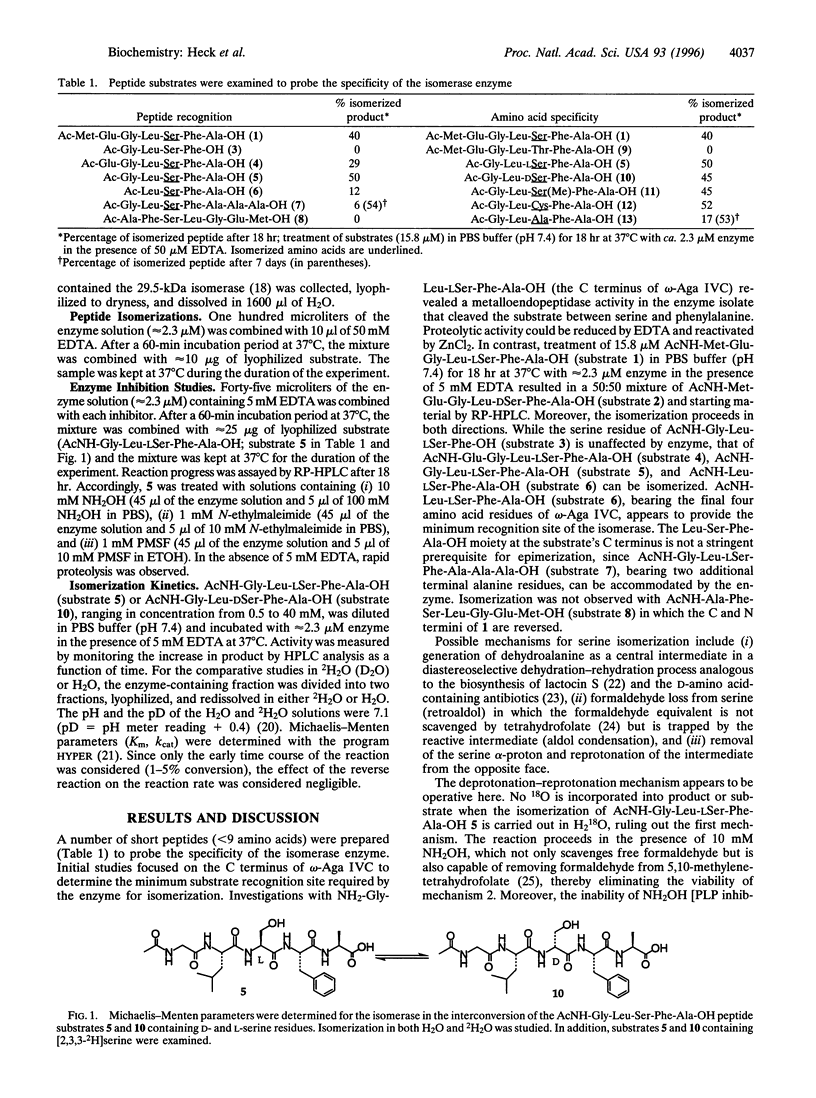
Abstract
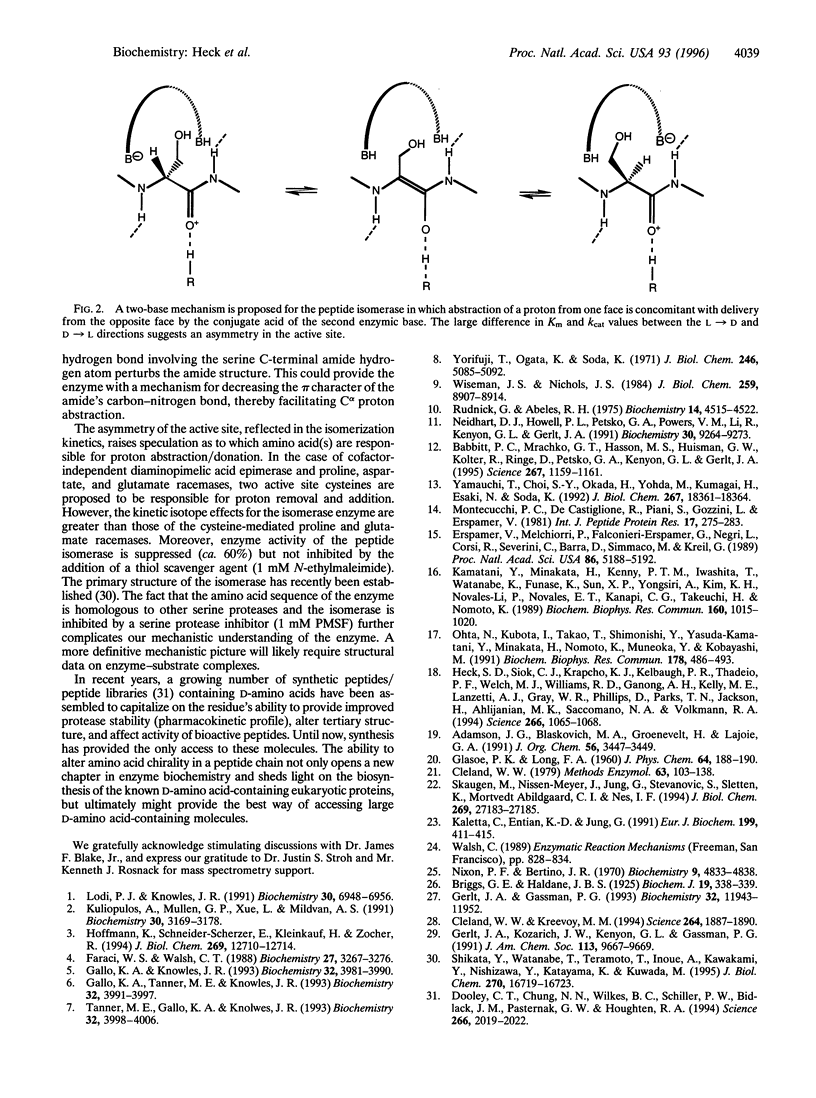
Since ribosomally mediated protein biosynthesis is confined to the L-amino acid pool, the presence of D-amino acids in peptides was considered for many years to be restricted to proteins of prokaryotic origin. Unicellular microorganisms have been responsible for the generation of a host of D-amino acid-containing peptide antibiotics (gramicidin, actinomycin, bacitracin, polymyxins). Recently, a series of mu and delta opioid receptor agonists [dermorphins and deltorphins] and neuroactive tetrapeptides containing a D-amino acid residue have been isolated from amphibian (frog) skin and mollusks. Amino acid sequences obtained from the cDNA libraries coincide with the observed dermorphin and deltorphin sequences, suggesting a stereospecific posttranslational amino acid isomerization of unknown mechanism. A cofactor-independent serine isomerase found in the venom of the Agelenopsis aperta spider provides the first major clue to explain how multicellular organisms are capable of incorporating single D-amino acid residues into these and other eukaryotic peptides. The enzyme is capable of isomerizing serine, cysteine, O-methylserine, and alanine residues in the middle of peptide chains, thereby providing a biochemical capability that, until now, had not been observed. Both D- and L-amino acid residues are susceptible to isomerization. The substrates share a common Leu-Xaa-Phe-Ala recognition site. Early in the reaction sequence, solvent-derived deuterium resides solely with the epimerized product (not substrate) in isomerizations carried out in 2H2O. Significant deuterium isotope effects are obtained in these reactions in addition to isomerizations of isotopically labeled substrates (2H at the epimerizeable serine alpha-carbon atom). The combined kinetic and structural data suggests a two-base mechanism in which abstraction of a proton from one face is concomitant with delivery from the opposite face by the conjugate acid of the second enzymic base.
Full text
PDF

Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Babbitt P. C., Mrachko G. T., Hasson M. S., Huisman G. W., Kolter R., Ringe D., Petsko G. A., Kenyon G. L., Gerlt J. A. A functionally diverse enzyme superfamily that abstracts the alpha protons of carboxylic acids. Science. 1995 Feb 24;267(5201):1159–1161. doi: 10.1126/science.7855594. [DOI] [PubMed] [Google Scholar]
- Briggs G. E., Haldane J. B. A Note on the Kinetics of Enzyme Action. Biochem J. 1925;19(2):338–339. doi: 10.1042/bj0190338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cleland W. W., Kreevoy M. M. Low-barrier hydrogen bonds and enzymic catalysis. Science. 1994 Jun 24;264(5167):1887–1890. doi: 10.1126/science.8009219. [DOI] [PubMed] [Google Scholar]
- Cleland W. W. Statistical analysis of enzyme kinetic data. Methods Enzymol. 1979;63:103–138. doi: 10.1016/0076-6879(79)63008-2. [DOI] [PubMed] [Google Scholar]
- Dooley C. T., Chung N. N., Wilkes B. C., Schiller P. W., Bidlack J. M., Pasternak G. W., Houghten R. A. An all D-amino acid opioid peptide with central analgesic activity from a combinatorial library. Science. 1994 Dec 23;266(5193):2019–2022. doi: 10.1126/science.7801131. [DOI] [PubMed] [Google Scholar]
- Erspamer V., Melchiorri P., Falconieri-Erspamer G., Negri L., Corsi R., Severini C., Barra D., Simmaco M., Kreil G. Deltorphins: a family of naturally occurring peptides with high affinity and selectivity for delta opioid binding sites. Proc Natl Acad Sci U S A. 1989 Jul;86(13):5188–5192. doi: 10.1073/pnas.86.13.5188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Faraci W. S., Walsh C. T. Racemization of alanine by the alanine racemases from Salmonella typhimurium and Bacillus stearothermophilus: energetic reaction profiles. Biochemistry. 1988 May 3;27(9):3267–3276. doi: 10.1021/bi00409a022. [DOI] [PubMed] [Google Scholar]
- Gallo K. A., Knowles J. R. Purification, cloning, and cofactor independence of glutamate racemase from Lactobacillus. Biochemistry. 1993 Apr 20;32(15):3981–3990. doi: 10.1021/bi00066a019. [DOI] [PubMed] [Google Scholar]
- Gallo K. A., Tanner M. E., Knowles J. R. Mechanism of the reaction catalyzed by glutamate racemase. Biochemistry. 1993 Apr 20;32(15):3991–3997. doi: 10.1021/bi00066a020. [DOI] [PubMed] [Google Scholar]
- Gerlt J. A., Gassman P. G. Understanding the rates of certain enzyme-catalyzed reactions: proton abstraction from carbon acids, acyl-transfer reactions, and displacement reactions of phosphodiesters. Biochemistry. 1993 Nov 16;32(45):11943–11952. doi: 10.1021/bi00096a001. [DOI] [PubMed] [Google Scholar]
- Heck S. D., Siok C. J., Krapcho K. J., Kelbaugh P. R., Thadeio P. F., Welch M. J., Williams R. D., Ganong A. H., Kelly M. E., Lanzetti A. J. Functional consequences of posttranslational isomerization of Ser46 in a calcium channel toxin. Science. 1994 Nov 11;266(5187):1065–1068. doi: 10.1126/science.7973665. [DOI] [PubMed] [Google Scholar]
- Hoffmann K., Schneider-Scherzer E., Kleinkauf H., Zocher R. Purification and characterization of eucaryotic alanine racemase acting as key enzyme in cyclosporin biosynthesis. J Biol Chem. 1994 Apr 29;269(17):12710–12714. [PubMed] [Google Scholar]
- Kaletta C., Entian K. D., Jung G. Prepeptide sequence of cinnamycin (Ro 09-0198): the first structural gene of a duramycin-type lantibiotic. Eur J Biochem. 1991 Jul 15;199(2):411–415. doi: 10.1111/j.1432-1033.1991.tb16138.x. [DOI] [PubMed] [Google Scholar]
- Kamatani Y., Minakata H., Kenny P. T., Iwashita T., Watanabe K., Funase K., Sun X. P., Yongsiri A., Kim K. H., Novales-Li P. Achatin-I, an endogenous neuroexcitatory tetrapeptide from Achatina fulica Férussac containing a D-amino acid residue. Biochem Biophys Res Commun. 1989 May 15;160(3):1015–1020. doi: 10.1016/s0006-291x(89)80103-2. [DOI] [PubMed] [Google Scholar]
- Kuliopulos A., Mullen G. P., Xue L., Mildvan A. S. Stereochemistry of the concerted enolization catalyzed by delta 5-3-ketosteroid isomerase. Biochemistry. 1991 Apr 2;30(13):3169–3178. doi: 10.1021/bi00227a003. [DOI] [PubMed] [Google Scholar]
- Lodi P. J., Knowles J. R. Neutral imidazole is the electrophile in the reaction catalyzed by triosephosphate isomerase: structural origins and catalytic implications. Biochemistry. 1991 Jul 16;30(28):6948–6956. doi: 10.1021/bi00242a020. [DOI] [PubMed] [Google Scholar]
- Montecucchi P. C., de Castiglione R., Piani S., Gozzini L., Erspamer V. Amino acid composition and sequence of dermorphin, a novel opiate-like peptide from the skin of Phyllomedusa sauvagei. Int J Pept Protein Res. 1981 Mar;17(3):275–283. doi: 10.1111/j.1399-3011.1981.tb01993.x. [DOI] [PubMed] [Google Scholar]
- Neidhart D. J., Howell P. L., Petsko G. A., Powers V. M., Li R. S., Kenyon G. L., Gerlt J. A. Mechanism of the reaction catalyzed by mandelate racemase. 2. Crystal structure of mandelate racemase at 2.5-A resolution: identification of the active site and possible catalytic residues. Biochemistry. 1991 Sep 24;30(38):9264–9273. doi: 10.1021/bi00102a019. [DOI] [PubMed] [Google Scholar]
- Nixon P. F., Bertino J. R. Inhibition of peptide chain initiation in Escherichia coli by hydroxylamine. Reaction of hydroxylamine with folate coenzymes. Biochemistry. 1970 Dec 8;9(25):4833–4838. doi: 10.1021/bi00827a001. [DOI] [PubMed] [Google Scholar]
- Ohta N., Kubota I., Takao T., Shimonishi Y., Yasuda-Kamatani Y., Minakata H., Nomoto K., Muneoka Y., Kobayashi M. Fulicin, a novel neuropeptide containing a D-amino acid residue isolated from the ganglia of Achatina fulica. Biochem Biophys Res Commun. 1991 Jul 31;178(2):486–493. doi: 10.1016/0006-291x(91)90133-r. [DOI] [PubMed] [Google Scholar]
- Rudnick G., Abeles R. H. Reaction mechanism and structure of the active site of proline racemase. Biochemistry. 1975 Oct 7;14(20):4515–4522. doi: 10.1021/bi00691a028. [DOI] [PubMed] [Google Scholar]
- Shikata Y., Watanabe T., Teramoto T., Inoue A., Kawakami Y., Nishizawa Y., Katayama K., Kuwada M. Isolation and characterization of a peptide isomerase from funnel web spider venom. J Biol Chem. 1995 Jul 14;270(28):16719–16723. doi: 10.1074/jbc.270.28.16719. [DOI] [PubMed] [Google Scholar]
- Skaugen M., Nissen-Meyer J., Jung G., Stevanovic S., Sletten K., Inger C., Abildgaard M., Nes I. F. In vivo conversion of L-serine to D-alanine in a ribosomally synthesized polypeptide. J Biol Chem. 1994 Nov 4;269(44):27183–27185. [PubMed] [Google Scholar]
- Tanner M. E., Gallo K. A., Knowles J. R. Isotope effects and the identification of catalytic residues in the reaction catalyzed by glutamate racemase. Biochemistry. 1993 Apr 20;32(15):3998–4006. doi: 10.1021/bi00066a021. [DOI] [PubMed] [Google Scholar]
- Wiseman J. S., Nichols J. S. Purification and properties of diaminopimelic acid epimerase from Escherichia coli. J Biol Chem. 1984 Jul 25;259(14):8907–8914. [PubMed] [Google Scholar]
- Yamauchi T., Choi S. Y., Okada H., Yohda M., Kumagai H., Esaki N., Soda K. Properties of aspartate racemase, a pyridoxal 5'-phosphate-independent amino acid racemase. J Biol Chem. 1992 Sep 15;267(26):18361–18364. [PubMed] [Google Scholar]
- Yorifuji T., Ogata K. Arginine racemase of Pseudomonas graveolens. I. Purification, crystallization, and properties. J Biol Chem. 1971 Aug 25;246(16):5085–5092. [PubMed] [Google Scholar]



