Abstract
Through linkage analysis, candidate gene approach, and genome-wide association studies (GWAS), many genetic susceptibility factors for substance dependence have been discovered, such as the alcohol dehydrogenase gene (ALDH2) for alcohol dependence (AD) and nicotinic acetylcholine receptor (nAChR) subunit variants on chromosomes 8 and 15 for nicotine dependence (ND). However, these confirmed genetic factors contribute only a small portion of the heritability responsible for each addiction. Among many potential factors, rare variants in those identified and unidentified susceptibility genes are supposed to contribute greatly to the missing heritability. Several studies focusing on rare variants have been conducted by taking advantage of next-generation sequencing technologies, which revealed that some rare variants of nAChR subunits are associated with ND in both genetic and functional studies. However, these studies investigated variants for only a small number of genes and need to be expanded to broad regions/genes in a larger population. This review presents an update on recently developed methods for rare-variant identification and association analysis and on studies focused on rare-variant discovery and function related to addictions.
Keywords: rare variants, next-generation sequencing, drug addiction
Introduction
Substance abuse and addiction pose significant threats to public health world-wide. According to World Health Organization estimates, there were 2 billion alcohol abusers, 1.3 billion tobacco users, and 230 million illicit-drugs users world-wide in 2004 [1]. In the United States, the harmful use of alcohol results in 2.5 million deaths each year, and cigarette smoking accounts for 30% of deaths from cancers and nearly 80% of deaths from chronic obstructive pulmonary disease (COPD) [2, 3]. Globally, more than 6 million people were killed by cigarette smoking, and about 0.7% of the global burden of diseases was attributable to illicit drugs world-wide, with the social cost of illicit substance use being in the region of 2% of GDP in those countries that have measured it [1].
Epidemiologic studies have found that many individuals become addicted to multiple drugs after the initiation of one drug [4, 5]. Evidence from family, twin, and adoption studies strongly implicate genetic factors in each step of addiction, including vulnerability to initiation, continued use, and propensity to become dependent [6]. Family study has shown siblings of drug-abuser and - dependent probands are at approximately a 1.7-fold higher risk of developing marijuana dependence, cocaine dependence, or habitual smoking than are siblings of non-dependent individuals [7]. Further, twin studies suggest shared environmental influence contributes more to the availability of and exposure to the substance, such as smoking initiation; however, genetic factors have greater effects on smoking progression to nicotine dependence (ND) [4]. Many large twin studies for alcohol-related behaviors have consistently shown that heritability of alcohol abuse and dependence ranges from 50% to 70% [8]. Meta-analysis of the twin studies shows that both genetics and environment are important in smoking-related behaviors, with an estimated average heritability of 0.50 for smoking initiation and 0.59 for ND [9]. Several family studies of illicit drug use estimate heritability ranging from 30%–80% [8, 10, 11]. Finally, it deserves to be mentioned that a heritability estimate is specific to the sample under study. Thus, the role of genetic influences may differ across samples, and heritability can be affected by many factors, such as sex, age, education, socioeconomic status, and cultural background.
Identifying the Genetic Risk Factors and GWAS
At least two linkage studies, Collaborative Study on the Genetics of Alcoholism (COGA), conducted on multigenerational pedigrees densely affected by alcoholism, and the NIAAA linkage study on a homogeneous population from a southwestern Native American tribe revealed that susceptibility loci on chromosome 4 increase the risk of alcohol dependence (AD) [12–14]. However, the two linkage peaks were not located in the same genomic region. The linkage peaks identified in COGA from the general US population were located close to chromosome 4q, the alcohol dehydrogenase (ADH) gene cluster, whereas the NIAAA study detected the linkage signal close to the GABRA2–GABRB1 cluster; both findings were confirmed in other, independent studies [6, 15–17]. Several GWAS studies have been conducted for AD, and only one GWAS for alcoholism reported two correlated intergenic single nucleotide polymorphisms (SNPs) (rs7590720, P = 9.72 × 10−9; rs1344694, P = 1.69 × 10−8) on chromosome 2q35 that reached genome-wide significance in the combined GWAS and follow-up replication studies [18]. Other investigators did not observe a significant association with AD. The ADH-encoding genes are the best-studied candidate genes for AD, and ADH1B, ADH1C, and ALDH2 coding variants are well characterized with the phenotype of AD, such as ADH1B Arg48His [19, 20], ADH1C Ile350Val [21], and ALDH2 Glu504Lys [22].
To identify susceptibility loci for ND, more than 20 genome-wide linkage analyses have been conducted among different populations with a variety of assessments of ND, including smoking initiation (SI), smoking quantity (SQ), Heaviness of Smoking Index (HSI), Fagerström Test for Nicotine Dependence (FTND), ever-smoking, habitual smoking, cigarettes per day (CPD), or maximum number of cigarettes smoked in a 24-h period [23]. Multiple regions, located on chromosomes 3–7, 9–11, 17, 20, and 22, have shown “significant” or “suggestive” linkage [23, 24]. Four regions, on chromosomes 9q, 10q, 11p, and 17p, have been replicated in at least four independent studies [25–29]. Eight regions, on chromosomes 1, 5, 10, 11, 12, 16, 20, and 22, have been nominated as significant loci for ND-related phenotypes by reaching either genome-wide significance or a theoretical linkage threshold [24, 28, 30–35]. Recent GWAS and candidate gene studies have identified several SNPs in the nAChR subunit genes, on human chromosomes 15q (CHRNA5-CHRNA3-CHRNB4) and 8p (CHRNA6-CHRNB3), that influence the risk of ND, as defined by FTND and CPD [36–45]. Further, an important nicotine metabolism gene, CYP2A6, was reported to be significantly associated with CPD in a recent GWAS [41]. In addition, candidate gene-based association studies revealed that DRD2/ANKK1 [46–48], NRXN1 [49, 50], DBH [51], and BDNF [51, 52] showed strong associations with CPD, FTND, or smoking initiation/cessation, results that have been replicated in at least two independent studies.
For illicit drugs, few linkage analyses have been conducted, and most of the results have not been replicated. The first linkage study of cannabis dependence was conducted in a linked set of family, twin, and adoption studies, which revealed suggestive linkage on chromosomes 3q and 9q for cannabis-dependence vulnerability [53]. Two loci, on chromosomes 16 and 19, were linked to severe cannabis use/antisocial subtype in a Native American community study [54]. For other illicit drugs, significant linkage peaks have been identified on chromosomes 9 and 12 for cocaine dependence [55], chromosome 17 for a heavy opioid-use cluster-defined trait [56] and on 14q for DSM-IV opioid dependence [57]. Li and Burmeister [6] summarized that regions on chromosomes 2–5, 7, 9–11, 13, 14, and 17 have independent evidence of “suggestive” or “significant” linkage, with regions on chromosomes 4, 5, 9, 10, 11, and 17 receiving the strongest support for harboring susceptibility genes for addictions to multiple drugs. Multiple candidate gene studies revealed OPRM1, OPRD1, and several other genes to be associated with opioid dependence [58–62]. A few GWAS studies have also been conducted on illicit drug addiction; however, none of these findings reached genome-wide significance [63–65]. Although genetic studies have been successful in identifying a number of common variants that showed significant association with substance abuse, these variants contributed only a small portion of the phenotypic variance related to substance abuse, suggesting further investigation is greatly needed to identify the unexplained phenotypic variance.
Common vs Rare Variants
During recent years, GWAS approaches have been utilized extensively to study complex human traits; however, most common variants identified with such an approach can explain only a small proportion of genetic variation, which has sparked an intense debate about the common disease– common variant hypothesis. For example, despite a type 2 diabetes study with a discovery sample of 10,128 and a replication sample of 53,975, the 18 common variants significantly associated with the disease seem to explain only about 6% of the higher risk of disease among relatives [66, 67]. A meta-analysis of schizophrenia GWAS that included 8,008 cases and 19,077 controls identified only seven significant SNPs, some in high linkage disequilibrium (LD) with each other and each with an odds ratio below 1.3, despite an estimated heritability of 80–85% for schizophrenia [68]. This “missing heritability problem” suggests that a few dozen loci with moderate effects and intermediate frequencies each explaining part of the disease risk in a population simply is not the case, as is typically observed in crosses or pedigrees. Since then, several new hypotheses have been proposed: 1) a large number of small-effect common variants across the entire allele frequency spectrum (the infinitesimal model) [69]; 2) a large number of large-effect rare variants (the rare-allele model) [70]; and 3) some combination of genotypic, environmental, and epigenetic interactions (the broad-sense heritability model) [71, 72].
The rare-variant hypothesis [67, 73] proposes that a significant proportion of the inherited susceptibility to relatively common human chronic diseases is attributable to the summation of the effects of a series of low-frequency dominantly and independently acting variants of several genes, each conferring a moderate but readily detectable increase in relative risk [74]. Such rare variants will mostly be population specific because of founder effects resulting from genetic drift [74]. Furthermore, evolutionary theory predicts that disease alleles should be rare. The disease-promoting variants will be prevented from drifting to a higher frequency in the population under purifying selection. The population data from exome sequencing showed that non-synonymous coding variants are significantly skewed toward low frequencies, which certainly can be explained by purifying selection. However, the rare-variant hypothesis has its limitations, as there is no evidence that the rare variants make a large contribution to the genetic variance yet to be detected in GWAS. Although the rare variants may not solve all the issues, they do provide a strong complement to the functional evidence, which cannot be explained by the common variants identified by most GWAS. As more and more subjects are being sequenced by whole-genome or exome sequencing, the cumulative excess of rare coding variants with substantial effect size remains to be discovered. Considerable resolution of the burden of deleterious rare variants will no doubt emerge in the next few years as whole-exome and whole-genome sequencing ramps up [75, 76].
Rare-Variant Discovery and Next-Generation Whole-Genome and Exome Sequencing
After completion of the HapMap and 1000 Genome Projects, millions of novel SNPs were deposited in public databases [77–79]. However, more than 95% of the variants discovered in the 1000 Genome Pilot Project are common (MAF ≥5%); low-frequency (1–5%) and rare (<1%) variants remain poorly characterized [78]. These variants are highly enriched for potentially functional mutations, such as protein code-changing variants. The second phase of the 1000 Genome Project finished more than 1000 genomes from 14 populations from Europe, East Asia, sub-Saharan Africa, and the Americas, which captured as many as 98% of accessible SNPs having a frequency of 1% in related populations. These data enable analysis of common and low-frequency variants in individuals from diverse, including admixed, populations [78]. The enormous low-coverage whole-genome sequencing data, deep-coverage exome sequencing data, and high-density SNP genotyping data allowed the identification of 10,000–50,000 potentially functional variants with MAF <5% per individual in each population [78].
Both whole-genome and exome sequencing have been successful in identifying potentially functional variants of low frequency. Whole-genome sequencing can be conducted easily through standard library preparation protocols based on available sequencing platforms. For the current next-generation sequencing technology, Illumina HiSeq2000 and Lifetech SOLiD 5500XL are the two most powerful high-throughput sequencing platforms. Illumina Hiseq2000 has the highest throughput (600 Gb/run) and lowest price for per gigabyte of data (~$100/Gb). Lifetech SOLiD 5500XL has flexibility to fit small to large whole-genome sequencing projects; however, the cost for high-coverage (~30×) whole-genome sequencing is still an obstacle for processing thousands of individuals. Low-coverage whole-genome sequencing has been adapted, as the simulation study showed that sequencing 3,000 individuals at 4× depth provides similar power to that of deep sequencing of >2000 individuals at 30× depth but requires only ~20% of the sequencing effort by assuming that disease-associated variants have a frequency of greater than 0.2% [80]. Low-coverage whole-genome sequencing can be used to build a reference panel for imputing additional samples and improving the coverage crossing the genome to increase the power.
Exome sequencing is an alternative approach to whole-genome sequencing that sequences only the coding and UTR regions. This is intended to identify functional variants likely to change amino acid and gene expression pattern. The cost for exome sequencing is much lower, as it sequences less than 2% of the whole genome. However, exome sequencing requires extra capture enrichment steps prior to the sequencing. Currently, there are two capture methods; i.e., array-based and probe-based. Array-based capture is the first-developed exome capture technology; it hybridizes targeted fragments to the oligonucleotides synthesized on the microarray. Nimblegen SeqCap is the only array-based capture exome enrichment kit. The limitations of this technology include the need for expensive hardware as well as the relative large amount of DNA needed (10–15 µg). The probe-based capture method was then developed by pooling custom oligonucleotides and conducting hybridization in solution, which not only simplifies the hybridization process but also requires much less DNA (~3 µg). The oligonucleotides used for hybridization can be either DNA or RNA. Agilent SureSelect, Illumina TrueSeq, and Lifetech TargetSeq are the three available probe-based exome capture kits. The Agilent SureSelect kit is an RNA probe-based capture that can be used on most high-throughput sequencing platform, whereas both Lifetech TargetSeq and Illumina TruSeq are DNA probe-based capture kits and can be used only on their own proprietary platforms.
Recently, Flanigan et al. [81] reported a comparison of the performance of three commercial exome capture kits based on the neuromuscular disorder (NMD) gene panels. Although 92%–94% of the known NMD exons were included in all three targeted regions, the actual capture results demonstrated that at best, 60% of these exons obtained 100% coverage. The best-performing kit, Agilent SureSelect (v3), captured an average of 92.7% of the bases in the NMD gene exons, but only 58% of the NMD isoforms had all exons captured at 100% (e.g., 42% of the NMD genes had at least one exon with one base insufficiently captured to make a genotyping call). Illumina TruSeq captured 89.2% of exons, but only 36.5% of the genes had 100% coverage; and Nimblegen v2 captured 90.5% of the genes, but only 46.3% of the genes had 100% sequence coverage.
Targeted Resequencing
Although the cost of next-generation sequencing has been substantially reduced within the last few years, the total cost can be still high when thousands of samples need to be sequenced at the genome/exome level to detect low-frequency or rare variants. An alternative approach is targeted resequencing of smaller regions or a few dozen genes, usually 10 Kb to 10 Mb. Prior to the adoption of next-generation sequencing, amplicon resequencing was conducted on a traditional automated capillary-based Sanger sequencing platform, such as ABI3730, which produces accurate genotype calls for each individual but with relatively low throughput. Next-generation sequencing, along with a variety of capture enrichment methods, provides a powerful tool for sequencing targeted regions at relatively low cost. Several technologies have been developed based on the targeted region size, capture enrichment methods, and sequencing technologies. Multiplex amplicon and DNA/RNA probe-based capture are the two most popular enrichment methods (Table 2). Multiplex amplicon sequencing can be applied to several genes and targeted regions < 500 kb at relatively low cost compared with capture enrichment, and three companies have this technology with different amplicon sizes: Lifetech (< 10 Kb), Illumina (< 500 Kb), and RainDance (< 10 Mb). The RainDance multiplex amplicon can provide targeted resequencing of as much as 10 Mb, although the cost is higher than that of other capture enrichment methods. However, RainDance does have an advantage in that there are many customized panels that have been widely adopted in the genetic diagnostics field, such as for cancer, autism spectrum disorder (ASD), and human leukocyte antigen identification [82, 83]. The sample preparation steps are highly automated and compatible with all sequencing platforms. For probe-based in-solution capture enrichment, Agilent HaloPlex/SureSelect (1Kb – 10Mb), Lifetech TargetSeq (100Kb – 10Mb), and Illumina TruSeq (500Kb – 10Mb) targeted resequencing kits are using the same technology as their whole-exome capture kits, just with smaller capture sizes.
Table 2.
Targeted Enrichment/Amplicon Resequencing Platform Comparison
| Application | Size | Agilent | Lifetech | Illumina | RainDance |
|---|---|---|---|---|---|
| Target size (sequencing platform) | 1–10 kb | HaloPlex (ILM, ION) | Ion Xpress (ION) | TruSeq (MSE, HSE) | RDT 1000 (ILM, ION, SOL, ROC) |
| 10–500 kb | TargetSeq (ION, SOL) | ||||
| 200 kb-10 Mb | SureSelect (ILM, SOL, ROC) | ||||
| Enrichment | Probe (< 10 Mb) | Amplicon (<10 kb) Probe (0.1–10 Mb) | Amplicon (<500 kb) Probe (0.5–25 Mb) | Amplicon (< 10 Mb) | |
| Application | Compatible with most high-throughput sequencing platforms; widely adopted | Work only with Ion Torrent or SOLiD system | Work only with MSE or HSE system | Many panels available: cancer, ASD, HLA, pharmacogenetic, etc. | |
| Cost | $$ | $ | $ | $$$ |
Abbreviations: ILM = ILLUMINA, SOL = SOLiD, ION = Ion Torrent, ROC = ROCHE, MSE = Illumina MiSeq, HSE = Illumina HiSeq.
The molecular inversion probe is similar to in-solution hybridization capture, as the only difference is capture by circularization, which has a higher capacity than the multiplex amplicon [84]. The OS-Seq is a new oligonucleotide selective-capture method involving capturing and sequencing of genomic targets on a sequencer’s solid-phase support, such as the Illumina flow cell, which overcomes the limitations of either in-solution or array-based capture, such as the capture efficiency. First, target-specific oligonucleotides (40mers) were synthesized using the same method utilized in the traditional microarray and then immobilized on the flow cell; these “primer probes” served as both capture probes and sequencing primers. Second, a single-adaptor library prepared from genomic DNA was added to the flow cell, where the desired targets were captured by the immobilized primer probes. Third, the captured library fragments were prepared for bridge amplification, clustered, and sequenced. The capture efficiency was much higher than with the in-solution capture methods (~90% vs. ~60%) [84]; however, this technology may require some extra front-end input to optimize probe design, as no commercial platform is available at present.
DNA pooling was proposed as a strategy for reducing the cost of large-scale genotypingbased disease association studies [85, 86]. However, the difficulty in measuring allele frequencies accurately from intensity data has limited the use of this strategy. Unlike pooled genotyping, pooled DNA sequencing not only provides digital allele counts for each variant but also can detect novel sequence variants. Several recent studies have demonstrated the potential of pooled sequencing using next-generation platforms for identifying disease-associated rare mutations [87, 88]. The pooling approach can significantly reduce the overall cost by sequencing pooling samples for variant discovery and following with high-throughput genotyping verification in the large set of samples.
In Sillico Rare-Variant Identification Using Genotype Imputation
Another solution for rare-variant initial identification is genotype imputation, as many large-scale genotyping and sequencing projects have released information on millions of variants to public databases, which provides an enormous resource for rare-variant discovery using the imputation approach. Genotype imputation allows the evaluation of association variants that are not directly genotyped based on the variants within each individual and reference population from HapMap and the 1000 Genomes Project. Imputation is useful not only for combining results from different studies conducted on different genotyping platforms but also for increasing the power of individual scans for both traditional GWAS and sequencing-based association analysis. Unlike the common variants, rare variants tend to be recent discoveries and to share shorter haplotype stretches, which gives a much higher discordance rate during imputation. The percentage of missingness also was significantly increased [89]. One of the largest human exome sequencing projects from the National Heart, Lung, and Blood Institute (NHLBI) reported that approximately 73% of all protein coding variants and approximately 86% of variants predicted to be deleterious arose in the past 5,000–10,000 years. Because of the nature of rare variants, the reference panel size used for imputation needs to be much larger than for common-variant imputation. A recent review reported that across all imputation panels and genotyping chips, the imputation error rate increases as the minor allele frequency decreases, which is in line with previous observations that rare SNPs are more difficult to tag than common SNPs [90]. Using a reference panel phased with trio information boosts imputation performance compared with a reference panel phased without trio information, and the combination of the CEU + YRI + JPT + CHB reference panel can improve the imputation performance and accuracy across all the populations when imputing genotypes at SNPs with MAF < 5% [89].
Although rare-variant imputation is a feasible approach, in addition to sequencing for rarevariant discovery, genotyping/sequencing validation is necessary after imputation. Multiple imputation programs have been developed. Detailed information about these programs is provided in the next section.
Rare-Variant Analysis and Statistical Methods
Sequencing Data Analysis, Variant Calling, and Imputation
After high-throughput sequencing, variant calling is essential to retrieve the rare variants accurately from either high-coverage exome sequencing or low-coverage whole-genome sequencing data. The variant-calling process pipeline has several components, including base calling, alignment, post-alignment processing, variant and genotype calling, and candidate variant filtering. The base calling processes all the raw image data to sequence data, which usually is performed by high-throughput sequencing software. A number of alignment, post-alignment processing, and variant-calling programs are available. The Burrow-Wheeler Aligner (BWA) provides accurate and fast alignment, allowing gapped alignment among all alignment programs [91]. Genome Analysis Toolkit (GATK) packages provide post-alignment processing, including local realignment around indels and quality recalibration [92]. Both Unified genotyper (GATK) and SAMtools [93] have built-in probabilistic approaches that facilitate variant calling with different coverage. The variant calling can be carried out in multiple individuals simultaneously. Targeted resequencing can be applied to the same variant-calling pipeline as the whole exome/genome sequencing. However, if the targeted resequencing is carried out with pooling samples, the pipeline requires different programs for variant calling after alignment and post-alignment processing. The Syzygy program [88] was developed especially for pooling sample resequencing, and the program computes the likelihood that the position contains a non-reference allele, using Bayes’ Rule and a parameter that specifies the number of chromosomes in the pool.
Moreover, genotype imputation can fully utilize the sequencing data to increase the availability of variants for association analysis and enhance the power of statistical analysis to facilitate the combination of results across studies using meta-analysis [89]. Multiple imputation programs have been developed. MaCH [94], MaCH-Admix (http://www.sph.umich.edu/csg/yli/software.html) [95], IMPUTE [96], IMPUTE2 (http://mathgen.stats.ox.ac.uk/impute/impute_v2.html#home) [97] are based on the extension of the HMM model originally developed as part of importance sampling schemes for simulating coalescent trees, modeling LD, and estimating recombination rates [89]. MaCH works by successively updating the phase of each individual’s genotype data conditional on the current haplotype estimates of all the other samples, and the MaCH-Admix, based on the MaCH, allows the user to decide to either to use an integrated model for estimating parameters (recombination rate and error rate) by program or give fixed parameters acquired from the reference panel only or both reference haplotypes and target genotypes [95]. However, IMPUTE and IMPUTE2 are using fixed estimates of mutation rates and recombination maps. Furthermore, MACH uses a random subset of sample haplotypes as templates, whereas IMPUTE2 uses a subset of haplotypes selected to be similar to the haplotypes of the individual currently being estimated. The IMPUTE2 strategy appears to permit greater improvement in accuracy, as sample size increases and model complexity (the number of states) is held constant [98]. Minimac is a low-memory, computationally efficient implementation of the MaCH algorithm for genotype imputation that is designed to work on phased genotypes and can handle reference panels with hundreds or even thousands of haplotypes, such as the 1000 Genomes Project [99]. A similar approach has been implanted in IMPUTE2. BEAGLE is based on a graphical model of a set of haplotypes. This method works iteratively by fitting the model to the current set of estimated haplotypes and then resampling new estimated haplotypes for each individual based on the model of fit; http://faculty.washington.edu/browning/beagle/beagle.html [100, 101]. This program can be used for both case-control and family-based study designs.
Rare-Variant Association Analysis Methods
Unlike the GWAS, the power of single-marker analysis is poor for rare-variant association analysis because of the extremely low frequency [102]. Impelled by growing interests, burden tests have been proposed for rare variants, and these methods often employ the ideas of pooling or collapsing multiple rare variants within a region, weighting and/or prioritizing rare variants based on the functional and other criteria, and distribution-based approaches [102–105]. Recent evidence suggests that multiple rare variants often act collectively on disease risks [87, 103, 106, 107], with the need for aggregate effects of low-frequency variants in order to increase the power.
The cohort allelic sum test (CAST) was the first method developed to collapse information on all rare variants within a region, for example, the exons of a gene, into a single dichotomous variable for each subject by indicating whether the subject has any rare variants within the region and then applying a univariate test [103]. However, this method does not easily accommodate covariates and consider weighting of the variants, which is also not compatible with quantitative traits. The combined multivariate and collapsing (CMC) test [102] extends the CAST by collapsing variants in subgroups according to allele frequencies and combining these subgroups using Hotelling’s T2 test, which controls Type I error well. However, the disadvantage of this approach is that the threshold is not easy to select for the variants in a biological meaningful way. Compared with the CAST method of assigning similar effects to all rare variants, weighting methods usually assign high priority to alleles based on their frequency in the control population, potential functional changes predicted by PolyPhen/SIFT, or other criteria. Weighted sum statistics (WSS) is the first method developed based on the weighting method by grouping variants according to function, and the permutation of disease status were applied among affected and unaffected individuals to test the excess of variants in the affected individuals [104]. The weight method does have limitations in that it applies much higher weights to very rare variants in some scenarios. Besides collapsing or weighting variants, the variable threshold test (VT) uses a variable allele frequency threshold instead of a fixed threshold, and then assesses the statistical significance by permutation testing with variable thresholds [108]. The above all burden test methods require either specification of thresholds for collapsing or the use of permutation to estimate the threshold. Permutation tests are computationally expensive, especially on the whole-genome scale, and are difficult to adjust for covariates because permutation requires independence of the genotype from the covariates.
The C-alpha test is a non-burden-based test and robust to the direction and magnitude of effect that compares the expected variant with the actual variance of the distribution of effect for the case-control data, which improves the power relative to the burden-based test methods [109]. However, the C-alpha method is not easy to adjust for covariates, such as controlling population stratification. Kernel-based methods are a non-burden test, such as the sequence kernel association test (SKAT), which aggregates individual variant-score test statistics with weights when SNP effects are modeled linearly instead of aggregating variants. The SKAT extends kernel machine-based tests for rare variants with more accurate asymptotic approximations in the tail distribution [110], and it is powerful when a genetic region has both protective and deleterious variants of many non-causal variants. In general, SKAT first checks each single variant causal direction, then generates a weight for each variant, which can avoid loss of analytical power. However, in an extreme case, SKAT may have less power than other software if all variants in a gene or region are truly causal and affect the phenotype in the same direction [110, 111]. The SKAT produces conservative Type I errors for small-sample case-control sequencing association studies, which could lead to power loss and are often observed in current exome sequencing studies [110, 112]. The SKAT-O was further improved [113] through maximizing power by adaptively using the data to optimally combine the burden test and the non-burden sequence kernel association (SKAT), which are computationally efficient and can easily be applied to GWAS studies. Recently, family-based SKAT (famSKAT) has been developed based on the framework of linear mixed effected model to extend SKAT for rare variant association analysis with quantitative traits for family data [114]. Moreover, several popular multivariate tests for GWA studies have been evaluated for rare-variant association, including the minimum of univariate P-values (UminP) [115], sum score [116], sum of squared score (SSU) [117], and weighted sum of squared score (SSUw) tests [111]. Although more methods are being developed, there is still no method available that can be fit to all scenarios for rare-variant association analysis. In a real case, various methods should be tested.
Example of Rare-Variant Discover In Addiction Studies: ND
Discovery of Rare Variants Related to ND
GWAS have identified common variations in the several nAChR subunit genes/clusters, such as CHRNA5-CHRNA3-CHRNB4 and CHRNA6-CHRNB3, which contribute to ND. However, the role of rare variation in the risk of ND in these nicotinic receptor genes has not been well studied. In Wessel et al’s study [118], eleven nAChR subunit genes were sequenced through the amplicon approach, and a total of 44 common and 129 rare SNPs (MAF < 5%) were identified and tested for association with the FTND score using data obtained from 430 individuals, 18 of whom were excluded because of the reduced completion rate. The cohort allelic sum test and the weighted sum statistic methods were used for a rare-variant association test only and the multivariate distance matrix regression method for both common and rare SNPs. Significant association was observed between the FTND score and common and rare SNP/SNVs in CHRNA5 and CHRNB2 and of rare SNVs in CHRNA4. Both common and rare SNP/SNVs from multiple nAChR subunit genes were associated with the FTND score in this sample of treatment-seeking smokers. A follow-up CHRNA4 rare-variant study resequenced exon 5 from more than 2000 individuals, including both European American (EA) and African American (AA) populations [119], and the association test suggested that rare variants in CHRNA4 confer protection against ND. Recently, a rare-variant study was reported through pooled sequencing of the coding regions and flanking sequence of CHRNA5, CHRNA3, CHRNB4, CHRNA6, and CHRNB3 through an amplicon approach in AA and EA nicotine-dependent smokers and smokers without symptoms of dependence [120]. The carrier status of individuals harboring rare missense variants at conserved sites in each of these genes was then compared in cases and controls to test for an association with ND. Missense variants discovered at conserved residues in CHRNB4 are associated with a lower risk of ND in AA and EA, with two variants (T375I and T91I) contributing most to this association [120].
The above rare-variant studies utilized amplicon approaches along with either Sanger or next-generation sequencing or both, which screened only a handful of genes. However, the linkage analysis, candidate gene, and GWAS studies also identified several other possibly important genes beyond the nAChR subunits. We recently developed a customized targeted capture panel of 32 genes (Supplementary Table 1), including both nAChR subunit genes and several neurotransmitter receptors and metabolism genes, which have been reported to be associated with ND from various studies [6, 121]. The Agilent SureSelect Capture panel (250 kb) includes the coding regions, UTR regions, and flanking sequence of these genes. A total of 400 samples (200 sib pairs) were selected from the Mid-South Tobacco family study and divided into 8 pools (50 samples/per pool) based on ethic group (EA and AA), smoking status (smoking and non-smoking), and FTND (light and heavy smokers). The concentrations of individual DNA samples were first measured using the Quant-iT™ dsDNA assay kit (Lifetech) and pooled in equimolar amounts as suggested by manufacturers, and then library preparation, targeted capture, and high-throughput sequencing (72 bp paired-end) were conducted, following by data analysis, including base quality recalibration and alignment using BWA and hg19 assembly build as the reference. Variant calling was conducted using Syzygy designed for a pooling approach.
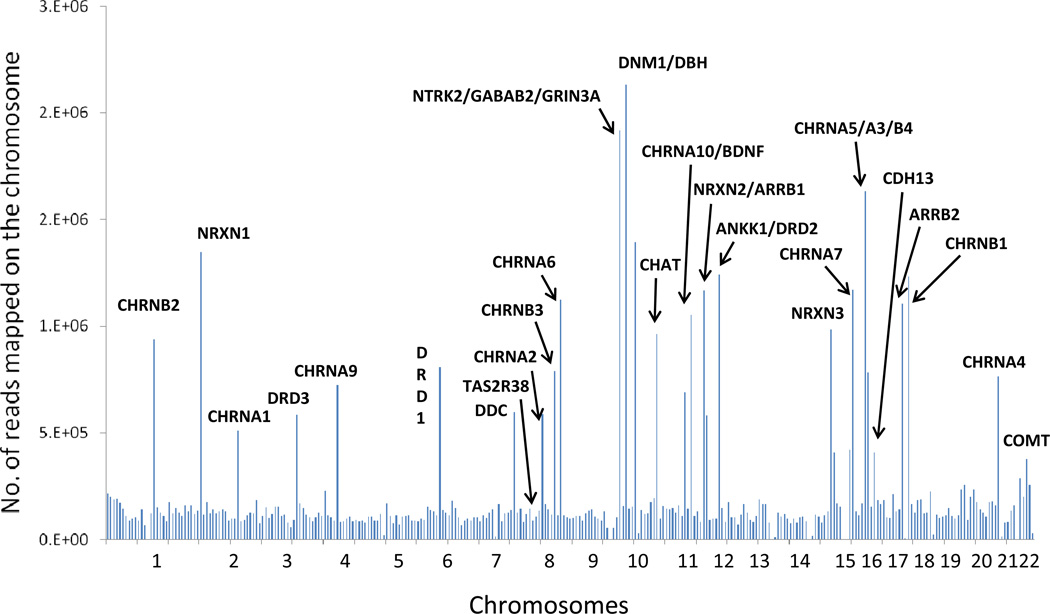
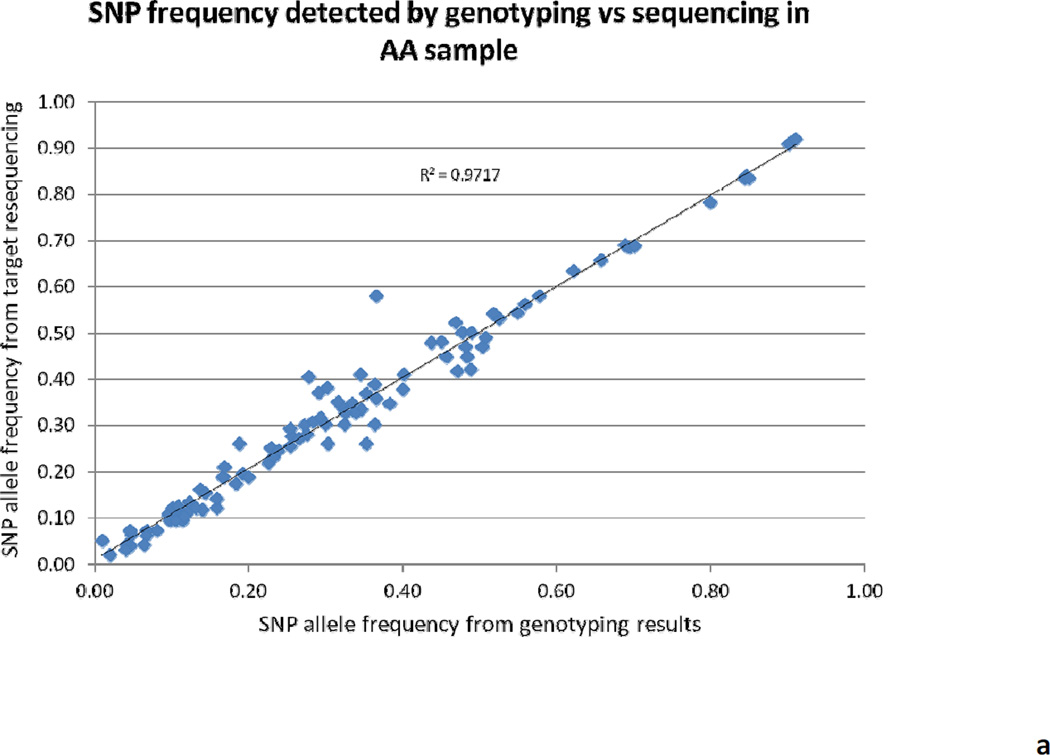
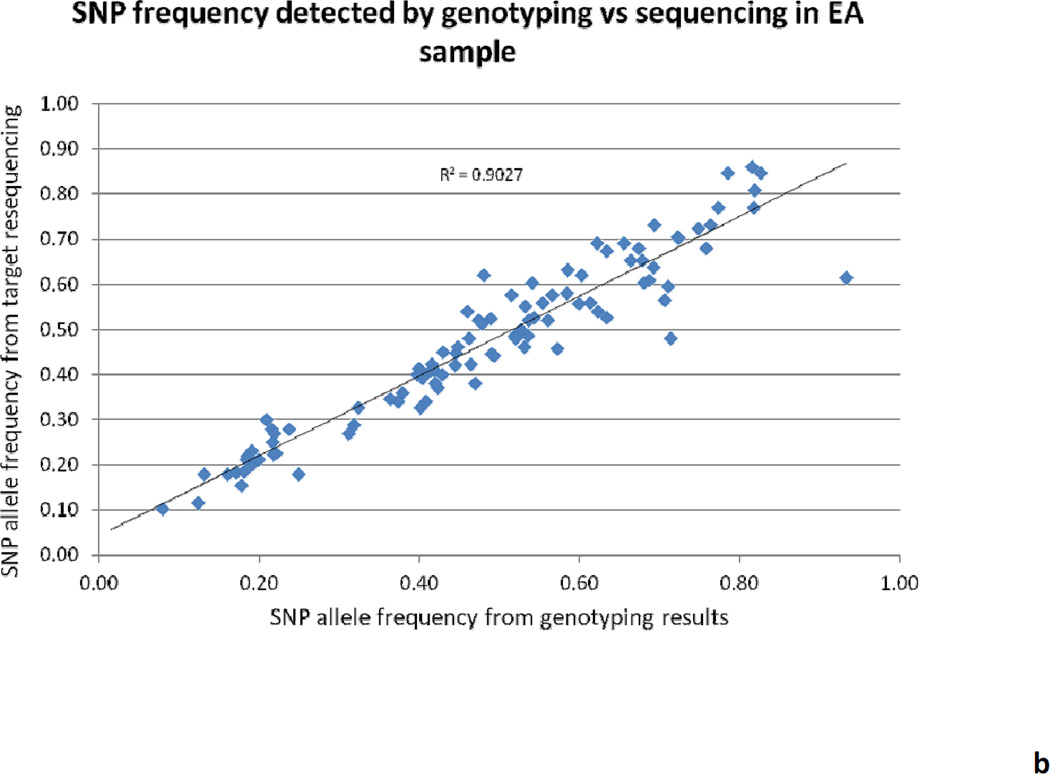
Overall, 62 Gb (868 million reads) of raw sequencing data was produced with an average of 108 million reads per pool. More than 80% of the raw sequencing data was mapped on the human genome (hg19) after filtering and mapping. A total of 147 million reads were mapped to the targeted regions, which is about 20% of the total mapped reads, and the entire targeted regions were 100% covered with a median coverage of 106× for each individual in the pool. The distribution of reads crossing the genome is shown in Figure 1. The minor allele frequency (MAF) was calculated for several common variants (MAF ≥ 0.05) from pooling the sequencing and then compared with our previous genotyping results based on ABI TaqMan assay; the correlation between these methods is 0.97 for AA samples and 0.90 for EA samples (Figure 2). The variants identified with a minimum MAF of 0.75% and minimum sequencing read counts of 500 were set up for variant selection. After removing the intron and synonymous variants, a total of 430 putative functional rare variants were identified from the eight pools and ranked according to the Polyphen and SIFT scores and variant frequency. Table 3 shows the summarized results of putative functional variants identified from each pool, including 28 premature stop codons, 212 damaging variants, and 190 tolerated variants. Several predicted-functional rare variants were selected for further validation in the Mid-South Tobacco Case/Control samples [45] and showed significant association with ND (Table 4), which is consistent with the previous report from Haller et al. [120].
Figure 1.
Figure 2.
Table 3.
Discovery of Putative Rare Variants from ND Candidate Gene Resequencing*
| Library/Pooled DNA Group | Synonymous | Non-synonymous | ||
|---|---|---|---|---|
| Tolerated | Damaging | Stop | ||
| African American Heavy Smokers | 197 | 97 | 129 | 22 |
| African American Heavy Smokers: Nonsmoker Sibling Controls | 166 | 82 | 59 | 5 |
| African American Light Smokers | 158 | 79 | 61 | 3 |
| African American Light Smoker: Nonsmoker Sibling Controls | 159 | 87 | 63 | 4 |
| European American Heavy Smokers | 99 | 62 | 51 | 5 |
| European American Heavy Smokers: Nonsmoker Sibling Controls | 103 | 61 | 46 | 3 |
| European American Light Smokers | 101 | 62 | 50 | 5 |
| European American Light Smokers: Nonsmoker Sibling Controls | 106 | 54 | 47 | 4 |
| Total | 190 | 212 | 28 | |
Number of variants shown in each row may overlap.
Table 4.
Significantly Associated Rare Variants in nACh Receptor Subunit Genes*
| SNP | A1 | American African | European American | Pooled | ||||||
|---|---|---|---|---|---|---|---|---|---|---|
| MAF (%) |
FTND | Q1 | MAF (%) |
FTND | Q1 | MAF (%) | FTND | Q1 | ||
| CHRNA3_H217Y | A | 0.05 | 0.192 | 0.681 | 0.22 | 0.139 | 0.0185 | 0.10 | 0.487 | 0.0123 |
| 0.192 | 0.681 | 0.139 | 0.0185 | 0.487 | 0.0123 | |||||
| CHRNB4_S140G | A | 4.25 | 0.0460 | 0.388 | 0.83 | 0.427 | 0.904 | 3.21 | 0.208 | 0.507 |
| 0.0408 | 0.274 | 0.427 | 0.904 | 0.200 | 0.398 | |||||
| CHRNB4_T91I | A | 0.73 | 0.429 | 0.484 | 3.58 | 0.0165 | 0.091 | 1.60 | 0.0015 | 0.128 |
| 0.429 | 0.484 | 0.0161 | 0.086 | 0.0016 | 0.128 | |||||
| CHRNB4_N41S | C | 1.40 | 0.246 | 0.890 | 0.22 | 0.136 | 0.035 | 1.04 | 0.190 | 0.653 |
| 0.246 | 0.890 | 0.136 | 0.035 | 0.190 | 0.653 | |||||
Note: Several low-frequency and rare variants within CHRNA5-CHRNA3-CHRNB4 discovered from targeted resequencing were selected for further validation on the TaqMan® OpenArray® Genotyping System (Life Technologies Inc.). The samples used in the replication study were from the Mid-South Tobacco Case-Control Study (MSTCC) population, which consists of 4,548 smokers and non-smokers aged 18 years or older of either African American (AA) (N = 3,161) or European American (EA) (N = 1,387) origin, who were recruited primarily from the city of Jackson, Mississippi during 2005–2011 [45]. Although questionnaires assessing various smoking-related behaviors were administered to each participant, only the Fagerström Test for Nicotine Dependence (FTND) and indexed CPD data were analyzed for this report. The association analysis was performed using a linear regression model by regressing FTND scores and indexed CPD on age and sex, in PLINK [130]. The FTND scores were continuous, ranging from 0 to 10, and indexed CPD categories were defined as 1: ≤10 CPD, 2: 11 to 20 CPD, 3: 21 to 30 CPD and 4: ≥31 CPD, as we did in our previous studies [38, 45]. Non-smokers were excluded from the regression model.
Function Studies of Rare Variants in ND
The majority of low-frequency and rare variants change the amino acid coding and offer the potential to study the function of these variants, which explains the phenotypic variance they cause. For nAChR subunit genes, in vitro electrophysiology has commonly been used to study the function of variants. The function of two CHRNB4 variants (T375I and T91I) and a missense variant in CHRNA3 (R37H) in strong LD with T91I were examined in vitro by an electrophysiology approach in HEK293 cells. The minor allele of each polymorphism increased the cellular response to nicotine (T375I: P = 0.01; T91I: P = 0.02; R37H: P = 0.003), but the largest effect on in vitro receptor activity was seen in the presence of both CHRNB4 T91I and CHRNA3 R37H (P = 2 × 10−6) [120].
In vitro function study does have its limitations; several knockout (KO) mice provide a chance to study the in vivo function of variants. KO mice with modified CHRNA5 [122] and CHRNB4 [123] subunits, as well as CHRNA3 KO [124] heterozygous knockout mice, facilitate the functional study of rare variants in the CHRNA5-A3-B4 gene clusters. Remarkably, A5 subunit knockdown in the medial habenula (MHb) did not alter the rewarding effects of nicotine but abolished the inhibitory effects of higher nicotine doses on brain reward systems [125]. Because the MHb projections extend almost exclusively to the interpeduncular nucleus (IPN), IPN activation was diminished in response to nicotine in CHRNA5-knockout mice, which further increased nicotine intake in rats with disruption of IPN signaling.
Taking it one step further for the in vivo function study, Hong et al. [126] reported the variant Asp398Asn in CHRNA5 is associated with the dorsal anterior cingulate-ventral/extended amygdala circuit through a resting-state functional connectivity (rsFC) approach, which decreases the intrinsic resting functional connectivity strength in this circuit. Although Asp398Asn is a common variant, it extended the functional study from in vitro to in vivo. Xie et al. [119] examined the effect of rare variants on in vivo nAchR binding using single-photon emission-computed tomography (SPECT) in a subsample of 139 subjects. One of the rare variants was associated with substantially greater nAchR availability in the brain than was seen in four age-matched individuals, suggesting that the variant alters nAchR availability. All the recent rare-variant functional studies related to ND focus on nAchR subunit genes; however, the rare variants of nAchR subunit genes may not explain all the phenotypic variance in ND. Several other genes that have been reported in ND GWAS [6, 121, 127, 128] and/or candidate studies need further investigation for potential functional rare variants associated with ND.
Conclusions
To date, all reported rare-variant studies related to substance abuse targeted only a handful of candidate genes. Almost all the recent studies were conducted on ND, but not on AD and other illicit drug dependences. Although these genes have been well investigated with confirmation of strong association with drug addiction based on the GWAS or candidate gene studies or both, unidentified gene/variants associated with drug addiction may still be missing. Whole-exome sequencing or largescale candidate genes targeted resequencing is necessary for studying unknown or less well studied genes/variants, especially the low-frequency and rare variants. However, rare-variant discovery requires extensive sequencing of much larger populations than is needed for common-variant identification based on the case-control studies design used for GWAS. Family-based designs may have some advantage over case-control studies when exploring the potential functional rare variants that change amino acid coding or gene expression pattern, as rare variants may be enriched in a few family samples that may increase the statistical power. Family-based designs are efficient for whole-genome sequencing because of the ability to impute the sequence of non-founders. Considering that linkage peaks identified in the family studies usually are not consistent with GWAS results, this suggests those linkage peaks likely harbor some rare variants that cannot be detected by GWAS because of the low frequency and lack of power. Taken together, these findings indicate that rare variants may have great potential to elucidate the unexplained contribution to the phenotypic variance for complex traits, but confirming this requires even greater efforts.
Supplementary Material
Table 1.
Recent GWAS/Candidate Studies in Drug Addiction
| Drug | Chr | Gene/Location | Variant(s) | Phenotype | References |
|---|---|---|---|---|---|
| Alcohol | 2 | 2q35 | rs7590720 rs1344694 |
AD | [18] |
| 4 | ADH1B | rs1229984 (R48H) | AD, esophageal cancer, upper aerodigestive-tract cancer | [19, 20] | |
| ADH1C | rs698 (I50V) | AD | [21] | ||
| 12 | ALDH2 | rs671 (E504K) | AD, alcoholic liver disease, cirrhosis, pancreatitis | [19, 22] | |
| 11 | ANKK1 | rs1800497 (Taq1A) | AD | [129] | |
| Nicotine | 2 | NRXN1 | rs10490162 | FTND | [49, 50] |
| 8 | CHRNB3 | rs6474412 | CPD, FTND, lung cancer | [41, 44, 48] | |
| CHRNA6 | rs2304297 | [41] | |||
| 9 | DBH | rs3025343 | Smoking cessation | [48, 51] | |
| 10 | 10q25 | rs1028936 rs1329650 |
CPD | [51] | |
| 11 | BDNF | rs6265 | Smoking initiation | [51, 52] | |
| ANKK1 | rs2734849 (Arg/His) rs4938012 |
FTND | [46, 47] | ||
| DRD2 | rs4245150 rs17602038 |
||||
| 15 | CHRNA5 | rs588765 rs578776 rs16969968 (D398N) rs6495308 rs55853698 rs2036527 |
Chronic pulmonary disease | [6, 127, 128] | |
| CHRNA3 | rs1051730 rs6495308 |
||||
| CHRNB4 | rs1996371 | ||||
| Nicotine | 19 | CYP2A6-CYP2B6 | rs4105144 | Chronic pulmonary disease | [41] |
| EGLN2 | rs3733829 | Chronic pulmonary disease | [51] | ||
| Opioid | 6 | OPRM1 | rs1799971 (A118G) | Opioid addiction Heroin addiction |
[58, 61–63] |
| rs1799972 (A6V) | |||||
| rs510769 | |||||
| 1 | OPRD1 | rs2236861 rs1042114 |
Opioid addiction Heroin addiction |
[58, 60, 63] |
The SNPs listed for AD and ND were reported either in the GWAS study, which reached genome-wide significance, or in the candidate gene-based association studies that are significant after correction and have been replicated in independent studies.
Acknowledgements
The preparation of this review was supported by NIH grant DA-012844 to MDL. The authors thank Dr. David Bronson for his excellent editing.
References
- 1.WHO. Report on the Global Tobacco Epidemic, 2008: The MPOWER package. Geneva: World Health Organization; 2008. [Google Scholar]
- 2.Mokdad AH, Marks JS, Stroup DF, Gerberding JL. Actual causes of death in the United States, 2000. JAMA. 2004;291:1238–1245. doi: 10.1001/jama.291.10.1238. [DOI] [PubMed] [Google Scholar]
- 3.Centers for Disease, C and Prevention. Smoking-attributable mortality, years of potential life lost, and productivity losses--United States, 2000–2004. MMWR. Morbidity and mortality weekly report. 2008;57:1226–1228. [PubMed] [Google Scholar]
- 4.Rhee SH, Hewitt JK, Young SE, Corley RP, Crowley TJ, Stallings MC. Genetic and environmental influences on substance initiation, and problem use in adolescents. Archives of general psychiatry. 2003;60:1256–1264. doi: 10.1001/archpsyc.60.12.1256. [DOI] [PubMed] [Google Scholar]
- 5.Palmer RHC, Young SE, Hopfer CJ, Corley RP, Stallings MC, Crowley TJ, Hewitt JK. Developmental epidemiology of drug use and abuse in adolescence and young adulthood: Evidence of generalized risk. Drug and Alcohol Dependence. 2009;102:78–87. doi: 10.1016/j.drugalcdep.2009.01.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Li MD, Burmeister M. New insights into the genetics of addiction. Nat Rev Genet. 2009;10:225–231. doi: 10.1038/nrg2536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Bierut LJ, Dinwiddie SH, Begleiter H, Crowe RR, Hesselbrock V, Nurnberger JI, Jr, Porjesz B, Schuckit MA, Reich T. Familial transmission of substance dependence: alcohol, marijuana, cocaine, and habitual smoking: a report from the Collaborative Study on the Genetics of Alcoholism. Archives of general psychiatry. 1998;55:982–988. doi: 10.1001/archpsyc.55.11.982. [DOI] [PubMed] [Google Scholar]
- 8.Agrawal A, Lynskey MT. Are there genetic influences on addiction: evidence from family, adoption and twin studies. Addiction. 2008;103:1069–1081. doi: 10.1111/j.1360-0443.2008.02213.x. [DOI] [PubMed] [Google Scholar]
- 9.Li MD, Cheng R, Ma JZ, Swan GE. A meta-analysis of estimated genetic and environmental effects on smoking behavior in male and female adult twins. Addiction. 2003;98:23–31. doi: 10.1046/j.1360-0443.2003.00295.x. [DOI] [PubMed] [Google Scholar]
- 10.Tsuang MT, Bar JL, Harley RM, Lyons MJ. The Harvard Twin Study of Substance Abuse: what we have learned. Harvard review of psychiatry. 2001;9:267–279. [PubMed] [Google Scholar]
- 11.Agrawal A, Lynskey MT, Hinrichs A, Grucza R, Saccone SF, Krueger R, Neuman R, Howells W, Fisher S, Fox L, et al. A genome-wide association study of DSM-IV cannabis dependence. Addiction biology. 2011;16:514–518. doi: 10.1111/j.1369-1600.2010.00255.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Long JC, Knowler WC, Hanson RL, Robin RW, Urbanek M, Moore E, Bennett PH, Goldman D. Evidence for genetic linkage to alcohol dependence on chromosomes 4 and 11 from an autosome-wide scan in an American Indian population. Am J Med Genet. 1998;81:216–221. doi: 10.1002/(sici)1096-8628(19980508)81:3<216::aid-ajmg2>3.0.co;2-u. [DOI] [PubMed] [Google Scholar]
- 13.Reich T, Edenberg HJ, Goate A, Williams JT, Rice JP, Van Eerdewegh P, Foroud T, Hesselbrock V, Schuckit MA, Bucholz K, et al. Genome-wide search for genes affecting the risk for alcohol dependence. Am J Med Genet. 1998;81:207–215. [PubMed] [Google Scholar]
- 14.Foroud T, Edenberg HJ, Goate A, Rice J, Flury L, Koller DL, Bierut LJ, Conneally PM, Nurnberger JI, Bucholz KK, et al. Alcoholism susceptibility loci: confirmation studies in a replicate sample and further mapping. Alcohol Clin Exp Res. 2000;24:933–945. [PubMed] [Google Scholar]
- 15.Cui WY, Seneviratne C, Gu J, Li MD. Genetics of GABAergic signaling in nicotine and alcohol dependence. Hum Genet. 2012;131:843–855. doi: 10.1007/s00439-011-1108-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Gelernter J, Kranzler HR. Genetics of alcohol dependence. Hum Genet. 2009;126:91–99. doi: 10.1007/s00439-009-0701-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Goldman D, Oroszi G, Ducci F. The genetics of addictions: uncovering the genes. Nat Rev Genet. 2005;6:521–532. doi: 10.1038/nrg1635. [DOI] [PubMed] [Google Scholar]
- 18.Treutlein J, Cichon S, Ridinger M, Wodarz N, Soyka M, Zill P, Maier W, Moessner R, Gaebel W, Dahmen N, et al. Genome-wide association study of alcohol dependence. Archives of general psychiatry. 2009;66:773–784. doi: 10.1001/archgenpsychiatry.2009.83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Li D, Zhao H, Gelernter J. Strong association of the alcohol dehydrogenase 1B gene (ADH1B) with alcohol dependence and alcohol-induced medical diseases. Biological psychiatry. 2011;70:504–512. doi: 10.1016/j.biopsych.2011.02.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Guo H, Zhang G, Mai R. Alcohol dehydrogenase-1B Arg47His polymorphism and upper aerodigestive tract cancer risk: a meta-analysis including 24,252 subjects. Alcoholism, clinical and experimental research. 2012;36:272–278. doi: 10.1111/j.1530-0277.2011.01621.x. [DOI] [PubMed] [Google Scholar]
- 21.Li D, Zhao H, Gelernter J. Further clarification of the contribution of the ADH1C gene to vulnerability of alcoholism and selected liver diseases. Hum Genet. 2012;131:1361–1374. doi: 10.1007/s00439-012-1163-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Li D, Zhao H, Gelernter J. Strong protective effect of the aldehyde dehydrogenase gene (ALDH2) 504lys (*2) allele against alcoholism and alcohol-induced medical diseases in Asians. Human genetics. 2012;131:725–737. doi: 10.1007/s00439-011-1116-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Li MD. Identifying susceptibility loci for nicotine dependence: 2008 update based on recent genome-wide linkage analyses. Hum Genet. 2008;123:119–131. doi: 10.1007/s00439-008-0473-0. [DOI] [PubMed] [Google Scholar]
- 24.Li MD, Ma JZ, Payne TJ, Lou XY, Zhang D, Dupont RT, Elston RC. Genome-wide linkage scan for nicotine dependence in European Americans and its converging results with African Americans in the Mid-South Tobacco Family sample. Mol Psychiatry. 2008;13:407–416. doi: 10.1038/sj.mp.4002038. [DOI] [PubMed] [Google Scholar]
- 25.Gelernter J, Liu X, Hesselbrock V, Page GP, Goddard A, Zhang H. Results of a genomewide linkage scan: support for chromosomes 9 and 11 loci increasing risk for cigarette smoking. American journal of medical genetics. Part B, Neuropsychiatric genetics : the official publication of the International Society of Psychiatric Genetics. 2004;128B:94–101. doi: 10.1002/ajmg.b.30019. [DOI] [PubMed] [Google Scholar]
- 26.Morley KI, Medland SE, Ferreira MA, Lynskey MT, Montgomery GW, Heath AC, Madden PA, Martin NG. A Possible Smoking Susceptibility Locus on Chromosome 11p12: Evidence from Sex-limitation Linkage Analyses in a Sample of Australian Twin Families. Behav Genet. 2006;36:87–99. doi: 10.1007/s10519-005-9004-0. [DOI] [PubMed] [Google Scholar]
- 27.Vink JM, Posthuma D, Neale MC, Eline Slagboom P, Boomsma DI. Genome-wide Linkage Scan to Identify Loci for Age at First Cigarette in Dutch Sibling Pairs. Behavior genetics. 2006;36:100–111. doi: 10.1007/s10519-005-9012-0. [DOI] [PubMed] [Google Scholar]
- 28.Saccone SF, Pergadia ML, Loukola A, Broms U, Montgomery GW, Wang JC, Agrawal A, Dick DM, Heath AC, Todorov AA, et al. Genetic linkage to chromosome 22q12 for a heavy-smoking quantitative trait in two independent samples. Am J Hum Genet. 2007;80:856–866. doi: 10.1086/513703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Loukola A, Broms U, Maunu H, Widen E, Heikkila K, Siivola M, Salo A, Pergadia ML, Nyman E, Sammalisto S, et al. Linkage of nicotine dependence and smoking behavior on 10q, 7q and 11p in twins with homogeneous genetic background. Pharmacogenomics J. 2008;8:209–219. doi: 10.1038/sj.tpj.6500464. [DOI] [PubMed] [Google Scholar]
- 30.Morabia A, Cayanis E, Costanza MC, Ross BM, Bernstein MS, Flaherty MS, Alvin GB, Das K, Morris MA, Penchaszadeh GK, et al. Association between lipoprotein lipase (LPL) gene and blood lipids: a common variant for a common trait? Genet Epidemiol. 2003;24:309–321. doi: 10.1002/gepi.10229. [DOI] [PubMed] [Google Scholar]
- 31.Wang D, Ma JZ, Li MD. Mapping and verification of susceptibility loci for smoking quantity using permutation linkage analysis. Pharmacogenomics J. 2005;5:166–172. doi: 10.1038/sj.tpj.6500304. [DOI] [PubMed] [Google Scholar]
- 32.Li MD, Payne TJ, Ma JZ, Lou XY, Zhang D, Dupont RT, Crews KM, Somes G, Williams NJ, Elston RC. A genomewide search finds major susceptibility Loci for nicotine dependence on chromosome 10 in african americans. Am J Hum Genet. 2006;79:745–751. doi: 10.1086/508208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Swan GE, Hops H, Wilhelmsen KC, Lessov-Schlaggar CN, Cheng LS, Hudmon KS, Amos CI, Feiler HS, Ring HZ, Andrews JA, et al. A genome-wide screen for nicotine dependence susceptibility loci. Am J Med Genet B Neuropsychiatr Genet. 2006;141:354–360. doi: 10.1002/ajmg.b.30315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Gelernter J, Panhuysen C, Weiss R, Brady K, Poling J, Krauthammer M, Farrer L, Kranzler HR. Genomewide linkage scan for nicotine dependence: identification of a chromosome 5 risk locus. Biological psychiatry. 2007;61:119–126. doi: 10.1016/j.biopsych.2006.08.023. [DOI] [PubMed] [Google Scholar]
- 35.Li MD, Sun D, Lou XY, Beuten J, Payne TJ, Ma JZ. Linkage and association studies in African- and Caucasian-American populations demonstrate that SHC3 is a novel susceptibility locus for nicotine dependence. Mol Psychiatry. 2007;12:462–473. doi: 10.1038/sj.mp.4001933. [DOI] [PubMed] [Google Scholar]
- 36.Bierut LJ, Stitzel JA, Wang JC, Hinrichs AL, Grucza RA, Xuei X, Saccone NL, Saccone SF, Bertelsen S, Fox L, et al. Variants in Nicotinic Receptors and Risk for Nicotine Dependence. Am J Psychiatry. 2008;165:1163–1171. doi: 10.1176/appi.ajp.2008.07111711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Li MD, Xu Q, Lou XY, Payne TJ, Niu T, Ma JZ. Association and interaction analysis of variants in CHRNA5/CHRNA3/CHRNB4 gene cluster with nicotine dependence in African and European Americans. Am J Med Genet B Neuropsychiatr Genet. 2010;153B:745–756. doi: 10.1002/ajmg.b.31043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li MD, Yoon D, Lee JY, Han BG, Niu T, Payne TJ, Ma JZ, Park T. Associations of variants in CHRNA5/A3/B4 gene cluster with smoking behaviors in a Korean population. PLoS One. 2010;5:e12183. doi: 10.1371/journal.pone.0012183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Liu JZ, Tozzi F, Waterworth DM, Pillai SG, Muglia P, Middleton L, Berrettini W, Knouff CW, Yuan X, Waeber G, et al. Meta-analysis and imputation refines the association of 15q25 with smoking quantity. Nature genetics. 2010;42:436–440. doi: 10.1038/ng.572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Saccone NL, Culverhouse RC, Schwantes-An TH, Cannon DS, Chen X, Cichon S, Giegling I, Han S, Han Y, Keskitalo-Vuokko K, et al. Multiple independent loci at chromosome 15q25.1 affect smoking quantity: a meta-analysis and comparison with lung cancer and COPD. PLoS genetics. 2010;6 doi: 10.1371/journal.pgen.1001053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Thorgeirsson TE, Gudbjartsson DF, Surakka I, Vink JM, Amin N, Geller F, Sulem P, Rafnar T, Esko T, Walter S, et al. Sequence variants at CHRNB3-CHRNA6 and CYP2A6 affect smoking behavior. Nat Genet. 2010;42:448–453. doi: 10.1038/ng.573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tobacco & Genetics. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nature genetics. 2010;42:441–447. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Truong T, Hung RJ, Amos CI, Wu X, Bickeboller H, Rosenberger A, Sauter W, Illig T, Wichmann HE, Risch A, et al. Replication of lung cancer susceptibility loci at chromosomes 15q25, 5p15, and 6p21: a pooled analysis from the International Lung Cancer Consortium. Journal of the National Cancer Institute. 2010;102:959–971. doi: 10.1093/jnci/djq178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Saccone NL, Schwantes-An TH, Wang JC, Grucza RA, Breslau N, Hatsukami D, Johnson EO, Rice JP, Goate AM, Bierut LJ. Multiple cholinergic nicotinic receptor genes affect nicotine dependence risk in African and European Americans. Genes, brain, and behavior. 2010;9:741–750. doi: 10.1111/j.1601-183X.2010.00608.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Cui WY, Wang S, Yang J, Yi SG, Yoon D, Kim YJ, Payne TJ, Ma JZ, Park T, Li MD. Significant association of CHRNB3 variants with nicotine dependence in multiple ethnic populations. Mol Psychiatry. 2013 doi: 10.1038/mp.2012.190. [DOI] [PubMed] [Google Scholar]
- 46.Gelernter J, Yu Y, Weiss R, Brady K, Panhuysen C, Yang BZ, Kranzler HR, Farrer L. Haplotype spanning TTC12 and ANKK1, flanked by the DRD2 and NCAM1 loci, is strongly associated to nicotine dependence in two distinct American populations. Hum Mol Genet. 2006;15:3498–3507. doi: 10.1093/hmg/ddl426. [DOI] [PubMed] [Google Scholar]
- 47.Huang W, Payne TJ, Ma JZ, Beuten J, Dupont RT, Inohara N, Li MD. Significant association of ANKK1 and detection of a functional polymorphism with nicotine dependence in an African-American sample. Neuropsychopharmacology. 2009;34:319–330. doi: 10.1038/npp.2008.37. [DOI] [PubMed] [Google Scholar]
- 48.Saccone NL, Saccone SF, Hinrichs AL, Stitzel JA, Duan W, Pergadia ML, Agrawal A, Breslau N, Grucza RA, Hatsukami D, et al. Multiple distinct risk loci for nicotine dependence identified by dense coverage of the complete family of nicotinic receptor subunit (CHRN) genes. Am J Med Genet B Neuropsychiatr Genet. 2009;150B:453–466. doi: 10.1002/ajmg.b.30828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bierut LJ, Madden PA, Breslau N, Johnson EO, Hatsukami D, Pomerleau OF, Swan GE, Rutter J, Bertelsen S, Fox L, et al. Novel genes identified in a high-density genome wide association study for nicotine dependence. Hum Mol Genet. 2007;16:24–35. doi: 10.1093/hmg/ddl441. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Nussbaum J, Xu Q, Payne TJ, Ma JZ, Huang W, Gelernter J, Li MD. Significant association of the neurexin-1 gene (NRXN1) with nicotine dependence in European- and African-American smokers. Hum Mol Genet. 2008;17:1569–1577. doi: 10.1093/hmg/ddn044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.TAG. Genome-wide meta-analyses identify multiple loci associated with smoking behavior. Nat Genet. 2010;42:441–447. doi: 10.1038/ng.571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Beuten J, Ma JZ, Payne TJ, Dupont RT, Quezada P, Huang W, Crews KM, Li MD. Significant association of BDNF haplotypes in European-American male smokers but not in European-American female or African-American smokers. Am J Med Genet B Neuropsychiatr Genet. 2005;139:73–80. doi: 10.1002/ajmg.b.30231. [DOI] [PubMed] [Google Scholar]
- 53.Hopfer CJ, Lessem JM, Hartman CA, Stallings MC, Cherny SS, Corley RP, Hewitt JK, Krauter KS, Mikulich-Gilbertson SK, Rhee SH, et al. A genome-wide scan for loci influencing adolescent cannabis dependence symptoms: evidence for linkage on chromosomes 3 and 9. Drug Alcohol Depend. 2007;89:34–41. doi: 10.1016/j.drugalcdep.2006.11.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Ehlers CL, Gilder DA, Gizer IR, Wilhelmsen KC. Heritability and a genome-wide linkage analysis of a Type II/B cluster construct for cannabis dependence in an American Indian community. Addiction biology. 2009;14:338–348. doi: 10.1111/j.1369-1600.2009.00160.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Gelernter J, Panhuysen C, Weiss R, Brady K, Hesselbrock V, Rounsaville B, Poling J, Wilcox M, Farrer L, Kranzler HR. Genomewide linkage scan for cocaine dependence and related traits: significant linkages for a cocaine-related trait and cocaine-induced paranoia. Am J Med Genet B Neuropsychiatr Genet. 2005;136B:45–52. doi: 10.1002/ajmg.b.30189. [DOI] [PubMed] [Google Scholar]
- 56.Gelernter J, Panhuysen C, Wilcox M, Hesselbrock V, Rounsaville B, Poling J, Weiss R, Sonne S, Zhao H, Farrer L, et al. Genomewide linkage scan for opioid dependence and related traits. Am J Hum Genet. 2006;78:759–769. doi: 10.1086/503631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lachman HM, Fann CS, Bartzis M, Evgrafov OV, Rosenthal RN, Nunes EV, Miner C, Santana M, Gaffney J, Riddick A, et al. Genomewide suggestive linkage of opioid dependence to chromosome 14q. Hum Mol Genet. 2007;16:1327–1334. doi: 10.1093/hmg/ddm081. [DOI] [PubMed] [Google Scholar]
- 58.Levran O, Awolesi O, Linzy S, Adelson M, Kreek MJ. Haplotype block structure of the genomic region of the mu opioid receptor gene. Journal of human genetics. 2011;56:147–155. doi: 10.1038/jhg.2010.150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Levran O, Yuferov V, Kreek MJ. The genetics of the opioid system and specific drug addictions. Human genetics. 2012;131:823–842. doi: 10.1007/s00439-012-1172-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Zhang H, Kranzler HR, Yang BZ, Luo X, Gelernter J. The OPRD1 and OPRK1 loci in alcohol or drug dependence: OPRD1 variation modulates substance dependence risk. Molecular psychiatry. 2008;13:531–543. doi: 10.1038/sj.mp.4002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Bart G, Heilig M, LaForge KS, Pollak L, Leal SM, Ott J, Kreek MJ. Substantial attributable risk related to a functional mu-opioid receptor gene polymorphism in association with heroin addiction in central Sweden. Molecular psychiatry. 2004;9:547–549. doi: 10.1038/sj.mp.4001504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Tan EC, Tan CH, Karupathivan U, Yap EP. Mu opioid receptor gene polymorphisms and heroin dependence in Asian populations. Neuroreport. 2003;14:569–572. doi: 10.1097/00001756-200303240-00008. [DOI] [PubMed] [Google Scholar]
- 63.Levran O, Londono D, O'Hara K, Nielsen DA, Peles E, Rotrosen J, Casadonte P, Linzy S, Randesi M, Ott J, et al. Genetic susceptibility to heroin addiction: a candidate gene association study. Genes, brain, and behavior. 2008;7:720–729. doi: 10.1111/j.1601-183X.2008.00410.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Nielsen DA, Ji F, Yuferov V, Ho A, He C, Ott J, Kreek MJ. Genome-wide association study identifies genes that may contribute to risk for developing heroin addiction. Psychiatric genetics. 2010;20:207–214. doi: 10.1097/YPG.0b013e32833a2106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nishizawa D, Fukuda K, Kasai S, Hasegawa J, Aoki Y, Nishi A, Saita N, Koukita Y, Nagashima M, Katoh R, et al. Genome-wide association study identifies a potent locus associated with human opioid sensitivity. Molecular psychiatry. 2012 doi: 10.1038/mp.2012.164. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Feinberg AP. Phenotypic plasticity and the epigenetics of human disease. Nature. 2007;447:433–440. doi: 10.1038/nature05919. [DOI] [PubMed] [Google Scholar]
- 67.Mackay TF, Stone EA, Ayroles JF. The genetics of quantitative traits: challenges and prospects. Nature reviews. Genetics. 2009;10:565–577. doi: 10.1038/nrg2612. [DOI] [PubMed] [Google Scholar]
- 68.Shi J, Levinson DF, Duan J, Sanders AR, Zheng Y, Pe'er I, Dudbridge F, Holmans PA, Whittemore AS, Mowry BJ, et al. Common variants on chromosome 6p22.1 are associated with schizophrenia. Nature. 2009;460:753–757. doi: 10.1038/nature08192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Visscher PM, Hill WG, Wray NR. Heritability in the genomics era--concepts and misconceptions. Nature reviews. Genetics. 2008;9:255–266. doi: 10.1038/nrg2322. [DOI] [PubMed] [Google Scholar]
- 70.Cirulli ET, Goldstein DB. Uncovering the roles of rare variants in common disease through whole-genome sequencing. Nature reviews. Genetics. 2010;11:415–425. doi: 10.1038/nrg2779. [DOI] [PubMed] [Google Scholar]
- 71.Feldman MW, Lewontin RC. The heritability hang-up. Science. 1975;190:1163–1168. doi: 10.1126/science.1198102. [DOI] [PubMed] [Google Scholar]
- 72.Eichler EE, Flint J, Gibson G, Kong A, Leal SM, Moore JH, Nadeau JH. Missing heritability and strategies for finding the underlying causes of complex disease. Nature reviews. Genetics. 2010;11:446–450. doi: 10.1038/nrg2809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Mackay TF. The genetic architecture of quantitative traits. Annual review of genetics. 2001;35:303–339. doi: 10.1146/annurev.genet.35.102401.090633. [DOI] [PubMed] [Google Scholar]
- 74.Gibson G. Rare and common variants: twenty arguments. Nature reviews. Genetics. 2011;13:135–145. doi: 10.1038/nrg3118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Lupski JR, Belmont JW, Boerwinkle E, Gibbs RA. Clan genomics and the complex architecture of human disease. Cell. 2011;147:32–43. doi: 10.1016/j.cell.2011.09.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Ng SB, Turner EH, Robertson PD, Flygare SD, Bigham AW, Lee C, Shaffer T, Wong M, Bhattacharjee A, Eichler EE, et al. Targeted capture and massively parallel sequencing of 12 human exomes. Nature. 2009;461:272–276. doi: 10.1038/nature08250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Abecasis GR, Altshuler D, Auton A, Brooks LD, Durbin RM, Gibbs RA, Hurles ME, McVean GA. A map of human genome variation from population-scale sequencing. Nature. 2010;467:1061–1073. doi: 10.1038/nature09534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Abecasis GR, Auton A, Brooks LD, DePristo MA, Durbin RM, Handsaker RE, Kang HM, Marth GT, McVean GA. An integrated map of genetic variation from 1,092 human genomes. Nature. 2012;491:56–65. doi: 10.1038/nature11632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Altshuler DM, Gibbs RA, Peltonen L, Altshuler DM, Gibbs RA, Peltonen L, Dermitzakis E, Schaffner SF, Yu F, Peltonen L, et al. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li Y, Sidore C, Kang HM, Boehnke M, Abecasis GR. Low-coverage sequencing: implications for design of complex trait association studies. Genome Res. 2011;21:940–951. doi: 10.1101/gr.117259.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Flanigan KM, Gastier-Foster J, Pyatt R, Rosales XQ, Thrush DL, Kneile K, Mendell JR, Kelly B, Newsom D, Hu P, et al. Comparison of commercially-available exome capture kits in the diagnosis of neuromuscular disorders. Neuromuscular Disorders. 2012;22:808. [Google Scholar]
- 82.Ewing CM, Ray AM, Lange EM, Zuhlke KA, Robbins CM, Tembe WD, Wiley KE, Isaacs SD, Johng D, Wang Y, et al. Germline mutations in HOXB13 and prostate-cancer risk. The New England journal of medicine. 2012;366:141–149. doi: 10.1056/NEJMoa1110000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Mondal K, Ramachandran D, Patel VC, Hagen KR, Bose P, Cutler DJ, Zwick ME. Excess variants in AFF2 detected by massively parallel sequencing of males with autism spectrum disorder. Human molecular genetics. 2012;21:4356–4364. doi: 10.1093/hmg/dds267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Myllykangas S, Buenrostro JD, Natsoulis G, Bell JM, Ji HP. Efficient targeted resequencing of human germline and cancer genomes by oligonucleotide-selective sequencing. Nature biotechnology. 2011;29:1024–1027. doi: 10.1038/nbt.1996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Sham P, Bader JS, Craig I, O'Donovan M, Owen M. DNA Pooling: a tool for large-scale association studies. Nat Rev Genet. 2002;3:862–871. doi: 10.1038/nrg930. [DOI] [PubMed] [Google Scholar]
- 86.Norton N, Williams NM, O'Donovan MC, Owen MJ. DNA pooling as a tool for large-scale association studies in complex traits. Annals of medicine. 2004;36:146–152. doi: 10.1080/07853890310021724. [DOI] [PubMed] [Google Scholar]
- 87.Nejentsev S, Walker N, Riches D, Egholm M, Todd JA. Rare variants of IFIH1, a gene implicated in antiviral responses, protect against type 1 diabetes. Science. 2009;324:387–389. doi: 10.1126/science.1167728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Calvo SE, Tucker EJ, Compton AG, Kirby DM, Crawford G, Burtt NP, Rivas M, Guiducci C, Bruno DL, Goldberger OA, et al. High-throughput, pooled sequencing identifies mutations in NUBPL and FOXRED1 in human complex I deficiency. Nature genetics. 2010;42:851–858. doi: 10.1038/ng.659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nature reviews. Genetics. 2010;11:499–511. doi: 10.1038/nrg2796. [DOI] [PubMed] [Google Scholar]
- 90.Frazer KA, Ballinger DG, Cox DR, Hinds DA, Stuve LL, Gibbs RA, Belmont JW, Boudreau A, Hardenbol P, Leal SM, et al. A second generation human haplotype map of over 3.1 million SNPs. Nature. 2007;449:851–861. doi: 10.1038/nature06258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Li H, Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25:1754–1760. doi: 10.1093/bioinformatics/btp324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.McKenna A, Hanna M, Banks E, Sivachenko A, Cibulskis K, Kernytsky A, Garimella K, Altshuler D, Gabriel S, Daly M, et al. The Genome Analysis Toolkit: a MapReduce framework for analyzing next-generation DNA sequencing data. Genome research. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Li H. A statistical framework for SNP calling, mutation discovery, association mapping and population genetical parameter estimation from sequencing data. Bioinformatics. 2011;27:2987–2993. doi: 10.1093/bioinformatics/btr509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Scott LJ, Mohlke KL, Bonnycastle LL, Willer CJ, Li Y, Duren WL, Erdos MR, Stringham HM, Chines PS, Jackson AU, et al. A genome-wide association study of type 2 diabetes in Finns detects multiple susceptibility variants. Science. 2007;316:1341–1345. doi: 10.1126/science.1142382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Liu EY, Li M, Wang W, Li Y. MaCH-admix: genotype imputation for admixed populations. Genetic epidemiology. 2013;37:25–37. doi: 10.1002/gepi.21690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Marchini J, Howie B, Myers S, McVean G, Donnelly P. A new multipoint method for genome-wide association studies by imputation of genotypes. Nature genetics. 2007;39:906–913. doi: 10.1038/ng2088. [DOI] [PubMed] [Google Scholar]
- 97.Howie BN, Donnelly P, Marchini J. A flexible and accurate genotype imputation method for the next generation of genome-wide association studies. PLoS Genet. 2009;5:e1000529. doi: 10.1371/journal.pgen.1000529. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Browning SR, Browning BL. Haplotype phasing: existing methods and new developments. Nature reviews. Genetics. 2011;12:703–714. doi: 10.1038/nrg3054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Howie B, Fuchsberger C, Stephens M, Marchini J, Abecasis GR. Fast and accurate genotype imputation in genome-wide association studies through pre-phasing. Nat Genet. 2012;44:955–959. doi: 10.1038/ng.2354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Browning SR, Browning BL. Rapid and accurate haplotype phasing and missing-data inference for whole-genome association studies by use of localized haplotype clustering. Am J Hum Genet. 2007;81:1084–1097. doi: 10.1086/521987. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Browning BL, Browning SR. A unified approach to genotype imputation and haplotype-phase inference for large data sets of trios and unrelated individuals. American journal of human genetics. 2009;84:210–223. doi: 10.1016/j.ajhg.2009.01.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Li B, Leal SM. Methods for detecting associations with rare variants for common diseases: application to analysis of sequence data. Am J Hum Genet. 2008;83:311–321. doi: 10.1016/j.ajhg.2008.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Morgenthaler S, Thilly WG. A strategy to discover genes that carry multi-allelic or mono-allelic risk for common diseases: a cohort allelic sums test (CAST) Mutation research. 2007;615:28–56. doi: 10.1016/j.mrfmmm.2006.09.003. [DOI] [PubMed] [Google Scholar]
- 104.Madsen BE, Browning SR. A groupwise association test for rare mutations using a weighted sum statistic. PLoS Genet. 2009;5:e1000384. doi: 10.1371/journal.pgen.1000384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Morris AP, Zeggini E. An evaluation of statistical approaches to rare variant analysis in genetic association studies. Genetic epidemiology. 2010;34:188–193. doi: 10.1002/gepi.20450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Cohen JC, Kiss RS, Pertsemlidis A, Marcel YL, McPherson R, Hobbs HH. Multiple rare alleles contribute to low plasma levels of HDL cholesterol. Science. 2004;305:869–872. doi: 10.1126/science.1099870. [DOI] [PubMed] [Google Scholar]
- 107.Fearnhead NS, Wilding JL, Winney B, Tonks S, Bartlett S, Bicknell DC, Tomlinson IP, Mortensen NJ, Bodmer WF. Multiple rare variants in different genes account for multifactorial inherited susceptibility to colorectal adenomas. Proceedings of the National Academy of Sciences of the United States of America. 2004;101:15992–15997. doi: 10.1073/pnas.0407187101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Price AL, Kryukov GV, de Bakker PI, Purcell SM, Staples J, Wei LJ, Sunyaev SR. Pooled association tests for rare variants in exon-resequencing studies. American journal of human genetics. 2010;86:832–838. doi: 10.1016/j.ajhg.2010.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Neale BM, Rivas MA, Voight BF, Altshuler D, Devlin B, Orho-Melander M, Kathiresan S, Purcell SM, Roeder K, Daly MJ. Testing for an unusual distribution of rare variants. PLoS genetics. 2011;7:e1001322. doi: 10.1371/journal.pgen.1001322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Wu Michael C, Lee S, Cai T, Li Y, Boehnke M, Lin X. Rare-Variant Association Testing for Sequencing Data with the Sequence Kernel Association Test. The American Journal of Human Genetics. 2011;89:82–93. doi: 10.1016/j.ajhg.2011.05.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Basu S, Pan W. Comparison of statistical tests for disease association with rare variants. Genet Epidemiol. 2011;35:606–619. doi: 10.1002/gepi.20609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Lin DY, Tang ZZ. A general framework for detecting disease associations with rare variants in sequencing studies. American journal of human genetics. 2011;89:354–367. doi: 10.1016/j.ajhg.2011.07.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Lee S, Emond MJ, Bamshad MJ, Barnes KC, Rieder MJ, Nickerson DA, Christiani DC, Wurfel MM, Lin X Team NGESP-ELP. Optimal unified approach for rare-variant association testing with application to small-sample case-control whole-exome sequencing studies. American journal of human genetics. 2012;91:224–237. doi: 10.1016/j.ajhg.2012.06.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Chen H, Meigs JB, Dupuis J. Sequence kernel association test for quantitative traits in family samples. Genet Epidemiol. 2013;37:196–204. doi: 10.1002/gepi.21703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Conneely KN, Boehnke M. So many correlated tests, so little time! Rapid adjustment of P values for multiple correlated tests. American journal of human genetics. 2007;81:1158–1168. doi: 10.1086/522036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Chapman J, Whittaker J. Analysis of multiple SNPs in a candidate gene or region. Genetic epidemiology. 2008;32:560–566. doi: 10.1002/gepi.20330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Pan W. Asymptotic tests of association with multiple SNPs in linkage disequilibrium. Genetic epidemiology. 2009;33:497–507. doi: 10.1002/gepi.20402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Wessel J, McDonald SM, Hinds DA, Stokowski RP, Javitz HS, Kennemer M, Krasnow R, Dirks W, Hardin J, Pitts SJ, et al. Resequencing of nicotinic acetylcholine receptor genes and association of common and rare variants with the Fagerstrom test for nicotine dependence. Neuropsychopharmacology : official publication of the American College of Neuropsychopharmacology. 2010;35:2392–2402. doi: 10.1038/npp.2010.120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Xie P, Kranzler HR, Krauthammer M, Cosgrove KP, Oslin D, Anton RF, Farrer LA, Picciotto MR, Krystal JH, Zhao H, et al. Rare nonsynonymous variants in alpha-4 nicotinic acetylcholine receptor gene protect against nicotine dependence. Biological psychiatry. 2011;70:528–536. doi: 10.1016/j.biopsych.2011.04.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Haller G, Druley T, Vallania FL, Mitra RD, Li P, Akk G, Steinbach JH, Breslau N, Johnson E, Hatsukami D, et al. Rare missense variants in CHRNB4 are associated with reduced risk of nicotine dependence. Human molecular genetics. 2012;21:647–655. doi: 10.1093/hmg/ddr498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Ho MK, Goldman D, Heinz A, Kaprio J, Kreek MJ, Li MD, Munafo MR, Tyndale RF. Breaking barriers in the genomics and pharmacogenetics of drug addiction. Clin Pharmacol Ther. 2010;88:779–791. doi: 10.1038/clpt.2010.175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Salas R, Orr-Urtreger A, Broide RS, Beaudet A, Paylor R, De Biasi M. The nicotinic acetylcholine receptor subunit alpha 5 mediates short-term effects of nicotine in vivo. Mol Pharmacol. 2003;63:1059–1066. doi: 10.1124/mol.63.5.1059. [DOI] [PubMed] [Google Scholar]
- 123.Salas R, Pieri F, De Biasi M. Decreased signs of nicotine withdrawal in mice null for the beta4 nicotinic acetylcholine receptor subunit. The Journal of neuroscience : the official journal of the Society for Neuroscience. 2004;24:10035–10039. doi: 10.1523/JNEUROSCI.1939-04.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Salas R, Cook KD, Bassetto L, De Biasi M. The alpha3 and beta4 nicotinic acetylcholine receptor subunits are necessary for nicotine-induced seizures and hypolocomotion in mice. Neuropharmacology. 2004;47:401–407. doi: 10.1016/j.neuropharm.2004.05.002. [DOI] [PubMed] [Google Scholar]
- 125.Fowler CD, Lu Q, Johnson PM, Marks MJ, Kenny PJ. Habenular alpha5 nicotinic receptor subunit signalling controls nicotine intake. Nature. 2011;471:597–601. doi: 10.1038/nature09797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Hong LE, Hodgkinson CA, Yang Y, Sampath H, Ross TJ, Buchholz B, Salmeron BJ, Srivastava V, Thaker GK, Goldman D, et al. A genetically modulated, intrinsic cingulate circuit supports human nicotine addiction. Proc Natl Acad Sci U S A. 2010;107:13509–13514. doi: 10.1073/pnas.1004745107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Bierut LJ. Convergence of genetic findings for nicotine dependence and smoking related diseases with chromosome 15q24-25. Trends Pharmacol Sci. 2010;31:46–51. doi: 10.1016/j.tips.2009.10.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Wang JC, Kapoor M, Goate AM. The genetics of substance dependence. Annual review of genomics and human genetics. 2012;13:241–261. doi: 10.1146/annurev-genom-090711-163844. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 129.Munafo MR, Matheson IJ, Flint J. Association of the DRD2 gene Taq1A polymorphism and alcoholism: a meta-analysis of case-control studies and evidence of publication bias. Mol Psychiatry. 2007;12:454–461. doi: 10.1038/sj.mp.4001938. [DOI] [PubMed] [Google Scholar]
- 130.Purcell S, Neale B, Todd-Brown K, Thomas L, Ferreira MA, Bender D, Maller J, Sklar P, de Bakker PI, Daly MJ, et al. PLINK: a tool set for whole-genome association and population-based linkage analyses. Am J Hum Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.