Abstract
The RAG proteins are comprised of core endonuclease domains and non-core regions that modulate endonuclease activity. Mutation or deletion of non-core RAG regions in humans causes immunodeficiency and altered TCR repertoire, and mice expressing core but not full-length Rag1 (Rag1C/C) or Rag2 (Rag2C/C) exhibit lymphopenia, reflecting impaired V(D)J recombination and lymphocyte development. Rag1C/C mice display reduced D-to-J and V-to-DJ rearrangements of TCRβ and IgH loci, while Rag2C/C mice show decreased V-to-DJ rearrangements and altered Vβ/VH repertoire. Since Vβs/VHs only recombine to DJ complexes, the Rag1C/C phenotype could reflect roles for non-core RAG1 regions in promoting recombination during only the D-to-J step or during both steps. Here, we demonstrate that a pre-assembled TCRβ gene, but not a pre-assembled DβJβ complex or the pro-survival BCL2 protein, completely rescues αβ T cell development in Rag1C/C mice. We find that Rag1C/C mice exhibit altered Vβ utilization in Vβ-to-DJβ rearrangements, increased usage of 3’Jα gene segments in Vα-to-Jα rearrangements, and abnormal changes in Vβ repertoire during αβ TCR selection. Inefficient Vβ/VH recombination signal sequences (RSSs) have been hypothesized to cause impaired V-to-DJ recombination on the background of a defective recombinase as in core-Rag mice. We show that replacement of the Vβ14 RSS with a more efficient RSS increases Vβ14 recombination and rescues αβ T cell development in Rag1C/C mice. Our data indicate that non-core RAG1 regions establish a diverse TCR repertoire by overcoming Vβ RSS inefficiency to promote Vβ recombination and αβ T cell development, and by modulating TCRβ and TCRα gene segment utilization.
Introduction
The lymphocyte-specific RAG1/RAG2 (1) endonuclease generates Ag receptor diversity by recombining germline variable (V), diversity (D), and joining (J) gene segments of T cell receptor (TCR) and immunoglobulin (Ig) loci. RAG cleavage between two participating gene segments and their adjacent RSSs yields DNA double strand breaks (DSBs) comprised of hairpin-sealed coding ends and blunt signal ends (1, 2). RAG proteins along with DNA damage response/repair proteins hold these DNA ends in a stable post-cleavage complex and facilitate their repair by non-homologous end-joining (NHEJ) factors (3, 4). The combination of possible V(D)J joining events and the imprecise manner by which coding ends are processed cooperate to generate Ag receptor diversity.
The RAG1 and RAG2 proteins are each comprised of core endonuclease domains, defined as the minimal sequences required for DNA cleavage in vitro, and non-core regions that modulate this activity (2, 5). RAG1 or RAG2 mutations that alter or delete non-core RAG1 or RAG2 region amino acids and reduce overall V(D)J recombinase activity cause Omenn Syndrome (OS) or other fatal severe combined immunodeficiencies associated with oligoclonal TCR repertoire and increased T cell mediated autoimmunity (6-9). However, the contribution of diminished recombinase activity to aberrant TCR repertoire and autoreactive T cells in OS patients remains undetermined (6). Rag1C/C and Rag2C/C mice each display reduced numbers of mature T and B cells, reflecting impaired lymphocyte development beyond the progenitor stages associated with reduced TCRβ and IgH recombination (10-12). While Rag1C/C mice display reduced levels of both D-to-J and V-to-DJ recombination of TCRβ and IgH loci (11), Rag2C/C mice show predominantly decreased V-to-DJ rearrangements associated with altered Vβ/VH usage (10, 12). Although no reductions in the levels of V-to-J recombination of Igκ or TCRα loci were discovered in either Rag1C/C or Rag2C/C mice, potential changes in the utilization of individual Igκ or TCRα gene segments have not been assayed (10-12). Considering that Vβs and VHs only recombine to DJ complexes, the Rag1C/C phenotype could reflect roles for non-core RAG1 regions in promoting recombination during only the D-to-J step or during both the D-to-J and V-to-DJ steps (11). Since Vβ and VH rearrangements are selectively impaired in Rag2C/C mice, it has been hypothesized that the Rag2C/C mouse phenotype is due to the interaction of a defective recombinase with unique features of Vβ/VH RSSs (10). In support of this notion, Vβ/VH RSSs that more closely resemble those of Dβ and Vλ/Vα/Vγ/Vδ segments are more efficiently cleaved by core-RAG proteins in vitro and their associated Vβ/VH segments are recombined and expressed at higher frequencies in Rag2C/C mice relative to normal mice (10). Yet, neither this decade-old model nor its prediction that Rag1C/C mice also would exhibit altered Vβ and VH utilization has been tested.
In humans and mice, αβ T cells develop in the thymus through a differentiation program that involves the ordered assembly, expression, and selection of TCR genes. TCRβ genes assemble through Dβ-to-Jβ and then Vβ-to-DβJβ rearrangements in CD4-CD8- “double-negative” (DN) thymocytes (13). The Dβ-to-Jβ recombination step initiates in c-Kit+CD25- DN1 cells and continues in c-Kit+CD25+ DN2 cells, while Vβ-to-DβJβ recombination occurs in c-Kit-CD25+ DN3 cells (13). Since TCRβ loci contain 31 Vβ segments (Trbv1 through Trbv31) and two Dβ-Jβ-Cβ clusters (Trbd1-Trbj1-Trbc1 and Trbd2-Trbj2-Trbc2) each with one Dβ and six functional Jβ segments, secondary Vβ rearrangements can occur to Dβ2Jβ2 (Trbd2Trbj2) complexes on alleles with primary Vβ rearrangements to assembled Dβ1Jβ1 (Trbd1Trbj1) complexes (14). Utilization of individual Vβ segments in primary Vβ-to-Dβ1Jβ1 rearrangements is biased independent of Vβ position (15). However, Vβ position relative to a pre-assembled VβDβJβ1Cβ1 gene can influence the usage of Vβ segments in secondary Vβ-to-Dβ2Jβ2 rearrangements (14, 16). Assembly and expression of a functional TCRβ gene generates TCRβ chains that pair with pre-Tα proteins to form pre-TCRs, which promote survival and differentiation, down-regulate RAG expression, and induce expression of the Cyclin D3 (Ccnd3) protein (17, 18). Ccnd3 drives proliferation as DN3 cells down-regulate RAG expression and differentiate into c-Kit-CD25- DN4 and then CD4+CD8+ “double-positive” (DP) thymocytes (17, 18). TCRα genes assemble through Vα-to-Jα rearrangements on both alleles in DP cells, where Vβ-to-DβJβ recombination is silenced (19, 20). The assembly and expression of a functional VαJαCα gene generates TCRα chains that can pair with TCRβ chains to generate αβ TCRs, which are selected based on interactions with thymic epithelial cells (17). Positive selection increases expression of αβ TCRs and promotes differentiation of DP thymocytes into CD4+CD8- or CD4-CD8+ “single-positive” thymocytes that emigrate from the thymus as mature naive αβ T cells (17). Since TCRα loci contain ~100 Vα and ~50 Jα segments, successive Vα-to-Jα rearrangements can occur until positive selection or until all Vα or Jα segments have been utilized (13, 19). Although pre-TCR selection and thymocyte expansion does not significantly change Vβ repertoire during DN-to-DP thymocyte differentiation (15), positive selection can substantially alter Vβ representation in αβ TCRs during DP-to-SP thymocyte development (21-27).
The mouse αβ T cell differentiation program provides a useful experimental model to elucidate roles for non-core RAG1 regions in promoting V(D)J recombination and controlling TCR gene repertoire. Rag1C/C mice exhibit reduced Dβ-Jβ and Vβ-DβJβ recombination in DN3 thymocytes and impaired DN3-to-DP thymocyte development from accumulation of cells at the DN3 stage (11). These phenotypes may arise from impaired Dβ-to-Jβ and/or Vβ-to-DβJβ recombination in the absence of non-core RAG1 regions. Yet, considering that RAG DSBs induce changes in the expression of proteins involved in cellular survival, lymphocyte differentiation, and Ag gene receptor selection (28), these phenotypes also may arise from impaired signaling in response to RAG DSBs induced in Rag1C/C DN3 thymocytes. To determine how non-core RAG1 regions promote TCRβ gene assembly and αβ T cell development, we have created and analyzed Rag1C/C mice containing a pre-assembled DβJβ complex or pre-assembled functional TCRβ gene, expressing the pro-survival EμBCL2 transgene, or with the 3’Dβ1 RSS in place of the Vβ14 RSS. We demonstrate that this TCRβ gene, but not the DβJβ complex or BCL2, completely rescues DN3-to-DP thymocyte development in Rag1C/C mice, indicating that the predominant function of non-core RAG1 regions in differentiating αβ T cells is to promote Vβ recombination. We show that Rag1C/C mice exhibit altered Vβ utilization in Vβ-to-DJβ rearrangements and that neither apoptosis of cells attempting Vβ recombination nor TCRβ-dependent expansion of DN3 thymocytes contributes to this abnormal Vβ repertoire. We detect increased usage of 3’Jα segments in Vα-to-Jα rearrangements and abnormal selection of the Vβ repertoire in Rag1C/C mice, revealing that non-core RAG1 regions also function during TCRα recombination in DP thymocytes. Finally, we show that the 3’Dβ1 RSS increases Vβ14 recombination and partially rescues αβ T cell development in Rag1C/C mice. Collectively, our data indicate that non-core RAG1 regions establish a diverse αβ TCR repertoire by overcoming Vβ RSS inefficiency to promote Vβ recombination and αβ T cell development and by modulating TCRβ and TCRα gene segment utilization.
Materials and Methods
Mice
Rag1C/C (11), EμBCL2 (29), Jβ1DJ/DJ (22), Vβ1NT/NT (14), and Ccnd3-/- (18) mice were obtained and utilized to generate the mice described in this study. The germline Vβ143’Dβ1RSS mice were generated by Cre-loxP-mediated gene-targeting using W4 mouse embryonic stem cells and the construct previously employed to make chimeric Vβ143’Dβ1RSS mice (21). All experimental mice were on a mixed 129SvEv and C57BL/6 background and were littermate or age-matched mice between 4-6 weeks of age. All experiments were conducted in accordance with national guidelines and approved by the Institutional Animal Care and Use Committee of the Children’s Hospital of Philadelphia.
Flow Cytometry
Single cell suspensions were stained with antibodies in PBS containing 2% BSA. All antibodies were purchased from BD Pharmingen. CD4 and CD8 analysis was performed using anti-CD4 (553653), anti-CD8 (553031), and anti-TCRβ (553174) antibodies. DN stage analysis was performed on lineage-negative cells stained with a mixture of PE-conjugated anti-CD4 (553653), anti-CD8a (553033), anti-TCRβ (553172), anti-TCRγ (553178), anti-B220 (553090), anti-CD19 (553786), anti-CD11b (553311), anti-CD11c (557401), anti-NK1.1 (553165), and anti-Ter119 (553673) antibodies in addition to anti-CD25 (552880) and anti-CD117 (553356) antibodies. Vβ analysis was performed using antibodies against TCRβ (553174) as well as Vβ5 (553189), Vβ6 (553192), Vβ8 (553861), Vβ10 (553285), Vβ14 (553258), and Streptavidin-FITC (554060). Data was acquired on a FACSCalibur (BD Biosciences, San Jose, CA) using CellQuest software (BD Biosciences) and analyzed using FlowJo software (Tree Star).
PCR
Genomic DNA from sorted DP thymocytes (on 100 ng and 1:5 serial dilutions) was subjected to long-range PCR with the use of primers and PCR conditions as described (30, 31).
Results
Transgenic BCL2 expression partially rescues early αβ T cell development in Rag1C/C mice
To determine whether impaired cellular survival in response to RAG DSBs causes impaired DN3-to-DP thymocyte development in Rag1C/C mice, we created and analyzed Rag1C/C mice containing the EμBCL2 transgene (EμBCL2:Rag1C/C mice) since expression of the anti-apoptotic BCL2 protein increases survival of DN3 cells attempting TCRβ rearrangements (32). Consistent with the pro-survival effect of BCL2, we detected ~3-fold increases in the numbers DN and DP thymocytes in EμBCL2:Rag1C/C mice compared to Rag1C/C mice, although the difference in DN thymocyte numbers did not reach statistical significance (Fig. 1 A and B). Similarly, we found equivalent numbers of DN3 and DN4 thymocytes in EμBCL2:Rag1C/C and Rag1C/C mice (Fig. 1 C and D). We also detected ~4.5-fold decreased numbers of total and DP thymocytes in EμBCL2:Rag1C/C mice relative to EμBCL2 mice (Fig. 1 A and B), indicating that BCL2 expression does not completely rescue impaired DN-to-DP thymocyte development in Rag1C/C mice. These observations indicate that BCL2 expression does not substantially enhance the survival of Rag1C/C DN3 thymocytes, yet has a more pronounced effect on promoting survival of DP thymocytes or cells during DN-to-DP thymocyte expansion and differentiation in Rag1C/C mice. Therefore, we conclude that reduced survival of DN3 thymocytes in response to RAG DSBs is not a major cause of the impaired TCRβ gene assembly and DN-to-DP thymocyte development in Rag1C/C mice.
FIGURE 1.
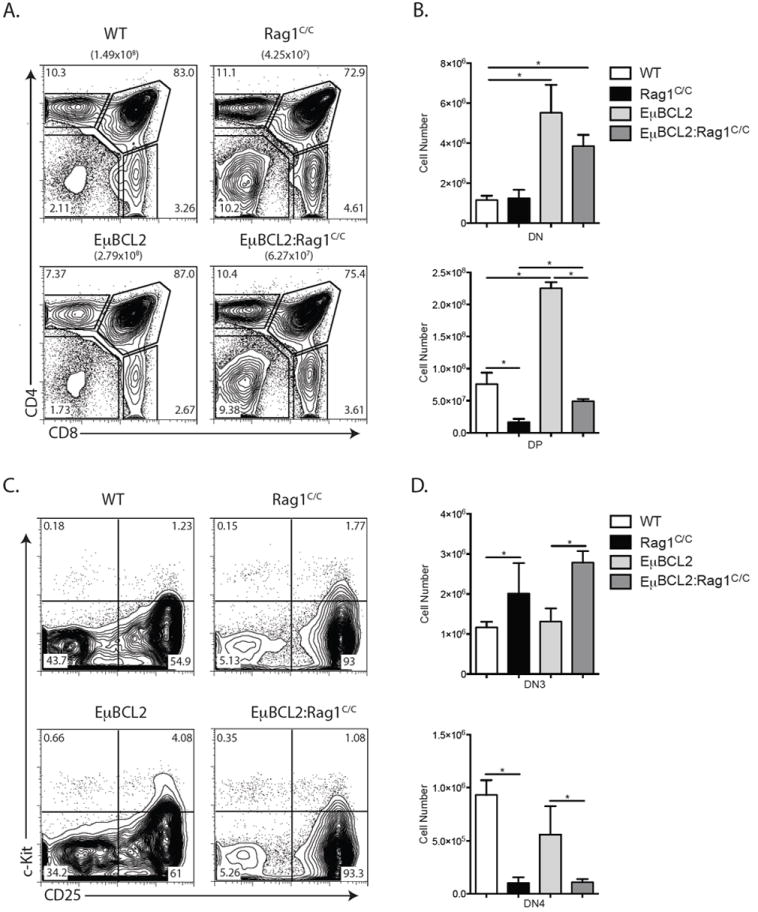
Transgenic BCL2 expression partially rescues αβ T cell development in Rag1C/C mice. A. Representative flow cytometry analysis of CD4 and CD8 expression on total thymocytes isolated from littermate or age-matched WT (n=3), Rag1C/C (n=3), EμBCL2 (n=3), or EμBCL2:Rag1C/C (n=3) mice. The average number of total thymocytes for mice of each genotype is indicated in parentheses, and the frequencies of cells in the DN, DP, CD4+ SP, and CD8+ SP quadrants are indicated on the plots. B. Graphs showing the average numbers of DN and DP thymocytes from mice of the indicated genotypes. Error bars are standard error of the mean. Lines with asterisks above indicate significant differences (p≤0.05). A and B. This experiment was independently performed three times, each time on one mouse of each genotype. C. Representative flow cytometry analysis of c-Kit and CD25 expression on DN thymocytes isolated from littermate or age-matched WT (n=3), Rag1C/C (n=3), EμBCL2 (n=3), or EμBCL2:Rag1C/C (n=3) mice. The frequencies of DN cells in the DN1, DN2, DN3, and DN4 quadrants are indicated on the plots. D. Graphs showing the average numbers of DN3 and DN4 thymocytes from mice of the indicated genotypes. Error bars are standard error of the mean. Lines with asterisks above indicate significant differences (p≤0.05). C and D. This experiment was independently performed three times, each time on one mouse of each genotype.
A pre-assembled TCRβ gene completely rescues early αβ T cell development in Rag1C/C mice
To determine potential functions of non-core Rag1 regions in promoting Dβ-to-Jβ recombination and Vβ-to-DJβ recombination, we generated and analyzed Rag1C/C mice that contain a pre-assembled DβJβ complex (Jβ1DJ) or pre-assembled functional TCRβ gene (Vβ1NT) on both TCRβ alleles. The Jβ1DJ allele contains a pre-assembled Dβ1Jβ1.1 (Trbd1Trbj1.1) complex, lacks Dβ2 (Trbd2) and Jβ2 (Trbj2) segments, and must recombine one of 35 Vβ segments to the Dβ1Jβ1.1 complex to promote αβ T lymphocyte development (22). The Vβ1NT allele contains a pre-assembled functional endogenous Vβ1Dβ1Jβ1.4Cβ1 (Trbv5Trbd1Trbj1.4Trbc1) gene that promotes αβ T cell development independent of TCRβ recombination (14). We detected ~2-fold more DP thymocytes in Rag1C/C Jβ1DJ/DJ mice relative to Rag1C/C mice (Fig. 2 A and B). However, the DP cell numbers in Rag1C/CJβ1DJ/DJ mice and Rag1C/C mice were 2-fold or more lower as compared to EμBCL2 and WT mice, respectively (Fig. 2 B versus 1 B). We also detected ~8-fold more DN4 cells in Rag1C/CJβ1DJ/DJ mice relative to Rag1C/C mice (Fig. 2 C and D), with DN4 thymocyte numbers in Rag1C/CJβ1DJ/DJ mice and Rag1C/C mice significantly lower than in EμBCL2 and WT mice, respectively (Fig. 2 D versus Fig 1 E). These data demonstrate that a pre-assembled DβJβ1 complex on both TCRβ alleles partially rescues DN3-to-DN4 and DN-to-DP thymocyte development in Rag1C/C mice. In Rag1C/CVβ1NT/NT mice relative to Rag1C/C mice, we observed greater numbers of DP (4-fold more) and DN4 (11-fold more) thymocytes (Fig. 2 E - H). Notably, the numbers of DP and DN4 thymocytes in Rag1C/CVβ1NT/NT mice were similar to those in Vβ1NT/NT and WT mice (Fig. 2 E - H), showing that a pre-assembled functional TCRβ gene completely rescues both DN3-to-DN4 and DN-to-DP thymocyte development in Rag1C/C mice. Therefore, our data indicate s that non-core RAG1 regions promote both Dβ-to-Jβ and Vβ-to-DJβ rearrangements and that reduced Vβ-to-DJβ recombination is the major cause of accumulation of cells at the DN3 stage and impaired DN-to-DP thymocyte development in Rag1C/C mice.
FIGURE 2.
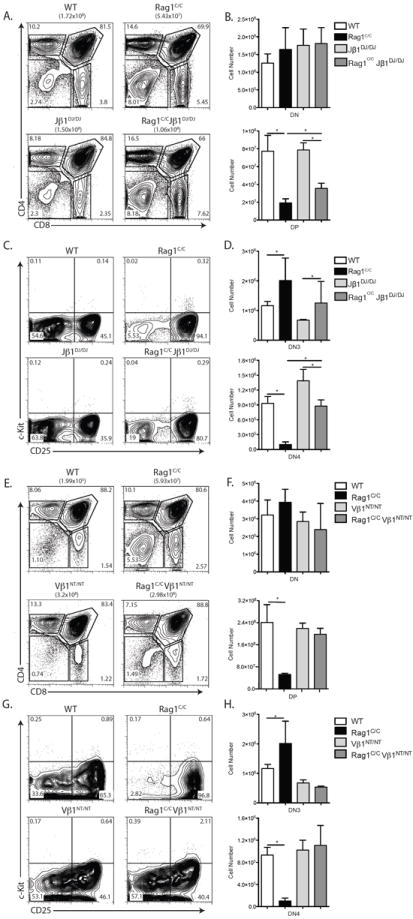
A pre-assembled TCRβ gene completely rescues early αβ T cell development in Rag1C/C mice. A. Representative flow cytometry analysis of CD4 and CD8 expression on total thymocytes isolated from littermate or age-matched WT (n=3), Rag1C/C (n=3), Jβ1DJ/DJ (n=3), and Rag1C/CJβ1DJ/DJ mice (n=3). The average number of total thymocytes for each genotype is indicated in parentheses, and the frequencies of cells in the DN, DP, CD4+ SP, and CD8+ SP quadrants are indicated on the plots. B. Graphs showing the average numbers of DN and DP thymocytes from mice of the indicated genotypes. Error bars are standard error of the mean. Lines with asterisks above indicate significant differences (p≤0.05). A and B. This experiment was independently performed three times, each time on one mouse of each genotype. C. Representative flow cytometry analysis of c-Kit and CD25 expression on DN thymocytes isolated from littermate or age-matched WT (n=3), Rag1C/C (n=3), Jβ1DJ/DJ (n=3), and Rag1C/CJβ1DJ/DJ (n=3) mice. The frequencies of DN cells in the DN1, DN2, DN3, and DN4 quadrants are indicated. D. Graphs showing the average numbers of DN3 and DN4 thymocytes from mice of the indicated genotypes. Error bars are standard error of the mean. Lines with asterisks above indicate significant differences (p≤0.05). C and D. This experiment was independently performed three times, each time on one mouse of each genotype. E. Representative flow cytometry analysis of CD4 and CD8 expression on total thymocytes isolated from littermate or age-matched WT (n=3), Rag1C/C (n=3), Vβ1NT/NT (n=6), and Vβ1NT/NTRag1C/C (n=6) mice. The average number of total thymocytes for each genotype is indicated in parentheses, and the frequencies of cells in the DN, DP, CD4+ SP, and CD8+ SP quadrants are indicated on the plots. F. Graphs showing the average numbers of DN and DP thymocytes from mice of the indicated genotypes. Error bars are standard error of the mean. The line with an asterisk above indicates a significant difference (p≤0.05). E and F. This experiment was independently performed three times, each time on at least mouse of each genotype. G. Representative flow cytometry analysis of c-Kit and CD25 expression on DN thymocytes isolated from littermate or age-matched WT (n=3), Rag1C/C (n=3), Vβ1NT/NT (n=3), and Vβ1NT/NTRag1C/C (n=3) mice. The frequencies of DN cells in the DN1, DN2, DN3, and DN4 quadrants are indicated. H. Graphs showing the average numbers of DN3 and DN4 thymocytes from mice of the indicated genotypes. Error bars are standard error of the mean. Lines with asterisks above indicate significant differences (p≤0.05). G and H. This experiment was independently performed three times, each time on one mouse of each genotype.
Rag1C/C mice exhibit altered Vβ utilization in primary and secondary Vβ rearrangements
The TCRβ locus architecture permits primary Vβ rearrangements to Dβ1Jβ1 complexes and then secondary Vβ rearrangements to Dβ2Jβ2 complexes, which occur on the Vβ1NT allele (14). Our current analysis of thymocyte development in Rag1C/CJβ1DJ/DJ and Rag1C/CVβ1NT/NT mice and our previous analysis of Vβ8 (Trbv13.1, Trbv13.2, and Trbv13.3) and Vβ10 (Trbv4) rearrangements in DN3 cells of Rag1C/C mice (11) demonstrate that primary Vβ-to-DJβ rearrangements are impaired in the absence of non-core Rag1 regions. However, these analyses cannot address potential function of non-core Rag1 regions in secondary Vβ rearrangements nor quantify relative usage of individual Vβ segments in Vβ-to-DβJβ rearrangements. Since DN-to-DP thymocyte differentiation does not significantly alter Vβ repertoire (15), the use of flow cytometry to monitor Vβ expression on TCRβintermediate DP thymocytes provides a more sensitive means than PCR to quantify relative Vβ usage in Vβ rearrangements (14, 15, 21). Thus, to determine the contributions of non-core Rag1 regions in Vβ utilization during primary Vβ-to-DβJβ rearrangements, we assayed Vβ5 (Trbv12.1 and Trbv12.2), Vβ6 (Trbv19), Vβ8, Vβ10, and Vβ14 (Trbv31) expression on DP thymocytes of Rag1C/CJβ1DJ/DJ and Jβ1DJ/DJ mice, as only primary Vβ rearrangements occur on the Jβ1DJ allele (22). We observed lower percentages of Vβ5+ (5-fold less) and Vβ10+ (~3-fold less) DP thymocytes in Rag1C/CJβ1DJ/DJ mice as compared to Jβ1DJ/DJ mice (Fig. 3 A and B). We detected increases in the percentages of Vβ8+ (2-fold more) and Vβ14+ (3-fold more) DP thymocytes, but no difference in the percentages of Vβ6+ DP cells, in Rag1C/CJβ1DJ/DJ mice relative to Jβ1DJ/DJ mice (Fig. 3 A and B). Since only secondary Vβ-to-DβJβ rearrangements involving Vβ10 occur on the Vβ1NT allele (14), we next quantified Vβ10 expression on DP thymocytes of Rag1C/CVβ1NT/NT and Vβ1NT/NT mice to evaluate whether non-core Rag1 regions promote such secondary Vβ rearrangements. We found a 7-fold lower percentage of Vβ10+ DP thymocytes in Rag1C/CVβ1NT/NT mice as compared to Vβ1NT/NT mice (Fig. 3 C and D). Collectively, these data indicate that non-core Rag1 regions affect Vβ utilization in primary and secondary Vβ rearrangements, at least on Jβ1DJ and Vβ1NT alleles.
FIGURE 3.
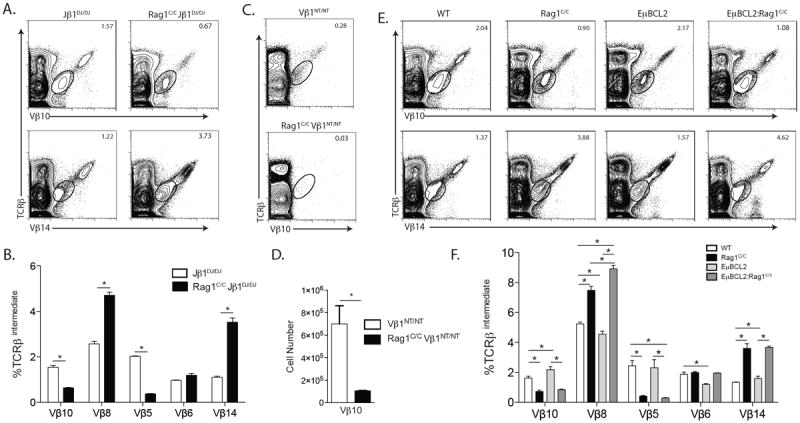
Rag1C/C mice exhibit altered Vβ utilization in primary and secondary Vβ rearrangements. A. Representative flow cytometry analysis of TCRβ and Vβ expression shown for Vβ10 or Vβ14 on total thymocytes isolated from littermate or age-matched Jβ1DJ/DJ (n=3) and Rag1C/CJβ1DJ/DJ (n=3) mice. The frequencies of cells in the depicted TCRβintermediate gate are indicated. B. Graph showing the average frequencies of TCRβintermediate cells expressing Vβ10, Vβ8, Vβ5, Vβ6, or Vβ14 in thymocytes from Jβ1DJ/DJ and Rag1C/CJβ1DJ/DJ mice. Error bars are standard error of the mean. Lines with asterisks above indicate significant differences (p≤0.05). A and B. This experiment was independently performed three times, each time on one mouse of each genotype. C. Representative flow cytometry analysis of TCRβ and Vβ10 expression on total thymocytes isolated from littermate or age-matched Vβ1NT/NT (n=6) and Vβ1NT/NTRag1C/C (n=6) mice. The frequencies of cells in the depicted TCRβintermediate gate are indicated. D. Graph showing the average number of TCRβintermediate cells expressing Vβ10 in thymocytes from Vβ1NT/NT and Vβ1NT/NTRag1C/C mice. Error bars are standard error of the mean. The line with an asterisk above indicates a significant difference (p≤0.05). C and D. This experiment was independently performed three times, each time on at least one mouse of each genotype. E. Representative flow cytometry analysis of TCRβ and Vβ10 or Vβ14 expression on total thymocytes isolated from littermate or age-matched WT (n=3), Rag1C/C (n=3), EμBCL2 (n=3), or EμBCL2:Rag1C/C (n=3) mice. The frequencies of cells in the depicted TCRβintermediate gate are indicated. F. Graph showing the average frequencies of TCRβintermediate cells expressing Vβ10, Vβ8, Vβ5, Vβ6, or Vβ14 in thymocytes from mice of the indicated genotypes. Error bars are standard error of the mean. Lines with asterisks above indicate significant differences (p≤0.05). E and F. This experiment was independently performed three times, each time on one mouse of each genotype.
To evaluate whether non-core Rag1 regions affect Vβ utilization in total Vβ rearrangements on normal TCRβ alleles, we quantified expression of Vβ5, Vβ6, Vβ8, Vβ10, and Vβ14 on DP thymocytes of Rag1C/C and WT mice. We detected lower percentages of Vβ5+ (~6-fold less) and Vβ10+ (~2-fold less) DP cells in Rag1C/C mice relative to WT mice (Fig. 3 E and F). We also detected ~2-fold increased percentage of Vβ8+ and ~3-fold increased percentage of Vβ14+ DP thymocytes, but no difference in the percentage of Vβ6+ DP cells, in Rag1C/C mice as compared to WT mice (Fig. 3 E and F). These data demonstrate that non-core Rag1 regions influence Vβ utilization in total Vβ rearrangements on normal TCRβ alleles.
While we did not observe a major role for impaired survival of Rag1C/C thymocytes in response to RAG DSBs during TCRβ recombination, altered survival of DN3 thymocytes during rearrangements of particular Vβ segments could influence Vβ repertoire in Rag1C/C mice. To investigate this possibility, we quantified the expression of Vβ5, Vβ6, Vβ8, Vβ10, and Vβ14 on DP thymocytes of EμBCL2:Rag1C/C and Rag1C/C mice. We detected no significant differences in the frequencies of Vβ6+, Vβ5+, Vβ10+ or Vβ14+ DP thymocytes between EμBCL2:Rag1C/C and Rag1C/C mice (Fig. 3 E and F), however we did observe a slightly higher frequency of Vβ8+ DP cells in EμBCL2:Rag1C/C mice as compared to Rag1C/C mice (Fig. 3 E and F). These data suggest that impaired survival of DN3 cells in response to RAG DSBs does not cause the altered relative representation of Vβ5, Vβ6, Vβ8, Vβ10, and Vβ14 on DP thymocytes of Rag1C/C mice.
While Vβ representation is not significantly altered during DN-to-DP thymocyte differentiation in mice with normal levels of Vβ rearrangements (15), altered expansion of DN cells expressing particular Vβ segments could contribute to the altered Vβ repertoire in DP thymocytes of Rag1C/C mice which have reduced numbers of thymocytes caused by impaired Vβ-to-DβJβ recombination. To assess this possibility, we generated and analyzed Rag1C/CCcnd3-/- mice since expression of TCRβ chains drive DN-to-DP thymocyte expansion through Ccnd3, and Ccnd3-/- mice exhibit normal Vβ repertoire in DP thymocytes (18, 33). We found equivalent numbers of DN thymocytes, but ~10-fold lower numbers of DP cells, in Rag1C/CCcnd3-/- mice as compared to Ccnd3-/- mice (Fig. 4 A and B), indicating that TCRβ-mediated DN-to-DP thymocyte development is profoundly impaired in Rag1C/CCcnd3-/- mice relative to Ccnd3-/- mice. Notably, the numbers of DP cells in Rag1C/CCcnd3-/- mice were reduced ~40-fold as compared to Rag1C/C mice versus ~17 fold for Ccnd3-/- mice relative to WT mice (Compare Fig. 4 B and Fig. 1 B). We also found a ~5-fold lower frequency of Vβ10+ DP cells but a ~2-fold increase frequency of Vβ14+ DP thymocytes in Rag1C/CCcnd3-/- mice as compared to Ccnd3-/- mice (Fig. 4 C and D). Considering that we observed similar increased and deceased frequencies of Vβ10+ and Vβ14+ DP thymocytes, respectively, in Rag1C/C mice relative to WT mice (Fig. 3 E and F), our data indicate that the altered representation of Vβ10 and Vβ14 on DP thymocytes of Rag1C/C mice is not caused by differences in TCRβ-mediated proliferation of DN cells expressing particular Vβ segments. Therefore, based on our quantification of Vβ expression on DP thymocytes of mice expressing wild-type or core-Rag1 proteins in combination with other genetic modifications, we conclude that Rag1 non-core regions control Vβ repertoire at the level of relative Vβ usage in primary and secondary Vβ-to-DβJβ rearrangements.
FIGURE 4.
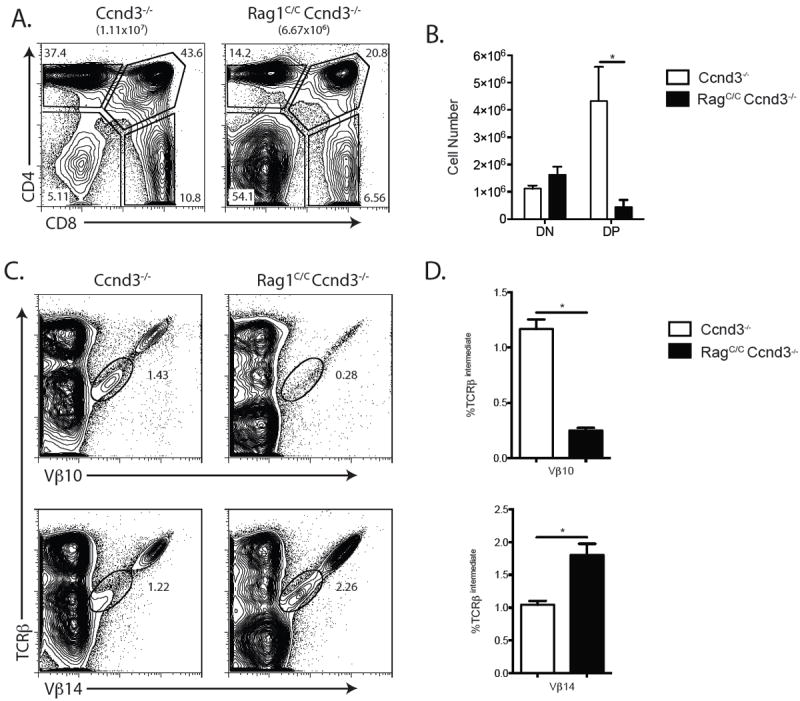
TCRβ recombination and TCRβ-mediated Ccnd3-dependent DN thymocyte proliferative expansion cooperate in αβ T cell development. A. Representative flow cytometry analysis of CD4 and CD8 expression on total thymocytes isolated from littermate or age-matched Ccnd3-/- (n=4) and Rag1C/CCcnd3-/- (n=5) mice. The average number of total thymocytes for each genotype is indicated in parentheses, and the frequencies of cells in the DN, DP, CD4+ SP, and CD8+ SP quadrants are indicated on the plots. B. Graph showing the average numbers of DN and DP thymocytes from Ccnd3-/- and Rag1C/CCcnd3-/- mice. Error bars are standard error of the mean. The line with an asterisk above indicates a significant difference (p≤0.05). A and B. This experiment was independently performed three times, each time on at least one mouse of each genotype. C. Representative flow cytometry analysis of TCRβ and Vβ10 or Vβ14 expression on total thymocytes isolated from littermate or age-matched Ccnd3-/- and Rag1C/CCcnd3-/- mice. The frequencies of cells in the depicted TCRβintermediate gate are indicated. D. Graph showing the average frequencies of TCRβintermediate cells expressing Vβ10 or Vβ14 in thymocytes from Ccnd3-/- (n=4) and Rag1C/CCcnd3-/- (n=5) mice. Error bars are standard error of the mean. Lines with asterisks above indicate significant differences (p≤0.05). C and D. This experiment was independently performed three times, each time on at least one mouse of each genotype.
Rag1C/C mice exhibit altered Jα utilization in Vα-to-Jα rearrangements and abnormal changes in Vβ repertoire during αβ TCR selection
In DP thymocytes of wild-type mice, TCRα recombination occurs on both alleles and normally involves successive rounds of Vα-to-Jα rearrangements on each allele (19). Although both TCRα alleles are recombined in mature αβ T cells of Rag1C/C mice (11), whether the absence of non-core Rag1 regions results in a modest reduction in Vα-to-Jα rearrangements in DP thymocytes is not known. Since reduced V(D)J recombinase activity in DP cells leads to increased representation of 5’Jαs and decreased representation of 3’Jαs in Vα-to-Jα rearrangements (34), we investigated whether Vα rearrangements in DP cells of Rag1C/C mice are similarly biased. For this purpose, we used PCR primers that hybridize to the Vα3 (Trav9-4) family of gene segments or to Jα61 (Traj61), Jα42 (Traj42), Jα17 (Traj17), or Jα4 (Traj4) to amplify Vα3-to-Jα rearrangements involving these 5’ (Jα61, Jα42) or 3’ (Jα17, Jα4) Jα gene segments from sort-purified DP cells of WT or Rag1C/C mice. The levels of PCR products representing Vα3 rearrangements to Jα61 and Jα42 were reduced in DP thymocytes from Rag1C/C mice relative to WT mice (Fig. 5 A). In contrast, the levels of PCR products representing Vα3 rearrangements to Jα17 and Jα4 were elevated in DP thymocytes from Rag1C/C mice as compared to WT mice (Fig. 5 A). These data reveal that loss of non-core Rag1 regions results in decreased usage of 5’Jαs and increased usage of 3’Jαs in Vα-to-Jα rearrangements. Although this biased targeting of Vα3 rearrangements toward 3’Jα gene segments is not consistent with diminished recombinase activity at the TCRα locus, our results demonstrate that non-core Rag1 regions control formation of the TCRα gene repertoire during Vα-to-Jα recombination in DP thymocytes.
FIGURE 5.
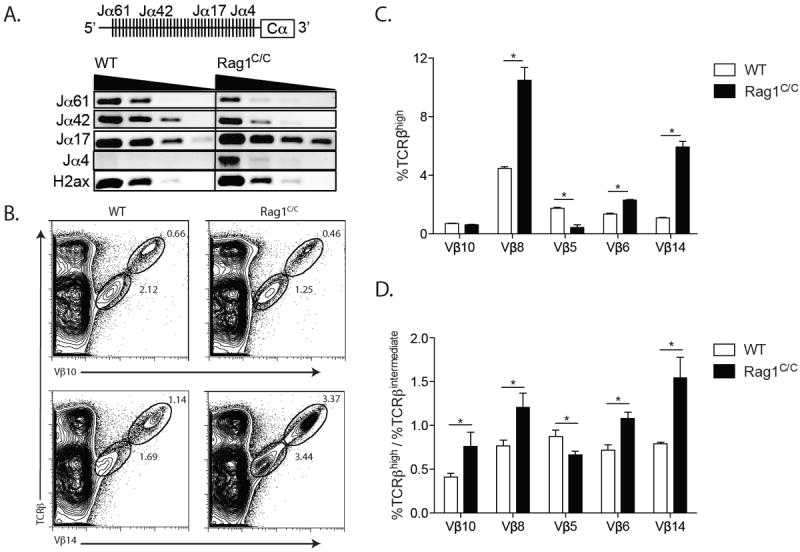
Rag1C/C mice exhibit altered Jα utilization in Vα-to-Jα rearrangements and abnormal changes in Vβ repertoire during αβ TCR selection. A. Schematic of the Jα-Cα region of the TCRα locus (Upper). Representative PCR analysis of Vα3 rearrangements to the indicated Jα gene segments or H2ax as a loading control on 5-fold dilutions of genomic DNA from sort-purified CD4+CD8+ WT (n=3) and Rag1C/C (n=3) thymocytes (Lower). B. Representative flow cytometry analysis of TCRβ and Vβ10 or Vβ14 expression on total thymocytes isolated from littermate or age-matched WT (n=3) and Rag1C/C (n=3) mice. The frequencies of cells in the depicted TCRβintermediate and TCRβhigh gates are indicated. C. Graph showing the average frequencies of TCRβhigh cells expressing Vβ10, Vβ8, Vβ5, Vβ6, or Vβ14 in thymocytes from mice of the indicated genotypes. Error bars are standard error of the mean. Lines with asterisks above indicate significant differences (p≤0.05). D. Graph showing the average ratios of the frequencies of TCRβhigh and TCRβintermediate cells in thymocytes from WT and Rag1C/C mice. Error bars are standard error of the mean. Lines with asterisks above indicate significant differences (p≤0.05). A - D. These experiments were independently performed three times, each time on one mouse of each genotype.
Since positive selection of αβ TCRs expressed on DP thymocytes shapes TCRβ repertoire (22-27, 35) and Jα repertoire is altered in DP thymocytes of Rag1C/C mice (Fig. 5 A), we investigated the impact of non-core Rag1 regions on Vβ repertoire during DP-to-SP thymocyte development. We detected ~4-fold lower frequencies of Vβ5+ TRCβhigh SP thymocytes and higher frequencies of Vβ6+ (~2-fold more) Vβ8+ (~2-fold more), and Vβ14+ (~5-fold more) TRCβhigh SP cells in Rag1C/C mice relative to WT mice (Fig. 5 B and C). Considering that we observed ~2-fold lower frequencies of Vβ10+ DP thymocytes and equivalent frequencies of Vβ6+ DP cells in Rag1C/C mice relative to WT mice (Fig. 3 E and F), these data indicate that positive selection of DP thymocytes alters Vβ repertoire differently in αβ TCRs of Rag1C/C and WT mice. To confirm this notion, we calculated the ratios of the frequencies of cells expressing Vβ5, Vβ6, Vβ8, or Vβ14 on TCRβhigh versus TCRβintermediate thymocytes in Rag1C/C and WT mice (Fig. 5 D). This analysis indicated enhanced selection for Vβ6+, Vβ8+, Vβ10+, and Vβ14+ cells and increased selection against Vβ5+ cells during positive selection of DP thymocytes in Rag1C/C mice as compared to WT mice (Fig. 5 D). Collectively, these data show that loss of non-core Rag1 regions leads to abnormal changes in Vβ repertoire during αβ TCR selection.
Replacement of the Vβ14 RSS with the more efficient 3’Dβ1 RSS increases Vβ14 recombination frequency and rescues αβ T cell development in Rag1C/C mice
We have shown that Rag1C/C mice exhibit impaired DN3-to-DP thymocyte development caused by reduced levels of Vβ-to-DJβ rearrangements, and that this impaired Vβ recombination is associated with altered recombination frequencies of individual Vβ segments. A similar phenotype in Rag2C/C mice lead to the hypothesis that conserved sequence features of Vβ/VH RSSs that make them inefficient relative to 3’Dβ and Vκ/Vλ/Vδ /Vα RSSs contributes to impaired V-to-DJ recombination on the background of a diminished recombinase (10). In support of this model, the Vβ8 RSS more closely resembles the consensus Vκ/Vλ/Vδ/Vα RSS than canonical Vβ RSSs, and Vβ8 exhibits a 1.8-fold higher rearrangement and expression in Rag1C/C and Rag2C/C mice compared to WT mice ((10); Fig. 3 E and F). In addition, the Vβ5 and Vβ10 RSSs more closely resemble the consensus Vβ/VH RSS than canonical Vβ RSSs, and Vβ5 and Vβ10 exhibit ~2-fold lower rearrangement and expression in Rag1C/C mice relative to WT mice (Fig. 3 E and F). However, the Vβ14 RSS more closely resembles the Vβ/VH RSS consensus than other Vβ RSSs, yet Vβ14 exhibits 2-fold higher recombination and expression in Rag1C/C mice relative to WT mice (Fig. 3 E and F). This latter finding suggests that factors in addition to Vβ RSS inefficiency may contribute the altered relative frequency of Vβ segments in Rag1C/C mice.
To directly test the prediction that Vβ RSS inefficiency causes impaired Vβ rearrangement in Rag1C/C mice, we sought to determine whether replacement of a Vβ RSS with a 3’Dβ or Vκ/Vλ/Vδ/Vα RSS increases recombination and expression of this Vβ to the same extent in Rag1C/C and WT mice. For this purpose, we established mice with gene-targeted replacement of the Vβ14 RSS with the 3’Dβ1 RSS on an otherwise normal TCRβ allele (Vβ143’Dβ1RSS/+ mice) since we previously showed in chimeric mice that this RSS replacement increases the frequencies of Vβ14 rearrangement and expression on a wild-type RAG background (21). We then made and analyzed in parallel Vβ143’Dβ1RSS/+ and Rag1C/CVβ143’Dβ1RSS/+ mice, as well as control Rag1C/C and WT mice. Consistent with our previous findings (21), we detected a ~20-fold increase in the frequency of Vβ14+ TCRβintermediate DP thymocytes in Vβ143’Dβ1RSS/+ mice relative to WT mice (Fig. 6 A and B). We observed a similar ~20-fold increase in the frequency of Vβ14+ TCRβintermediate DP thymocytes in Rag1C/CVβ143’Dβ1RSS/+ mice relative to Rag1C/C mice (Fig. 6 A and B), revealing that sequence of the RSS attached to Vβ14 is a major determinant of the frequency of Vβ14 rearrangement and expression in both WT and Rag1C/C mice. Therefore, our data provide direct support for the decade-old model that sequence features of Vβ/VH RSSs that renders them less efficient than 3’Dβ and Vκ/Vλ/Vδ/Vα RSSs contributes to impaired V rearrangements on the background of a defective recombinase as in Rag2C/C and Rag1C/C mice. Yet, we still detected ~2-fold greater frequencies of Vβ14 rearrangements and expression in Rag1C/CVβ143’Dβ1RSS/+ mice as compared to Vβ143’Dβ1RSS/+ mice (Fig. 6 A and B) as we observed between Rag1C/C and WT mice (Fig 3 E and F), highlighting that non-core Rag1 regions may modulate Vβ usage in Vβ rearrangements through mechanisms in addition to their function with inefficient Vβ RSSs.
FIGURE 6.
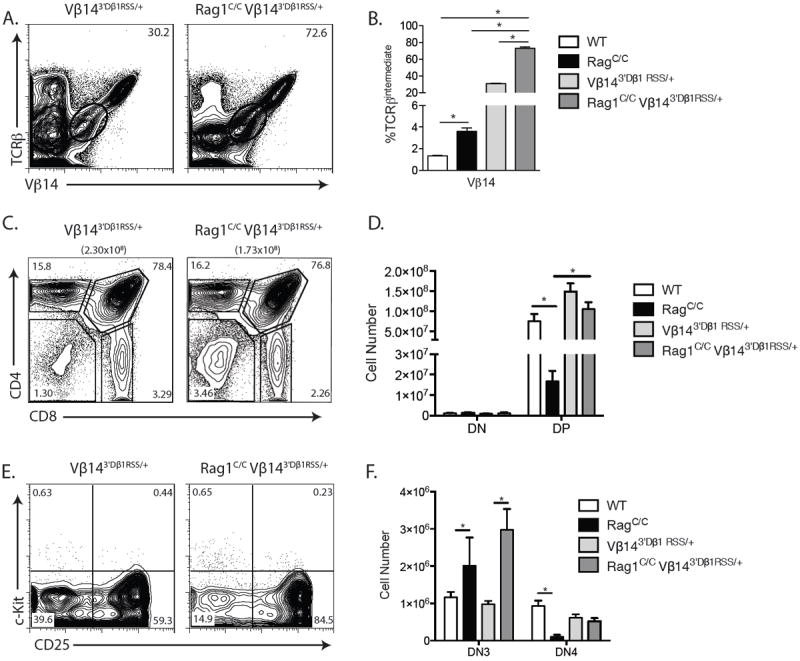
Replacement of the Vβ14 RSS with the more efficient 3’Dβ1 RSS increases Vβ14 recombination frequency and rescues αβ T cell development in Rag1C/C mice.A. Representative flow cytometry analysis of TCRβ and Vβ14 expression on total thymocytes isolated from littermate or age-matched Vβ143’Dβ1RSS/+ (n=3) and Rag1C/CVβ143’Dβ1RSS/+ (n=3) mice. The frequencies of cells in the depicted TCRβintermediate gates are indicated. B. Graph showing the average frequencies of TCRβintermediate cells expressing Vβ14 in thymocytes from mice of the indicated genotypes. Significant differences of *p<0.05 indicated. A and B. This experiment was independently performed three times, each time on at least one mouse of each genotype. C. Representative flow cytometry analysis of CD4 and CD8 expression on total thymocytes isolated from littermate or age-matched Vβ143’Dβ1RSS/+ (n=3) and Rag1C/CVβ143’Dβ1RSS/+ (n=3) mice. The average number of total thymocytes for each genotype is indicated in parentheses, and the frequencies of DN, DP, CD4+ SP, and CD8+ SP cell populations are indicated on the plots. D. Graph showing the average number of DN and DP cells from mice of the indicated genotypes. Error bars are standard error of the mean. Significant differences of *p<0.05 indicated. C and D. This experiment was independently performed three times, each time on at least one mouse of each genotype. E. Representative flow cytometry analysis of c-Kit and CD25 expression on DN thymocytes isolated from littermate or age-matched Vβ143’Dβ1RSS/+ (n=3) and Rag1C/CVβ143’Dβ1RSS/+ (n=3) mice with the frequencies of DN1, DN2, DN3, and DN4 cell populations indicated. F. Graph showing the average numbers of DN3 and DN4 cells from mice of the indicated genotypes. Error bars are standard error of the mean. Significant differences of *p<0.05 indicated. This experiment was independently performed three times on a total of three mice of each genotype.C and D. This experiment was independently performed three times, each time on at least one mouse of each genotype.
Since the 3’Dβ1 RSS replacement drives increased Vβ14 rearrangement in Rag1C/C DN3 cells, we next investigated the effect of this single RSS replacement on the impaired DN3-to-DP thymocyte development of Rag1C/C mice. We found ~5-fold greater numbers of total and DP thymocytes in Rag1C/CVβ143’Dβ1RSS/+ mice as compared to Rag1C/C mice (Fig. 6 C and D), with no significant differences in total and DP cell numbers or the percentages of thymocytes at the DP stage among Rag1C/CVβ143’Dβ1RSS/+, Vβ143’Dβ1RSS/+, and WT mice (Fig. 6 D and E). While DN cell numbers were similar among all four genotypes (Fig. 6 D), the percentages of thymocytes at the DN stage were lower in Rag1C/CVβ143’Dβ1RSS/+ mice compared to Rag1C/C mice (Fig. 6 C), yet higher in Rag1C/CVβ143’Dβ1RSS/+ mice relative to Vβ143’Dβ1RSS/+ and WT mice (Fig. 6 C). Consistent with these observations, we detected increased numbers of DN4 cells and an increased percentage of DN thymocytes at the DN4 stage in Rag1C/CVβ143’Dβ1RSS/+ mice as compared to Rag1C/C mice (Fig. 6 E), but lower values for these two parameters in Rag1C/CVβ143’Dβ1RSS/+ mice relative to Vβ143’Dβ1RSS/+ and WT mice (Fig. 6 E). These data indicate that replacement of the RSS of only one of the 20 functional Vβ segments with the more efficient 3’Dβ1 RSS partially rescues DN3-to-DN4 thymocyte development and completely rescues DN-to-DP thymocyte development in Rag1C/C mice. Therefore, we conclude that non-core Rag1 regions drive αβ T cell development by overcoming inherent inefficiencies of Vβ RSSs to promote Vβ rearrangements.
Discussion
We have taken a genetic approach with Rag1C/C mice to determine how non-core RAG1 regions promote TCRβ gene assembly and αβ T cell development. The reduced levels of Dβ-Jβ and Vβ-DβJβ recombination in DN3 thymocytes and impaired DN-to-DP thymocyte development from accumulation of cells at the DN3 stage of Rag1C/C mice could arise from decreased efficiency of TCRβ recombination and/or survival of DN3 thymocytes in response to RAG DSBs. Our observation that expression of the anti-apoptotic BCL2 protein does not substantially enhance the survival of Rag1C/C DN3 cells and only minimally rescues thymocyte development in Rag1C/C mice indicates that preventing death of DN3 cells in response to RAG DSBs is not the major means through which non-core RAG1 regions promote TCRβ recombination and αβ T cell development. Our finding that a pre-assembled DβJβ complex partially rescues thymocyte development in Rag1C/C mice confirms the hypothesis that non-core RAG1 regions promote αβ T cell development in part by stimulating Dβ-to-Jβ recombination (11), and also provides unequivocal evidence that non-core RAG1 regions drive Vβ-to-DJβ recombination. Combined with these data, our observation that a pre-assembled TCRβ gene completely rescues thymocyte development in Rag1C/C mice proves that non-core RAG1 regions promote TCRβ gene assembly and αβ T cell development by stimulating Vβ recombination. However, because Dβ-to-Jβ recombination precedes Vβ-to-DβJβ recombination, our data cannot differentiate the relative contribution of non-core RAG1 regions in each of these steps of TCRβ gene assembly in driving αβ T cell development.
In contrast to Rag1C/C mice, which we have shown exhibit substantial defects in both Dβ-to-Jβ and Vβ-to-DJβ recombination and altered Vβ repertoire, Rag2C/C mice have a major impairment in only Vβ-to-DJβ recombination that is associated with altered Vβ usage (10). Since Rag2C/C mice also have a reduction in overall recombinase activity in thymocytes but no apparent defect in TCRα recombination, it was proposed that the interaction of a diminished recombinase with RSS sequences unique to Vβs may cause the Vβ-to-DJβ recombination defect, rather than loss of specific properties of the non-core RAG2 region in stimulating Vβ rearrangements (10). Our findings that a pre-assembled DβJβ complex or functional TCRβ gene each rescues αβ T cell development to some extent cannot distinguish between impaired TCRβ gene assembly caused by diminished recombinase activity versus by the loss of specific functions of Rag1 non-core regions that stimulate Dβ-to-Jβ and/or Vβ-to-DJβ recombination. However, our demonstration that replacement of the Vβ14 RSS with the more efficient 3’Dβ1 RSS dramatically increases the frequency of Vβ14 rearrangement in Rag1C/C thymocytes and partially rescues thymocyte development proves that sequence features of the Vβ14 RSS that distinguishes it from the 3’Dβ1 RSS contributes to impaired Vβ-to-DJβ recombination in Rag1C/C mice. Notably, in the presence or absence of this Vβ14 RSS replacement, we have shown that loss of non-core Rag1 regions results in increased Vβ14 rearrangements, indicating that the interaction of a defective recombinase with inefficient Vβ RSSs is not the sole cause of impaired Vβ recombination in Rag1C/C mice. Furthermore, our discovery that Vα3 rearrangements are biased toward 3’Jα segments in DP thymocytes of Rag1C/C mice is opposite the prior observation of increased usage of 5’Jαs in mice with diminished recombinase activity in DP thymocytes (34). Therefore, we conclude that specific activities of non-core RAG1 regions modulate Vβ and Jα gene segment utilization during V(D)J recombination.
How might non-core RAG1 regions modulate Vβ and Jα usage? Both Vβ-to-DJβ and Vα-to-Jα recombination are driven by TCR enhancers and promoters that enable RAG2 binding to histone H3 proteins tri-methylated on lysine 4 located over D/J segments and also render D/J RSSs nucleosome-free and accessible for RAG cleavage (36-38). The RAG1/RAG2 proteins in D/J recombination centers capture accessible V RSSs to mediate V-to-(D)J rearrangements (39). Changes in locus topology that place V segments near D/J segments are likely critical for the formation of synaptic complexes between V and D/J RSSs (39, 40), which may dissociate more often than they are cleaved by RAG to assemble V(D)J joins (21, 39). The relative usage of individual Vβ and Jα gene segments in rearrangements should be determined by accessibility of their RSSs, frequencies at which they enter synaptic complexes, efficiencies of their flanking RSSs, and physical forces that destabilize pre-cleavage synaptic complexes (21, 39, 41). Since differences in TCRα locus conformation have been observed between DP cells that lack Rag1 protein or express a cleavage-defective mutant Rag1 protein (42), non-core RAG1 regions may promote or preserve changes in TCRβ and TCRα locus topology that, respectively, help assemble or stabilize synaptic complexes for some Vβ and Jα segments over others. The non-core RAG1 regions contain a Really Interesting New Gene domain with E3 ubiquitin ligase activity (43, 44) and other sequences that bind the RING E3 ubiquitin ligase complex VprBP/DDB1/Cul4A/Roc1 (45) The RAG1 RING domain catalyzes histone H3 monoubiquitylation, which reduces core histone binding and promotes transcription of genes (46-48), and RAG1 mutations that reduce this E3 ubiquitin ligase activity impair V(D)J recombination (46, 47, 49). While targets of VprBP/DDB1/Cul4A/Roc1 in the context of V(D)J recombination are not known, deletion of VprBP initiating in pro-B cells causes a block in B cell development at this stage that correlates with greater impairment of VH-to-DHJH recombination than DH-to-JH recombination (45). Differences in accessibility and transcription among germline Vβ and Jα gene segments are observed in DN and DP thymocytes, respectively (13, 20, 50), while changes in TCRα locus topology control Jα usage in Vα-to-Jα rearrangements (51). Thus, non-core RAG1 regions could modulate Vβ and Jα usage by ubiquitylating H3 histones or other proteins to remove nucleosomes from some Vβ and Jα RSSs more than others and/or similarly function to modify proteins that organize TCRβ and TCRα locus topology. The increased utilization of 3’Jα segments in Rag1C/C mice suggests that non-core RAG1 regions might inhibit successive Vα-to-Jα rearrangements. Although transcription from Vα promoters stimulates such rearrangements to shape TCRα repertoire (50) mechanisms that suppress Vα-to-Jα recombination and provide DP thymocytes time to express and select TCRα genes are not known. Considering that the RAG1 RING domain promotes RAG1 polyubiquitylation (43), and ubiquitylation targets proteins for proteasomal degradation and changes in cellular localization, our data is consistent with a regulatory role of the RAG1 non-core region in constraining RAG1 protein expression to ensure time for assembled TCRα genes to be expressed and selected before initiation of further Vα-to-Jα recombination. Another possibility is that RAG1-mediated ubiquitylation of histones cooperates with ATM-dependent histone H2A ubiquitlyation (52) to transiently inhibit accessibility and transcription of germline Vα gene segments in response to RAG DSBs induced during Vα-to-Jα rearrangements. However, the biased utilization of 3’Jα segments in Rag1C/C mice could result from ability of the core-RAG1 recombinase to directly recognize downstream Jα segments without proceeding through the normal 5’ to 3’ gradient of replacement rearrangements. Another possibility is that the altered TCRβ repertoire in Rag1C/C mice results in fewer αβ TCRs that can undergo positive selection, increasing successive TCRα rearrangements and resulting in increased representation of downstream Jα segments. In this later scenario, the altered Jα usage of Rag1C/C mice would be the result of altered RAG activity at the TCRβ locus rather than at the TCRα locus.
Generation, selection, and expression of a broad αβ TCR and IgH/IgL repertoires are critical for effective adaptive immunity. RAG1 or RAG2 mutations that diminish RAG endonuclease activity cause inefficient TCR gene assembly, reduced numbers of αβ T cells beyond the progenitor stage, restricted TCRβ and TCRα repertoires, and immunodeficiency (6-9), revealing that efficient V(D)J recombination is critical for generation of αβ TCR diversity. TCRβ-mediated, Ccnd3-dependent thymocyte expansion contributes to αβ TCR diversity by allowing multiple chances for each unique TCRβ gene assembled in DN thymocyte to be selected with a different TCRα chain in DP cells (18). Our demonstration that impaired TCRβ recombination is the predominant cause of reduced DP thymocyte numbers in Rag1C/C mice and that DP cell numbers are lower in Rag1C/CCcnd3-/- mice than in Rag1C/C and Ccnd3-/- mice indicates that TCRβ recombination efficiency and TCRβ-mediated thymocyte expansion cooperate to generate αβ TCR diversity. A polymorphism in the human VκA2 RSS impairs VκA2 recombination efficiency, reduces VκA2 representation in the Vκ repertoire, and confers susceptibility to Haemophilus Influenzae (53), revealing that alterations to the mechanisms that control utilization of individual V gene segments can have deleterious consequences. Our study indicates that, similar to loss of the non-core Rag2 region (10), absence of non-core RAG1 regions decreases Vβ-to-Jβ recombination and alters the primary Vβ repertoire generated during Vβ-to-DJβ recombination. Our study also reveals that the loss of non-core RAG1 regions leads to abnormal changes in Vβ repertoire during αβ TCR selection, which could arise from altered Jα usage in TCRα rearrangements and/or impaired signaling of gene expression changes in response to RAG DSBs in DP thymocytes. Regardless, our findings suggest that non-core regions of RAG1 may have co-evolved with the non-core RAG2 domain and Vβ RSS sequences to protect host organisms from pathogens by promoting sufficient TCRβ gene diversity and advantageous representation of individual Vβ segments in the naive αβ TCR repertoire.
Acknowledgments
These experiments were supported by a Leukemia and Lymphoma Society Scholar Award and NIH grants CA125195 and CA136470 (C.H.B.).
Footnotes
The authors have no conflicts of interest to disclose.
References
- 1.Fugmann SD. RAG1 and RAG2 in V(D)J recombination and transposition. Immunol Res. 2001;23:23–39. doi: 10.1385/IR:23:1:23. [DOI] [PubMed] [Google Scholar]
- 2.Schatz DG, Swanson PC. V(D)J recombination: mechanisms of initiation. Annu Rev Genet. 2011;45:167–202. doi: 10.1146/annurev-genet-110410-132552. [DOI] [PubMed] [Google Scholar]
- 3.Lieber MR. The mechanism of double-strand DNA break repair by the nonhomologous DNA end-joining pathway. Annu Rev Biochem. 79:181–211. doi: 10.1146/annurev.biochem.052308.093131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Helmink BA, Sleckman BP. The response to and repair of RAG-mediated DNA double-strand breaks. Annu Rev Immunol. 30:175–202. doi: 10.1146/annurev-immunol-030409-101320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Sadofsky MJ. Recombination-activating gene proteins: more regulation, please. Immunol Rev. 2004;200:83–89. doi: 10.1111/j.0105-2896.2004.00164.x. [DOI] [PubMed] [Google Scholar]
- 6.Wong SY, Roth DB. Murine models of Omenn syndrome. J Clin Invest. 2007;117:1213–1216. doi: 10.1172/JCI32214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Villa A, Sobacchi C, Notarangelo LD, Bozzi F, Abinun M, Abrahamsen TG, Arkwright PD, Baniyash M, Brooks EG, Conley ME, Cortes P, Duse M, Fasth A, Filipovich AM, Infante AJ, Jones A, Mazzolari E, Muller SM, Pasic S, Rechavi G, Sacco MG, Santagata S, Schroeder ML, Seger R, Strina D, Ugazio A, Valiaho J, Vihinen M, Vogler LB, Ochs H, Vezzoni P, Friedrich W, Schwarz K. V(D)J recombination defects in lymphocytes due to RAG mutations: severe immunodeficiency with a spectrum of clinical presentations. Blood. 2001;97:81–88. doi: 10.1182/blood.v97.1.81. [DOI] [PubMed] [Google Scholar]
- 8.Notarangelo LD. Primary immunodeficiencies. J Allergy Clin Immunol. 2010;125:S182–194. doi: 10.1016/j.jaci.2009.07.053. [DOI] [PubMed] [Google Scholar]
- 9.Villa A, Marrella V, Rucci F, Notarangelo LD. Genetically determined lymphopenia and autoimmune manifestations. Curr Opin Immunol. 2008;20:318–324. doi: 10.1016/j.coi.2008.02.001. [DOI] [PubMed] [Google Scholar]
- 10.Liang HE, Hsu LY, Cado D, Cowell LG, Kelsoe G, Schlissel MS. The “dispensable” portion of RAG2 is necessary for efficient V-to-DJ rearrangement during B and T cell development. Immunity. 2002;17:639–651. doi: 10.1016/s1074-7613(02)00448-x. [DOI] [PubMed] [Google Scholar]
- 11.Dudley DD, Sekiguchi J, Zhu C, Sadofsky MJ, Whitlow S, DeVido J, Monroe RJ, Bassing CH, Alt FW. Impaired V(D)J recombination and lymphocyte development in core RAG1-expressing mice. J Exp Med. 2003;198:1439–1450. doi: 10.1084/jem.20030627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Akamatsu Y, Monroe R, Dudley DD, Elkin SK, Gartner F, Talukder SR, Takahama Y, Alt FW, Bassing CH, Oettinger MA. Deletion of the RAG2 C terminus leads to impaired lymphoid development in mice. Proc Natl Acad Sci U S A. 2003;100:1209–1214. doi: 10.1073/pnas.0237043100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Krangel MS. Mechanics of T cell receptor gene rearrangement. Curr Opin Immunol. 2009;21:133–139. doi: 10.1016/j.coi.2009.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Brady BL, Oropallo MA, Yang-Iott KS, Serwold T, Hochedlinger K, Jaenisch R, Weissman IL, Bassing CH. Position-dependent silencing of germline Vss segments on TCRss alleles containing preassembled VssDJssCss1 genes. J Immunol. 2010;185:3564–3573. doi: 10.4049/jimmunol.0903098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Wilson A, Marechal C, MacDonald HR. Biased V beta usage in immature thymocytes is independent of DJ beta proximity and pT alpha pairing. J Immunol. 2001;166:51–57. doi: 10.4049/jimmunol.166.1.51. [DOI] [PubMed] [Google Scholar]
- 16.Steinel NC, Brady BL, Carpenter AC, Yang-Iott KS, Bassing CH. Posttranscriptional silencing of VbetaDJbetaCbeta genes contributes to TCRbeta allelic exclusion in mammalian lymphocytes. J Immunol. 2010;185:1055–1062. doi: 10.4049/jimmunol.0903099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.von Boehmer H, Melchers F. Checkpoints in lymphocyte development and autoimmune disease. Nat Immunol. 2009;11:14–20. doi: 10.1038/ni.1794. [DOI] [PubMed] [Google Scholar]
- 18.Sicinska E, Aifantis I, Le Cam L, Swat W, Borowski C, Yu Q, Ferrando AA, Levin SD, Geng Y, von Boehmer H, Sicinski P. Requirement for cyclin D3 in lymphocyte development and T cell leukemias. Cancer Cell. 2003;4:451–461. doi: 10.1016/s1535-6108(03)00301-5. [DOI] [PubMed] [Google Scholar]
- 19.Krangel MS, Carabana J, Abbarategui I, Schlimgen R, Hawwari A. Enforcing order within a complex locus: current perspectives on the control of V(D)J recombination at the murine T-cell receptor alpha/delta locus. Immunol Rev. 2004;200:224–232. doi: 10.1111/j.0105-2896.2004.00155.x. [DOI] [PubMed] [Google Scholar]
- 20.Chen F, Rowen L, Hood L, Rothenberg EV. Differential transcriptional regulation of individual TCR V beta segments before gene rearrangement. J Immunol. 2001;166:1771–1780. doi: 10.4049/jimmunol.166.3.1771. [DOI] [PubMed] [Google Scholar]
- 21.Wu C, Bassing CH, Jung D, Woodman BB, Foy D, Alt FW. Dramatically increased rearrangement and peripheral representation of Vbeta14 driven by the 3’Dbeta1 recombination signal sequence. Immunity. 2003;18:75–85. doi: 10.1016/s1074-7613(02)00515-0. [DOI] [PubMed] [Google Scholar]
- 22.Carpenter AC, Yang-Iott KS, Chao LH, Nuskey B, Whitlow S, Alt FW, Bassing CH. Assembled DJ beta complexes influence TCR beta chain selection and peripheral V beta repertoire. J Immunol. 2009;182:5586–5595. doi: 10.4049/jimmunol.0803270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Blackman MA, Marrack P, Kappler J. Influence of the major histocompatibility complex on positive thymic selection of V beta 17a+ T cells. Science. 1989;244:214–217. doi: 10.1126/science.2784868. [DOI] [PubMed] [Google Scholar]
- 24.Kappler JW, Roehm N, Marrack P. T cell tolerance by clonal elimination in the thymus. Cell. 1987;49:273–280. doi: 10.1016/0092-8674(87)90568-x. [DOI] [PubMed] [Google Scholar]
- 25.Kappler JW, Staerz U, White J, Marrack PC. Self-tolerance eliminates T cells specific for Mls-modified products of the major histocompatibility complex. Nature. 1988;332:35–40. doi: 10.1038/332035a0. [DOI] [PubMed] [Google Scholar]
- 26.MacDonald HR, Lees RK, Schneider R, Zinkernagel RM, Hengartner H. Positive selection of CD4+ thymocytes controlled by MHC class II gene products. Nature. 1988;336:471–473. doi: 10.1038/336471a0. [DOI] [PubMed] [Google Scholar]
- 27.Wade T, Bill J, Marrack PC, Palmer E, Kappler JW. Molecular basis for the nonexpression of V beta 17 in some strains of mice. J Immunol. 1988;141:2165–2167. [PubMed] [Google Scholar]
- 28.Bredemeyer AL, Helmink BA, Innes CL, Calderon B, McGinnis LM, Mahowald GK, Gapud EJ, Walker LM, Collins JB, Weaver BK, Mandik-Nayak L, Schreiber RD, Allen PM, May MJ, Paules RS, Bassing CH, Sleckman BP. DNA double-strand breaks activate a multi-functional genetic program in developing lymphocytes. Nature. 2008;456:819–823. doi: 10.1038/nature07392. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Strasser A, Harris AW, Cory S. bcl-2 transgene inhibits T cell death and perturbs thymic self-censorship. Cell. 1991;67:889–899. doi: 10.1016/0092-8674(91)90362-3. [DOI] [PubMed] [Google Scholar]
- 30.Guo J, Hawwari A, Li H, Sun Z, Mahanta SK, Littman DR, Krangel MS, He YW. Regulation of the TCRalpha repertoire by the survival window of CD4(+)CD8(+) thymocytes. Nat Immunol. 2002;3:469–476. doi: 10.1038/ni791. [DOI] [PubMed] [Google Scholar]
- 31.Riegert P, Gilfillan S. A conserved sequence block in the murine and human TCR J alpha region: assessment of regulatory function in vivo. J Immunol. 1999;162:3471–3480. [PubMed] [Google Scholar]
- 32.Brady BL, Rupp LJ, Bassing CH. Requirement for dicer in survival of proliferating thymocytes experiencing DNA double-strand breaks. J Immunol. 2013;190:3256–3266. doi: 10.4049/jimmunol.1200957. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Brady BL, Bassing CH. Differential regulation of proximal and distal Vbeta segments upstream of a functional VDJbeta1 rearrangement upon beta-selection. J Immunol. 187:3277–3285. doi: 10.4049/jimmunol.1101079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yannoutsos N, Wilson P, Yu W, Chen HT, Nussenzweig A, Petrie H, Nussenzweig MC. The role of recombination activating gene (RAG) reinduction in thymocyte development in vivo. J Exp Med. 2001;194:471–480. doi: 10.1084/jem.194.4.471. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Speiser DE, Lees RK, Hengartner H, Zinkernagel RM, MacDonald HR. Positive and negative selection of T cell receptor V beta domains controlled by distinct cell populations in the thymus. J Exp Med. 1989;170:2165–2170. doi: 10.1084/jem.170.6.2165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ji Y, Little AJ, Banerjee JK, Hao B, Oltz EM, Krangel MS, Schatz DG. Promoters, enhancers, and transcription target RAG1 binding during V(D)J recombination. J Exp Med. 2010;207:2809–2816. doi: 10.1084/jem.20101136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kondilis-Mangum HD, Cobb RM, Osipovich O, Srivatsan S, Oltz EM, Krangel MS. Transcription-dependent mobilization of nucleosomes at accessible TCR gene segments in vivo. J Immunol. 2010;184:6970–6977. doi: 10.4049/jimmunol.0903923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Osipovich O, Cobb RM, Oestreich KJ, Pierce S, Ferrier P, Oltz EM. Essential function for SWI-SNF chromatin-remodeling complexes in the promoter-directed assembly of Tcrb genes. Nat Immunol. 2007;8:809–816. doi: 10.1038/ni1481. [DOI] [PubMed] [Google Scholar]
- 39.Schatz DG, Ji Y. Recombination centres and the orchestration of V(D)J recombination. Nat Rev Immunol. 2011;11:251–263. doi: 10.1038/nri2941. [DOI] [PubMed] [Google Scholar]
- 40.Jhunjhunwala S, van Zelm MC, Peak MM, Murre C. Chromatin architecture and the generation of antigen receptor diversity. Cell. 2009;138:435–448. doi: 10.1016/j.cell.2009.07.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bassing CH, Whitlow S, Mostoslavsky R, Yang-Iott K, Ranganath S, Alt FW. Vbeta cluster sequences reduce the frequency of primary Vbeta2 and Vbeta14 rearrangements. Eur J Immunol. 2008;38:2564–2572. doi: 10.1002/eji.200838347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chaumeil J, Micsinai M, Ntziachristos P, Deriano L, Wang JM, Ji Y, Nora EP, Rodesch MJ, Jeddeloh JA, Aifantis I, Kluger Y, Schatz DG, Skok JA. Higher-order looping and nuclear organization of Tcra facilitate targeted rag cleavage and regulated rearrangement in recombination centers. Cell Rep. 2013;3:359–370. doi: 10.1016/j.celrep.2013.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Jones JM, Gellert M. Autoubiquitylation of the V(D)J recombinase protein RAG1. Proc Natl Acad Sci U S A. 2003;100:15446–15451. doi: 10.1073/pnas.2637012100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Yurchenko V, Xue Z, Sadofsky M. The RAG1 N-terminal domain is an E3 ubiquitin ligase. Genes Dev. 2003;17:581–585. doi: 10.1101/gad.1058103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Kassmeier MD, Mondal K, Palmer VL, Raval P, Kumar S, Perry GA, Anderson DK, Ciborowski P, Jackson S, Xiong Y, Swanson PC. VprBP binds full-length RAG1 and is required for B-cell development and V(D)J recombination fidelity. EMBO J. 2011;31:945–958. doi: 10.1038/emboj.2011.455. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jones JM, Bhattacharyya A, Simkus C, Vallieres B, Veenstra TD, Zhou M. The RAG1 V(D)J recombinase/ubiquitin ligase promotes ubiquitylation of acetylated, phosphorylated histone 3.3. Immunol Lett. 2011;136:156–162. doi: 10.1016/j.imlet.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 47.Grazini U, Zanardi F, Citterio E, Casola S, Goding CR, McBlane F. The RING domain of RAG1 ubiquitylates histone H3: a novel activity in chromatin-mediated regulation of V(D)J joining. Mol Cell. 2010;37:282–293. doi: 10.1016/j.molcel.2009.12.035. [DOI] [PubMed] [Google Scholar]
- 48.Wright DE, Wang CY, Kao CF. Histone ubiquitylation and chromatin dynamics. Front Biosci (Landmark Ed) 17:1051–1078. doi: 10.2741/3973. [DOI] [PubMed] [Google Scholar]
- 49.Simkus C, Anand P, Bhattacharyya A, Jones JM. Biochemical and folding defects in a RAG1 variant associated with Omenn syndrome. J Immunol. 2007;179:8332–8340. doi: 10.4049/jimmunol.179.12.8332. [DOI] [PubMed] [Google Scholar]
- 50.Abarrategui I, Krangel MS. Germline transcription: a key regulator of accessibility and recombination. Adv Exp Med Biol. 2009;650:93–102. doi: 10.1007/978-1-4419-0296-2_8. [DOI] [PubMed] [Google Scholar]
- 51.Seitan VC, Hao B, Tachibana-Konwalski K, Lavagnolli T, Mira-Bontenbal H, Brown KE, Teng G, Carroll T, Terry A, Horan K, Marks H, Adams DJ, Schatz DG, Aragon L, Fisher AG, Krangel MS, Nasmyth K, Merkenschlager M. A role for cohesin in T-cell-receptor rearrangement and thymocyte differentiation. Nature. 476:467–471. doi: 10.1038/nature10312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Shanbhag NM, Rafalska-Metcalf IU, Balane-Bolivar C, Janicki SM, Greenberg RA. ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell. 141:970–981. doi: 10.1016/j.cell.2010.04.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nadel B, Tang A, Lugo G, Love V, Escuro G, Feeney AJ. Decreased frequency of rearrangement due to the synergistic effect of nucleotide changes in the heptamer and nonamer of the recombination signal sequence of the V kappa gene A2b, which is associated with increased susceptibility of Navajos to Haemophilus influenzae type b disease. J Immunol. 1998;161:6068–6073. [PubMed] [Google Scholar]


