Abstract
NFIL3 (nuclear factor, IL-3 regulated) is a transcription factor that regulates multiple immunologic functions. In myeloid cells, NFIL3 is IL-10 inducible, and has a key role as a repressor of IL-12p40 transcription. NFIL3 is a susceptibility gene for the human inflammatory bowel diseases. Here we describe spontaneous colitis in Nfil3−/− mice. Mice lacking both Nfil3 and Il10 (NIDKO) had severe early-onset colitis, suggesting NFIL3 and IL-10 independently regulate mucosal homeostasis. Lymphocytes were necessary for colitis, as Nfil3/Rag1 double knockout (NRDKO) mice were protected from disease. However, NRDKO mice adoptively transferred with wild type CD4+ T cells developed severe colitis compared to Rag1−/− recipients, suggesting that colitis was linked to defects in innate immune cells. Colitis was abrogated in Nfil3/Il12b double-deficient mice, identifying Il12b dysregulation as a central pathogenic event. Finally, germ-free Nfil3−/− mice do not have colonic inflammation. Thus, NFIL3 is a microbiota-dependent, IL-10-independent regulator of mucosal homeostasis via IL-12p40.
Keywords: Cells: Monocytes/Macrophages, Diseases: Autoimmunity, Molecules: Cytokines, Processes: Inflammation, Tissues: Mucosa
INTRODUCTION
Genetic variants that confer susceptibility to the human inflammatory bowel diseases (IBD), Crohn’s disease (CD) and ulcerative colitis (UC) highlight the importance of innate immune interactions with the enteric microbiota in controlling inflammation (1). A consequence of innate immune activation in IBD is the over-production of, and maladaptive responses to, pro-inflammatory cytokines (2). In this context of human IBD and mouse models of IBD, IL-12 and IL-23 have been shown in multiple contexts to have a central pathogenic role (3–7). IL-12 and IL-23 are heterodimeric cytokines composed of p40-p35 and p40-p19 subunits, respectively. Macrophages and dendritic cells activated by microbes and microbial products are the main cell types that produce IL-12 and IL-23. While IL-12 is decisive in regulating lineage commitment to Th1 development, IL-23 is essential for the maturation and stability of Th17 cells (8, 9). Several genes in the IL-12 and IL-23 expression and signaling pathways, notably IL23R, JAK2, STAT3, and IL12B are associated with CD and UC susceptibility (10, 11).
A key step in the normal homeostatic regulation of IL-12 and IL-23 from activated innate cells is inhibition by IL-10 derived from autocrine-paracrine signaling from activated myeloid cells, or from lymphocytes, especially regulatory T cells (12, 13). IL-12 and IL-23 are not the only IL-10 targets; in fact many pro-inflammatory genes implicated in IBD, including TNF and IL1B, are regulated by IL-10 by a variety of mechanisms (14, 15). However, in the context of intestinal mucosal homeostasis, the IL-10-mediated inhibitory pathway that tonically regulates excessive IL-12 and IL-23 is a central pathway. Defects in the production of or the responsiveness to IL-10 exert profound homeostatic effects in the mucosal immune system (12, 16, 17). Polymorphisms in IL10 confer susceptibility to UC while complete loss-of-function mutations in the IL-10 receptors cause a rare but severe form of very early onset of IBD (18, 19). Importantly, ablation of IL-12 and IL-23 significantly ameliorates IBD in the IL-10-deficient (Il10−/−) mouse model, confirming the essential role of the IL-10 inhibitory pathway that controls IL-12 and IL-23 (20–22).
The basic leucine zipper transcription factor NFIL3 (Nuclear factor, IL-3 regulated, also known as E4BP4) has recently been implicated as a regulator of multiple aspects of immune function. NFIL3 is essential for the development of all natural-killer (NK) cells (23, 24) and CD8α dendritic cells (DCs) (25), cytokine production by Th2 cells (26, 27), and IgE class-switching in B cells (28). IL-10 was reported to regulate NFIL3 mRNA expression over a decade ago (29). Following from that observation, we recently reported that NFIL3 is an essential transcriptional repressor of IL-12p40 in murine macrophages (30, 31). NFIL3 controls Il12b transcription by binding to an enhancer located ~10 kb upstream of the canonical Il12b promoter (31). An NFIL3 binding element has also been characterized in the proximal Il12b promoter region (30). Myeloid cells deficient in NFIL3 overproduce IL-12p40, however, they remain responsive to IL-10-mediated inhibition, arguing that NFIL3 is a necessary component of the IL-10 pathway but alone is not sufficient to block IL-12p40 production (30, 31). We also found that NFIL3 participates in host enteric microbial interactions in vivo and the enteric microbiota and IL-10 are co-factors for NFIL3 expression in the intestine. We found that colonic CD11b+ LPMC isolated from Nfil3−/− mice, like bone marrow-derived myeloid cells, made higher amounts of Il12b mRNA than WT CD11b+ colonic macrophages. In human IBD, intestinal CD14+ macrophages are major producers of pro-inflammatory cytokines including IL-12 family members (5). We found that NFIL3 expression was lower in CD14+ LPMC isolated from patients with CD and UC compared with non-inflammatory controls undergoing surgical resection (30). Our previous studies also concluded that intestinal NFIL3 expression inversely correlates with higher amounts IL-12p40 in mice and humans (30). Importantly, recent meta-analysis of CD and UC genome-wide association scans identified NFIL3 as a susceptibility gene for human inflammatory bowel disease (IBD) (32).
Collectively, these studies raised the possibility that NFIL3 would be involved in normal homeostatic regulation of the intestine. Here, we characterize a severe form of spontaneous colitis in Nfil3−/− mice that correlated with increased IL-12p40 in serum and colonic tissue. Colitis was dependent on Il12b-driven innate and adaptive immune interactions. These results implicate NFIL3 as a central determinant of mucosal immune homeostasis and coupled with the recent human genetic association, an important pathway to elucidate the pathogenesis of IBD.
MATERIALS AND METHODS
Mice
Nfil3−/− mice were generated by gene-targeting strategies as reported (28) and bred with C57BL/6 mice for at least 10 generations. Germ free (GF) Nfil3−/− mice were Caesarian derived as previously described (33) and were maintained according to standard techniques in the University of North Carolina National Gnotobiotic Resource Center. WT, Il10−/−, and Rag1−/− mice were obtained from Jackson Laboratory (Bar Harbor, ME). Il10−/− and Rag1−/− mice were crossed with Nfil3−/− mice to generate mice deficient in both genes (Nfil3−/−Il10−/−, NIDKO or Nfil3−/−Rag1−/−, NRDKO). All experimental mice were genotyped by polymerase chain reaction (PCR) screening before tissue collection, and age-matched littermates were used as controls. Mice were evaluated for colonic inflammation at the age of 12–16 weeks. Another strain of NFIL3-deficient mice (24) was maintained at St. Jude Children’s Research Hospital. All animal experiments were in accordance with protocols approved by the Institutional Animal Care and Use Committee of the University of North Carolina School of Medicine, the University of Iowa Carver College of Medicine and St. Jude Children’s Research Hospital.
Reagents
LPS was purchased from InvivoGen (San Diego, CA). M-CSF was obtained from Peprotech Inc (Rocky Hill, NJ).
Cell isolation
BMDMs were cultured as described (34). Mouse colonic lamina propria mononuclear cells (LPMCs) were isolated by an enzymatic method and separated into CD11b+ cells using MACS anti-CD11b microbeads (Miltenyi Biotec, Auburn, CA) (12).
Quantitative RT-PCR
Quantitative real-time RT-PCR was performed as described (35). Primer sequences are available upon request.
Colonic tissue explant cultures and histology
Colonic tissue explant cultures were performed as previously described (35). Supernatants were collected after 24 hours for cytokine ELISA. Cytokine values were normalized to dry weight of the colonic tissue explants (pg/ml/100 mg). Slides were prepared for H&E staining and histological analysis was performed by a pathologist blinded to the study groups (LBB) using criteria reported previously (36).
Flow cytometry
Flow cytometric analysis was performed as previously described (12). Anti-CD3, anti-CD4, anti-NKp46, anti-retinoid-acid receptor-related orphan receptor gamma t (RORχt), anti-forkhead box P3 (Foxp3), anti-IL-17A, and anti-IFN-γ antibodies were purchased from eBioscience (San Diego, California, USA). For intracellular staining, Cytofix/Cytoperm Kit (BD PharMingen) was used according to the manufacturer’s instructions. For cytokine staining, LPMCs were stimulated with phorbol myristate acetate (100 ng/ml) and ionomycin (1 μg/ml) (Sigma) in the presence of monensin (Golgistop; BD PharMingen) for 4 hours. Results were analyzed by Beckman-Coulter (Dako) CyAn ADP flow cytometer and Summit (version 4.3) software.
Immunohistochemistry
Mouse colonic tissues were harvested, swiss-rolled, and fixed with 4% paraformaldehyde (Sigma, St. Louis, MO) overnight. Fixed tissues were then embedded in OCT compound (Sakura, Tokyo, Japan) and kept at −80°C. Frozen tissue sections (7 μm) were first blocked for 30 min at room temperature with Blocking Reagent (PerkinElmer, Waltham, MA) and then incubated overnight at 4°C with primary antibodies as described below. Following overnight incubation, sections were washed and then incubated with HRP-conjugated secondary antibody for 1h at room temperature. After washing, sections were treated with tyramide-fluorescein (PerkinElmer), followed by counterstaining with DAPI (Sigma). All stained sections were analyzed by confocal microscope using either the 10× or the 20× objective (TCS SP2, Leica, Wetzlar, Germany). Images were processed with Adobe Photoshop software. The following antibodies were used: anti-CD11c and anti-F4/80 from BD Bioscience (Franklin Lakes, NJ), anti-CD3, anti-NKp46, and anti-B220 from eBioscience (San Diego, CA), HRP-conjugated donkey anti-rat IgG, and HRP-conjugated donkey anti-Armenian hamster IgG (H+L) from Jackson Immunoresearch (West Grove, PA).
ELISA
IL-12p40 concentration was determined by sandwich ELISA (BD Biosciences, San Jose, CA). IL-22 ELISA kit was obtained from eBioscience (San Diego, CA). IL-17A, IL-10, IFN-γ, and TNF-α were measured using the MILLIPLEX MAP Cytokine Kit (Millipore, Billerica, MA).
Induction of colitis by adoptive transfer of T cells
Splenocytes from WT mice were enriched for CD4+ cells by positive selection on MACS columns with anti-CD4 microbeads (Miltenyi Biotec). Rag1−/− or Nfil3−/−Rag1−/− (NRDKO) mice were i.p. injected with 1×106 CD4+ cells. Recipients and control mice were observed for clinical signs of colitis and body weight changes. Clinical score was recorded four weeks post-transfer as previously described (37) (wasting: 0, no wasting; 1, 0.1–10% loss of initial total body weight; 2, >10% loss of initial total body weight; diarrhea: 0, none; 1, soft stool; 2, watery and/or bloody; hunching/bristled fur/skin lesions: 0, unchanged; 1, any positive sign; rectum prolapse: 0, absent; 1, present).
Germ free to conventionalized transition of mice
8–10-week-old GF WT and Nfil3−/− mice were transitioned to conventionalized housing. To re-colonize the GF mice with conventionalized microbiota, each cage was given bedding from the same donor conventionalized WT cage. Mice were sacrificed after 2 weeks and monitored for colitis as described above.
Statistical analysis
Statistical significance for data subsets was assessed by the two-tailed Student’s t test unless specifically described. A p<0.05 was considered significant.
RESULTS
Nfil3−/− mice develop spontaneous Th1/Th17-mediated chronic colitis
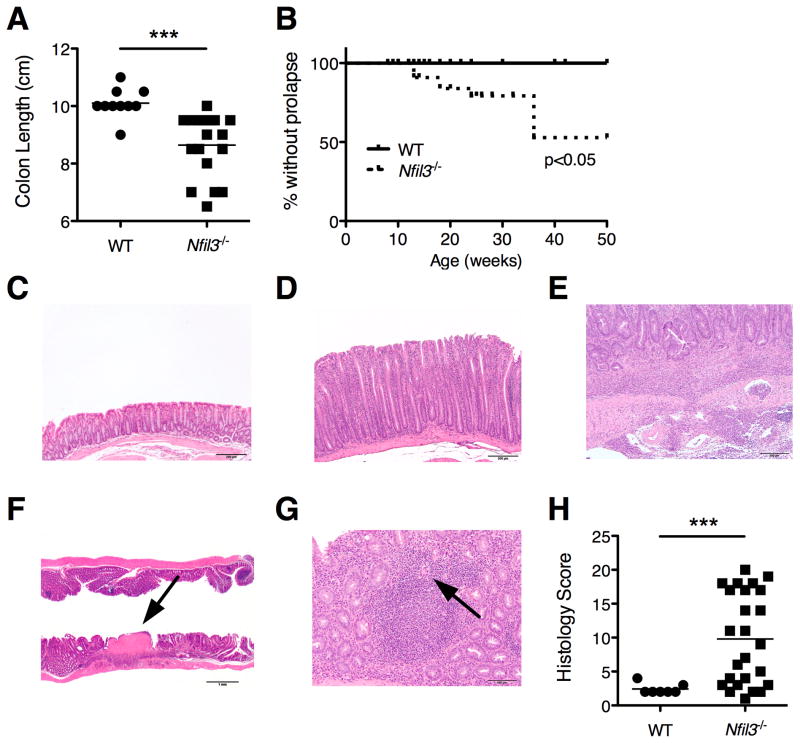
On necropsy, colons from 12-week-old Nfil3−/− mice were thickened and foreshortened with lack of formed fecal pellets. Colon length was significantly shorter in Nfil3−/− compared to WT mice (Fig. 1A). Approximately 20% of Nfil3−/− mice developed rectal prolapse by 20 weeks and 50% by 36 weeks of age (Fig. 1B).
Figure 1. Nfil3−/− mice develop spontaneous colitis.
(A) Colon length of 12–18 week-old Nfil3−/− (n = 14) and age-matched WT (n = 8) mice was measured. *, p<0.05 versus WT. (B) Prevalence of rectal prolapse was plotted (WT, n = 25; Nfil3−/−, n = 41). p<0.05 versus WT. (C) Histological severity (WT, n = 4; Nfil3−/−, n = 14) was scored based on Berg’s score. **, p<0.005 versus WT. (D, E) Colonic epithelium, H&E (20× magnification). Photomicrographs of colonic mucosa taken at the same magnification compared to WT mouse colons (D) demonstrate significant mucosal hypertrophy with crypt elongation and decreased goblet cells in the Nfil3−/− (E) mouse colons. (F) Colon, H&E (40× magnification). Inflammation in Nfil3−/− colons was characterized by multifocal aggregates of inflammatory cells that extended transmurally to the submucosa (G) Colon, H&E (4× magnification). Areas of ulceration (arrow) were also occasionally observed. (H) Colon, H&E (40× magnification). In Nfil3−/− the inflammation in the lamina propria was occasionally organized into de novo lymphoid follicles and occasional multinucleated giant cells (arrow) were observed.
Quantitative assessment of colonic histology revealed markedly increased frequency and severity of inflammation in Nfil3−/− mice compared to WT mice (Fig. 1C). As in other models of spontaneous colitis, histological severity was variable and penetrance of disease was incomplete (Fig. 1H Compared to colons from WT mice (Fig. 1D), significant elongation of colonic crypts was observed colons from Nfil3−/− mice (Fig. 1E). Inflammation was characterized by multifocal aggregates of inflammatory cells that were confined to the mucosa as well as extending transmurally to the submucosa (Fig. 1F). We also observed areas of ulceration (Fig. 1G, arrow). Inflammation in the lamina propria was occasionally organized into de novo lymphoid follicles, and multinucleated giant cells were detected (Fig. 1H, arrow). Quantitative assessment of colonic histological severity (n = 25, 12–16 week old Nfil3−/− mice) was significantly higher compared to age-matched WT mice (n = 7), but the penetrance of colitis was incomplete (Fig. 1C).
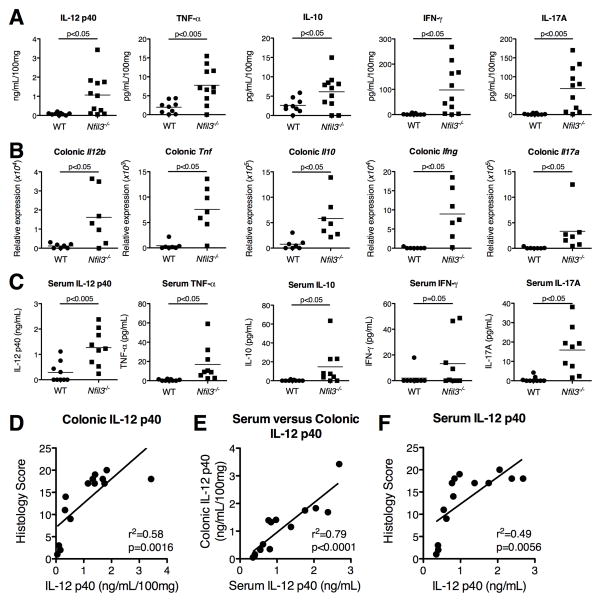
Colonic tissue explants from Nfil3−/− mice secreted significantly elevated amounts of pro-inflammatory cytokines IL-12p40, TNF-α, IFNγ and IL-17A compared to WT colonic explants (Fig. 2A). Quantitative real-time RT-PCR from colonic tissue also demonstrated increased mRNA expression for these cytokines (Fig. 2B). Serum IL-12p40, TNF-α, IFNγ and IL-17A were also increased in Nfil3−/− mice (Fig. 2C). Increased colonic and serum IL-10 was also detected, suggesting that the loss NFIL3-dependent pathways cannot be compensated by increased IL-10 expression (Fig. 2A, B, C). Moreover, serum IL-12p40 positively correlated with colonic IL-12p40 expression (Fig. 2D) and histologic severity of inflammation (Fig. 2E, F). Consequently, serum IL-12 p40 is a biomarker for histological severity of colitis in this model.
Figure 2. Characterization of cytokine profile in the colon and serum in Nfil3−/− mice.
(A) Colonic explants (WT, n = 9; Nfil3−/−, n = 11) were assayed for spontaneous secretion of cytokines. Colons were isolated from WT and Nfil3−/− mice, cleaned, and processed for intestinal tissue explant cultures as described in Experimental Procedures. Tissue fragments (approximately 0.05 g dry weight) were incubated in 1 ml of RPMI 1640 supplemented with 1% antibiotics and 10% FBS. Supernatants were collected after 24 h and spontaneous secretion of cytokines was measured by Multiplex. Values were normalized to dry weight of the colonic explants (pg/ml/100 mg). (B) Colonic mRNA from WT (n = 7) and Nfil3−/− (n = 7) mice was extracted and expression of Il12b, Tnf, Ifng, Il17 and Il10 mRNA was assessed by real-time RT-PCR. Results are expressed as relative expression normalized to β-actin. (C) Serum cytokine concentration in WT (n = 9) and Nfil3−/− (n = 9) mice was measured by Multiplex. (D) Correlation of serum and colonic tissue explant IL-12p40 in 12–18 week old Nfil3−/− mice (n = 11). R2=0.8883, p<0.0001. (E) Correlation of histological score and colonic tissue explant IL-12p40 in 12–18 week old Nfil3−/− mice (n = 11). R2=0.625, p<0.005. (F) Correlation of serum IL-12p40 and histological score in 12–18 week old Nfil3−/− mice (n = 11). R2=0.7914, p<0.0005.
Colonic macrophages are the main source of inflammatory cytokines in Nfil3−/− mice
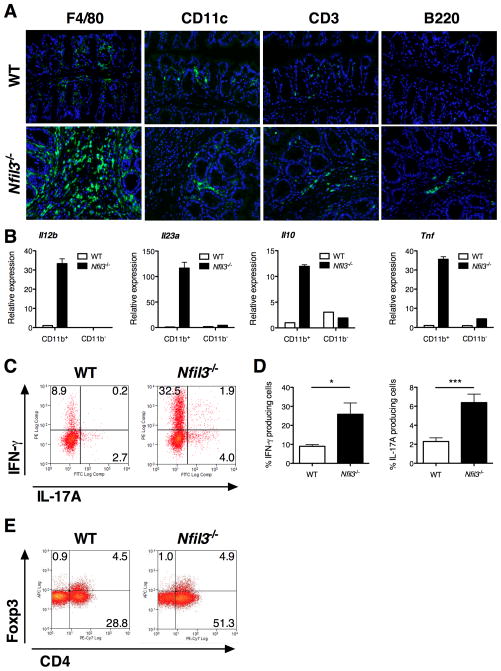
By immunohistochemical analyses, CD3+, B220+. and F4/80+ cells were increased in inflamed colonic lamina propria of Nfil3−/− mice compared to WT mice, with the most striking increase in F4/80+ cells (Fig. 3A). There was no significant increase in CD11c+ dendritic cell (DC) populations. B220+ plasma cells were localized inside and outside of secondary lymphoid follicles in Nfil3−/− mice in contrast to WT colons where the vast majority of B220+ cells localized within lymphoid structures. CD11b+ lamina propria mononuclear cells (LPMC) are comprised of colonic macrophages (F4/80+ CD11c− or CD11clow) and DCs (CD11c+ F4/80−) (38). We therefore isolated and analyzed colonic CD11b+ LPMCs from colons of Nfil3−/− as a representative colonic macrophage population. Nfil3−/− CD11b+ colonic LPMCs expressed higher Il12b, Il23a, Tnf, and Il10 mRNA compared to WT CD11b+ colonic LPMCs (Fig. 3B). Furthermore, these cytokine mRNAs were predominantly expressed in CD11b+ LPMCs, whereas expression in the CD11b− population was limited.
Figure 3. Characterization of increased inflammatory cell infiltration in the colon of Nfil3−/− mice.
(A) Fluorescence immunohistochemistry in colonic tissues from WT and Nfil3−/− mice was performed for F4/80, CD11c, CD3, and B220 (Green). Specimens were counterstained with DAPI (Blue). (B) Expression relative to β-actin of Il12b, Il23a, Tnf and Il10 transcripts in CD11b+ and CD11b− LPMC from WT and Nfil3−/− mice. Results represent 3 independent experiments. (C) LPMCs were isolated from WT and Nfil3−/− colons and stimulated with phorbol myristate acetate (100 ng/ml) and ionomycin (1 mg/ml) in the presence of monensin for 4 hours. Representative patterns for intracellular IFN-g and IL-17 in gated CD3+CD4+ lymphocytes from 3 independent experiments are shown. (D) Quantification of IFN-γ- and IL-17A-positive cells in CD3+CD4+ lymphocytes from WT and Nfil3−/− LPMCs. Data are presented as percentage of IFN-γ- or IL-17A-positive cells in gated CD3+CD4+ lymphocytes (mean ± SEM from 3 independent experiments). *, p<0.05; ***, p<0.0005 versus WT. (E) Representative patterns for intracellular FoxP3 staining in gated CD3+CD4+ lymphocytes from 3 independent experiments are shown.
Localization of IFNγ and IL-17A expression in CD3+CD4+ colonic lamina propria T cells was examined by intracellular cytokine staining in WT and Nfil3−/− mice. Both IFN-γ- and IL-17A-producing CD4+ T cells are significantly increased in Nfil3−/− mice (Fig. 3C, D). The percentage of Foxp3+CD4+ regulatory T cells did not differ between WT and Nfil3−/− colonic LPMCs (Fig. 3E).
Nfil3−/− mice have decreased numbers of colonic NK cells and lymphoid tissue inducer cells (LTi)
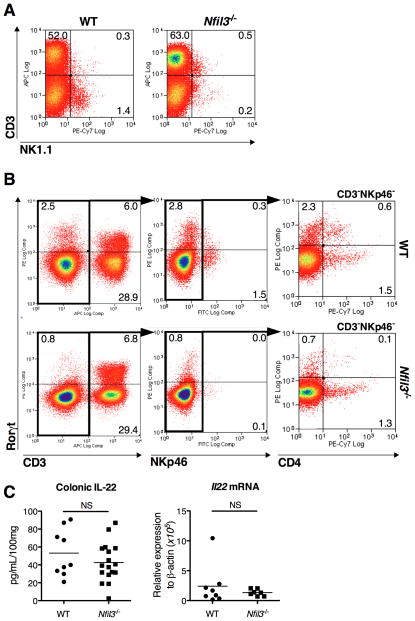
Nfil3−/− mice lack peripheral NK1.1+ NK cells (23, 24). However it is unclear whether these mice also lack intestinal NK cells, which express NKp46 (39). The lamina propria CD3−NK1.1+ NK cell population was significantly decreased, while CD3+NK1.1+ NK T cells were similar in number in Nfil3−/− compared to WT mice (Fig. 4A). The transcription factor retinoid-acid receptor-related orphan receptor gamma t (RORγt) is implicated in NK marker-expressing mucosal lymphoid cell development and function (39, 40). Therefore, RORγt expression was assessed in the colonic NKp46+ cell population. Both RORγt+ and RORγt− CD3−NKp46+ colonic lamina propria ILC populations were significantly decreased in number in Nfil3−/− compared to WT mice (Fig. 4B). Lymphoid tissue inducer (LTi) cells, defined as CD3− RORγt+, are required for the development of secondary lymphoid tissue in the gut (41). In contrast to CD3+RORγt+ cells that include IL-17A-producing T cells, the CD3− RORγt+ LTi population was significantly decreased in Nfil3−/− compared to WT mice. Both CD4+ and CD4− populations were decreased among CD3−NKp46−RORγt+ LTi cells in Nfil3−/− colons, even though de novo colonic mucosal lymphoid structures are present in Nfil3−/− mice (Fig. 1H). RORγt+ CD3−NKp46+ mucosal lymphoid cells are a major source of the homeostatic cytokine IL-22 (NK22 cells) (39, 40). Therefore, expression and production of colonic IL-22 was examined in Nfil3−/− mice. By contrast to other pro- and anti-inflammatory (IL-10) cytokines (Fig. 2A, B), IL-22 production in colonic explant culture and Il22 mRNA expression were not increased in Nfil3−/− colonic tissue (Fig. 4C).
Figure 4. Nfil3−/− mice show decreased mucosal NK cells and LTi subpopulation.
(A) CD4+ Colonic LPMCs were isolated and analyzed for CD3 and NK1.1. Lymphocytes were gated by forward and side scatter and CD45 expression. (B) Colonic LPMCs were isolated and analyzed for CD3, NKp46 and CD4 in combination with intracellular RORγt by flow cytometry. Lymphocytes were gated by forward- and side-scatter and further gated in CD3-negative cells (middle), CD3-negative NKp46-negative cells (right). (C) Colonic explant cultures were analyzed for IL-22 expression by ELISA (WT, n = 6; Nfil3−/−, n = 7) and colonic tissue was analyzed for Il22 expression by RT-PCR (WT, n = 6; Nfil3−/−, n = 7). NS, not significant versus WT.
IL-10 and NFIL3 independently but cooperatively regulate IL-12p40 in BMDMs and in vivo
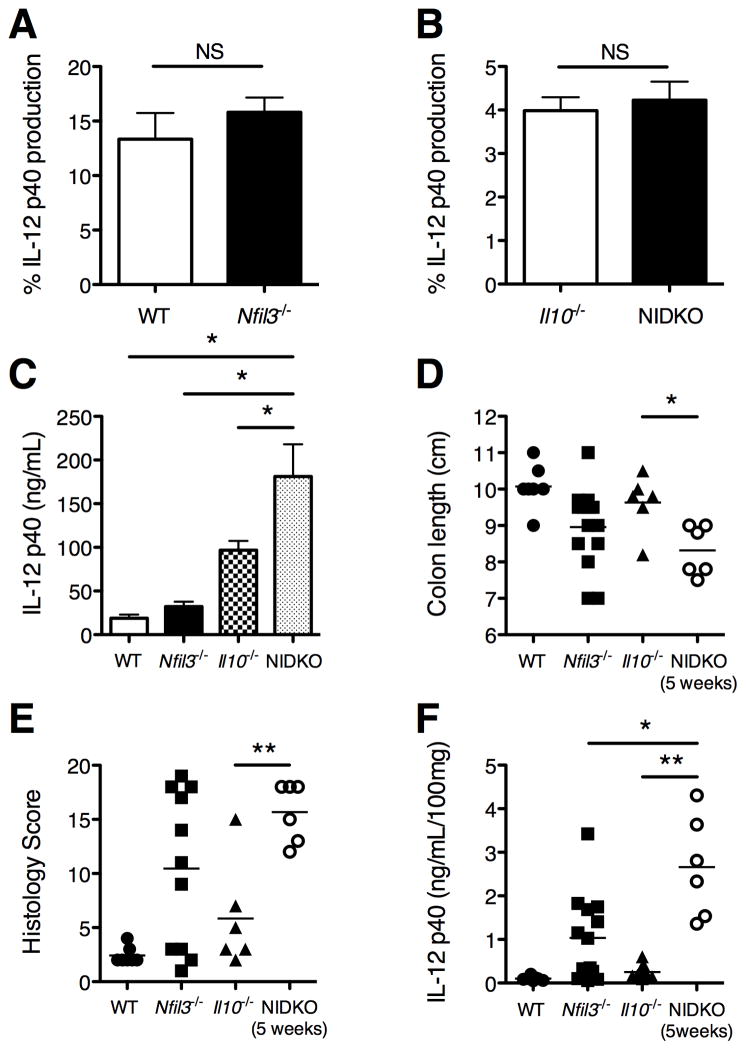
We have previously shown that IL-10 is required for LPS-induced NFIL3 expression in BMDMs and that NFIL3 partly mediates IL-10 inhibition of Il12b (30, 31). Moreover, Il10−/− mice demonstrated impaired induction of Nfil3 in the colon by the microbiota (30). However, inhibition of Il12b by NFIL3 does not require IL-10, since overexpression of NFIL3 in Il10−/− BMDMs effectively inhibited IL-12p40 (30). Therefore, whether IL-10 and NFIL3 independently regulate IL-12p40 in cells and in vivo was next addressed. The inhibitory effect of IL-10 on IL-12p40 in WT and Nfil3−/− BMDMs was compared. Recombinant IL-10 inhibited LPS-induced IL-12p40 to a similar extent in WT and Nfil3−/− BMDMs (Fig. 5A). We next generated Nfil3 × Il10 double knockout (NIDKO) mice, and hypothesized that colitis would be similar in severity in NIDKO mice compared to single knockouts if NFIL3 and IL-10 functions are completely dependent on each other. IL-12p40 production from activated macrophages was dramatically increased in NIDKO BMDMs compared to either single knockout cells (Fig. 5B). However, there was no difference in IL-10 mediated IL-12p40 inhibition in Il10−/− and NIDKO BMDMs (Fig. 5C), suggesting that inhibition of IL-12p40 by IL-10 does not require NFIL3. These data were ascertained independently with BMDMs from two different strains of NIDKO and single knockout mice derived at UNC and St. Jude Children’s Research Hospital.
Figure 5. IL-10 and NFIL3 independently regulate IL-12p40 in BMDMs and in vivo.
(A) WT and Nfil3−/− BMDMs were stimulated with LPS (10 ng/ml) in the presence or absence of IL-10 (1 ng/ml). Supernatants were harvested 24 hours post-LPS stimulation and analyzed for IL-12p40 by ELISA. Results are presented as percent IL-12p40 by LPS in the presence of IL-10 relative to LPS alone (mean ± SEM from 4 independent experiments). NS, not significant versus WT. (B) WT, Nfil3−/−, Il10−/−, and Nfil3/Il10−/− double knockout (NIDKO) BMDMs were stimulated with LPS (10 ng/ml) for 24 hours and IL-12p40 in the supernatants were analyzed by ELISA. Results are presented as mean ± SEM from at least 4 independent experiments. *, p<0.05 versus NIDKO. (C) Il10−/− and NIDKO BMDMs were stimulated with LPS (10 ng/ml) in the presence or absence of IL-10 (1 ng/ml) for 24 hours and analyzed for IL-12p40 by ELISA. Results are presented as percent IL-12p40 by LPS in the presence of IL-10 relative to LPS alone (mean ± SEM from 4 independent experiments). NS, not significant versus Il10−/−. (D) Quantitative histological scores from WT (n = 4), Nfil3−/− (n = 14), Il10−/− (n = 6) and NIDKO (n = 6) mice were given. **, p<0.005 versus NIDKO. (E) Comparison of spontaneous secretion of IL-12p40 by colonic explants from 12–18 week old WT (n = 7), Nfil3−/− (n = 13), Il10−/− (n = 6) and 5-week-old NIDKO (n = 6) mice. Values were normalized to dry weight of the colonic explants (ng/ml/100 mg). *, p<0.05; **, p<0.005 versus NIDKO. (F) Colon lengths from WT (n = 8), Nfil3−/− (n = 14), Il10−/− (n = 6) and NIDKO (n = 6) mice were measured. *, p<0.05 versus NIDKO.
Importantly, NIDKO mice had severe colitis with 100% penetrance (Fig. 5D) and higher colonic IL-12p40 expression (Fig. 5E) at 5 weeks of age (when these mice were euthanized due to severity of illness) compared to single knockout mice at 12–16 weeks of age. All double knockout mice developed severe diarrhea and 50% (3/6) developed rectal prolapse by 5 weeks of age. By comparison, 7 and 12 percent of Il10−/− (n=28, data not shown) and Nfil3−/− (n=41, see Fig. 1B), respectively, developed rectal prolapse in mice at 16 weeks of age. Colon length was shorter in NIDKO mice compared to Nfil3−/− and Il10−/− mice (Fig. 5F). Interestingly, histological colonic inflammation was more severe in Nfil3−/− mice than Il10−/− mice on a C57BL/6 background (Fig. 5D). A separate colony of NIDKO mice generated at St. Jude Children’s Research Hospital confirmed these findings; even though the St. Jude Nfil3−/− mice do not get colitis, combined loss of IL-10 and NFIL3 caused a dramatic acceleration in colitis. These findings demonstrate that NFIL3 and IL-10 coordinately but independently regulate mucosal homeostasis.
Nfil3+/+ T cells are sufficient to induce colitis in Nfil3/Rag1 double knockout mice
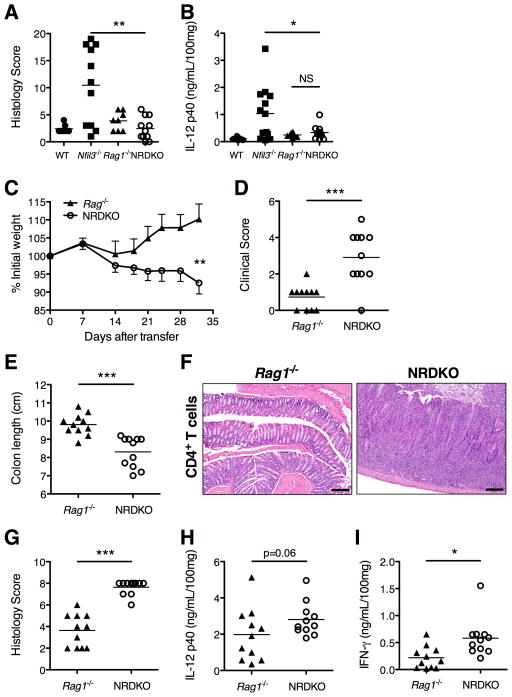
In an adoptive transfer model of colitis in Rag1−/− recipients, Motomura et al., demonstrated that naive Nfil3−/− CD4+T cells were potent in colitis induction potential compared to WT CD4+ T cells (27), mediated by defective IL-10 production from Nfil3−/− T cells. However, since IL-10 production was increased in Nfil3−/− colons (Fig. 2A, B, C), other mechanisms underlie colitis development in the Nfil3−/− background. Therefore, Nfil3−/− mice were crossed to Rag1−/− mice (NRDKO) to study whether lymphocytes are necessary for the colitis development in Nfil3−/− mice. NRDKO mice had no clinical signs of colitis (data not shown). Colonic histological scores (Fig. 6A) and colonic IL-12p40 (Fig. 6B) were comparable to WT and Rag−/− mice, showing that lymphocytes are necessary for colitis development.
Figure 6. Lymphocytes are necessary for the development of colitis in Nfil3−/− mice.
(A) Nfil3/Rag1 double knockout (NRDKO) mice were generated and histological scores were assigned at 16 weeks of age (WT, n = 4; Nfil3−/−, n = 14; Rag1−/−, n = 8; NRDKO, n = 11). **, p<0.005 versus NRDKO. (B) Comparison of spontaneous secretion of IL-12p40 by colonic explants from WT (n = 7), Nfil3−/− (n = 13), Rag1−/− (n = 8) and NRDKO (n = 11) mice. *, p<0.05; NS, not significant versus NRDKO. (C–I) Rag1−/− (n = 7) and NRDKO (n = 6) mice were given 1×106 unfractionated CD4+ T cells by i.p. injection and development of colitis was monitored over 4 weeks. (C) Body weight expressed as a percentage of the total body weight on Day 0 of the T cell transfer was measured. ***, p<0.0005 versus Rag1−/−. (D) Clinical scores were measured at time of sacrifice as described in Experimental Procedures. ***, p<0.0005 versus Rag1−/−. (E) Colon lengths were measured. ***, p<0.0005 versus Rag1−/−. (F) Colon, H&E (10× magnification). Representative photomicrographs from Rag1−/− and NRDKO mice given unfractionated CD4+ T cells. (G) Quantitative histological scores were given. ***, p<0.0005 versus Rag1−/−. (H) Comparison of spontaneous secretion of IL-12p40 by colonic explants from Rag1−/− (n = 7) and NRDKO (n = 6) mice given unfractionated CD4+ T cells. *, p<0.05 versus Rag1−/−. (I) Comparison of spontaneous secretion of IFN-γ by colonic explants. ***, p<0.0005 versus Rag1−/−.
We next asked whether innate-T cell interactions are necessary for the development of colitis in Nfil3−/− mice. WT (Nfil3+/+) unfractionated CD4+ T cells were adoptively transferred into Rag1−/− or NRDKO mice. Unfractionated CD4+ cells did not cause severe pathology when transferred to Rag1 single knockout recipients (Fig. 6C–G), as previously described (42). However, NRDKO recipients develop severe colitis clinically (Fig. 6C, D, E) and histologically (Fig. 6F, G), correlating with increased intestinal IL-12p40 (Fig. 6H) and IFN-γ (Fig. 6I). These results suggest that T cells are necessary for the development of colitis in Nfil3−/− mice; however the lack of NFIL3 in accessory cell populations is essential for intestinal inflammation. Furthermore, transferred Tregs in the unfractionated CD4+ population cannot rescue disease when NFIL3 is absent in non-lymphocyte cells.
Nfil3/Il12b double knockout mice do not develop colitis
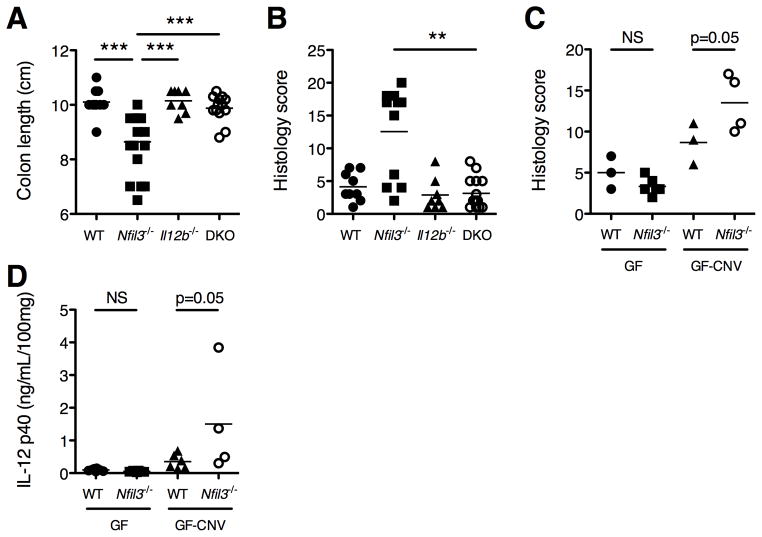
Genetic deletion of Il12b rescues several forms of intestinal inflammatory disease because IL-12 and IL-23 production is ablated (20, 43). Therefore, a key test of the relationship between NFIL3 and IL-12p40 is to determine if loss of Il12b rescues colitis in Nfil3−/− mice. We therefore generated Nfil3 × Il12b double knockout (DKO) mice and observed that these mice did not develop colitis up to 4 months of age (Fig. 7A, B), confirming that increased IL-12p40 production in Nfil3−/− mice is responsible for the development of colitis and of the inflammatory T cell phenotype.
Figure 7. Development of colitis in Nfil3−/− mice requires the microbiota and is driven by IL-12p40.
(A,B) Nfil3/Il12b double knockout (DKO) mice were generated and monitored for colitis development. (A) Colon lengths were measured at 16 weeks of age (WT, n = 8; Nfil3−/−, n = 14; Il12b−/−, n = 8; DKO, n = 13). ***, p<0.0005 versus Nfil3−/−. (B) Histological scores were assigned at 16 weeks of age (WT, n = 4; Nfil3−/−, n = 14; Il12b−/−, n = 8; DKO, n = 13). **, p<0.005 versus DKO. (C,D) Germ free (GF) Nfil3−/− were generated and monitored for colitis (WT, n = 12; Nfil3−/−, n = 6). Additionally, GF WT (n = 6) and Nfil3−/− (n = 4) were transitioned to conventional housing (GF to CNV) and monitored for colitis development for 14 days. (C) Histological scores were assigned. **, p<0.005 versus GF-CNV WT. (D) Comparison of spontaneous secretion of IL-12p40 by colonic explants. *, p<0.05 versus GF-CNV WT.
Colitis development in Nfil3−/− mice is dependent on the enteric microbiota
Nfil3−/− mice in our colony at the University of North Carolina develop significant spontaneous colitis, but mice from the founder colony at the University of Iowa (28) and a colony at St. Jude Children’s Research Hospital (31) do not develop colitis. Indeed, in a histologic analysis of 14 Nfil3−/− mice 6–8 months in age from the University of Iowa and St. Jude Children’s Research Hospital, no colitis was detected (data not shown), whereas 10 age-matched mice from the UNC colony manifest histologic colitis (data not shown). We assayed for Helicobacter spp. in the feces of Iowa and UNC mice. Non-H. hepaticus (H. hepaticus is routinely screened by both facilities) Helicobacter spp. were present in both colonies (data not shown), leading to the conclusion that other components of the intestinal microbiota are necessary for colitis development (although interactions of other enteric microbes with Helicobacter spp. are possible).
To definitively confirm the microbiota dependence for colitis development, Nfil3−/− mice were derived germ free (GF). Importantly, GF Nfil3−/− mice did not manifest colitis at 6 months of age (Fig. 7C, left) and colonic explant cultures produce low levels of IL-12p40 (Fig. 7D, left). However, upon transition to conventional housing (GF-CNV), Nfil3−/− mice demonstrated histologic colitis (Fig. 7C, right), and colonic explant cultures produced increased IL-12p40 (Fig. 7D, right) at 14 days, compared to similarly transitioned GF-CNV WT mice.
DISCUSSION
Here we report that NFIL3 deficiency, akin to IL-10 deficiency (17), results in the development of spontaneous colitis in mice. Immunohistochemistry showed that F4/80+ macrophages are predominant amongst the infiltrating inflammatory cells in the colons of inflamed Nfil3−/− mice. Colonic inflammation in Nfil3−/− mice correlated with increased colonic expression of innate derived cytokines TNF and IL-12p40, both central to human IBD pathogenesis. Moreover, colonic CD11b+ cells, comprising mainly macrophages, are the major source of increased IL-12p40 in Nfil3−/− mice. Increased expression of IFN-γ and IL-17 in Nfil3−/− mice is consistent with the hypothesis that NFIL3 deficiency leads to a pathogenic inflammatory Th1 and Th17 immune response. This inflammatory T cell phenotype is driven by innate interactions with the enteric microbiota, as Nfil3−/− mice crossed with Il12b−/− mice do not develop colitis. Two recent papers reported that NFIL3 regulates Th2 cytokine production in T cells but does not affect Th1 and Th17 cytokines (26, 27), providing further evidence that the lack of NFIL3 in T cells does not directly explain the increased Th1/Th17 response in Nfil3−/− mice.
The enteric microbiota and IL-10 are co-factors for NFIL3 expression in the intestine (29, 30). Indeed, we found colonic CD11b+ LPMC isolated from Nfil3−/− mice produced higher amounts of Il12b mRNA than WT CD11b+ colonic macrophages. In human IBD, intestinal CD14+ macrophages are the main producers of pro-inflammatory cytokines including IL-12 family members. We previously demonstrated that NFIL3 expression was lower in CD14+ LPMC isolated from patients with CD and UC compared with non-inflammatory controls undergoing surgical resection (30), and intestinal NFIL3 expression inversely correlated with higher amounts IL-12p40 in mice and humans (30). Increased colonic expression of IL-10 was detected in Nfil3−/− mice, suggesting that even in the presence of abundant IL-10, the loss of the downstream NFIL3 pathway cannot be overcome by increased local amounts of IL-10. This notion is consistent with our data showing that IL-10 and NFIL3 mediate cooperative but independent functions in macrophages and in vivo; IL-10 potently inhibited Il12b to the same extent in WT, Nfil3−/−, and Nfil3−/− × Il10−/− macrophages arguing that IL-10-dependent, NFIL3-independent pathways can regulate IL-12p40 production. Furthermore, Nfil3−/− × Il10−/− mice developed severe colitis at an early age, demonstrating non-overlapping homeostatic functions of NFIL3 and IL-10. However, the NFIL3-independent pathways are not sufficient to prevent colitis in mice able to express IL-10.
Although T cells were necessary for colitis development, the critical role of innate immune contributions in directing the inflammatory T cell response was conclusively demonstrated through several lines of experimental evidence. First, Nfil3−/− × Il12b−/− mice were protected from colitis. Moreover, Nfil3−/− × Rag1−/− mice developed worse colitis than Rag1−/− mice when adoptively transferred with unfractionated WT CD4+ T cells. NFIL3 was shown to be essential for NK cell development; therefore, Nfil3−/− mice lack peripheral NK1.1+ NK cells (23, 24). Recently, Lang et al., demonstrated that the presence of NK cells limited T cell expansion in vivo, and Nfil3−/− mice failed to regulate the expansion of IFNγ-producing CD8+ cells (44). Thus it is possible that systemic NK cell deficiency leads to an insufficient regulation of T cell responses in Nfil3−/− mice. We demonstrated that colonic lamina propria from Nfil3−/− mice show a severe paucity of both RORγt+ and RORγt− NKp46+ mucosal NK cells, and it was recently demonstrated that Nfil3−/− mice lack intraepithelial NKp46+ ILC1 in the small intestine (45). Therefore, it is also possible that a local mucosal NK cell deficiency plays a role in the pathogenesis of Nfil3−/− colitis that requires further analysis. Interestingly, amongst multiple colonic cytokines we examined in this study, IL-22 was the only cytokine that was not increased. IL-22 is largely produced by RORγt+NKp46+ cells in the gut (NK22 cells) and is implicated in intestinal immune homeostasis (39, 46). Therefore, lack of IL-22 induction may reflect the inability of Nfil3−/− mice to properly respond to and resolve inflammation through IL-22. Further study is needed to unravel the role of mucosal NK cells and IL-22 in the development of spontaneous colitis in Nfil3−/− mice. Taken together, our study suggests that lack of NFIL3 in non-lymphocyte populations is essential for the development of colitis in Nfil3−/− mice through subsequent Th1/Th17-biased acquired immune responses. Macrophages and mucosal NK cells may play key roles in this process, and an imbalance in IL-12p40 and IL-22 may be factors bridging innate immune dysfunction and T cell polarization described in this study.
Colitis in Nfil3−/− mice is dependent on the presence and composition of the enteric microbiota. Nfil3−/− mice demonstrated incomplete penetrance of colitis in the UNC facility, and there was a marked difference in penetrance and severity of disease between institutions maintaining the same strain of Nfil3−/− mice (University of Iowa) (28) or at St. Jude Children’s Research Hospital where a different Nfil3−/− mouse is maintained (24, 31). Importantly, the four described NFIL3-deficient mice (23, 24, 27, 28) have the same phenotypes, arguing that the microbiota at a given location is a key non-genetic factor in determining whether colitis develops. Many of the currently available murine models of IBDs also exhibit incomplete penetrance and variable severity of disease (47). Hence, compositional differences between enteric microbiota are the most plausible environmental determinants for the development of colitis in different models at different sites. Importantly, our GF Nfil3−/− mice do not manifest colitis at 6 months of age, and colonic explant cultures produce low levels of IL-12p40, conclusively demonstrating a role for the microbiota in colitis pathogenesis in this model. Non-H. hepaticus (H. hepaticus is routinely screened by all facilities) Helicobacter spp. were ubiquitously present in the UNC and Iowa colonies, leading to the conclusion that other components of the intestinal microbiota are necessary for colitis development, although interactions of distinct enteric microbes with Helicobacter spp. leading to colitis are possible. Moreover, components of the enteric microbiota that colonize Nfil3−/− (and by inference WT) mice at the UNC facility not present in the Iowa facility are colitogenic in Nfil3−/− mice. Further studies to characterize the enteric microbial communities in WT and in Nfil3−/− mice at different facilities with different colitis penetrance are likely to provide important insights into gene-environment relationships relevant to human IBD pathogenesis.
Absence of colitis development in other institutions also raised a critical question for the essential role for Nfil3 in the development of colitis. Therefore, we also generated Nfil3/Il10 DKO in St. Jude Children’s Research Hospital. The DKO demonstrate more severe and more consistent colitis than the Il10−/− counterparts as we saw at UNC, despite the absence of clinically apparent colitis in Nfil3 singles (data not shown). The DKOs also needed to be euthanized at an early age due to severity of disease with very high prevalence of rectal prolapse. This clear contrast of penetrance between Il10/Nfil3 DKO (complete penetrance) and singles (incomplete) across the different institutions elegantly shows the compensating/interacting roles of IL-10 and NFIL3, and provides a model for elucidating multi-hit mechanisms by multiple IBD susceptibility genes.
Several genes in pathways regulating the expression and/or function of the cytokines IL-10, IL-12 and IL-23 contribute to IBD susceptibility (IL10, IL23R, IL12B, IL12RB1, STAT3, JAK2, TYK2, HCK) (10, 18, 32). Work from our group and others have shown that other IBD susceptibility genes are functionally embedded in these pathways (IFNG, IRF1, REL, CEBPB) (32, 48–51). Moreover, the NFIL3 risk polymorphism is relatively common, present in 70% of IBD cases; thus, approximately 50% of cases are homozygous for the risk allele and 10% homozygous for the protective allele. Hence, we speculate that NFIL3 may be one gene of many that affects IL12B expression in human IBD, with polymorphisms leading to an incremental increase in IBD susceptibility. Moreover, there may be rare genetic variants of NFIL3, akin to recently described IL-10 and IL-10 receptor polymorphisms, which underlie a severe clinical phenotype such as very early onset IBD (19).
Collectively, our results show that excessive IL-12p40 production in Nfil3−/− colonic macrophages in response to the enteric microbiota is a key trigger that leads to an exaggerated intestinal Th1/Th17 response, resulting in colitis development. As NFIL3 is a human IBD susceptibility gene, the Nfil3−/− mouse is a new model to understand how a genetic defect in a transcriptional repressor in innate immune cells leads to dysregulated T cell inflammatory responses to the enteric microbiota. Furthermore, NFIL3 may be a useful therapeutic target for human IBD given its central homeostatic functions in the intestines.
Acknowledgments
SOURCE SUPPORT: This work was supported by grants from the National Institutes of Health (NIH): RO1 DK54452 (SEP), P40 RR018603 (National Gnotobiotic Rodent Resource Center), P30 DK034987 (Center for Gastrointestinal Biology and Disease; Immunotechnologies, Gnotobiotic and Histology Cores), and by grants from American Gastroenterological Association’s Research Scholars Award (SZS), Crohn’s and Colitis Foundation of America Research Fellowship Award (TK, KM and NM), Uehara Memorial Foundation (TK).
ABBREVIATIONS
- BMDMs
bone marrow derived macrophages
- DC
dendritic cells
- DKO
double knockout
- Foxp3
forkhead box P3
- IBD
inflammatory bowel diseases
- LPMCs
lamina propria mononuclear cells
- LTi
lymphoid tissue inducer
- NFIL3
nuclear factor, IL-3 regulated
- RORγt
retinoid-acid receptor-related orphan receptor gamma t
- WT
wild-type
Footnotes
Author contributions were as follows: Designed research (TK, KM, SEP); Performed research (TK, ECS, SMR, TN, NM, LBB, BH); Analyzed data (TK, ECS, SMR, TN, NM, LBB, JVG, PJM, SEP); Contributed new reagents/analytic tools (PBR, MK); Wrote the paper (TK, SEP); Revised paper (JVG, PBR, MK, SZS, PJM).
The authors have no conflict of interests to declare.
References
- 1.Rioux JD, Xavier RJ, Taylor KD, Silverberg MS, Goyette P, Huett A, Green T, Kuballa P, Barmada MM, Datta LW, Shugart YY, Griffiths AM, Targan SR, Ippoliti AF, Bernard EJ, Mei L, Nicolae DL, Regueiro M, Schumm LP, Steinhart AH, Rotter JI, Duerr RH, Cho JH, Daly MJ, Brant SR. Genome-wide association study identifies new susceptibility loci for Crohn disease and implicates autophagy in disease pathogenesis. Nat Genet. 2007;39:596–604. doi: 10.1038/ng2032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Xavier RJ, Podolsky DK. Unravelling the pathogenesis of inflammatory bowel disease. Nature. 2007;448:427–434. doi: 10.1038/nature06005. [DOI] [PubMed] [Google Scholar]
- 3.Camoglio L, Juffermans NP, Peppelenbosch M, te Velde AA, ten Kate FJ, van Deventer SJ, Kopf M. Contrasting roles of IL-12p40 and IL-12p35 in the development of hapten-induced colitis. Eur J Immunol. 2002;32:261–269. doi: 10.1002/1521-4141(200201)32:1<261::AID-IMMU261>3.0.CO;2-X. [DOI] [PubMed] [Google Scholar]
- 4.Hue S, Ahern P, Buonocore S, Kullberg MC, Cua DJ, McKenzie BS, Powrie F, Maloy KJ. Interleukin-23 drives innate and T cell-mediated intestinal inflammation. J Exp Med. 2006;203:2473–2483. doi: 10.1084/jem.20061099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kamada N, Hisamatsu T, Okamoto S, Chinen H, Kobayashi T, Sato T, Sakuraba A, Kitazume MT, Sugita A, Koganei K, Akagawa KS, Hibi T. Unique CD14 intestinal macrophages contribute to the pathogenesis of Crohn disease via IL-23/IFN-gamma axis. J Clin Invest. 2008;118:2269–2280. doi: 10.1172/JCI34610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kobayashi T, Okamoto S, Hisamatsu T, Kamada N, Chinen H, Saito R, Kitazume MT, Nakazawa A, Sugita A, Koganei K, Isobe K, Hibi T. IL23 differentially regulates the Th1/Th17 balance in ulcerative colitis and Crohn’s disease. Gut. 2008;57:1682–1689. doi: 10.1136/gut.2007.135053. [DOI] [PubMed] [Google Scholar]
- 7.Neurath MF, Fuss I, Kelsall BL, Stuber E, Strober W. Antibodies to interleukin 12 abrogate established experimental colitis in mice. J Exp Med. 1995;182:1281–1290. doi: 10.1084/jem.182.5.1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.McGeachy MJ, Chen Y, Tato CM, Laurence A, Joyce-Shaikh B, Blumenschein WM, McClanahan TK, O’Shea JJ, Cua DJ. The interleukin 23 receptor is essential for the terminal differentiation of interleukin 17-producing effector T helper cells in vivo. Nat Immunol. 2009;10:314–324. doi: 10.1038/ni.1698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Trinchieri G, Pflanz S, Kastelein RA. The IL-12 family of heterodimeric cytokines: new players in the regulation of T cell responses. Immunity. 2003;19:641–644. doi: 10.1016/s1074-7613(03)00296-6. [DOI] [PubMed] [Google Scholar]
- 10.Cho JH, Weaver CT. The genetics of inflammatory bowel disease. Gastroenterology. 2007;133:1327–1339. doi: 10.1053/j.gastro.2007.08.032. [DOI] [PubMed] [Google Scholar]
- 11.Zenewicz LA, Abraham C, Flavell RA, Cho JH. Unraveling the genetics of autoimmunity. Cell. 2010;140:791–797. doi: 10.1016/j.cell.2010.03.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kamada N, Hisamatsu T, Okamoto S, Sato T, Matsuoka K, Arai K, Nakai T, Hasegawa A, Inoue N, Watanabe N, Akagawa KS, Hibi T. Abnormally differentiated subsets of intestinal macrophage play a key role in Th1-dominant chronic colitis through excess production of IL-12 and IL-23 in response to bacteria. J Immunol. 2005;175:6900–6908. doi: 10.4049/jimmunol.175.10.6900. [DOI] [PubMed] [Google Scholar]
- 13.Murai M, Turovskaya O, Kim G, Madan R, Karp CL, Cheroutre H, Kronenberg M. Interleukin 10 acts on regulatory T cells to maintain expression of the transcription factor Foxp3 and suppressive function in mice with colitis. Nat Immunol. 2009;10:1178–1184. doi: 10.1038/ni.1791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Fiorentino DF, Zlotnik A, Mosmann TR, Howard M, O’Garra A. IL-10 inhibits cytokine production by activated macrophages. J Immunol. 1991;147:3815–3822. [PubMed] [Google Scholar]
- 15.Smallie T, Ricchetti G, Horwood NJ, Feldmann M, Clark AR, Williams LM. IL-10 inhibits transcription elongation of the human TNF gene in primary macrophages. J Exp Med. 2010;207:2081–2088. doi: 10.1084/jem.20100414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Kaser A, Zeissig S, Blumberg RS. Genes and environment: how will our concepts on the pathophysiology of IBD develop in the future? Dig Dis. 2010;28:395–405. doi: 10.1159/000320393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kuhn R, Lohler J, Rennick D, Rajewsky K, Muller W. Interleukin-10-deficient mice develop chronic enterocolitis. Cell. 1993;75:263–274. doi: 10.1016/0092-8674(93)80068-p. [DOI] [PubMed] [Google Scholar]
- 18.Anderson CA, Boucher G, Lees CW, Franke A, D’Amato M, Taylor KD, Lee JC, Goyette P, Imielinski M, Latiano A, Lagace C, Scott R, Amininejad L, Bumpstead S, Baidoo L, Baldassano RN, Barclay M, Bayless TM, Brand S, Buning C, Colombel JF, Denson LA, De Vos M, Dubinsky M, Edwards C, Ellinghaus D, Fehrmann RS, Floyd JA, Florin T, Franchimont D, Franke L, Georges M, Glas J, Glazer NL, Guthery SL, Haritunians T, Hayward NK, Hugot JP, Jobin G, Laukens D, Lawrance I, Lemann M, Levine A, Libioulle C, Louis E, McGovern DP, Milla M, Montgomery GW, Morley KI, Mowat C, Ng A, Newman W, Ophoff RA, Papi L, Palmieri O, Peyrin-Biroulet L, Panes J, Phillips A, Prescott NJ, Proctor DD, Roberts R, Russell R, Rutgeerts P, Sanderson J, Sans M, Schumm P, Seibold F, Sharma Y, Simms LA, Seielstad M, Steinhart AH, Targan SR, van den Berg LH, Vatn M, Verspaget H, Walters T, Wijmenga C, Wilson DC, Westra HJ, Xavier RJ, Zhao ZZ, Ponsioen CY, Andersen V, Torkvist L, Gazouli M, Anagnou NP, Karlsen TH, Kupcinskas L, Sventoraityte J, Mansfield JC, Kugathasan S, Silverberg MS, Halfvarson J, Rotter JI, Mathew CG, Griffiths AM, Gearry R, Ahmad T, Brant SR, Chamaillard M, Satsangi J, Cho JH, Schreiber S, Daly MJ, Barrett JC, Parkes M, Annese V, Hakonarson H, Radford-Smith G, Duerr RH, Vermeire S, Weersma RK, Rioux JD. Meta-analysis identifies 29 additional ulcerative colitis risk loci, increasing the number of confirmed associations to 47. Nature genetics. 2011;43:246–252. doi: 10.1038/ng.764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Glocker EO, Kotlarz D, Boztug K, Gertz EM, Schaffer AA, Noyan F, Perro M, Diestelhorst J, Allroth A, Murugan D, Hatscher N, Pfeifer D, Sykora KW, Sauer M, Kreipe H, Lacher M, Nustede R, Woellner C, Baumann U, Salzer U, Koletzko S, Shah N, Segal AW, Sauerbrey A, Buderus S, Snapper SB, Grimbacher B, Klein C. Inflammatory bowel disease and mutations affecting the interleukin-10 receptor. N Engl J Med. 2009;361:2033–2045. doi: 10.1056/NEJMoa0907206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davidson NJ, Hudak SA, Lesley RE, Menon S, Leach MW, Rennick DM. IL-12, but not IFN-gamma, plays a major role in sustaining the chronic phase of colitis in IL-10-deficient mice. J Immunol. 1998;161:3143–3149. [PubMed] [Google Scholar]
- 21.Kobayashi M, Kweon MN, Kuwata H, Schreiber RD, Kiyono H, Takeda K, Akira S. Toll-like receptor-dependent production of IL-12p40 causes chronic enterocolitis in myeloid cell-specific Stat3-deficient mice. J Clin Invest. 2003;111:1297–1308. doi: 10.1172/JCI17085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yen D, Cheung J, Scheerens H, Poulet F, McClanahan T, McKenzie B, Kleinschek MA, Owyang A, Mattson J, Blumenschein W, Murphy E, Sathe M, Cua DJ, Kastelein RA, Rennick D. IL-23 is essential for T cell-mediated colitis and promotes inflammation via IL-17 and IL-6. J Clin Invest. 2006;116:1310–1316. doi: 10.1172/JCI21404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gascoyne DM, Long E, Veiga-Fernandes H, de Boer J, Williams O, Seddon B, Coles M, Kioussis D, Brady HJ. The basic leucine zipper transcription factor E4BP4 is essential for natural killer cell development. Nat Immunol. 2009;10:1118–1124. doi: 10.1038/ni.1787. [DOI] [PubMed] [Google Scholar]
- 24.Kamizono S, Duncan GS, Seidel MG, Morimoto A, Hamada K, Grosveld G, Akashi K, Lind EF, Haight JP, Ohashi PS, Look AT, Mak TW. Nfil3/E4bp4 is required for the development and maturation of NK cells in vivo. J Exp Med. 2009;206:2977–2986. doi: 10.1084/jem.20092176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kashiwada M, Pham NL, Pewe LL, Harty JT, Rothman PB. NFIL3/E4BP4 is a key transcription factor for CD8alpha dendritic cell development. Blood. 2011;117:6193–6197. doi: 10.1182/blood-2010-07-295873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kashiwada M, Cassel SL, Colgan JD, Rothman PB. NFIL3/E4BP4 controls type 2 T helper cell cytokine expression. EMBO J. 2011;30:2071–2082. doi: 10.1038/emboj.2011.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Motomura Y, Kitamura H, Hijikata A, Matsunaga Y, Matsumoto K, Inoue H, Atarashi K, Hori S, Watarai H, Zhu J, Taniguchi M, Kubo M. The transcription factor E4BP4 regulates the production of IL-10 and IL-13 in CD4+ T cells. Nat Immunol. 2011;12:450–459. doi: 10.1038/ni.2020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kashiwada M, Levy DM, McKeag L, Murray K, Schroder AJ, Canfield SM, Traver G, Rothman PB. IL-4-induced transcription factor NFIL3/E4BP4 controls IgE class switching. Proc Natl Acad Sci U S A. 2010;107:821–826. doi: 10.1073/pnas.0909235107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lang R, Patel D, Morris JJ, Rutschman RL, Murray PJ. Shaping gene expression in activated and resting primary macrophages by IL-10. J Immunol. 2002;169:2253–2263. doi: 10.4049/jimmunol.169.5.2253. [DOI] [PubMed] [Google Scholar]
- 30.Kobayashi T, Matsuoka K, Sheikh SZ, Elloumi HZ, Kamada N, Hisamatsu T, Hansen JJ, Doty KR, Pope SD, Smale ST, Hibi T, Rothman PB, Kashiwada M, Plevy SE. NFIL3 is a regulator of IL-12 p40 in macrophages and mucosal immunity. J Immunol. 2011;186:4649–4655. doi: 10.4049/jimmunol.1003888. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Smith AM, Qualls JE, O’Brien K, Balouzian L, Johnson PF, Schultz-Cherry S, Smale ST, Murray PJ. A distal enhancer in Il12b is the target of transcriptional repression by the STAT3 pathway and requires the basic leucine zipper (B-ZIP) protein NFIL3. J Biol Chem. 2011;286:23582–23590. doi: 10.1074/jbc.M111.249235. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Jostins L, Ripke S, Weersma RK, Duerr RH, McGovern DP, Hui KY, Lee JC, Schumm LP, Sharma Y, Anderson CA, Essers J, Mitrovic M, Ning K, Cleynen I, Theatre E, Spain SL, Raychaudhuri S, Goyette P, Wei Z, Abraham C, Achkar JP, Ahmad T, Amininejad L, Ananthakrishnan AN, Andersen V, Andrews JM, Baidoo L, Balschun T, Bampton PA, Bitton A, Boucher G, Brand S, Buning C, Cohain A, Cichon S, D’Amato M, De Jong D, Devaney KL, Dubinsky M, Edwards C, Ellinghaus D, Ferguson LR, Franchimont D, Fransen K, Gearry R, Georges M, Gieger C, Glas J, Haritunians T, Hart A, Hawkey C, Hedl M, Hu X, Karlsen TH, Kupcinskas L, Kugathasan S, Latiano A, Laukens D, Lawrance IC, Lees CW, Louis E, Mahy G, Mansfield J, Morgan AR, Mowat C, Newman W, Palmieri O, Ponsioen CY, Potocnik U, Prescott NJ, Regueiro M, Rotter JI, Russell RK, Sanderson JD, Sans M, Satsangi J, Schreiber S, Simms LA, Sventoraityte J, Targan SR, Taylor KD, Tremelling M, Verspaget HW, De Vos M, Wijmenga C, Wilson DC, Winkelmann J, Xavier RJ, Zeissig S, Zhang B, Zhang CK, Zhao H, Silverberg MS, Annese V, Hakonarson H, Brant SR, Radford-Smith G, Mathew CG, Rioux JD, Schadt EE, Daly MJ, Franke A, Parkes M, Vermeire S, Barrett JC, Cho JH. Host-microbe interactions have shaped the genetic architecture of inflammatory bowel disease. Nature. 2012;491:119–124. doi: 10.1038/nature11582. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.MacDonald TT, Carter PB. Contact sensitivity in the germ-free mouse. J Reticuloendothel Soc. 1978;24:287–293. [PubMed] [Google Scholar]
- 34.Xiong H, Zhu C, Li F, Hegazi R, He K, Babyatsky M, Bauer AJ, Plevy SE. Inhibition of interleukin-12 p40 transcription and NF-kappaB activation by nitric oxide in murine macrophages and dendritic cells. J Biol Chem. 2004;279:10776–10783. doi: 10.1074/jbc.M313416200. [DOI] [PubMed] [Google Scholar]
- 35.Hegazi RA, Rao KN, Mayle A, Sepulveda AR, Otterbein LE, Plevy SE. Carbon monoxide ameliorates chronic murine colitis through a heme oxygenase 1-dependent pathway. J Exp Med. 2005;202:1703–1713. doi: 10.1084/jem.20051047. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Berg DJ, Zhang J, Weinstock JV, Ismail HF, Earle KA, Alila H, Pamukcu R, Moore S, Lynch RG. Rapid development of colitis in NSAID-treated IL-10-deficient mice. Gastroenterology. 2002;123:1527–1542. doi: 10.1053/gast.2002.1231527. [DOI] [PubMed] [Google Scholar]
- 37.Maillard MH, Cotta-de-Almeida V, Takeshima F, Nguyen DD, Michetti P, Nagler C, Bhan AK, Snapper SB. The Wiskott-Aldrich syndrome protein is required for the function of CD4(+)CD25(+)Foxp3(+) regulatory T cells. J Exp Med. 2007;204:381–391. doi: 10.1084/jem.20061338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Denning TL, Wang YC, Patel SR, Williams IR, Pulendran B. Lamina propria macrophages and dendritic cells differentially induce regulatory and interleukin 17-producing T cell responses. Nat Immunol. 2007;8:1086–1094. doi: 10.1038/ni1511. [DOI] [PubMed] [Google Scholar]
- 39.Satoh-Takayama N, Vosshenrich CA, Lesjean-Pottier S, Sawa S, Lochner M, Rattis F, Mention JJ, Thiam K, Cerf-Bensussan N, Mandelboim O, Eberl G, Di Santo JP. Microbial flora drives interleukin 22 production in intestinal NKp46+ cells that provide innate mucosal immune defense. Immunity. 2008;29:958–970. doi: 10.1016/j.immuni.2008.11.001. [DOI] [PubMed] [Google Scholar]
- 40.Sanos SL, Bui VL, Mortha A, Oberle K, Heners C, Johner C, Diefenbach A. RORgammat and commensal microflora are required for the differentiation of mucosal interleukin 22-producing NKp46+ cells. Nat Immunol. 2009;10:83–91. doi: 10.1038/ni.1684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ivanov II, Diehl GE, Littman DR. Lymphoid tissue inducer cells in intestinal immunity. Curr Top Microbiol Immunol. 2006;308:59–82. doi: 10.1007/3-540-30657-9_3. [DOI] [PubMed] [Google Scholar]
- 42.Powrie F, Leach MW, Mauze S, Caddle LB, Coffman RL. Phenotypically distinct subsets of CD4+ T cells induce or protect from chronic intestinal inflammation in C. B-17 scid mice. Int Immunol. 1993;5:1461–1471. doi: 10.1093/intimm/5.11.1461. [DOI] [PubMed] [Google Scholar]
- 43.Tozawa K, Hanai H, Sugimoto K, Baba S, Sugimura H, Aoshi T, Uchijima M, Nagata T, Koide Y. Evidence for the critical role of interleukin-12 but not interferon-gamma in the pathogenesis of experimental colitis in mice. J Gastroenterol Hepatol. 2003;18:578–587. doi: 10.1046/j.1440-1746.2003.03024.x. [DOI] [PubMed] [Google Scholar]
- 44.Lang PA, Lang KS, Xu HC, Grusdat M, Parish IA, Recher M, Elford AR, Dhanji S, Shaabani N, Tran CW, Dissanayake D, Rahbar R, Ghazarian M, Brustle A, Fine J, Chen P, Weaver CT, Klose C, Diefenbach A, Haussinger D, Carlyle JR, Kaech SM, Mak TW, Ohashi PS. Natural killer cell activation enhances immune pathology and promotes chronic infection by limiting CD8+ T-cell immunity. Proc Natl Acad Sci U S A. 2011 doi: 10.1073/pnas.1118834109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Fuchs A, Vermi W, Lee JS, Lonardi S, Gilfillan S, Newberry RD, Cella M, Colonna M. Intraepithelial type 1 innate lymphoid cells are a unique subset of IL-12- and IL-15-responsive IFN-gamma-producing cells. Immunity. 2013;38:769–781. doi: 10.1016/j.immuni.2013.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zenewicz LA, Yancopoulos GD, Valenzuela DM, Murphy AJ, Stevens S, Flavell RA. Innate and adaptive interleukin-22 protects mice from inflammatory bowel disease. Immunity. 2008;29:947–957. doi: 10.1016/j.immuni.2008.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Mahler M, Leiter EH. Genetic and environmental context determines the course of colitis developing in IL-10-deficient mice. Inflamm Bowel Dis. 2002;8:347–355. doi: 10.1097/00054725-200209000-00006. [DOI] [PubMed] [Google Scholar]
- 48.Plevy SE, Gemberling JH, Hsu S, Dorner AJ, Smale ST. Multiple control elements mediate activation of the murine and human interleukin 12 p40 promoters: evidence of functional synergy between C/EBP and Rel proteins. Mol Cell Biol. 1997;17:4572–4588. doi: 10.1128/mcb.17.8.4572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Sanjabi S, Hoffmann A, Liou HC, Baltimore D, Smale ST. Selective requirement for c-Rel during IL-12 P40 gene induction in macrophages. Proceedings of the National Academy of Sciences of the United States of America. 2000;97:12705–12710. doi: 10.1073/pnas.230436397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Sheikh SZ, Matsuoka K, Kobayashi T, Li F, Rubinas T, Plevy SE. Cutting Edge: IFN-{gamma} Is a Negative Regulator of IL-23 in Murine Macrophages and Experimental Colitis. J Immunol. 2010;184:4069–4073. doi: 10.4049/jimmunol.0903600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Sheikh SZ, Kobayashi T, Matsuoka K, Onyiah JC, Plevy SE. Characterization of an interferon-stimulated response element (ISRE) in the Il23a promoter. The Journal of biological chemistry. 2011;286:1174–1180. doi: 10.1074/jbc.M110.147884. [DOI] [PMC free article] [PubMed] [Google Scholar]