Abstract
RifDH10, the dehydratase domain from the terminal module of the rifamycin polyketide synthase, catalyzed the stereospecific syn dehydration of the model substrate (2S,3S)-2-methyl-3-hydroxypentanoyl-RifACP10, resulting in exclusive formation of (E)-2-methyl-2-pentenoyl-RifACP10. RifDH10 did not dehydrate any of the other three diastereomeric, RifACP10-bound, diketide thioester substrates. On the other hand, when EryACP6, from the sixth module of the erythromycin polyketide synthase, was substituted for RifACP10, RifDH10 stereospecifically dehydrated only (2R,3R)-2-methyl-3-hydroxypentanoyl-EryACP6 to give exclusively (E)-2-methyl-2-pentenoyl-EryACP6, with no detectable dehydration of any of the other three diastereomeric, EryACP6-bound, diketides. An identical alteration in substrate diastereospecificity was observed for the corresponding N-acetylcysteamine or pantetheine thioester analogues, regardless of acyl chain length or substitution pattern. Incubation of (2RS)-2-methyl-3-ketopentanoyl-RifACP10 with the didomain reductase-dehydratase RifKR10-RifDH10 yielded (E)-2-methyl-2-pentenoyl-RifACP10, the expected product of syn dehydration of (2S,3S)-2-methyl-3-hydroxypentanoyl-RifACP10, while incubation with the corresponding EryACP6-bound substrate, (2RS)-2-methyl-3-ketopentanoyl-EryACP6, gave only the reduction product (2S,3S)-2-methyl-3-hydroxypentanoyl-EryACP6 with no detectable dehydration. These results establish the intrinsic syn dehydration stereochemistry and substrate diastereoselectivity of RifDH10 and highlight the critical role of the natural RifACP10 domain in chaperoning the proper recognition and processing of the natural ACP-bound undecaketide substrate. The 1.82 Å-resolution structure of RifDH10 revealed the atomic resolution details of the active site and allowed modeling of the syn-dehydration of the (2S,3S)-2-methyl-3-hydroxyacyl-RifACP10 substrate. These results suggest that generation of the characteristic cis double bond of the rifamycins occurs after formation of the full-length RifACP10-bound acyclic trans-unsaturated undecaketide intermediate, most likely during the subsequent macrolactamization catalyzed by the amide synthase RifF.
Polyketides are an enormously diverse group of microbial natural products, many of which are important antibacterial, antifungal, immunosuppressive, and anticancer agents. The multimodular polyketide synthases (PKSs) that assemble complex polyketides are structurally and mechanistically related to the metazoan fatty acid synthases (FASs).1–3 While FASs condense 2-carbon acetate units and reduce newly-formed β-ketoacyl groups in an iterative fashion, multimodular PKSs exploit an assembly-line logic in which 5 to 25 modules (sets of enzymes similar to those in an FAS) each perform a decarboxylative chain-elongation reaction that adds successive malonyl, methylmalonyl, or ethylmalonyl building blocks to the growing polyketide and processing reactions that control the oxidation level and stereochemistry of the substituents attached to the α- and β-positions of the newly extended chain.
Type I PKS modules contain at least three independently folded catalytic domains: 1) an acyltransferase (AT) that selects a unique malonyl-, methylmalonyl-, or ethylmalonyl-CoA chain extender from the cellular acyl-CoA pool, 2) an acyl carrier protein (ACP) that acquires the extender unit from the AT through a flexible, 18-Å phosphopantetheine arm, and 3) a β-ketoacyl-ACP synthase (ketosynthase, KS) that catalyzes the decarboxylative chain-elongation reaction between a growing polyketide transferred to the KS by the upstream module and the ACP-tethered α-carboxyacyl extender unit. (The closely related trans-AT PKS modules lack an integrated AT domain).4 Most modules also contain one or more processing domains, such as a ketoreductase (KR) that stereoselectively catalyzes the NADPH-dependent reduction of the β-keto group and can stereospecifically set the configuration of a neighboring α-substituent, a dehydratase (DH) that eliminates the resulting β-hydroxyl group as well as an α-proton to form an α,β-enoyl double bond, and an enoylreductase (ER) that catalyzes the stereoselective, NADPH-dependent reduction of the double bond. During polyketide assembly, growing chains remain covalently bound through a thioester linkage either to the phosphopantetheinyl thiol of an ACP or, transiently, to the active site cysteine residue of the KS domain, with the relatively mobile ACP domains shuttling the attached substrates among processing domains and to downstream modules. In most PKSs. the release of the fully elongated and processed polyketide is catalyzed by a dedicated thioesterase (TE) domain located at the C-terminal end of the ultimate module in the synthase, resulting in either macrolactonization or hydrolysis.
The vast majority of the more than 2000 known complex polyketides contain one or more double bonds, most of which have trans (E) geometry. A minor but nonetheless significant fraction of complex polyketides also harbor one or more cis (Z) double bonds (Supporting Information, Figure S1). How these cis double bonds are installed in polyketides such as the protein phosphatase inhibitors fostriecin5, 6 and phoslactomycin,7 the anti-angiogenic agent borrelidin,8 the microtubule stabilizer epothilone,9 the microtubule polymerization inhibitor curacin A,10 and the rifamycin family of antibacterials11–13 remains largely unknown. Reynolds and coworkers have recently established that Plm1, the first module of the phoslactomycin PKS, most likely produces cis-3-cyclohexylpropenoate and that the PlmKR1 domain generates a (3S)-hydroxyacyl thioester intermediate that serves as the substrate for the PlmDH1 domain.14, 15
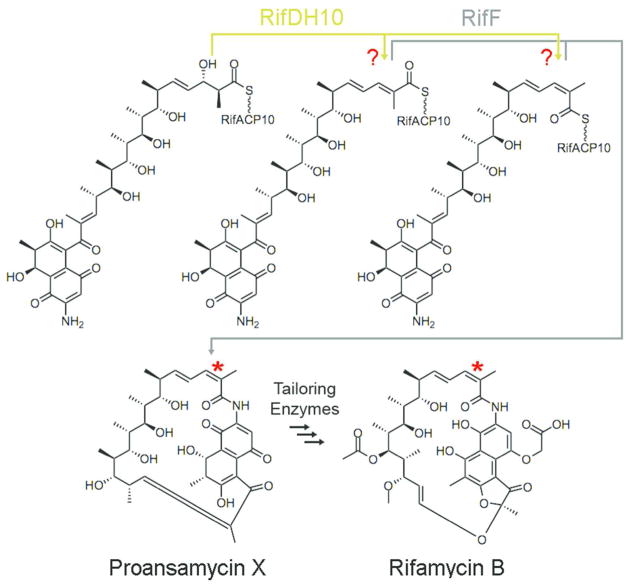
The ansamycin antibiotic rifamycin B (Figure 1) is produced by the actinomycete Amycolatopsis mediterranei. The semisynthetic derivatives rifapentine, rifampicin, and rifabutin inhibit the RNA polymerase of mycobacteria and are clinically prescribed to tuberculosis patients, with rifampicin listed by the World Health Organization as one of the five essential anti-tuberculosis drugs.16 Rifamycins are distinguished from the more common macrolide and polyene polyketides by several unique structural features that include an aminonaphthoquinone, a macrolactam ring, and a trisubstituted cis-double bond.17 The rifamycin biosynthetic gene cluster harbors a 10-module PKS that extends a 3-amino-5-hydroxybenzoate (AHBA) starter unit with 2 acetates and 8 propionates.11–13 The rifamycin PKS lacks the typical TE domain fused to the terminal module 10. Instead, release of the fully-processed, RifACP10-bound undecaketide is catalyzed by the separately-encoded amide synthase, RifF. Although two research teams have independently reported that mutation or deletion of rifF leads to release of acyclic 2-methyl-2-enoyl undecaketides, these investigators reached contradictory conclusions regarding the geometry of the 15,16-double bond (rifamycin numbering) of this abortive product.18–20 The geometry of the double bond in the fully-processed RifACP10-undecaketide intermediate produced by the rifamycin PKS, which serves as the actual substrate for cyclization by amide synthase RifF, therefore remains unsettled, nor is it known whether RifDH10 itself or the cyclase RifF sets the characteristic cis double bond geometry of the rifamycins (Figure 1).
Figure 1.
RifDH10 dehydrates a RifACP10-bound (2S,3S)-2-methyl-3-hydroxyacyl undecaketide generated by the paired ketoreductase domain RifKR10 in the terminal module 10 of the rifamycin PKS. The amide synthase RifF catalyzes the macrolactamization of the acyclic undecaketide product to the rifamycin B precursor, proansamycin X, either before or after isomerization to form the characteristic cis double bond.
Neither sequence nor structural alignments have revealed any features that correlate DH protein sequence with the trans or cis geometry of the double bond in the resultant product.21 For example, the active site of EryDH4, which catalyzes the formation of a trisubstituted trans (E) double bond,22, 23 is structurally equivalent to that established for the DH domains of the curacin PKS subunits CurF and CurH, one of which is thought to generate a cis double bond.10 DH-containing modules that generate trans 3double bonds are often paired with KR domains that produce 3R-hydroxyacyl products. Such KR domains are readily recognized by their characteristic amino acid sequence fingerprints.24–27 On the other hand, there is only limited circumstantial evidence correlating KR domains that generate 3S-hydroxyacyl intermediates with the generation of cis double bonds by their paired DH domains.21 For example, KR domains may provide 3S-hydroxyacyl substrates for the DH domains of modules 2 and 3 of the fostriecin PKS that generate cis double bond diketide and triketide intermediates,6 as well as for the DH domains of modules 1 and 2 of the phoslactomycin PKS,14, 15, 28 module 4 of the epothilone PKS,29 the CurG module of the curacin PKS,10 and module 10 of the rifamycin PKS.11–13 Whether modules that contain both an A-type KR and a DH actually generate cis double bonds, however, is still largely untested, except in the above-mentioned case of phoslactomycin module 1.14, 15 As a further complication, the formation of some cis double bonds can involve post-PKS transformations such as dehydration7 or double-bond isomerization.30
We have recently established that RifKR10, the KR domain from the tenth module of the rifamycin PKS, as well as RifKR7 from the seventh module of the same PKS, each mediate the stereospecific epimerization/reduction of (2R)-2-methyl-3-ketoacyl-ACP substrates to give exclusively (2S,3S)-2-methyl-3-hydroxyacyl-ACP products.31 The RifKR10-generated product must serve as the substrate for the paired dehydratase domain of module 10, RifDH10. Although the (3S)-configuration of this 3-hydroxyacyl substrate might superficially suggest the formation of a cis-2-methyl-2-enoyl-ACP product, this geometry cannot be generated by syn dehydration of a (2S,3S)-2-methyl-3-hydroxyacyl intermediate. In fact, all DH domains that have been stereochemically characterized to date, including EryDH4 from the erythromycin synthase,23 NanDH2 from the nanchangmycin synthase,32 and TylDH2 from the tylactone synthase,27 display strict specificity for the diastereomeric (2R,3R)-2-methyl-3-hydroxyacyl-ACP substrates and catalyze completely stereospecific syn dehydration to give the corresponding trans-(E)-2-methyl-2-enoyl-ACP products. In light of not only its unusual substrate stereochemistry but also the uncertainty in the actual cis or trans geometry of its natural undecaketide product, the RifDH10-catalyzed dehydration reaction presents an intriguing and important mechanistic, stereochemical, and biosynthetic puzzle.
We now report the expression and purification of the recombinant RifDH10 domain, the determination of its protein structure to 1.82 Å resolution, and the demonstration that RifDH10 catalyzes the diastereospecific syn dehydration of (2S,3S)-2-methyl-3-hydroxypentanoyl-RifACP10 to give exclusively the corresponding trans-(E)-2-methyl-2-pentenoyl thioester product. Unexpectedly, the natural diastereospecificity of RifDH10 is reversed when the dehydratase is incubated with thioester substrates that are tethered to the non-cognate ACP domain EryACP6 or to the corresponding pantetheinyl or N-acetylcysteamine (NAC) analogues, resulting in the anomalous syn dehydration of only the diastereomeric (2R,3R)-2-methyl-3-hydroxypentanoyl thioesters to give the corresponding (E)-2-methyl-2-pentenoyl thioester products.
Experimental Procedures
Materials
Isopropylthio-β-D-galactopyranoside (IPTG) was purchased from Invitrogen. All other chemical reagents were purchased from Sigma-Aldrich and utilized without further purification. Ni-NTA affinity resin was purchased from Qiagen. Amicon Ultra Centrifugal Filter Units (Amicon Ultra-15, 30,000 MWCO) were purchased from Millipore. DNA primers were synthesized by Integrated DNA Technologies. Recombinant Ery[KS6][AT6], EryACP6, RifKR7, RifKR10, TylKR1, EryKR1, EryKR6, PicTE, and Sfp were each expressed and purified as previously described.31, 33–35 Reference standards for chiral GC-MS analysis of methyl (2S,3S)-2-methyl-3-hydroxypentanoate (4a), methyl (2R,3R)-2-methyl-3-hydroxypentanoate (4b), methyl (2R,3S)-2-methyl-3-hydroxypentanoate (4c), and methyl (2S,3R)-2-methyl-3-hydroxypentanoate (4d), were each prepared synthetically or chemoenzymatically as previously described.31, 36 (E)-2-Methyl-2-pentenoic acid (3) was purchased from Sigma-Aldrich and used to prepare (Z)-2-methyl-2-pentenoic acid as previously described.23 Propionyl-N-acetylcysteamine thioester and (2S,3R)-2-methyl-3-hydroxy-pentanoyl-SNAC was prepared as described.36 The synthesis of acyl-S-NAC and acyl-S-pantetheine thioester substrates and products is described in the Supporting Information. (2RS)-2-Methyl-3-keto-pentanoyl-CoA was prepared by Dr. Lynne Collett Pilcher as previously described,31, 37–39 purified by reverse phase C-18 HPLC, and characterized by 1H NMR and ESI-MS. Synthetic genes encoding recombinant domains RifDH10 and RifACP10, optimized for expression in E. coli, were designed and prepared by DNA2.0 and supplied in the vector pJ201.
Methods
General methods were as previously described.36, 40 Growth media and conditions used for E. coli and standard methods for handling E. coli in vivo and in vitro were those described previously, unless otherwise noted.40 All DNA manipulations were performed following standard procedures.40 DNA sequencing was carried out at the U. C. Davis Sequencing Facility, Davis, CA. All proteins were handled at 4 °C unless otherwise stated. Protein concentrations were determined according to the method of Bradford,41 using Hewlett Packard 8452A Diode Array or Thermo Evolution Array UV/Vis spectrophotometers with bovine serum albumin as the standard. SDS-PAGE and DNA gels were imaged and analyzed with a Bio-Rad ChemiDoc MP System. A Bio-Rad FX-Pro Plus Molecular Imager was utilized for radio-TLC analysis. GC-MS analysis was performed on a GC–MS Hewlett-Packard Series 2 GC-MSD, 70 eV EI in positive ion mode using a capillary CP-Chirasil-Dex CB column (25 m × 0.32 mm) from Agilent Technologies. For resolution and analysis of (E)-2-methyl-2-pentenoic acid (3) and (Z)-2-methyl-2-pentenoic acid, three different programs were used: Method 1 - initial oven temperature of 50 °C for 1 min, increase at 7.5 °C/min up to 200 °C; Method 2 - initial oven temperature of 50 °C for 1 min, increase at 15 °C/min up to 200 °C; Method 3 - initial oven temperature of 65 °C for 1 min, increase at 0.5 °C/min up to 100 °C, then 20 °C/min up to a final temperature 200 °C. For resolution and analysis of the 4 diastereomers of methyl 2-methyl-3-hydroxypentanoate (4) the following program was used: Method 4 – initial oven temperature of 50 °C for 1 min, increase at 1 °C/min up to 90 °C, followed by 20 °C/min up to 200 °C. A Thermo LXQ equipped with Surveyor HPLC system and Waters Symmetry C18 column (2.1 mm × 50 mm, 3.5 μm) was utilized for HPLC-ESI-MS analysis in positive ion mode.
General cloning strategy
The design of rifamycin module 10 domain boundaries for cloning of RifDH10, RifDH10-RifKR10, and RifACP10-NusA is described in the Supporting Information (Figures S3–S5). Synthetic genes encoding RifDH10, RifDH10-RifKR10, and RifACP10-NusA,42 optimized for expression in E. coli and flanked by suitable restriction sites, were ligated into pET28a and the resultant plasmids were used to transform the expression host E. coli BL21(DE3).
RifDH10 expression and purification
The synthetic gene for RifDH10, flanked by 5′-NdeI and 3′-XhoI sites, was ligated into pET28a. E. coli BL21(DE3) transformed with the RifDH10 expression plasmid was inoculated into LB media containing 50 mg/L kanamycin at 37 °C, grown to OD600 0.4, and induced with 0.5 mM IPTG. After 12 h at 15 °C, cells were collected by centrifugation and resuspended in lysis buffer (0.5 M NaCl, 10% (v/v) glycerol, 0.1 M HEPES, pH 7.5). Following sonication, cell debris was removed by centrifugation (30,000g for 30 min). The supernatant was poured over a column of Nickel-NTA resin (Thermoscientific), which was then washed with 50 mL lysis buffer containing 15 mM imidazole and eluted with 5 mL lysis buffer containing 150 mM imidazole. RifDH10 was further purified using a Superdex 200 gel filtration column (GE Healthcare Life Sciences) equilibrated with 150 mM NaCl, 10 mM HEPES, pH 7.5. The eluted protein (~100 mg of purified RifDH10 from 2 L of culture) was concentrated to 15 mg/mL in the equilibration buffer and stored at −80 °C until needed. SDS-PAGE analysis of purified His6RifDH10 protein showed an Mr 34,900 (calc MW 34,109 Da) (Figure S6).
apo-RifACP10-NusA fusion protein
The synthetic gene for RifACP10, flanked by 5′-NdeI and 3′-XhoI sites, was initially ligated into the NdeI/XhoI sites of pET28a. Since the resultant recombinant RifACP10 was obtained only as insoluble inclusion bodies when expressed in E. coli BL21(DE3), we constructed the corresponding RifACP10-NusA protein. The synthetic DNA encoding RifACP10 was PCR-amplified from the RifACP10 construct using the primer pair pET28a-FP (5′-ATGGGCAGCAGCCATCATCAT-3′) and RifACP10-EcoRI-RP (5′-TATTTAAAATTCAGAATTCCAGCAGCTCATTCAAGTG-TGC-3′; EcoRI site in italics). The resultant amplified DNA was digested with NdeI and EcoRI and ligated into the previously described plasmid,31 RifKR10-NusA–pET28, that had been digested with NdeI and EcoRI so as to excise the DNA segment encoding RifKR10 and to replace it with DNA encoding RifACP10, thus giving apo-RifACP10-NusA-pET28 which was used to transform E. coli BL21(DE3). Recombinant His6-tag-apo-RifACP10-NusA, harboring an HRV 3C protease cleavage site between the RifACP10 and the C-terminal NusA, was expressed in soluble form by growth in Terrific Broth or in Super Broth until an OD600 of 0.4–0.8. Protein expression was induced by addition of 0.2 mM IPTG and the cell culture was continuously grown for additional 16–18 h at 18 °C overnight. The cells were harvested by centrifugation at 4,200g for 20 min and the cell pellet was collected and stored at −80 °C. The frozen cells were thawed at RT and dissolved and lysed on ice for 30 min in starting buffer (500 mM NaCl, 50 mM phosphate, pH 7.8) containing 1 mg/ml of lysozyme, followed by sonication. Cell supernatant and the pellet were separated by centrifugation at 23,000g for 30 min and the supernatant was loaded on a pre-charged Ni-NTA column. The column was washed with starting buffer and then washing buffer (300 mM NaCl, 50 mM phosphate, pH 7.6, 10 mM imidazole) and proteins were eluted from the Ni-column by elution buffer (150 mM NaCl, 50 mM phosphate, pH 7.5, 150 mM imidazole, 10 % glycerol). The eluted fractions were collected, concentrated with an Amicon MWCO 30,000 filter, and buffer was exchanged with a PD-10 column to 150 mM NaCl, 50 mM phosphate, pH 7.5, 10 % glycerol, yielding ~200 mg of His6-tag-apo-RifACP10-NusA per 2 L culture. SDS-PAGE analysis of purified His6-tag-apo-RifACP10-NusA protein showed an Mr 70,900 (calc MW 68,955 Da) (Figure S6).
RifDH10-KR10 fusion protein
DNA encoding the synthetic gene for RifKR10 was amplified from the previously described expression construct31 using the primer pair RifKR10-NdeI-BamHI-FP (5′-ATTTTCGAATTACATATGATGAATTTTAAAGGATCCCTGTACCGCGTCGACTG-3′; NdeI site in bold and BamHI site in italics) and pET-28a XhoI RP (5′-TCGGGCTTTGTTAGCAGC-3′). The amplified DNA fragment was digested with NdeI and XhoI followed by ligation into pET-28a vector to give RifKR10-NdeI-BamHI-pET28a. DNA encoding RifDH10 was amplified from the above-described expression construct using primer pair RifDH10-PAGA-BamHI-RP (5′-TAATTCGAAAATGGATCCTGCACCCGCTGGACCCGCTGCAGTGGTC-3′; BamHI site in italics) and pET28a-FP (5′-ATGGGCAGCAGCCATCATCAT-3′) followed by DNA restriction by NdeI and BamHI. The resultant DNA fragment was ligated into NdeI/BamHI-digested plasmid RifKR10-NdeI-BamHI-pET28a. The His6-tag-RifDH10-KR10 was expressed in E. coli BL21(DE3) and purified by the same procedures used to express the His6-tag-apo-RifACP10-NusA protein, then further purified by size exclusion chromatography, yielding 16 mg of partially purified His6-tag-RifDH10-KR10 didomain protein per 2 L culture. SDS-PAGE analysis of partially purified His6-tag-RifDH10-KR10 protein gave Mr 80,100 (calc MW 80,150 Da) (Figure S6).
Incubation of RifDH10 with (2S,3S)-2-methyl-3-hydroxypentanoyl-RifACP10-NusA
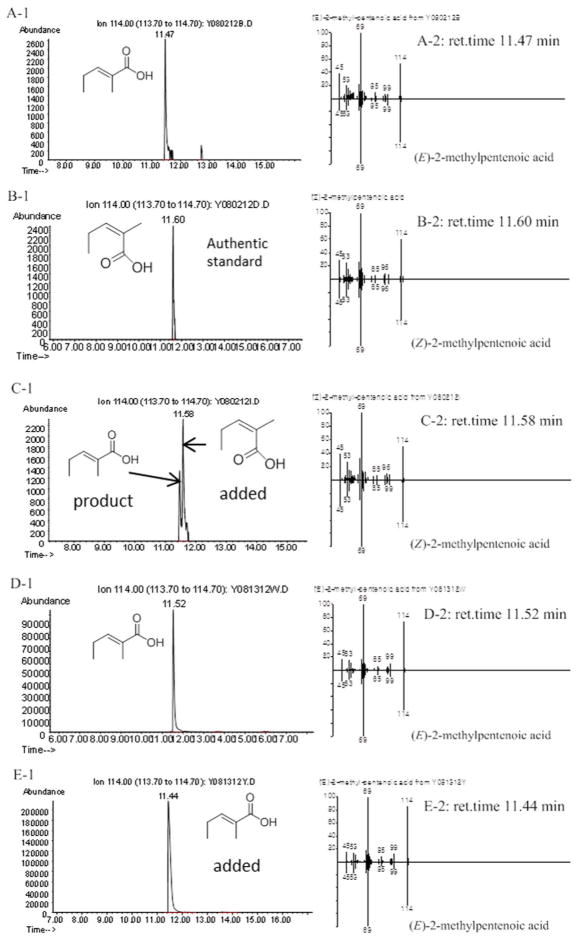
A mixture of 500 μM (2RS)-2-methyl-3-ketopentanoyl-CoA, 40 μM Sfp (surfactin phosphopantetheinyl transferase),43 and 400 μM apo-RifACP10-NusA were added to the reaction buffer (150 mM NaCl, 50 mM NaH2PO4, pH 7.2) containing 20 mM MgCl2 and 4 mM DTT. After incubation at 37 °C for 1 h, 300 μM RifKR7, 300 μM RifDH10 and 2 mM NADPH were added and the mixture was incubated for an additional 1 h at RT. The final reaction volume was 500 μL. The RifACP10-NusA was hydrolytically released by incubation with 100 μM PicTE for 10 min at RT. The reaction mixtures were acidified with 1 M HCl to pH < 3, and extracted with ethyl acetate. After solvent evaporation, the product was dissolved in 100 μL methanol and unsaturated diketide was analyzed by chiral GC-MS and direct comparison with authentic standards of (E)-2-methyl-2-pentenoic acid and (Z)-2-methyl-2-pentenoic acid, using both Methods 1 and 2 (Figure 3).23 In control experiments carried out in the absence of RifDH10, 20 μL TMS-diazomethane was added to the hydrolyzed organic extract and the derived diketide methyl ester was analyzed by chiral GC-MS and direct comparison with authentic methyl (2S,3S)-2-methyl-3-hydroxypentanoate, using Method 4 (Figure S7). As previously reported, direct comparison with authentic standards of 4a–4b was essential since the observed retention time on the chiral column varied slightly from run to run, although the order of elution never changed.31, 36
Figure 3.
Chiral GC-MS analysis (Method 1) of the incubation of (2RS)-2-methyl-3-ketopentanoyl-RifACP10-NusA with RifKR7 and RifDH10. A and D: (E)-2-Methyl-2-pentenoic acid (3) from RifKR7-catalyzed reduction of (2RS)-2-methyl-3-ketopentanoyl-RifACP10-NusA followed by dehydration by RifDH10. B: (Z)-2-methylpentenoic acid authentic standard. C: Co-injection of A with B. E: Co-injection of (E)-2-methyl-2-pentenoic acid standard with D. A-1, B-1, C-1, D-1 and E-1: Extracted ion current (XIC) at m/z 114 (base peak). A-2, B-2, C-2, D-2 and E-2: mass spectra of selected peak, upper half, observed spectrum, lower half, inverted mass spectrum of reference standard.
Incubations with TylKR1, EryKR6, and EryKR1 in place of RifKR7 were also carried out so as to generate the corresponding RifACP10-bound intermediates (2R,3R)-1b, (2R,3S)-1c, and (2S,3R)-1d. The derived hydrolysis products were analyzed by GC-MS and direct comparison with authentic standards of each diastereomer of methyl 2-methyl-3-hydroxypentanoate (4a–4d) (Method 4) and both (E)-2-methyl-2-pentenoic acid (3) and (Z)-2-methyl-2-pentenoic acid (Method 2), as described for the incubations using RifKR7 (Figures S8–S10).
Incubation of recombinant RifDH10-RifKR10 didomain with chemoenzymatically generated (2RS)-2-methyl-3-ketopentanoyl-RifACP10
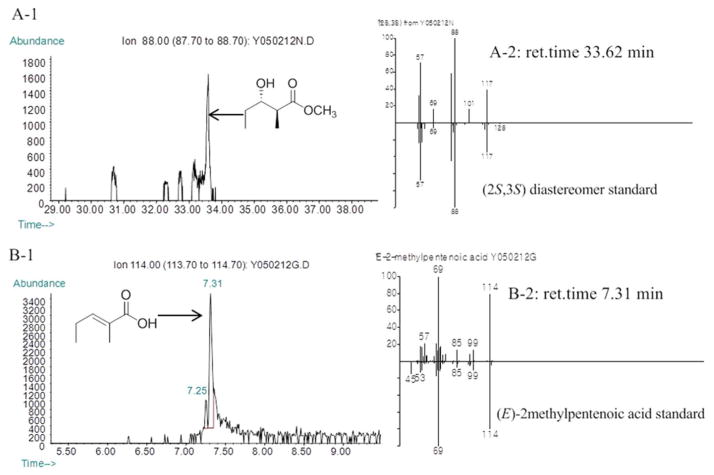
A mixture of 500 μM (2RS)-2-methyl-3-keto-pentanoyl-CoA, 40 μM Sfp, and 400 μM apo-RifACP10-NusA was added to the reaction buffer (150 mM NaCl, 50 mM NaH2PO4, pH 7.2) containing 20 mM MgCl2 and 4 mM DTT. After incubation at 37 °C for 1 h, 300 μM RifDH10-RifKR10 and 2 mM NADPH were added and the mixture was incubated for an additional 1 h at RT. The final reaction volume of each assay was 500 μL. The diketide acid was hydrolytically released from RifACP10-NusA by incubation with 100 μM PicTE for 10 min at RT. The reaction mixtures were acidified with 1 M HCl to pH < 3, and extracted with ethyl acetate. After solvent evaporation, the product was dissolved in 100 μL methanol and the formation of unsaturated (E)-2-methyl-2-pentenoic acid was confirmed by GC-MS (Method 2) and direct comparison with authentic standards of 3 and (Z)-2-methyl-2-pentenoic acid. For the detection of reduced diketide product, 20 μL TMS-diazomethane was added and the derived diketide methyl ester, methyl (2S,3S)-2-methyl-3-hydroxypentanoate (4a), was identified by chiral GC-MS (Method 4) and direct comparison with authentic standards (Figure 4).
Figure 4.
Chiral GC-MS analysis of the incubation of (2RS)-2-methyl-3-ketopentanoyl-RifACP10-NusA with RifDH10-KR10 (Methods 2 and 4). Left panels: A1. Extracted ion current (XIC) at m/z 88 (base peak for 4a) (Method 4); B-1 and C-1. Extracted ion current (XIC) at m/z 114 (base peak for 3) (Method 2). Right panels, mass spectra of selected peaks, upper half, observed spectrum, lower half, inverted mass spectrum of reference standard. A. (2S,3S)-4a produced by RifDH10-KR10. B. (E)-2-Methylpentenoic acid (3) produced by RifDH10-KR10.
Incubation of recombinant RifDH10 with chemoenzymatically generated (E)-2-methyl-2-pentenoyl-RifACP10 and with (E)-2-methylpentenoyl-EryACP6
A mixture of 500 μM (E)-2-methyl-2-pentenoyl-CoA, 40 μM Sfp, and either 400 μM apo-RifACP10-NusA or 200 μM apo-EryACP6 was added to the reaction buffer (150 mM NaCl, 50 mM NaH2PO4, pH 7.2) containing 20 mM MgCl2 and 4 mM DTT. After incubation at 37 °C for 1 h, 300 μM RifDH10 was added and the mixture was incubated for an additional 1 h at RT. The final reaction volume of each assay was 500 μL. The RifACP10-NusA or EryACP6 was hydrolytically released by incubation with 100 μM PicTE for 10 min at RT. The reaction mixtures were acidified with 1 M HCl to pH <3, and extracted with ethyl acetate. After solvent evaporation, the product was dissolved in 100 μL methanol and 20 μL TMS-diazomethane was added. The derived diketide methyl esters were analyzed as described above by chiral GC-MS and direct comparison with authentic standards (Figures 5 [RifACP10] and S16 [EryACP6]). In a control incubation without added RifDH10 domain, the chemoenzymatically-generated 2-methyl-2-pentenoyl-RifACP10-NusA was treated with HRV 3C protease to remove the C-terminal NusA and the MW of the resulting 2-methyl-2-pentenoyl-RifACP10 was verified by LC-ESI(+)-MS, which gave an observed MW of 13144 Da (calcd for [M+H]+ ion of 2-methyl-2-pentenoyl-RifACP10, 13143.9) (Figure S11). LC-ESI(+)-MS was also used to confirm the formation of 2-methyl-2-pentenoyl-ACP6 (6), MW (observed) 11728 (calc. 11727.7 Da), compared to the peak for holo-eryACP6, MW (observed) 11632 (calc. 11631.6 Da), with the difference of 96 amu matching that expected for addition of the 2-methyl-2-pentenoyl moiety. Additional control incubations established that the isomeric (Z)-2-methylpentenoyl-EryACP6 was not a substrate for RifDH10.
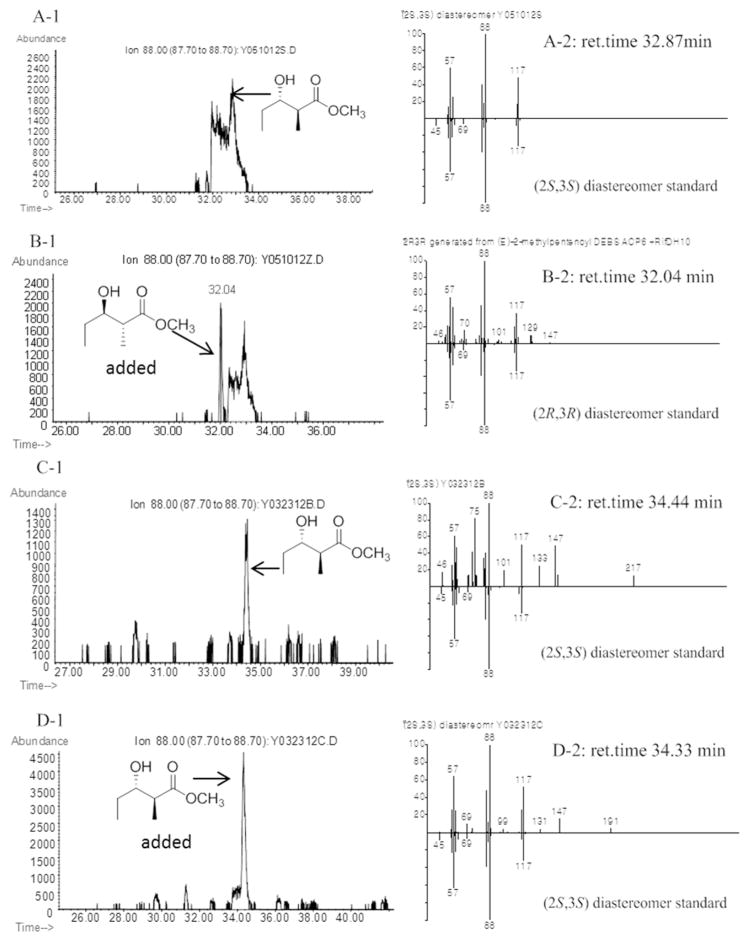
Figure 5.
Chiral GC-MS analysis (Method 4) of the incubation of (E)-2-methyl-2-pentenoyl-RifACP10-NusA with RifDH10. Left panels: Extracted ion current (XIC) at m/z 88 (base peak). Right panels, mass spectra of selected peaks corresponding to diastereomers of methyl 2-methyl-3-hydroxypentanoate, upper half, observed spectrum, lower half, inverted mass spectrum of reference standard. A and C. (2S,3S)-4a generated by RifDH10. B. A plus (2R,3R)-4b. D. C plus (2S,3S)-4a. Detailed analysis of the full mass spectra of all peaks detected in Panel A1 between 31.00 and 34.00 min showed only the presence of the single (2S,3S)-4a.
TLC-phosphorimaging assay of the incubation of recombinant RifDH10 with reconstituted Ery[KS6][AT6], EryACP6, and recombinant KR domains
Radio-TLC assay of the conversion of propionyl-SNAC to 2-methyl-3-hydroxypentanoic acids and 2-methyl-2-pentenoic acid by coupled incubation with reconstituted Ery[KS6][AT6] and EryACP6 in combination with EryKR6, TylKR1, EryKR1, or RifKR7 and RifDH10 was carried out as previously described.23, 31, 36 For each incubation, 5 mM propionyl-SNAC and 40 μM Ery[KS6][AT6] were added to the reaction buffer (50 mM NaH2PO4, pH 7.2) containing 2.5 mM TCEP and the mixture was incubated at RT. After 1 h, 200 μM of the holo-EryACP6, 300 μM methylmalonylCoA containing a trace amount of [2-14C]-methylmalonylCoA, 2 mM NADPH and 300 μM of the individual KR domains both with or without 300 μM RifDH10 were added to the reaction mixture which was then incubated for an additional 1 h. The final reaction volume of each assay was 125 μL. The reaction was quenched and the diketide acids were hydrolytically released from EryACP6 by addition of 0.5 M NaOH (40 % by volume) followed by incubation at 65 °C for 20 min. After adjustment to pH <3 with 1 M HCl and extraction with ethyl acetate, the concentrated organic extracts were dissolved in 15 μL ethyl acetate and spotted on a silica gel TLC plate, which was developed with 1:9 dichloromethane:ethyl acetate containing 0.1% AcOH. The dried TLC plate was then visualized by phosphorimaging over 48 h (Figure S12).
Incubation of recombinant RifDH10 or RifDH10-RifKR10 didomain with reconstituted Ery[KS6][AT6], EryACP6, and TylKR1
The incubation procedures for the in situ generation of (2R,3R)-2-methyl-3-hydroxyacyl-EryACP6 derivatives were based on those previously described.31, 36 In a typical assay, 5 mM propionyl-SNAC or 5 mM (2S,3R)-2-methyl-3-hydroxy-pentanoyl-SNAC was pre-incubated with 40 μM Ery[KS6][AT6] in 50 mM NaH2PO4, pH 7.2 containing 2.5 mM tris-2-carboxyethylphosphine (TCEP) at RT. After 1 h, 200 μM of holo-EryACP6, 300 μM methylmalonylCoA, 2 mM NADPH, 300 μM recombinant TylKR1, and 300 μM recombinant RifDH10 were added to the reaction mixture which was then incubated for an additional 1 h (Figure S13). In a second experiment, 300 μM RifDH10-KR10 didomain was added instead of the TylKR1 and RifDH10 pair (Figure S14). The final reaction volume of each assay was 500 μL. The reaction was quenched and EryACP6 was hydrolytically released from the product by addition of 0.5 M NaOH (40 % by volume) and incubation at 65 °C for 20 min. Alternatively, EryACP6 was enzymatically released by incubation with 100 μM PicTE for 10 min at RT. The reaction mixtures were acidified with 1 M HCl to pH < 3, and then extracted with ethyl acetate. After solvent evaporation, the product was dissolved in 100 μL of ethyl acetate or methanol and analyzed by chiral GC/MS (Method 3) (Figures S13 and S14). TMS-diazomethane (20 μL) was added to convert recovered 2-methyl-3-hydroxypentanoates to the derived 2-methyl-3-hydroxypentanoate methyl esters which were analyzed by chiral GC-MS and direct comparison with synthetic standards (Method 4).36 For the GC-MS analysis of methyl (2E,4R,5R)-2,4-dimethyl-5-hydroxy-2-heptenoate, the temperature program of Method 3 was used (Figure S15).
Incubation of RifDH10 with acyl-S-NAC or pantetheine thioesters
RifDH10 (30 μM) was incubated overnight with 10 mM of each of the chemoenzymatically prepared 2-methyl-3-hydroxyacyl or 2,3-unsaturated enoyl-S-NAC or pantetheine thioester analogues in a 25 μL solution containing 150 mM NaCl, 10% (v/v) glycerol, 150 mM HEPES pH 7.5. After 16 h the reaction mixture was injected onto a C18 reversed-phase HPLC column and monitored at λ 200–600 nm (100% water with 0.1% TFA to 100% MeOH with 0.1% TFA, 30 min). All products were characterized by mass spectrometry and comparison with authentic samples as appropriate (Figures S17–S24).
RifDH10 catalyzed dehydration of (2R,3R)-2-methyl-3-hydroxy-butanoyl-S-pantetheine and subsequent acyl transthioesterification to NAC
trans-2-Methyl-2-butenoyl-S-pantetheine (11) (20 mg) was incubated overnight in 150 mM NaCl, 150 mM HEPES pH 7.5, 10% (v/v) glycerol, and 1 mg/mL RifDH10 in a total volume of 1 mL at 22 °C. After 16 h the reaction mixture was injected onto a C18 reversed-phase HPLC column and monitored at λ 200–600 nm (100% water with 0.1% TFA to 100% MeOH with 0.1% TFA, 15 min). A peak unobserved in a control reaction without RifDH10 with λmax 232 nm was collected and concentrated. The concentrated fractions yielded 5 mg of the hydrated product, (2R,3R)-2-methyl-3-hydroxybutanoyl-S-pantetheine (12b). To confirm its stereochemistry, 3 mg of (2R,3R)-2-methyl-3-hydroxybutanoyl-S-pantetheine (12b) was resuspended in 5 mL of 50 mM NaHCO3 (aq.) and 50 mM of NAC and stirred overnight. The reaction mixture was injected on a C18 reversed-phase HPLC column, and monitored at λ 200–600 nm (100% water with 0.1% TFA to 100% MeOH with 0.1% TFA, 15 min). A peak unobserved in a control reaction without NAC with λmax 232 nm was collected and concentrated. The compound ran identically to authentic (2R,3R)-2-methyl-3-hydroxybutanoyl-S-NAC (9b) on a ChiralCel OC-H column (250 × 4.6 mm) on a Beckman Coulter HPLC system equipped with a 20 μL loop with an isocratic flow rate of 0.6 mL/min using 1:24 ethanol/hexanes as the mobile phase (Figure S24).44 Compounds were observed by 235 nm UV-absorbance. Synthetic standards, prepared as previously described, were provided by Shawn Piasecki.44
Crystallization and structure determination
Crystals of RifDH10 grew over a period of 2 days to 1 week by sitting drop vapor diffusion at 22 °C. Drops were formed by mixing 3 μL protein solution (15 mg/mL RifDH10, 150 mM NaCl, 10 mM HEPES pH 7.5) with 1 μL crystallization buffer (1.1 M sodium citrate, 0.1 M HEPES pH 6.8). Crystals were soaked briefly in crystallization buffer with 15% (v/v) glycerol before being frozen in liquid nitrogen. The diffraction data, collected at ALS Beamline 5.0.2, were processed by iMosflm and scaled with SCALA from the CCP4 suite.45 The structure was solved to 1.82 Å resolution by molecular replacement with PhaserMR46 using EryDH4 (PDB code: 3EL6) as the search model.21 The model generated from the molecular replacement solution was refined with Coot47 and Refmac5.48 Aside from the N- and C-termini, only loop residues 232–242 did not show in the electron density maps.
Results
Expression and purification of RifDH10 and RifACP10
To investigate the mechanism and structure of RifDH10, we used a synthetic gene with codons optimized for expression in E. coli to express the RifDH10 domain as a discrete recombinant protein corresponding to the region from A2569 to G2872 of RifE (EMBL AAC01714.1; UniProt ID O54593; residues herein numbered 1–304), based on alignment with the previously described standalone EryDH4 domain21, 23 and flanked by the deduced C-terminal and N-terminal boundaries, respectively, of the RifAT10 and RifKR10 domains12, 13 (Figures S3 and S4). After ligation of the synthetic gene into pET28a and transformation into E. coli BL21(DE3), the expressed recombinant RifDH10 carrying an N-terminal His6-tag was purified by Ni(II)-affinity chromatography and analyzed for purity and Mr by SDS-PAGE (Figure S6). Attempted expression of recombinant RifACP10 corresponding to the region from L3324 to L3409 of RifE, using a synthetic RifACP10 gene with codons optimized for expression in E. coli (Figure S5), gave only insoluble inclusion bodies. Instead, apo-RifACP10-NusA (hereafter referred to simply as apo-RifACP10) could be obtained as a soluble fusion protein carrying an N-terminal His6-tag and a C-terminal NusA.42. The resultant protein was readily purified by Ni(II)-affinity chromatography (Figure S6).
Stereochemistry of RifDH10-catalyzed dehydration of 2-methyl-3-hydroxyacyl-RifACP10
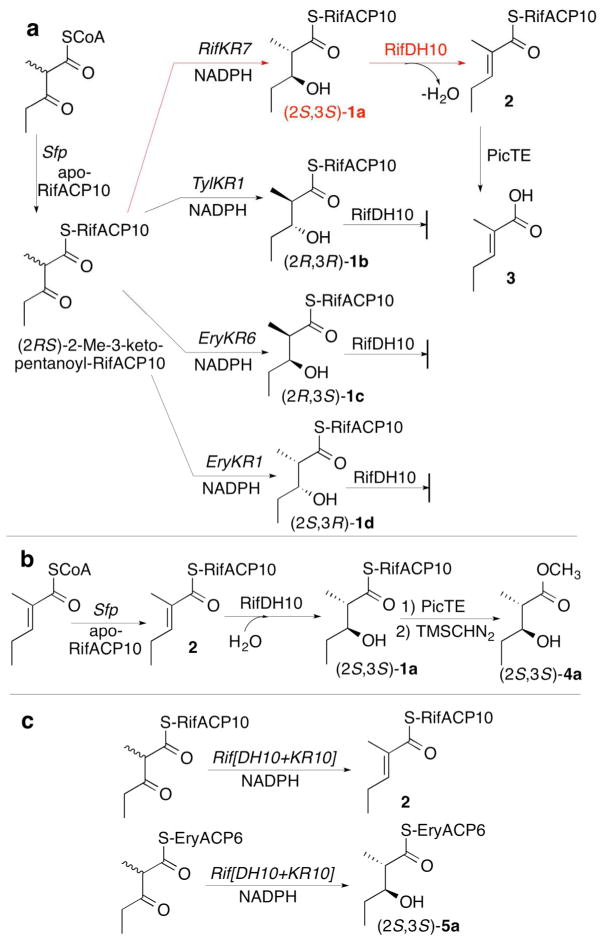
We have recently reported that (2S,3S)-2-methyl-3-hydroxypentanoyl-RifACP10 (1a) can be prepared by stereospecific reduction of (2RS)-2-methyl-3-ketopentanoyl-RifACP10 with either RifKR7 or RifKR10 in the presence of NADPH (Figures 2a and S7).31 The requisite RifACP10-bound 3-keto acylthioester was generated in situ by chemoenzymatic incubation of (2RS)-2-methyl-3-ketopentanoyl-CoA with apo-RifACP10 and the phosphopantetheinyl transferase Sfp.31 Incubation of (2S,3S)-1a with RifDH10 gave as the exclusive dehydration product (E)-2-methyl-2-pentenoyl-RifACP10 (2), as established by enzymatic hydrolysis with PicTE, the thioesterase domain from the picromycin synthase, followed by GC-MS analysis and direct comparison of the derived (E)-2-methyl-2-pentenoate (3) with an authentic sample of 3, as well as with the isomeric (Z)-2-methyl-2-pentenoic acid (Figures 2a and 3).23 Omission of RifDH10 resulted in exclusive formation of (2S,3S)-1a, as confirmed by PicTE-catalyzed hydrolysis and chiral GC-MS analysis of the derived methyl ester (2S,3S)-4a, which was identical in both retention time and mass spectrum by direct comparison with authentic 4a, as previously observed (Figure S7).31 By contrast, incubations of RifDH10 under identical conditions with any of the chemoenzymatically prepared diastereomeric substrates, (2R,3R)-1b (TylKR1), (2R,3S)-1c (EryKR6), or (2S,3R)-1d (EryKR1),27, 36, 49 resulted at most in generation of only very minor quantities (<5–10%) of 3, most likely due to the previously reported31 generation of minor amounts of contaminating (2S,3S)-1a by TylKR1 and EryKR1 upon incubation with (2RS)-2-methyl-3-ketopentanoyl-RifACP10 (Figures S8–S10).
Figure 2.
RifDH10-catalyzed dehydration/hydration of RifACP10-bound substrates a) Dehydration of (2S,3S)-2-methyl-3-hydroxypentanoyl-RifACP10 (1a) b) Hydration of (E)-2-methyl-2-pentenoyl-RifACP10 (2). c) Incubation of Rif[DH10+KR10] with RifACP10- and EryACP6-bound 2-methyl-3-ketoacyl thioesters.
We have previously established that RifKR10 has identical stereospecificity to RifKR7, reducing (2RS)-2-methyl-3-ketopentanoyl-RifACP10 to the corresponding (2S,3S)-2-methyl-3-hydroxypentanoyl-RifACP10 (1a).31 To confirm that the observed stereochemistry of the coupled RifDH10-catalyzed dehydration reaction is not influenced by the origin of the (2S,3S)-specific ketoreductase domain, we prepared a RifDH10-RifKR10 fusion protein. Incubation of Rif[DH][KR10] with (2RS)-2-methyl-3-ketopentanoyl-RifACP10 and NADPH gave (E)-2-methyl-2-pentenoyl-RifACP10 (2), as established by GC-MS analysis of the derived (E)-2-methyl-2-pentenoic acid (3) (Figures 2c and 4). Analysis of the methyl ester derived from the reduced diketide intermediate 1a confirmed the expected formation of exclusively (2S,3S)-4a.
Syn-stereochemistry of RifDH10-catalyzed hydration of (E)-2-methyl-2-pentenoyl-RifACP10
In a complementary experiment, we also investigated the reverse RifDH10-catalyzed hydration of (E)-2-methyl-2-pentenoyl-RifACP10 (2). Incubation of chemoenzymatically prepared (E)-2-methyl-2-pentenoyl-RifACP10 (2) resulted in stereospecific syn-hydration to give exclusively (2S,3S)-2-methyl-3-hydroxypentanoyl-RifACP10 (1a), as confirmed by PicTE hydrolysis, methylation, and chiral GC-MS detection of methyl (2S,3S)-4a (Figures 2b and 5).
Stereochemistry of RifDH10-catalyzed dehydration of 2-methyl-3-hydroxyacyl thioester analogues
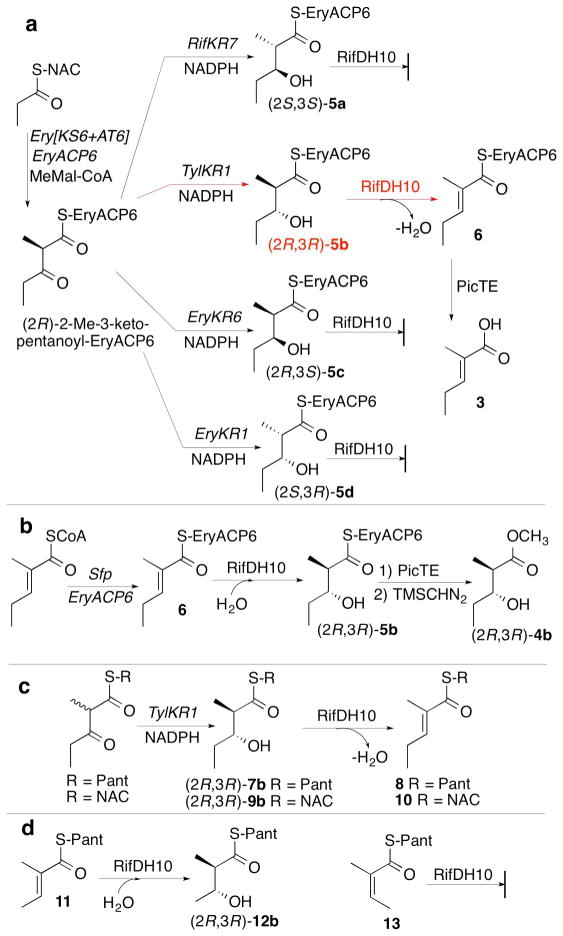
Although these results are all congruent with the previously established (2S,3S) stereospecificity of RifKR10,31 which generates the natural substrate for RifDH10, the observed syn dehydration of a (2S,3S)-2-methyl-3-hydroxyacyl-ACP substrate by RifDH10 to give the corresponding (E)-2-methyl-2-enoylacyl-ACP product is biochemically unprecedented. Thus all other mechanistically characterized DH domains, catalyze syn dehydration of only the diastereomeric (2R,3R)-2-methyl-3-hydroxyacyl-ACP substrates to give (E)-2-methyl-2-enoyl-ACP products,23, 32 while a (3S)-hydroxyacyl intermediate has been implicated in the formation of a cis-enoyl thioester in the biosynthesis of phoslactomycin (although the overall stereochemistry of the latter dehydration has not been determined).15 To explore any influence of the RifACP10 on the overall stereochemistry of the RifDH10-catalyzed reaction, we therefore also examined the stereospecificity of RifDH10-catalyzed dehydration of diketide thioester substrates that are covalently tethered to alternative thioester analogues of RifACP10. To our surprise, we found that replacement of RifACP10 by the heterologous EryACP6 domain, obtained from module 6 of the erythromycin PKS, resulted in an unexpected complete alteration in the diastereospecifity of RifDH10, without any effect on either the intrinsic syn stereospecificity of the dehydration reaction or the trans geometry of the resulting unsaturated product (Figure 6). Thus although RifDH10 was unable to dehydrate chemoenzymatically prepared (2S,3S)-2-methyl-3-hydroxyacyl-EryACP6 (5a), as established by TLC-phosphorimaging assay (Figure S12), RifDH10 did catalyze the syn dehydration of the diastereomeric anti-(2R,3R)-2-methyl-3-hydroxyacyl-EryACP6 (5b), which could be generated in situ by coupled incubation of the Ery[KS6][AT6] didomain from module 6 of the erythromycin PKS, EryACP6, and TylKR1 (from the first module of the tylactone PKS) with propionyl-SNAC, methylmalonyl-CoA, and NADPH, as previously described.23, 27, 36, 49 The reaction gave exclusively (E)-2-methyl-2-pentenoyl-EryACP6 (6), whose structure and stereochemistry were established by hydrolysis with 0.5 M NaOH for 20 min at 65 °C and GC-MS analysis of the derived methyl ester 3 (Figure 6a and S13). Similarly, an analogous incubation in which (2S,3R)-2-methyl-3-hydroxypentanoyl-SNAC was substituted for propionyl-SNAC gave exclusively (E)-(4R,5R)-2,4-dimethyl-5-hydroxy-2-heptenoyl-ACP6, as established by GC-MS analysis of the derived methyl ester (Figure S15). On the other hand, RifDH10 did not dehydrate either of the diastereomeric EryACP6-bound syn-thioesters, (2R,3S)-5c or (2S,3R)-5d (Figures 6a and S12). RifDH10 also catalyzed the reverse syn hydration of (E)-2-methyl-2-pentenoyl-EryACP6 (6), chemoenzymatically prepared in situ, as previously described23 to give methyl (2R,3R)-2-methyl-3-hydroxypentanoyl-EryACP6 (5b), whose structure and stereochemistry were established by PicTE-catalyzed hydrolysis and methylation, followed by chiral GC-MS analysis and direct comparison of the derived methyl ester (2R,3R)-4b with authentic standards of each of the four diastereomeric diketide methyl esters (Figures 6b and S16).
Figure 6.
RifDH10-catalyzed dehydration/hydration of acyl thioester analogues. a) Dehydration of (2R,3R)-2-methyl-3-hydroxypentanoyl-EryACP6 (5b). b) Hydration of (E)-2-methyl-2-pentenoyl-EryACP6 (6). c) Dehydration of (2R,3R)-2-methyl-3-hydroxypentanoyl-S-Pant (7b) and –S-NAC (9b) analogues. d) Incubation of RifDH10 with (E)-11 and (Z)-13.
A parallel series of incubations of RifDH10 with the corresponding pantetheine and NAC thioesters also resulted in syn dehydration/hydration of only the anomalous (2R,3R)-2-methyl-3-hydroxyacyl thioester diastereomers and their corresponding (E)-2-methyl-2-enoylacyl thioesters (Figures 6c and S17-S23). Thus, incubation of RifDH10 with (2R,3R)-2-methyl-3-hydroxypentanoyl-S-pantetheine (7b), prepared by diastereospecific reduction of (2RS)-2-methyl-3-ketopentanoyl-S-pantetheine with TylKR126, 44, 50 and a coupled enzymatic NADPH-regenerating system,44 gave exclusively (E)-2-methyl-2-pentenoyl-S-pantetheinate (8), as established by HPLC analysis and direct comparison with authentic synthetic standards (Figures 6c, S19, and S20). In agreement with this result, incubation of RifDH10 with (E)-2-methyl-2-butenoyl-S-pantetheine (11) yielded exclusively the syn-hydration product (2R,3R)-2-methyl-3-hydroxybutanoyl-S-pantetheine (12b) (Figures 6d, S22, and S24). The isomeric (Z)-2-methyl-2-butenoyl-S-pantetheine (13) did not undergo RifDH10-catalyzed hydration. RifDH10 did not dehydrate the diastereomeric diketide S-pantetheinates (2S,3S)-7a, (2R,3S)-7c, or (2R,3S)-2-methyl-3-hydroxypentanoyl-S-pantetheine (7d). Analogous results were also obtained using the corresponding NAC thioester analogues. Thus RifDH10 catalyzed the syn dehydration of only (2R,3R)-2-methyl-3-hydroxypentanoyl-S-NAC (9b) to give the corresponding (E)-2-methyl-2-pentenoyl-S-NAC (10) (Figure 6c and S19). We also examined the effects of acyl chain length and substitution pattern, observing that RifDH10 catalyzed the reversible dehydration of 3R-hydroxybutanoyl-S-pantetheine (Figures S21 and S24) and also catalyzed the hydration of (E,E)-2,4-hexadienoyl-S-pantetheine (Figure S23).
Incubation of the Rif[DH10][KR10] fusion protein with chemoenzymatically prepared (2RS)-2-methyl-3-ketopentanoyl-EryACP6 gave only the corresponding product of reduction by RifKR10, (2S,3S)-2-methyl-3-hydroxypentanoyl-EryACP6 (5a), with no detectable dehydration product, in contrast to the processing of the RifACP10-bound substrate, which resulted in both reduction and dehydration to give the corresponding (E)-2-methyl-3-pentenoyl-RifACP10 product, as described above. In like manner, although Rif[DH1][KR10] produced alcohol (2S,3S)-5a by diastereospecific reduction of (2R)-2-methyl-3-ketopentanoyl-EryACP6, generated in situ by decarboxylative condensation of propionyl-SNAC and methylmalonyl-CoA catalyzed by Ery[KS6][AT6] plus EryACP6, none of the corresponding dehydration product could be detected (Figures 2c and S14).
Structure of RifDH10
To better understand the protein structural basis for the dehydration reaction catalyzed by RifDH10, its structure was determined to 1.82-Å resolution (Figure 7 and Table 1). RifDH10 is structurally highly homologous to the other previously solved DH domains, including EryDH4 and the four DH domains from CurF, CurH, CurJ, and CurK of the curacin PKS (PDB Codes 3EL6, 3KG6, 3KG7, 3KG8, and 3KG9, respectively; rmsd 1.2 Å, 1.8 Å, 2.0 Å, 2.3 Å, and 1.7 Å, respectively).10, 21 RifDH10 possesses a characteristic double hotdog fold and dimerizes through an interface largely created by its ~25 N-terminal residues (Figure 7a). The catalytic dyad, comprised of a histidine (H50) and an aspartic acid (D220), almost perfectly superposes with the dyads of each of the other DH structures (Figure 7b). A hydrogen-bonding network beginning with the phenolic hydroxyl of Y164 may increase the pKa of the active site aspartate thereby allowing it to act as a general acid for the dehydration reaction. A similar hydrogen-bonding network is conserved in five of the six known DH structures, which employ either a glutamine or histidine (H224 in RifDH10) to bridge the network between Y164 and the catalytic aspartate. The exception is the CurH DH, in which an arginine occupies the corresponding site.
Figure 7.

The 1.82 Å-resolution structure of RifDH10. a) The 2Fo-Fc electron density map (contoured at 1.8 σ) shows a water molecule, bound to the catalytic histidine (H50) and aspartate (D220), as well as a nearly invariant Y174, that is representative of the water molecule eliminated through a syn-dehydration reaction. H224 and Y164 may help increase the pKa of D220 through a hydrogen-bond network. b) Stereodiagram of the superposition of the active sites of RifDH10, EryDH4, CurF DH, CurH DH, CurJ DH, and CurK DH, showing the catalytic His (H) and Asp (D) residues.
Table 1.
Crystallographic Data and Refinement Statistics
| Data collection | |
|---|---|
| Space group | P32 |
| Cell dimensions, a, b, c (Å) | 77.2, 77.2, 97.9 |
| Resolution (Å) | 50–1.82 |
| Rmerge | 0.075 (0.807) |
| I/σ(I) | 10.6 (2.0) |
| No. of reflections | 55,103 (4,134) |
| Completeness (%) | 99.1 (99.9) |
| Redundancy | 5.3 (4.8) |
| Wilson B value (Å2) | 40.1 |
|
| |
| Refinement | |
| Resolution (Å) | 50–1.82 |
| No. of reflections | 55,103 (4,134) |
| Rwork/Rfree | 0.211/0.254 |
| No. of atoms | 4444 |
| Protein | 4134 |
| Water | 310 |
| Average B factors (Å2) | |
| Protein | 40 |
| Water | 46 |
| RMS deviations | |
| Bond lengths (Å) | 0.003 |
| Bond angles (°) | 0.759 |
| Ramachandran Statistics (%) | |
| Preferred Regions | 95.8 |
| Allowed Regions | 4.2 |
| Outliers | 0.0 |
Values in parentheses refer to the highest resolution shell (1.92 -1.82 Å)
Discussion
RifDH10 is only the second PKS DH domain for which both the protein structure and the stereospecificity of the dehydration reaction have been experimentally determined.23 Consistent with the close similarity in overall protein architecture between RifDH10 and EryDH4, as well as the essentially identical relative positioning of their conserved active site histidine and aspartate residues, we have established that both dehydratases catalyze syn elimination of water to give trans-unsaturated, trisubstituted carboxylic acid thioesters. On the other hand, these two closely related dehydratases actually catalyze the dehydration of effectively enantiomeric syn-2-methyl-3-hydroxyacyl substrates. Catalysis of each dehydration reaction is believed to involve deprotonation of C-2 of the substrate by the imidazole sidechain of H50 which acts as the general base, with either stepwise or concerted elimination of water promoted by protonation of the C-3 hydroxyl by the carboxylic acid side chain of D220 (Figure 7b). These conserved active site residues have been shown to be essential not only in EryDH4,51 but in the dehydratase domain of animal FAS52 and in the corresponding Type II FabZ dehydratase proteins of bacterial fatty acid biosynthesis as well.53 The topological relationship of the active site histidine and aspartate residues is consistent only with net syn elimination of water from the substrate. Crotonase (enoyl-CoA hydratase), a mechanistically closely related enzyme from the fatty acid beta-oxidation pathway, although differing from PKS and FAS DH domains in both active site residues and overall protein structure, has also been shown to catalyze reversible concerted syn addition of water to trans-enoyl-CoA substrates.39, 54
In spite of all these mechanistic and structural similarities to other characterized DH domains, RifDH10 is unique in catalyzing the diastereospecific syn dehydration of a (2S,3S)-2-methyl-3-hydroxyacyl-RifACP10 substrate. This is in contrast to the demonstrated specificity of EryDH4, TylDH2, and NanDH2, all of which utilize only (2R,3R)-2-methyl-3-hydroxyacyl-ACP substrates, as well as the specificity of the DH domain of yeast FAS for 3R-hydroxyacyl-ACP substrates.55 Intriguingly, the intrinsic preference of RifDH10 for the (2S,3S)-2-methyl-3-hydroxyacyl thioesters is only evident when the substrate is tethered to its native acyl carrier protein, RifACP10. By contrast, RifDH10-catalyzed dehydration of the corresponding non-cognate EryACP6, as well as the NAC and pantetheine thioester analogues is specific for the corresponding (2R,3R)-2-methyl-3-hydroxyacyl diastereomers, while in no case do either the (2R,3S)- or (2S,3R)-2-methyl-3-hydroxyacyl derivatives undergo dehydration. This reversal of substrate specificity is unprecedented. Although several EryKS domains have been shown to exhibit strong preferences for their cognate ACP domains in both intramodular condensation reactions and intermodular chain transfers,35, 56, 57 and the stereoselectivity of KR-catalyzed reductions can require or be enhanced by tethering of 3-ketoacyl thioester substrates to an ACP domain,36, 50 this is the first example of the complete reversal of the diastereospecificity of a biochemical reaction based on the nature of the thioester conjugate.
Several interlocking lines of argument support the biosynthetic relevance of the observed strict preference for the RifACP10-linked, (2S,3S)-2-methyl-3-hydroxyacyl thioester substrate: 1) RifACP10 is derived from the same parent module of the rifamycin synthase as is RifDH10 and therefore must represent the native carrier for dehydration of the natural ACP-bound acyclic undecaketide substrate within Rif module 10. 2) RifKR10, the paired KR domain that is also harbored in Rif module 10, generates exclusively the (2S,3S)-2-methyl-3-hydroxyacyl thioester, independent of the nature or origin of the attached ACP domain. Under natural conditions, therefore, RifDH10 should encounter only the (2S,3S)-2-methyl-3-hydroxyacyl undecaketide which will be tethered to RifACP10. 3) While most of the substrate analogues that have been tested in vitro with RifDH10 are diketide derivatives, longer chain acyl thioester substrates have no effect on the observed stereospecificity of RifDH10. 4) It highly unlikely that the in vitro results are artifacts and that the natural reaction on the undecaketide substrate would generate a cis double bond based on both the highly conserved active site geometry of RifDH10, including the precise positioning of the catalytic histidine and aspartate residues in a topology that has been shown to catalyze syn dehydration by several other DH domains, as well as the exclusive formation and rehydration of trans-α,β-unsaturated trisubstituted enoyl thioesters. 5) If these in vitro results were simply the spurious consequence of the use of a deconstructed in vitro enzyme system, one would expect to see a simple degradation in overall diastereospecificity, rather than the observed complete reversal of the diastereospecificity that is cleanly correlated with the use of RifDH10 within its natural context.
The syn stereochemistry of the DH-catalyzed dehydration reaction is thus an intrinsic catalytic property that is rooted in the topological placement of the active site histidine and aspartate residues. This reaction stereochemistry should not be influenced by simple variations in the structure and stereochemistry of the substrate. While it is not unusual to observe degradation in substrate stereospecificity with the use of alternative substrates, to our knowledge it is unprecedented to encounter a complete reversal in the apparent diastereospecificity that is a consequence of a change in the nature of the thioester conjugate. Although both the (2S,3S)-, and (2R,3R)-2-methyl-3-hydroxyacyl thioesters can be modeled into the RifDH10 active site, with productive alignments of both the syn-related C2 proton and the C3 hydroxyl group with the corresponding H50 and D220 residues, the attached pantetheinyl moieties would be differently positioned in each case (Figure 8). While there is as yet no protein structure of a DH–ACP complex, several lines of evidence have supported the postulated existence of strong interactions between DH domains and their cognate ACP domains. Both pull-down and surface plasmon resonance experiments have shown a high affinity of the Helicobacter pylori ACP (HpACP) for the HpFabZ protein, with a Kd of 10 nM.58 Similarly, in silico protein ligand docking and molecular dynamics simulations have been used to model complexes involving specific interactions of EryDH4 and holo-EryACP4.59 We surmise that substitution by the non-cognate EryACP6, which has only 43% sequence identity with RifACP10, or by the simpler NAC and pantetheine analogues, results in the loss of critical RifDH10-RifACP10 interactions that are essential for proper positioning of the tethered substrate within the active site, thereby permitting syn dehydration of the anomalously-bound, diastereomeric (2R,3R)-2-methyl-3-hydroxyacyl thioester analogues.
Figure 8.
RifDH10-catalyzed dehydration. a) syn-Dehydration of a (2S,3S)-2-methyl-3-hydroxyacyl thioester substrate generates a trans (E) double bond. b) Stereodiagram showing (2S,3S)-2-methyl-3-hydroxyacyl thioester substrate modeled into the active site of RifDH10. See Figure S2 for modeling of diastereomeric thioesters into the active site.
Our results strongly indicate that during the course of rifamycin biosynthesis the combined action of the RifKR10 and RifDH10 domains from the terminal module of the rifamycin PKS naturally generates an acyclic (E)-2-methyl-2-enoyl undecaketide. We therefore suggest that isomerization of the (E)-2,3-double bond of the initially formed RifACP10-bound undecaketide intermediate to the corresponding cis geometry that is characteristic of both the cyclized intermediate proansamycin X and the ultimately derived rifamycin B probably most likely occurs during the subsequent macrolactamization reaction that is catalyzed by the discrete amide synthase RifF.
Supplementary Material
Acknowledgments
Instrumentation and technical assistance at the University of Texas were provided by Dr. Art Monzingo and the Macromolecular Crystallography Facility, with financial support from the College of Natural Sciences, the Office of the Executive Vice President and Provost, and the Institute for Cellular and Molecular Biology, the University of Texas at Austin. The Berkeley Center for Structural Biology is supported in part by the National Institutes of Health, National Institute of General Medical Sciences, and the Howard Hughes Medical Institute. The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the U.S. Department of Energy under Contract No. DE-AC02-05CH11231. Joshua F. Detelich helped synthesize several compounds used in this work.
Funding Sources
Financial support was provided by Welch Foundation Grant F-1712 (A.T.K.) and National Institutes of Health, NIGMS Grants GM106112 (A.T.K.) and GM022172 (D.E.C.).
Abbreviations
- ACP
acyl carrier protein
- AT
acyltransferase
- DH
dehydratase
- ER
enoylreductase
- Ery
6-deoxyerythonolide B synthase
- FAS
fatty acid synthase
- GC-MS
gas chromatography – mass spectrometry
- KR
ketoreductase
- KS
ketosynthase
- NAC
N-acetylcysteamine
- Nan
nanchangmycin synthase
- PKS
polyketide synthase
- Rif
rifamycin synthase
- TE
thioesterase
- Tyl
tylactone synthase
Footnotes
Accession Codes
The atomic coordinates and crystallographic structure factors for RifDH10 have been deposited in the Protein Data Bank (www.pdb.org) with accession code PDB ID 4LN9.
Structures of polyketides containing cis double bonds, modeling of reduced diketides into the RifDH10 active site, design of recombinant RifDH10 and RifACP10, GC-MS data for incubations of RifACP10- and EryACP6-bound substrates with RifDH10, LC-MS data for incubations of S-pantetheine and S-NAC thioesters with RifDH10, and preparation of S-pantetheine thioesters. This material is available free of charge via the Internet at http://pubs.acs.org.
References
- 1.Smith S, Tsai SC. The type I fatty acid and polyketide synthases: a tale of two megasynthases, Nat. Prod Rep. 2007;24:1041–1072. doi: 10.1039/b603600g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sherman DH, Smith JL. Clearing the skies over modular polyketide synthases, ACS Chem. Biol. 2006;1:505–509. doi: 10.1021/cb600376r. [DOI] [PubMed] [Google Scholar]
- 3.Cane DE. Programming of erythromycin biosynthesis by a modular polyketide synthase. J Biol Chem. 2010;285:27517–27523. doi: 10.1074/jbc.R110.144618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Piel J. Biosynthesis of polyketides by trans-AT polyketide synthases, Nat. Prod Rep. 2010;27:996–1047. doi: 10.1039/b816430b. [DOI] [PubMed] [Google Scholar]
- 5.Lewy DS, Gauss CM, Soenen DR, Boger DL. Fostriecin: chemistry and biology, Curr. Med Chem. 2002;9:2005–2032. doi: 10.2174/0929867023368809. [DOI] [PubMed] [Google Scholar]
- 6.Kong R, Liu X, Su C, Ma C, Qiu R, Tang L. Elucidation of the biosynthetic gene cluster and the post-PKS modification mechanism for fostriecin in Streptomyces pulveraceus, Chem. Biol. 2013;20:45–54. doi: 10.1016/j.chembiol.2012.10.018. [DOI] [PubMed] [Google Scholar]
- 7.Palaniappan N, Alhamadsheh MM, Reynolds KA. cis-Delta(2,3)-double bond of phoslactomycins is generated by a post-PKS tailoring enzyme. J Am Chem Soc. 2008;130:12236–12237. doi: 10.1021/ja8044162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Olano C, Wilkinson B, Sanchez C, Moss SJ, Sheridan R, Math V, Weston AJ, Brana AF, Martin CJ, Oliynyk M, Mendez C, Leadlay PF, Salas JA. Biosynthesis of the angiogenesis inhibitor borrelidin by Streptomyces parvulus Tu4055: cluster analysis and assignment of functions, Chem. Biol. 2004;11:87–97. doi: 10.1016/j.chembiol.2003.12.018. [DOI] [PubMed] [Google Scholar]
- 9.Tang L, Shah S, Chung L, Carney J, Katz L, Khosla C, Julien B. Cloning and heterologous expression of the epothilone gene cluster. Science. 2000;287:640–642. doi: 10.1126/science.287.5453.640. [DOI] [PubMed] [Google Scholar]
- 10.Akey DL, Razelun JR, Tehranisa J, Sherman DH, Gerwick WH, Smith JL. Crystal structures of dehydratase domains from the curacin polyketide biosynthetic pathway. Structure. 2010;18:94–105. doi: 10.1016/j.str.2009.10.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Schupp T, Toupet C, Engel N, Goff S. Cloning and sequence analysis of the putative rifamycin polyketide synthase gene cluster from Amycolatopsis mediterranei, FEMS Microbiol. Lett. 1998;159:201–207. doi: 10.1111/j.1574-6968.1998.tb12861.x. [DOI] [PubMed] [Google Scholar]
- 12.August PR, Tang L, Yoon YJ, Ning S, Muller R, Yu TW, Taylor M, Hoffmann D, Kim CG, Zhang X, Hutchinson CR, Floss HG. Biosynthesis of the ansamycin antibiotic rifamycin: deductions from the molecular analysis of the rif biosynthetic gene cluster of Amycolatopsis mediterranei S699, Chem. Biol. 1998;5:69–79. doi: 10.1016/s1074-5521(98)90141-7. [DOI] [PubMed] [Google Scholar]
- 13.Tang L, Yoon YJ, Choi CY, Hutchinson CR. Characterization of the enzymatic domains in the modular polyketide synthase involved in rifamycin B biosynthesis by Amycolatopsis mediterranei. Gene. 1998;216:255–265. doi: 10.1016/s0378-1119(98)00338-2. [DOI] [PubMed] [Google Scholar]
- 14.Alhamadsheh MM, Palaniappan N, Daschouduri S, Reynolds KA. Modular polyketide synthases and cis double bond formation: establishment of activated cis-3-cyclohexylpropenoic acid as the diketide intermediate in phoslactomycin biosynthesis, J. Am. Chem Soc. 2007;129:1910–1911. doi: 10.1021/ja068818t. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bonnett SA, Whicher JR, Papireddy K, Florova G, Smith JL, Reynolds KA. Structural and stereochemical analysis of a modular polyketide synthase ketoreductase domain required for the generation of a cis-alkene, Chem. Biol. 2013;20:772–783. doi: 10.1016/j.chembiol.2013.04.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.World Health Organization. Treatment of Tuberculosis Guidelines. 4. WHO Press; Geneva, Switzerland: 2010. Standard Treatment Regimens; pp. 29–30. [PubMed] [Google Scholar]
- 17.Hertweck C. The biosynthetic logic of polyketide diversity, Angew. Chem Int Ed Engl. 2009;48:4688–4716. doi: 10.1002/anie.200806121. [DOI] [PubMed] [Google Scholar]
- 18.Stratmann A, Toupet C, Schilling W, Traber R, Oberer L, Schupp T. Intermediates of rifamycin polyketide synthase produced by an Amycolatopsis mediterranei mutant with inactivated rifF gene. Microbiol. 1999;145:3365–3375. doi: 10.1099/00221287-145-12-3365. [DOI] [PubMed] [Google Scholar]
- 19.Yu TW, Shen Y, Doi-Katayama Y, Tang L, Park C, Moore BS, Richard Hutchinson C, Floss HG. Direct evidence that the rifamycin polyketide synthase assembles polyketide chains processively, Proc. Natl Acad Sci U S A. 1999;96:9051–9056. doi: 10.1073/pnas.96.16.9051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Floss HG, Yu TW. Lessons from the rifamycin biosynthetic gene cluster, Curr. Opin Chem Biol. 1999;3:592–597. doi: 10.1016/s1367-5931(99)00014-9. [DOI] [PubMed] [Google Scholar]
- 21.Keatinge-Clay A. Crystal structure of the erythromycin polyketide synthase dehydratase. J Mol Biol. 2008;384:941–953. doi: 10.1016/j.jmb.2008.09.084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Donadio S, McAlpine JB, Sheldon PJ, Jackson M, Katz L. An erythromycin analog produced by reprogramming of polyketide synthesis, Proc. Natl Acad Sci U S A. 1993;90:7119–7123. doi: 10.1073/pnas.90.15.7119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valenzano CR, You YO, Garg A, Keatinge-Clay A, Khosla C, Cane DE. Stereospecificity of the dehydratase domain of the erythromycin polyketide synthase. J Am Chem Soc. 2010;132:14697–14699. doi: 10.1021/ja107344h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Reid R, Piagentini M, Rodriguez E, Ashley G, Viswanathan N, Carney J, Santi DV, Hutchinson CR, McDaniel R. A model of structure and catalysis for ketoreductase domains in modular polyketide synthases. Biochemistry. 2003;42:72–79. doi: 10.1021/bi0268706. [DOI] [PubMed] [Google Scholar]
- 25.Caffrey P. Conserved amino acid residues correlating with ketoreductase stereospecificity in modular polyketide synthases. ChemBioChem. 2003;4:654–657. doi: 10.1002/cbic.200300581. [DOI] [PubMed] [Google Scholar]
- 26.Keatinge-Clay AT. A tylosin ketoreductase reveals how chirality is determined in polyketides, Chem. Biol. 2007;14:898–908. doi: 10.1016/j.chembiol.2007.07.009. [DOI] [PubMed] [Google Scholar]
- 27.Castonguay R, Valenzano CR, Chen AY, Keatinge-Clay A, Khosla C, Cane DE. Stereospecificity of ketoreductase domains 1 and 2 of the tylactone modular polyketide synthase. J Am Chem Soc. 2008;130:11598–11599. doi: 10.1021/ja804453p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Palaniappan N, Kim BS, Sekiyama Y, Osada H, Reynolds KA. Enhancement and selective production of phoslactomycin B, a protein phosphatase IIa inhibitor, through identification and engineering of the corresponding biosynthetic gene cluster. J Biol Chem. 2003;278:35552–35557. doi: 10.1074/jbc.M305082200. [DOI] [PubMed] [Google Scholar]
- 29.Tang L, Ward S, Chung L, Carney JR, Li Y, Reid R, Katz L. Elucidating the mechanism of cis double bond formation in epothilone biosynthesis, J. Am. Chem Soc. 2004;126:46–47. doi: 10.1021/ja030503f. [DOI] [PubMed] [Google Scholar]
- 30.Vergnolle O, Hahn F, Baerga-Ortiz A, Leadlay PF, Andexer JN. Stereoselectivity of isolated dehydratase domains of the borrelidin polyketide synthase: implications for cis double bond formation. Chembiochem. 2011;12:1011–1014. doi: 10.1002/cbic.201100011. [DOI] [PubMed] [Google Scholar]
- 31.You YO, Khosla C, Cane DE. Stereochemistry of reductions catalyzed by methyl-epimerizing ketoreductase domains of polyketide synthases, J. Am. Chem Soc. 2013;135:7406–7409. doi: 10.1021/ja4014776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Guo X, Liu T, Valenzano CR, Deng Z, Cane DE. Mechanism and stereospecificity of a fully saturating polyketide synthase module: nanchangmycin synthase module 2 and its dehydratase domain. J Am Chem Soc. 2010;132:14694–14696. doi: 10.1021/ja1073432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Chen AY, Schnarr NA, Kim CY, Cane DE, Khosla C. Extender unit and acyl carrier protein specificity of ketosynthase domains of the 6-deoxyerythronolide B synthase. J Am Chem Soc. 2006;128:3067–3074. doi: 10.1021/ja058093d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen AY, Cane DE, Khosla C. Structure-based dissociation of a type I polyketide synthase module, Chem. Biol. 2007;14:784–792. doi: 10.1016/j.chembiol.2007.05.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kim CY, Alekseyev VY, Chen AY, Tang Y, Cane DE, Khosla C. Reconstituting modular activity from separated domains of 6-deoxyerythronolide B synthase. Biochemistry. 2004;43:13892–13898. doi: 10.1021/bi048418n. [DOI] [PubMed] [Google Scholar]
- 36.Valenzano CR, Lawson RJ, Chen AY, Khosla C, Cane DE. The biochemical basis for stereochemical control in polyketide biosynthesis. J Am Chem Soc. 2009;131:18501–18511. doi: 10.1021/ja908296m. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wu N, Tsuji SY, Cane DE, Khosla C. Assessing the balance between protein-protein interactions and enzyme- substrate interactions in the channeling of intermediates between polyketide synthase modules. J Am Chem Soc. 2001;123:6465–6474. doi: 10.1021/ja010219t. [DOI] [PubMed] [Google Scholar]
- 38.Haapalainen AM, Merilainen G, Pirila PL, Kondo N, Fukao T, Wierenga RK. Crystallographic and kinetic studies of human mitochondrial acetoacetyl-CoA thiolase: The importance of potassium and chloride ions for its structure and function. Biochemistry. 2007;46:4305–4321. doi: 10.1021/bi6026192. [DOI] [PubMed] [Google Scholar]
- 39.Agnihotri G, Liu HW. Enoyl-CoA hydratase. reaction, mechanism, and inhibition. Bioorg Med Chem. 2003;11:9–20. doi: 10.1016/s0968-0896(02)00333-4. [DOI] [PubMed] [Google Scholar]
- 40.Sambrook J, Fritsch EF, Maniatis T. Molecular Cloning, A Laboratory Manual. 2. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1989. [Google Scholar]
- 41.Bradford M. A rapid and sensitive method for the quantitation of microgram quantitites of protein utilizing the principle of protein-dye binding, Anal. Biochem. 1976;72:248–254. doi: 10.1006/abio.1976.9999. [DOI] [PubMed] [Google Scholar]
- 42.De Marco V, Stier G, Blandin S, de Marco A. The solubility and stability of recombinant proteins are increased by their fusion to NusA, Biochem. Biophys Res Commun. 2004;322:766–771. doi: 10.1016/j.bbrc.2004.07.189. [DOI] [PubMed] [Google Scholar]
- 43.Weinreb PH, Quadri LE, Walsh CT, Zuber P. Stoichiometry and specificity of in vitro phosphopantetheinylation and aminoacylation of the valine-activating module of surfactin synthetase. Biochemistry. 1998;37:1575–1584. doi: 10.1021/bi9719859. [DOI] [PubMed] [Google Scholar]
- 44.Piasecki SK, Taylor CA, Detelich JF, Liu J, Zheng J, Komsoukaniants A, Siegel DR, Keatinge-Clay AT. Employing modular polyketide synthase ketoreductases as biocatalysts in the preparative chemoenzymatic syntheses of diketide chiral building blocks, Chem. Biol. 2011;18:1331–1340. doi: 10.1016/j.chembiol.2011.07.021. [DOI] [PubMed] [Google Scholar]
- 45.Collaborative Computational Project N. The CCP4 suite: programs for protein crystallography, Acta crystallographica. Section D, Biological crystallography. 1994;50:760–763. doi: 10.1107/S0907444994003112. [DOI] [PubMed] [Google Scholar]
- 46.McCoy AJ, Grosse-Kunstleve RW, Adams PD, Winn MD, Storoni LC, Read RJ. Phaser crystallographic software. J Appl Crystallogr. 2007;40:658–674. doi: 10.1107/S0021889807021206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Emsley P, Lohkamp B, Scott WG, Cowtan K. Features and development of Coot, Acta crystallographica. Section D, Biological crystallography. 2010;66:486–501. doi: 10.1107/S0907444910007493. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Murshudov GN, Vagin AA, Dodson EJ. Refinement of macromolecular structures by the maximum-likelihood method, Acta crystallographica. Section D, Biological crystallography. 1997;53:240–255. doi: 10.1107/S0907444996012255. [DOI] [PubMed] [Google Scholar]
- 49.Castonguay R, He W, Chen AY, Khosla C, Cane DE. Stereospecificity of ketoreductase domains of the 6-deoxyerythronolide B synthase. J Am Chem Soc. 2007;129:13758–13769. doi: 10.1021/ja0753290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Siskos AP, Baerga-Ortiz A, Bali S, Stein V, Mamdani H, Spiteller D, Popovic B, Spencer JB, Staunton J, Weissman KJ, Leadlay PF. Molecular basis of Celmer’s rules: stereochemistry of catalysis by isolated ketoreductase domains from modular polyketide synthases, Chem. Biol. 2005;12:1145–1153. doi: 10.1016/j.chembiol.2005.08.017. [DOI] [PubMed] [Google Scholar]
- 51.Bevitt DJ, Staunton J, Leadlay PF. Mutagenesis of the dehydratase active site in the erythromycin-producing polyketide synthase, Biochem. Soc Trans. 1993;21:30S. doi: 10.1042/bst021030s. [DOI] [PubMed] [Google Scholar]
- 52.Pasta S, Witkowski A, Joshi AK, Smith S. Catalytic residues are shared between two pseudosubunits of the dehydratase domain of the animal fatty acid synthase, Chem. Biol. 2007;14:1377–1385. doi: 10.1016/j.chembiol.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 53.Kimber MS, Martin F, Lu Y, Houston S, Vedadi M, Dharamsi A, Fiebig KM, Schmid M, Rock CO. The structure of (3R)-hydroxyacyl-acyl carrier protein dehydratase (FabZ) from Pseudomonas aeruginosa, J. Biol. Chem. 2004;279:52593–52602. doi: 10.1074/jbc.M408105200. [DOI] [PubMed] [Google Scholar]
- 54.Bahnson BJ, Anderson VE. Crotonase-catalyzed beta-elimination is concerted: a double isotope effect study. Biochemistry. 1991;30:5894–5906. doi: 10.1021/bi00238a013. [DOI] [PubMed] [Google Scholar]
- 55.Sedgwick B, Morris C, French SJ. Stereochemical course of dehydration catalyzed by the yeast fatty acid synthetase. J C S Chem Commun. 1978:193–194. [Google Scholar]
- 56.Kapur S, Lowry B, Yuzawa S, Kenthirapalan S, Chen AY, Cane DE, Khosla C. Reprogramming a module of the 6-deoxyerythronolide B synthase for iterative chain elongation, Proc. Natl Acad Sci U S A. 2012;109:4110–4115. doi: 10.1073/pnas.1118734109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Guo X, Liu T, Deng Z, Cane DE. Essential role of the donor acyl carrier protein in stereoselective chain translocation to a fully reducing module of the nanchangmycin polyketide synthase. Biochemistry. 2012;51:879–887. doi: 10.1021/bi201768v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Liu W, Du L, Zhang L, Chen J, Shen X, Jiang H. Helicobacter pylori acyl carrier protein: expression, purification, and its interaction with beta-hydroxyacyl-ACP dehydratase, Protein Expr. Purif. 2007;52:74–81. doi: 10.1016/j.pep.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 59.Anand S, Mohanty D. Modeling holo-ACP:DH and holo-ACP:KR complexes of modular polyketide synthases: a docking and molecular dynamics study, BMC Struct. Biol. 2012;12:10. doi: 10.1186/1472-6807-12-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.