compared with controls (Fig. 3B; population I/II 21.4% ± 2.2 vs. 7.0% ± 1.3, p < 0.0001). In addition, the cell surface expression of the murine terminal erythroid differentiation marker, Ter119, is lower on Pit1-deleted Ter119+ cells compared with controls (the geometric mean fluorescent intensity is over sevenfold lower than control mice; data not shown), suggesting that the cells are closer to the pro-erythroblast stage of differentiation. Splenic analyses show analogous findings (Fig. 3C). The absolute numbers of bone marrow CD71highCD44highTer119+FSChigh cells are similar in deleted and control mice, whereas the absolute numbers of more terminally differentiated erythroid precursors are markedly reduced in deleted mice, which is consistent with a block in erythroid differentiation around the proerythroblast stage. Hematopoietic colony assays showed similar findings (Supplementary Figure E5, online only, available at www.exphem.org). In addition, we demonstrated increased apoptosis in Pit1-deleted Ter119+ erythroid cells by annexin V staining (apoptosis was defined as annexin V+7AAD−/7AAD− cells, 17.9% ± 2.7 [n = 3 Pit1-deleted mice] vs. 8.0% ± 0.4 [n = 3 control mice]; p = 0.02, mean ± SEM, two-tailed Student t test; data not shown). We did not observe a difference in phospho-JNK between whole bone marrow from Pit1-deleted and control mice (data not shown), which has been reported in PiT1-depleted human cell lines and immortalized murine cells and purported to account for their increased sensitivity to the proapoptotic activity of tumor necrosis factor α [12].
Figure 3.
Pit1-deleted mice have abnormal erythropoiesis characterized by a block in terminal erythroid maturation, increased apoptosis, and dyserythro-poiesis. (A) Cytospin preparations of whole bone marrow cells from Pit1-deleted mice show dyserythropoiesis. Thin arrows indicate mitotic figures, and the block arrow indicates nuclear blebbing. The number of erythroid precursors with either nuclear blebbing or mitotic figures was increased on cytospin preparations from Pit1-deleted mice compared with control mice (3.5% ± 0.4% vs. 1.4% ± 0.2%; Pit1-deleted, n = 3; controls, n = 3; p = 0.02). The percentage of morphologic atypia was determined by visually counting 500 cells per cytospin preparation. Flow cytometric analyses of bone marrow (B) and spleen (C) from a representative control, Pit1-heterozygously-deleted (Pit1-het–deleted), and Pit1-deleted mouse immunostained with antibodies against CD71, CD44, and Ter119. CD71+ cells are gated in the lower panel of (B) and plotted as CD44 versus forward scatter (FSC) to define the terminal stages of erythroid differentiation: proerythroblasts and basophilic erythroblasts (I,II, CD71highCD44highFSChigh), polychromatophilic erythroblasts (III, CD71highCD44intermediateFSCintermediate), orthochromatophilic erythroblasts, and other maturing erythroid precursors (IV, CD71highCD44lowFSClow). Population V includes reticulocytes and mature red blood cells (CD71lowTer119high). Absolute marrow mononuclear cell numbers × 106 per mouse are shown (control, n = 9; Pit1-het, n = 7; Pit1-deleted, n = 12). To enrich for erythroid precursors, B220+, Gr1+, and Mac1+ cells were excluded from the analyses. The p values of all comparisons between the same individual control and Pit1-deleted populations, and of all comparisons between the same individual Pit1-het and Pit1-deleted populations are less than 0.002, unless indicated (populations I/II). The p values of all comparisons between the same individual control and Pit1-het-deleted populations are ≥0.10.
To investigate whether the observed anemia, neutrope-nia, and reduced numbers of marrow mononuclear cells reflect an effect of Pit1 deletion on the development of hematopoietic stem and progenitor cells, we performed additional flow cytometric analyses [24]. The number of KLS+ cells (IL7Rα−c-Kit+Lin−Sca-1+, which includes hematopoietic stem cells) was nonsignificantly reduced in deleted mice compared with controls (Fig. 4A). In addition, the number of common lymphoid progenitors (CLPs; IL7Rα+c-Kit+Lin−Sca-1+), common myeloid progenitors (CMPs), megakaryocytic-erythroid progenitors (MEPs; Lin−c-KithighSca-1−CD16/CD32lowCD34−), and granulocyte-macrophage progenitors (GMPs, Lin−c-Kithigh Sca-1−CD16/CD32highCD34−) were comparable in deleted and control mice (Fig. 4B, Supplementary Table E2, online only, available at www.exphem.org). Of note, deleted mouse marrow contained an expanded population of Lin−c-KithighSca-1−CD16/CD32highCD34− cells (asterisk in Fig. 4B), absent in control marrow. We further characterized these cells by morphologic review and additional im-munostaining and flow cytometric analyses designed to define specific cell populations in the mouse myeloeryth-roid hierarchy (PreMegE, Pre CFU-E, CFU-E, MkP, PreGM, and GMP) [18]; the precise lineage of this population remains unknown. We hypothesize that these cells represent erythroid precursors (Pre CFU-E) with aberrant expression of the myeloid antigen, CD16/CD32 (Supplementary Figure E6, online only, available at www.exphem.org).
Figure 4.
Pit1-deleted mice have normal numbers of early hematopoietic progenitors. (A) KLS+ cells (IL7Rα−c-Kit+Lin−Sca-1+). Numbers represent absolute numbers of cells − 104 per mouse; p = 0.6. (B) Bone marrow cells from a representative control and Pit1-deleted mouse stained for common myeloid progenitor (CMP, c-KithighLin−Sca-1−CD16/ CD32lowCD34+), megakaryocyte-erythroid progenitor (MEP, c-Kithigh-Lin−Sca-1−CD16/CD32lowCD34−) and granulocyte-macrophage progenitor (GMP; c-KithighLin−Sca-1−CD16/CD32highCD34+) cell surface markers. Numbers represent percentage of cells in gate shown, which did not differ between deleted and control animals apart from CMP (p = 0.03). Absolute numbers of each population were not significantly different. Control mice, n = 2; Pit1-deleted mice, n = 5. The number of CLPs (IL7Rα+c-Kit+Lin−Sca-1+) were similar in deleted and control mice (19.0 ± 1.4 × 103 [Pit1-deleted mice, n = 3] vs. 20.1 ± 8.1 × 103 [control mice, n = 3]; p = 0.9). Mean ± SEM, two-tailed Student t test.
During our analyses, we noticed that B220+ bone marrow cells (B lymphocytes at various stages of development) are reduced in Pit1-deleted animals compared with controls (3.0 × 106 ± 1.1 vs. 12.3 × 106 ± 1.2; n = 5 for both; p < 0.001, mean ± SEM, Student t test). This observation prompted us to quantify peripheral blood lymphocyte subsets. The peripheral blood mirrored the bone marrow find-ings and showed a reduced number of B lymphocytes (Fig. 5A). To define the stages of bone marrow B cell development impacted by a lack of PiT1, we evaluated B cell development flow cytometrically based on the expression of the cell surface markers B220, CD43, BP-1, CD24, and CD19. This analysis divides B cell progenitors into pre-pro B, early pro-B, late pro-B, and early pre-B cells, so-called Hardy fractions A, B, C, and C′ [19,20,25]. The absolute numbers of all these B cell progenitors were reduced in Pit1-deleted mice compared with controls (Fig. 5B, top). Furthermore, within early B cell progenitors (B220+CD43+), the proportions of pre-pro B (BP-1−CD24−) and early pro-B cells (BP-1−CD24+) are comparable between deleted and control mice, whereas the proportion of early pre-B cells (BP-1+CD24+) is dramatically reduced in deleted marrow (Fig. 5B, bottom). In addition, the percentage of CD19+ cells among early pro-B cells, which defines less (CD19−) and more (CD19+) differentiated cells [19,26], is markedly reduced (Fig. 5C). In total, our data suggest that B cell development is impaired between the CLP to pre-pro B cell stage and, additionally, is severely impaired at the early pro-B cell stage.
Figure 5.
Pit1-deleted mice have a defect in B cell development. (A) Peripheral blood cells were immunostained with antibodies against CD19 and B220. Absolute cell numbers were obtained by multiplying the percentage of cells in each population measured by flow cytometry with the lymphocyte count obtained by complete blood cell count. Peripheral blood CD19+ B cell numbers are similar to B220+ B cell numbers. Control mice, n ≥ 3; Pit1-deleted mice, n ≥ 6. (B) Flow cytometric analyses of bone marrow B cell progenitors from a representative control and Pit1-deleted mouse immunostained with antibodies against B220, CD43, BP-1, CD24, and CD19. Analyses include B220+CD43+ B cell progenitors plotted as BP-1 versus CD24 (flow plots). The relative percentages of B220+CD43+ B cell progenitors which mark as early pro-B (B220+CD43+BP-1−CD24+), and early pre-B (BP-1+CD24high) cells. (C) Percent of early pro-B cells expressing CD19. Analysis includes B220+CD43+BP-1−CD24+cells. Control mice, n = 3; Pit1-deleted mice, n = 3. All data represent mean ± SEM, two-tailed Student t test.
Importantly, the hematologic phenotype is intrinsic to the hematopoietic system because lethally irradiated mice transplanted with PiT1flox/flox;MxCre marrow and treated with poly(I)poly(C) to delete Pit1 specifically in engrafted cells also develop severe anemia, B cell lymphopenia, mild neutropenia, and thrombocytosis (Table 1). Together, our data suggest that PiT1 function is needed at multiple stages of hematopoiesis and additionally is uniquely required for terminal erythroid differentiation and pro-B cell development.
Table 1.
Complete blood counts in mice lacking PiT1 in the bone marrow and control micea
| Control (n = 3) | Pit1-deleted (n = 4) | p value | |
|---|---|---|---|
| WBC (1,000/μL) | 12.49 ± 1.05 | 6.05 ± 0.85 | 5.0 × 1033 |
| Neutrophils (1,000/μL) | 2.69 ± 0.24 | 1.56 ± 0.16 | 0.01 |
| Lymphocytes (1,000/μL) | 9.56 ± 1.15 | 4.26 ± 0.87 | 0.01 |
| Hemoglobin (g/dL) | 13.00 ± 1.34 | 6.42 ± 1.17 | 0.01 |
| MCV (fL) | 40.17 ± 0.73 | 37.57 ± 0.28 | 0.01 |
| MCH (pg) | 13.47 ± 0.27 | 13.40 ± 0.20 | 0.85 |
| Platelets (1,000/μL) | 685.00 ± 203.72 | 1658.50 ± 195.59 | 0.02 |
| B220+ (1,000/μL)b | 4.85 ± 0.44 | 1.12 ± 0.30 | <0.001 |
MCH = mean corpuscular hemoglobin; MCV = mean corpuscular volume.
Pep3b recipient mice were transplanted with Pit1wild-type/wild-type;MxCre or Pit1flox/flox;MxCre bone marrow and after stable engraftment, treated with poly(I) poly(C) to delete the floxed-allele. Data were obtained 4 weeks after poly(I)poly(C) treatment.
B220+ numbers calculated from CD45.2 donor-positive cells (determined by flow).
Serum phosphate and proerythroblast–basophilic erythroblast sodium-dependent phosphate uptake are not altered in Pit1-deleted mice
In humans, severe hypophosphatemia is reported to cause intravascular hemolysis [27,28], presumably by affecting red blood cell membrane deformability. Hyperphosphate-mia has been implicated as one cause for recombinant human erythropoietin–resistant anemia in a subset of chronic renal failure patients (with adequate iron status). These mostly correlative studies link serum phosphate concentration and parathyroid hormone (PTH) levels to erythropoietin dose [29–31]. It is unclear how PTH affects erythropoiesis [32–35]. The anemia in Pit1-deleted mice and mice lacking PiT1 only in hematopoietic cells (transplant setting) occurs in the absence of changes in serum phosphate or calcium concentrations and thus, is likely independent of changes in PTH (Supplementary Tables E3 and E4, online only, available at www.exphem.org).
In mature (enucleated) red blood cells, 75% of phosphate uptake occurs via the anion transporter, band 3; 20% occurs via a sodium-dependent transport mechanism, and 5% of uptake is mediated by a pathway linearly dependent on extracellular phosphate concentration [36]. In primary erythroid precursors, sodium-dependent phosphate uptake and the protein expression of sodium-dependent phosphate importers has not been reported. We found that Pit1, Pit2, and Npt2c are the only sodium-dependent phosphate importers expressed in wild type murine whole bone marrow and sorted proerythroblasts and basophilic erythroblasts (Supplementary Figure E7A, online only, available at www.exphem.org). Because PiT1 functions as a sodium-dependent phosphate import protein, we performed phosphate uptake studies using H333PO4 as an indicator. Pit1-deleted and control whole bone marrow and sorted proerythroblasts and basophilic erythroblasts, like MEFs derived from Pit1 null embryos [13], have equivalent total and sodium-dependent phosphate uptake (Fig. 6). Deleted cells demonstrate a fourfold upregulation of Pit2 mRNA expression (Supplementary Figure E7B, online only, available at www.exphem.org), which could account for the maintained sodium-dependent phosphate uptake. Thus, the hematopoietic phenotype of Pit1-deleted mice is not caused by altered cellular phosphate uptake.
Figure 6.
Total and sodium-dependent phosphate uptakes are comparable in Pit1-deleted and control mice proerythroblasts and basophilic erythroblasts. Phosphate uptake in flow cytometrically sorted proerythroblasts and basophilic erythroblasts (populations I and II in Fig. 3B) from deleted and control mice was measured in the presence of sodium chloride (total phosphate uptake) or choline chloride (sodium-independent phosphate uptake). Sample cells were derived from flow cytometrically sorted pooled whole bone marrow of three control mice or pooled whole bone marrow of two Pit1-deleted mice. Data represent mean ± SEM, two-tailed Student t test.
Pit1-deleted hematopoietic cells demonstrate a defect in cell cycle progression
Recent studies in nonhematopoietic cell lines implicate PiT1 in mediating cellular proliferation [11,12] which prompted us to evaluate the cell cycle profiles of CD71+CD44high-FSChigh erythroid cells from Pit1-deleted and control mice. The majority of deleted and control (consistent with published data [37]) proerythroblasts and basophilic erythroblasts are in S-G2/M phases of the cell cycle (Fig. 7A, center left panels). Because a DNA content-based cell cycle profile data cannot discern S from G2/M, we performed in vivo BrdU labeling studies 30 and 90 min after injection of BrdU to better delineate the cell cycle. After short labeling, the DNA content of the majority of BrdU-labeled deleted and control proerythroblasts and basophilic erythroblasts is less than 4N, suggesting that the cells recently entered S phase from G1 phase (Fig. 7A, right panels, 30 min). In addition, the fold-increase in geometric mean fluorescence of S phase over G1 phase cells (FIGF) is higher in deleted cells compared to control cells (deleted 21 vs. control 15, FIGF ratio 1.4). This suggests that DNA synthesis rates are not reduced in Pit1-deleted cells. Because the frequencies of BrdU-positive cells (deleted/control = 1.3) are also comparable, the cells likely enter S phase from G1 phase similarly. After 90-min labeling compared to after 30-min labeling, there are almost no unlabeled cells with DNA content of 4N from control mice (17.5% reduced to 4.1%). Alternatively, in deleted mice, the frequency of unlabeled cells with 4N DNA content increased from 6.4% to 21.1%, which is consistent with a delay in exiting G2 and M phases. Consistent with this conclusion, after 90-min labeling, the FIGF ratio is lower (deleted/control = 0.63; Fig. 7A, far right panels, 90 min), reflecting faster overall cell cycling of control cells. In summary, the Pit1-deleted cells have intact G1 to S entry and a delay in exiting G2 and M phases, resulting in overall slower cell cycling. Importantly, we also observed a defect in cell cycle progression in early pro-B cells and hematopoietic progenitors (Fig. 7B and C), which is consistent with a generalized defect in multiple hematopoi-etic cell types.
Figure 7.
Pit1-deleted hematopoietic populations have impaired cell cycle progression. (A) Cell cycle profiles (left) of proerythroblasts and basophilic erythroblasts (B220−Gr1−CD71+FSChigh) and BrdU cell cycle analyses (right) of proerythroblasts and basophilic erythroblasts. Our cell cycle profiles methods lyse terminally differentiated cells. For BrdU analyses, the percentages of cells in each phase of the cell cycle are shown. Mice were injected with BrdU, and bone marrow was harvested 30 and 90 minutes after injection for analyses. S phase BrdU median fluorescence intensity is commonly used as a measure of the S phase DNA synthesis rate. Because the baseline fluorescence is not uniform between mice, the fold-increase over G1 in geometric mean fluorescence, which reflects baseline fluorescence, is cited as a measure of DNA synthesis in the text. Repeated experiment showed similar results. (B) Cell cycle profiles of early pro-B cells. (C) Cell cycle profiles of KLS+ hematopoietic progenitors (c-Kit+Lin−Sca-1−); all n = 3.
Discussion
In this study, we show that mice lacking PiT1 develop a profound underproduction anemia characterized by mild macrocytosis, dyserythropoiesis, increased apoptosis and a near complete block in terminal erythroid differentiation around the proerythroblast stage. The animals also have a severe B cell deficiency, which primarily reflects a block in pro-B cell development, mild neutropenia, and thrombo-cytosis. The severity of the anemia and B cell lymphopenia are striking and suggest that PiT1 serves a fundamental and stage-specific role in these lineages.
In addition, Pit1-deleted mice have a general hemato-poietic cell defect in cell cycle progression, which is consistent with a role for PiT1 in governing cellular proliferation. This observation has been reported previously in nonhematopoietic cell types. In HeLa and hepatic cell lines, knockdown of PiT1 expression (and not PiT2) by RNA interference decreased cellular proliferation, delayed cell cycle progression, and increased apoptosis [11,12]. The proliferative defect did not correct with over-expression of PiT2, although this restored cellular phosphate uptake to nearly wild-type levels. Moreover, the proliferative defect was corrected by expressing either wild type PiT1 or a mutant PiT1 lacking sodium phosphate import function, but which traffics normally to the cell membrane, establishing that the effect on proliferation is specific to PiT1 knockdown and independent of phosphate transport activity through PiT1 [12]. Studies of MEFs derived from a PiT1 null embryo showed similar results and established that the proliferative potential of cells correlated with their levels of Pit1 mRNA expression [13]. In vivo data also support our observation that PiT1 functions in cellular proliferation. Tumor growth in nude mice injected with HeLa cells expressing shRNA against PiT1 is reduced compared with tumor growth in animals injected with HeLa cells expressing a control scramble shRNA [11]. In line with a role for PiT1 in regulating cellular proliferation, overexpression of human PiT1 (and not human PiT2) in two density-inhibited murine cell lines increased proliferation and increased sensitivity to fetal bovine serum-induced transformation [38].
Furthermore, we determined that cellular phosphate uptake in Pit1-deleted and control proerythroblasts and basophilic erythroblasts is comparable. Similarly, in both the PiT1 knockdown and overexpression models, cellular phosphate uptake did not differ between experimental and control cells [11,38]. Thus, although we suspect that the defect in terminal erythroid differentiation (and likely B cell development) results from an impairment in cellular proliferation, it seems unlikely that this is a direct consequence of phosphate deprivation.
During erythropoiesis, proliferation and terminal differentiation are mechanistically linked [39,40]. Similarly, early pro-B cells undergo a proliferative burst [38] concurrently with a critical differentiation step (interleukin 7 [IL-7] receptor-mediated heavy chain immunoglobulin gene rearrangement). Mice with targeted deletion of IL-7 or its receptor, like Pit1-deleted mice, have a severe block in B cell development at the pro-B cell stage [41,42]. Considering our results within this context, we speculate that the severe anemia and B cell lymphopenia in mice lacking PiT1 results from dysregulation in a lineage- and stage-specific coupling of proliferation with terminal differentiation; proerythroblast/basophilic erythroblasts and proB cell differentiation are particularly vulnerable to perturbations in the cell cycle. We recognize that Cre mediates recombination of the floxed-allele in other hematopoietic cells and nonhematopoietic cells and suspect that proliferation in these cells is also impacted by a lack of PiT1. This would explain the mild neutropenia we observed. However, we argue that the animals die of an anemia before clinically manifesting a defect in another cell type. Because deletion of the floxed allele is incomplete in the thymus and peripheral blood T cells, our model does not address the role of PiT1 in this lineage. Interestingly, mice expressing a hypo-morphic allele of c-Myb, a transcription factor that both promotes and represses proliferation and differentiation at multiple stages of hematopoiesis, share several phenotypic features with Pit1-deleted mice, including macrocytic anemia, thrombocytosis, and a block in B cell development at the pro-B cell stage [43]. In addition, c-Myb enables proB cells to respond to IL-7 [44]. Determining how PiT1 affects proliferation and differentiation at multiple stages of hematopoiesis and whether c-Myb–regulated pathways inform PiT1 biology remain open to future research.
Importantly, the hematologic phenotype of mice lacking PiT1 mimics the hematopoietic findings in low-grade human myelodysplastic syndromes (MDS) in which cell cycle defects may also play a pathophysiologic role [45]. The ineffective and dysplastic erythropoiesis, B cell lym-phopenia, and mild neutropenia observed in Pit1-deleted mice are also variably present in low-grade MDS [46–48]. As the human and feline orthologs of PiT1 function as retroviral receptors for gibbon ape leukemia virus (GALV) [6] and feline leukemia virus, subgroup, B (FeLV-B) [9], and retroviral infection downregulates function of its receptor, a pathologic link between PiT1 dysfunction and MDS is further supported by the observation that GALV and FeLV-B infection are associated with leukemia and MDS in nonhuman primates and cats [49–51]. The role of PiT1 in human hematopoiesis is unknown. Recently, Yoshida et al. [52] reported a point mutation predicted to be causative in SLC20A1, which encodes PiT1, in a patient with low-grade MDS. Given this report, the hematologic findings in mice lacking PiT1, and the close protein sequence homology between murine and human PiT1 [53], PiT1 may also function in human hematopoiesis and be required for terminal erythroid differentiation and B cell development. Future studies will address these possibilities.
Supplementary Material
Acknowledgments
We thank Janis L. Abkowitz for her ongoing mentorship, Yan Wang for early management of our mouse colony, Keith Loeb and Sue Knoblaugh for reviewing murine pathology slides, and John Byon for methodological advice on primary human erythroid culture.
This work was supported by National Institutes of Health grants P30 CA147883 and R01 HL062329, and National Heart, Lung, and Blood Institute grant T32 HL07828.
Footnotes
SBK, LL, MSB, and MC designed and performed research and analyzed data. SBK wrote the manuscript. CG contributed reagents and intellectually to the research.
Supplementary data related to this article can be found online at http://dx.doi.org/10.1016/j.exphem.2013.01.004.
Conflict of interest disclosure
No financial interest/relationships with financial interest relating to the topic of this article have been declared.
References
- 1.Lau WL, Festing MH, Giachelli CM. Phosphate and vascular calcifi-cation: Emerging role of the sodium-dependent phosphate co-transporter PiT-1. Thromb Haemost. 2010;104:464–470. doi: 10.1160/TH09-12-0814. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Berndt T, Kumar R. Phosphatonins and the regulation of phosphate homeostasis. Annu Rev Physiol. 2007;69:341–359. doi: 10.1146/annurev.physiol.69.040705.141729. [DOI] [PubMed] [Google Scholar]
- 3.Bergwitz C, Juppner H. Regulation of phosphate homeostasis by PTH, vitamin D, and FGF23. Annu Rev Med. 2010;61:91–104. doi: 10.1146/annurev.med.051308.111339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Murer H, Hernando N, Forster I, Biber J. Regulation of Na/Pi transporter in the proximal tubule. Annu Rev Physiol. 2003;65:531–542. doi: 10.1146/annurev.physiol.65.042902.092424. [DOI] [PubMed] [Google Scholar]
- 5.Sabbagh Y, O’Brien SP, Song W, et al. Intestinal npt2b plays a major role in phosphate absorption and homeostasis. J Am Soc Nephrol. 2009;20:2348–2358. doi: 10.1681/ASN.2009050559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.O’Hara B, Johann SV, Klinger HP, et al. Characterization of a human gene conferring sensitivity to infection by gibbon ape leukemia virus. Cell Growth Differ. 1990;1:119–127. [PubMed] [Google Scholar]
- 7.Miller DG, Miller AD. A family of retroviruses that utilize related phosphate transporters for cell entry. J Virol. 1994;68:8270–8276. doi: 10.1128/jvi.68.12.8270-8276.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kavanaugh MP, Miller DG, Zhang W, et al. Cell-surface receptors for gibbon ape leukemia virus and amphotropic murine retrovirus are inducible sodium-dependent phosphate symporters. Proc Natl Acad Sci USA. 1994;91:7071–7075. doi: 10.1073/pnas.91.15.7071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Takeuchi Y, Vile RG, Simpson G, O’Hara B, Collins MK, Weiss RA. Feline leukemia virus subgroup B uses the same cell surface receptor as gibbon ape leukemia virus. J Virol. 1992;66:1219–1222. doi: 10.1128/jvi.66.2.1219-1222.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Khoshniat S, Bourgine A, Julien M, Weiss P, Guicheux J, Beck L. The emergence of phosphate as a specific signaling molecule in bone and other cell types in mammals. Cell Mol Life Sci. 2011;68:205–218. doi: 10.1007/s00018-010-0527-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Beck L, Leroy C, Salaun C, Margall-Ducos G, Desdouets C, Fried-lander G. Identification of a novel function of PiT1 critical for cell proliferation and independent of its phosphate transport activity. J Biol Chem. 2009;284:31363–31374. doi: 10.1074/jbc.M109.053132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Salaun C, Leroy C, Rousseau A, Boitez V, Beck L, Friedlander G. Identification of a novel transport-independent function of PiT1/SL-C20A1 in the regulation of TNF-induced apoptosis. J Biol Chem. 2010;285:34408–34418. doi: 10.1074/jbc.M110.130989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Beck L, Leroy C, Beck-Cormier S, et al. The phosphate transporter PiT1 (Slc20a1) revealed as a new essential gene for mouse liver development. PLoS One. 2010;5:e9148. doi: 10.1371/journal.pone.0009148. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Festing MH, Speer MY, Yang HY, Giachelli CM. Generation of mouse conditional and null alleles of the type III sodium-dependent phosphate cotransporter PiT-1. Genesis. 2009;47:858–863. doi: 10.1002/dvg.20577. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wu H, Liu X, Jaenisch R, Lodish HF. Generation of committed erythroid BFU-E and CFU-E progenitors does not require erythropoi-etin or the erythropoietin receptor. Cell. 1995;83:59–67. doi: 10.1016/0092-8674(95)90234-1. [DOI] [PubMed] [Google Scholar]
- 16.Chen K, Liu J, Heck S, Chasis JA, An X, Mohandas N. Resolving the distinct stages in erythroid differentiation based on dynamic changes in membrane protein expression during erythropoiesis. Proc Natl Acad Sci USA. 2009;106:17413–17418. doi: 10.1073/pnas.0909296106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Socolovsky M, Nam H-S, Fleming MD, Haase VH, Brugnara C, Lod-ish HF. Ineffective erythropoiesis in Stat5a−/ −5b−/ − mice due to decreased survival of early erythroblasts. Blood. 2001;98:3261–3273. doi: 10.1182/blood.v98.12.3261. [DOI] [PubMed] [Google Scholar]
- 18.Pronk CJ, Rossi DJ, Mansson R, et al. Elucidation of the phenotypic, functional, and molecular topography of a myeloerythroid progenitor cell hierarchy. Cell Stem Cell. 2007;1:428–442. doi: 10.1016/j.stem.2007.07.005. [DOI] [PubMed] [Google Scholar]
- 19.Hardy RR, Kincade PW, Dorshkind K. The protean nature of cells in the B lymphocyte lineage. Immunity. 2007;26:703–714. doi: 10.1016/j.immuni.2007.05.013. [DOI] [PubMed] [Google Scholar]
- 20.Wen R, Chen Y, Bai L, et al. Essential role of phospholipase C gamma 2 in early B-cell development and Myc-mediated lymphomagenesis. Mol Cell Biol. 2006;26:9364–9376. doi: 10.1128/MCB.00839-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kennedy DW, Abkowitz JL. Kinetics of central nervous system micro-glial and macrophage engraftment: analysis using a transgenic bone marrow transplantation model. Blood. 1997;90:986–993. [PubMed] [Google Scholar]
- 22.Jono S, McKee MD, Murry CE, et al. Phosphate regulation of vascular smooth muscle cell calcification. Circ Res. 2000;87:E10–E17. doi: 10.1161/01.res.87.7.e10. [DOI] [PubMed] [Google Scholar]
- 23.Kuhn R, Schwenk F, Aguet M, Rajewsky K. Inducible gene targeting in mice. Science. 1995;269:1427–1429. doi: 10.1126/science.7660125. [DOI] [PubMed] [Google Scholar]
- 24.Barlow JL, Drynan LF, Hewett DR, et al. A p53-dependent mechanism underlies macrocytic anemia in a mouse model of human 5q-syndrome. Nat Med. 2010;16:59–66. doi: 10.1038/nm.2063. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Hardy RR, Carmack CE, Shinton SA, Kemp JD, Hayakawa K. Resolution and characterization of pro-B and pre-pro-B cell stages in normal mouse bone marrow. J Exp Med. 1991;173:1213–1225. doi: 10.1084/jem.173.5.1213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hardy RR, Hayakawa K. B cell development pathways. Annu Rev Im-munol. 2001;19:595–621. doi: 10.1146/annurev.immunol.19.1.595. [DOI] [PubMed] [Google Scholar]
- 27.Jacob HS, Amsden T. Acute hemolytic anemia with rigid red cells in hypophosphatemia. N Engl J Med. 1971;285:1446–1450. doi: 10.1056/NEJM197112232852602. [DOI] [PubMed] [Google Scholar]
- 28.Melvin JD, Watts RG. Severe hypophosphatemia: a rare cause of intra-vascular hemolysis. Am J Hematol. 2002;69:223–224. doi: 10.1002/ajh.10071. [DOI] [PubMed] [Google Scholar]
- 29.Diskin CJ, Stokes TJ, Dansby LM, Radcliff L, Carter TB. Can acidosis and hyperphosphataemia result in increased erythropoietin dosing in haemodialysis patients? Nephrology (Carlton) 2006;11:394–399. doi: 10.1111/j.1440-1797.2006.00674.x. [DOI] [PubMed] [Google Scholar]
- 30.Kalantar-Zadeh K, Lee GH, Miller JE, et al. Predictors of hyporespon-siveness to erythropoiesis-stimulating agents in hemodialysis patients. Am J Kid Dis. 2009;53:823–834. doi: 10.1053/j.ajkd.2008.12.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Trunzo JA, McHenry CR, Schulak JA, Wilhelm SM. Effect of parathy-roidectomy on anemia and erythropoietin dosing in end-stage renal disease patients with hyperparathyroidism. Surgery. 2008;144:915–918. doi: 10.1016/j.surg.2008.07.026. discussion 919. [DOI] [PubMed] [Google Scholar]
- 32.Urena P, Eckardt KU, Sarfati E, et al. Serum erythropoietin and eryth-ropoiesis in primary and secondary hyperparathyroidism: Effect of parathyroidectomy. Nephron. 1991;59:384–393. doi: 10.1159/000186596. [DOI] [PubMed] [Google Scholar]
- 33.Wu SG, Jeng FR, Wei SY, et al. Red blood cell osmotic fragility in chronically hemodialyzed patients. Nephron. 1998;78:28–32. doi: 10.1159/000044878. [DOI] [PubMed] [Google Scholar]
- 34.Meytes D, Bogin E, Ma A, Dukes PP, Massry SG. Effect of parathyroid hormone on erythropoiesis. J Clin Invest. 1981;67:1263–1269. doi: 10.1172/JCI110154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rao DS, Shih MS, Mohini R. Effect of serum parathyroid hormone and bone marrow fibrosis on the response to erythropoietin in uremia. N Engl J Med. 1993;328:171–175. doi: 10.1056/NEJM199301213280304. [DOI] [PubMed] [Google Scholar]
- 36.Shoemaker DG, Bender CA, Gunn RB. Sodium-phosphate cotransport in human red blood cells. Kinetics and role in membrane metabolism. J Gen Physiol. 1988;92:449–474. doi: 10.1085/jgp.92.4.449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Pop R, Shearstone JR, Shen Q, et al. A key commitment step in erythropoiesis is synchronized with the cell cycle clock through mutual inhibition between PU.1 and S-phase progression. PLoS Biol. 2010:8. doi: 10.1371/journal.pbio.1000484. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Byskov K, Jensen N, Kongsfelt IB, et al. Regulation of cell proliferation and cell density by the inorganic phosphate transporter PiT1. Cell Div. 2012;7:7. doi: 10.1186/1747-1028-7-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sankaran VG, Orkin SH, Walkley CR. Rb intrinsically promotes erythropoiesis by coupling cell cycle exit with mitochondrial biogenesis. Genes Dev. 2008;22:463–475. doi: 10.1101/gad.1627208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Inoue S, Yokota M, Nakada K, Miyoshi H, Hayashi J. Pathogenic mitochondrial DNA-induced respiration defects in hematopoietic cells result in anemia by suppressing erythroid differentiation. FEBS Lett. 2007;581:1910–1916. doi: 10.1016/j.febslet.2007.03.092. [DOI] [PubMed] [Google Scholar]
- 41.von Freeden-Jeffry U, Vieira P, Lucian LA, McNeil T, Burdach SE, Mur-ray R. Lymphopenia in interleukin (IL)-7 gene-deleted mice identifies IL-7 as a nonredundant cytokine. J Exp Med. 1995;181:1519–1526. doi: 10.1084/jem.181.4.1519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Peschon JJ, Morrissey PJ, Grabstein KH, et al. Early lymphocyte expansion is severely impaired in interleukin 7 receptor-deficient mice. J Exp Med. 1994;180:1955–1960. doi: 10.1084/jem.180.5.1955. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Sandberg ML, Sutton SE, Pletcher MT, et al. c-Myb and p300 regulate hematopoietic stem cell proliferation and differentiation. Dev Cell. 2005;8:153–166. doi: 10.1016/j.devcel.2004.12.015. [DOI] [PubMed] [Google Scholar]
- 44.Greig KT, de Graaf CA, Murphy JM, et al. Critical roles for c-Myb in lymphoid priming and early B-cell development. Blood. 2010;115:2796–2805. doi: 10.1182/blood-2009-08-239210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Boultwood J, Wainscoat JS. Gene silencing by DNA methylation in haematological malignancies. Br J Haematol. 2007;138:3–11. doi: 10.1111/j.1365-2141.2007.06604.x. [DOI] [PubMed] [Google Scholar]
- 46.Hsu AP, Sampaio EP, Khan J, et al. Mutations in GATA2 are associated with the autosomal dominant and sporadic monocytopenia and mycobacterial infection (MonoMAC) syndrome. Blood. 2011;118:2653–2655. doi: 10.1182/blood-2011-05-356352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Vardiman JW, Thiele J, Arber DA, et al. The 2008 revision of the World Health Organization (WHO) classification of myeloid neoplasms and acute leukemia: rationale and important changes. Blood. 2009;114:937–951. doi: 10.1182/blood-2009-03-209262. [DOI] [PubMed] [Google Scholar]
- 48.Li X, Xu F, He Q, Wu L, Zhang Z, Chang C. Comparison of immunological abnormalities of lymphocytes in bone marrow in myelodys-plastic syndrome (MDS) and aplastic anemia (AA) Intern Med. 2010;49:1349–1355. doi: 10.2169/internalmedicine.49.3477. [DOI] [PubMed] [Google Scholar]
- 49.Riedel N, Hoover EA, Gasper PW, Nicolson MO, Mullins JI. Molecular analysis and pathogenesis of the feline aplastic anemia retrovirus, feline leukemia virus C-Sarma. J Virol. 1986;60:242–250. doi: 10.1128/jvi.60.1.242-250.1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Jarrett O. Pathogenicity of feline leukemia virus is commonly associated with variant viruses. Leukemia. 1992;6(Suppl 3):153S–154S. [PubMed] [Google Scholar]
- 51.Tzavaras T, Stewart M, McDougall A, et al. Molecular cloning and characterization of a defective recombinant feline leukaemia virus associated with myeloid leukaemia. J Gen Virol. 1990;71(Pt 2):343–354. doi: 10.1099/0022-1317-71-2-343. [DOI] [PubMed] [Google Scholar]
- 52.Yoshida K, Sanada M, Shiraishi Y, et al. Frequent pathway mutations of splicing machinery in myelodysplasia. Nature. 2011;478:64–69. doi: 10.1038/nature10496. [DOI] [PubMed] [Google Scholar]
- 53.Farrell KB, Tusnady GE, Eiden MV. New structural arrangement of the extracellular regions of the phosphate transporter SLC20A1, the receptor for gibbon ape leukemia virus. J Biol Chem. 2009;284:29979–29987. doi: 10.1074/jbc.M109.022566. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.