Significance
The bacterium Sinorhizobium meliloti establishes an agriculturally and ecologically important nitrogen-fixing symbiosis with leguminous plants. During symbiosis, the bacterial cells undergo drastic cellular differentiation and alter their cell cycle regulation such that they become highly polyploid. Cell cycle research has been limited in S. meliloti because there has been no method to generate synchronous cell populations. Here we describe a robust method to synchronize S. meliloti and present a global analysis of S. meliloti cell cycle gene expression. The results of this study suggest that the S. meliloti cell cycle transcriptional regulatory network, especially the regulon of the master regulator CtrA, has adapted specifically to fit its lifestyle both within the soil and its legume host.
Keywords: cell cycle regulation, symbiosis, alpha-proteobacteria
Abstract
In α-proteobacteria, strict regulation of cell cycle progression is necessary for the specific cellular differentiation required for adaptation to diverse environmental niches. The symbiotic lifestyle of Sinorhizobium meliloti requires a drastic cellular differentiation that includes genome amplification. To achieve polyploidy, the S. meliloti cell cycle program must be altered to uncouple DNA replication from cell division. In the α-proteobacterium Caulobacter crescentus, cell cycle-regulated transcription plays an important role in the control of cell cycle progression but this has not been demonstrated in other α-proteobacteria. Here we describe a robust method for synchronizing cell growth that enabled global analysis of S. meliloti cell cycle-regulated gene expression. This analysis identified 462 genes with cell cycle-regulated transcripts, including several key cell cycle regulators, and genes involved in motility, attachment, and cell division. Only 28% of the 462 S. meliloti cell cycle-regulated genes were also transcriptionally cell cycle-regulated in C. crescentus. Furthermore, CtrA- and DnaA-binding motif analysis revealed little overlap between the cell cycle-dependent regulons of CtrA and DnaA in S. meliloti and C. crescentus. The predicted S. meliloti cell cycle regulon of CtrA, but not that of DnaA, was strongly conserved in more closely related α-proteobacteria with similar ecological niches as S. meliloti, suggesting that the CtrA cell cycle regulatory network may control functions of central importance to the specific lifestyles of α-proteobacteria.
The α-proteobacteria class includes bacteria adapted to a wide range of lifestyles and environments (1). To thrive in their specific ecological niches, α-proteobacteria have developed specialized cellular differentiation programs. For example, Sinorhizobium meliloti undergoes a drastic cellular differentiation during its ecologically and agriculturally important nitrogen-fixing symbiosis with Medicago, Melilotus, and Trigonella legume hosts (2–4). S. meliloti initially elicits nodules on the roots of compatible legumes and then invades these nodules through host-derived infection threads. The bacteria proliferate within the infection thread as it extends and, after reaching the interior of the developing nodule, bacteria at the tip of the infection thread are endocytosed individually into compartments termed “symbiosomes.” Within this compartment, S. meliloti undergoes a striking cellular differentiation to become a nitrogen-fixing bacteroid. This differentiation involves an alteration of the bacterial cell cycle, as not only are cell size and membrane permeability altered in bacteroids but multiple equivalents of the tripartite S. meliloti genome accumulate (5, 6). A large family of defensin-like nodule-specific cysteine-rich (NCR) peptides have been recently discovered to play key roles in orchestrating this differentiation process, however their molecular mechanism is largely unknown (7, 8). Work described by Penterman et al. in ref. 9 suggests that these NCR peptides may act in part by altering the transcriptional profiles of key cell cycle regulators and remodeling the transcriptome to favor symbiosis.
Several lines of evidence suggest that modulation of the S. meliloti cell cycle is critically important for the cellular differentiation during symbiosis. For example, it has been shown that altering the expression of genes central to S. meliloti cell cycle processes (i.e., ftsZ, dnaA, minE, and ccrM) produces bacteroid-like polyploid cells (10–13) and that mutation of conserved cell cycle regulators (cbrA, cpdR1, and divJ) blocks bacteroid formation and symbiosis (14–16). Furthermore, in the α-proteobacterium Caulobacter crescentus, the cellular differentiation program governing morphological and replicative asymmetry in progeny cells is genetically integrated with the cell cycle (17). This is achieved partially through coordinate expression of genes involved in cell cycle processes and cellular differentiation in a cell cycle phase-dependent manner (18). The transcriptional regulatory proteins at the top of this genetic network include the response regulator, CtrA, which modulates morphological and replicative asymmetry, and the DNA replication initiation protein DnaA (19–21).
Because the regulatory factors that govern the C. crescentus cell cycle are highly conserved in α-proteobacteria, the C. crescentus paradigm of transcriptional control of cell cycle progression has been postulated to also be conserved in most α-proteobacteria (22, 23). However, this hypothesis does not readily explain how a strictly conserved cell cycle regulatory circuit could accommodate the extremely variable lifestyles and cellular differentiation processes found in this diverse group of bacteria. To date, it has been difficult to test whether the C. crescentus paradigm of cell cycle regulation is conserved in S. meliloti on a global scale because no method existed to obtain synchronized cultures. Although single-gene studies have indicated that many cell cycle regulators including CtrA, DnaA, CcrM, DivJ, GcrA, and PleC are essential and/or functionally conserved in S. meliloti, our understanding of the role of transcriptional regulation in the S. meliloti cell cycle and the link between the cell cycle and cellular differentiation during symbiosis has been limited (11, 12, 24, 25).
Here we describe an efficient method for the synchronization of S. meliloti cell populations via nutrient downshift and present a microarray based gene expression analysis of the S. meliloti cell cycle. This analysis identified 462 genes exhibiting strong periods of up-regulation and down-regulation during the S. meliloti cell cycle. These genes include conserved cell cycle regulators as well as large number of genes involved in motility, attachment, and cell division. Importantly, these data firmly support the extension of the general model of C. crescentus cell cycle-regulated transcription to S. meliloti and possibly other α-proteobacteria. We also identified 126 cell cycle-regulated transcripts common to both C. crescentus and S. meliloti, which may represent a conserved core set of cell cycle-regulated genes in α-proteobacteria. However, the large divergence in cell cycle-regulated transcripts, as well as in the major differences between the predicted regulons of CtrA and DnaA between C. crescentus and S. meliloti, indicates that this network has specifically evolved in each species to facilitate their unique cellular differentiation programs and lifestyles.
Results
Synchronization of S. meliloti Cell Populations via Nutrient Downshift.
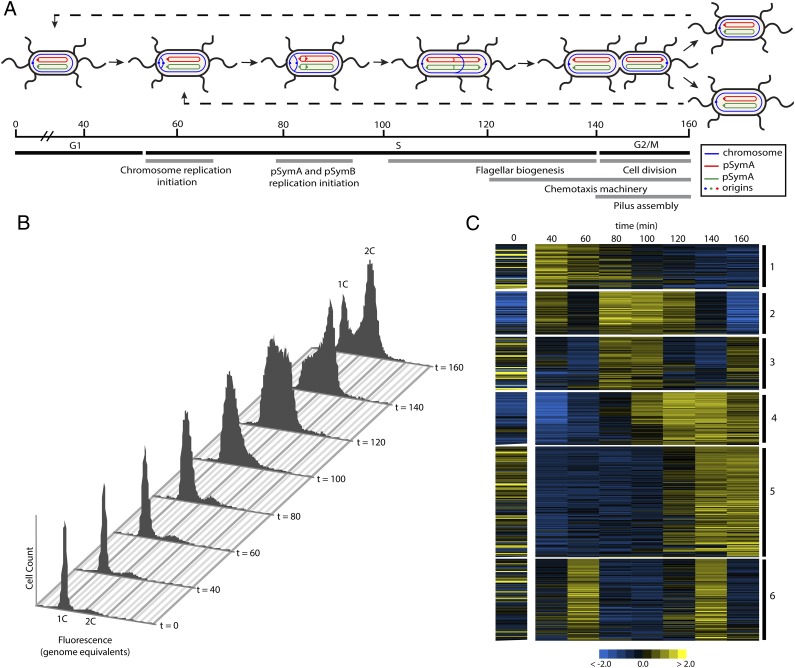
Previous attempts to use the C. crescentus method of differential centrifugation to isolate G1-arrested daughter cells (26) from S. meliloti cultures proved unsuccessful. However, an effective method for synchronizing populations of Escherichia coli involves growing cells in conditions that promote the highly conserved stringent response, which induces a G1 arrest (27). The stringent response of S. meliloti is stimulated by both carbon and nitrogen starvation and is dependent on ppGpp synthesis by the enzyme RelSm (28, 29). To test if stringent conditions would produce G1-arrested S. meliloti cells, early log phase Rm1021 cells were transferred to medium lacking preferred carbon and nitrogen sources (28). DNA content per cell was monitored via flow cytometry and after 4.5 h, ∼95% of the cells had one copy of their genome (1C) (Fig. 1 A and B). Arrested cells were pelleted and resuspended in rich medium, and after 40–50 min S. meliloti cells initiated DNA replication in unison and proceeded to replicate their genomes (Fig. 1 A and B). The synchronized cells reached mid-S phase by 100–120 min and completed replication by 140 min. At 160 min, cells underwent division and the 1C peak reemerged. Synchronous growth was thus maintained through one full cell cycle (Fig. 1B). This cell cycle progression is illustrated in Fig. 1A.
Fig. 1.
Synchronization of S. meliloti and microarray analysis of cell cycle-regulated gene expression. (A) Illustration of S. meliloti cell cycle progression. The timing of cell cycle events is based on expression patterns of specific genes as described in results. S. meliloti is peritrichously flagellated and divides asymmetrically (23, 56). (B) FACS profiles from cell synchronization. At t = 0, 95% of cells are arrested in G1. At t = 60, replication has initiated and S phase starts. Cells have completed S phase at t = 140 and divide at t = 160. (C) Heatmap of the clustering of 462 cell cycle-regulated transcripts. Clusters are indicated on the right. Each row represents a single gene. The color scale bar corresponding to log-fold expression is included at the bottom of the heatmap.
Microarray Analysis of Synchronized S. meliloti Cultures Identifies 462 Cell Cycle-Regulated Transcripts.
To test whether cell cycle-regulated gene expression was conserved in S. meliloti, gene expression was monitored in synchronized cultures of S. meliloti as they progressed through the cell cycle (18). RNA was isolated from cultures arrested in G1 (t = 0) and at 20-min intervals starting at the end of G1 (t = 40) until cell division (t = 160). We directly compared the expression of 6,046 S. meliloti genes in distinct phases of the cell cycle with the expression of these genes in early-log phase asynchronous culture via two-color microarray analysis. An SD cutoff was applied to the expression data to identify S. meliloti genes whose transcription varies significantly as a function of the cell cycle. Average replicate log ratio values of each gene for each time point were used to calculate the SD for all S. meliloti genes. These SD values were compared with SD values generated from random permuted gene expression profiles and genes with statistically significant SDs were chosen (30). To minimize the effect of the stringent response on variance, only values from the 40–160 time points were used to calculate the SD of expression for each gene.
Our analysis identified 462 genes whose expression varies as a function of the cell cycle (Dataset S1). The majority of cell cycle-regulated genes (320) are located on the chromosome, whereas 107 and 36 are located on the symbiotic megaplasmids pSymB and pSymA, respectively. Cell cycle-regulated transcription thus controls 9.5% of chromosomal genes, 6.8% of pSymB genes, and 2.7% pSymA genes. These data suggest that the chromosome is enriched for genes with cell cycle-regulated expression, an inference that was verified by a hypergeometric test (P < 0.001). The especially low percentage of genes demonstrating cell cycle-regulated transcription on pSymA compared with the chromosome and pSymB could be a result of pSymA being a more recently acquired, less domesticated replicon than pSymB (6, 31).
A fuzzy c-means clustering algorithm was used to group cell cycle-regulated genes with similar expression patterns to identify groups of genes involved in similar cell cycle functions (32, 33). This analysis yielded a total of six gene clusters that are displayed in Fig. 1C. Many genes with similar functions clustered together, especially the flagellar and chemotaxis machinery found in clusters 4 and 5, respectively. Importantly, each cluster represents a specific period of transcriptional activation and repression during the cell cycle—similar to the pattern of cell cycle gene expression observed in C. crescentus, (18). These data clearly demonstrate that temporally regulated gene expression during the cell cycle is conserved between S. meliloti and C. crescentus, which supports the extension of this paradigm to other α-proteobacteria.
Many S. meliloti Genes Exhibit Peak Expression Corresponding with the Timing of Their Cellular Function.
Fig. 2A illustrates the expression profiles of genes involved in DNA replication, recombination, repair, and chromosome segregation; genes denoted by an asterisk did not have a strong enough variance in their expression to be included in our list of cell cycle-regulated transcripts. Most of the genes encoding components of the DNA replication machinery did not exhibit strong cell cycle-regulated expression, which could mean that the activity of these factors is regulated posttranscriptionally or that the cell is sensitive to small differences in the expression of a particular gene. For example, transcription of dnaA, which did not make our cutoff, shows a slight up-regulation at 60 min corresponding with the start of S phase observed via flow cytometry (Figs. 1B and 2A). The calculated SD of the expression of the chromosome segregation genes parA and parB, which show early up-regulation in the heat map, was also not large enough to pass the stringent cutoff required by our analysis. However, our analysis did reveal cell cycle-regulated expression of the repABC genes, which govern the replication and segregation of pSymA and pSymB and thus play important roles in coordinating the replication of S. meliloti’s tripartite genome (34). These genes fall into cluster 3 with peak up-regulation occurring in early S phase between t = 80 and t = 100 (Fig. 1B). Specifically, the expression of repC1 and repC2 (which are required for the initiation of replication of pSymB and pSymA, respectively) occurred after the observed peak expression of dnaA, suggesting that initiation of the megaplasmid origins may follow that of the chromosomal origin instead of occurring simultaneously (illustrated in Fig. 1A). This phenomenon has precedence in bacteria with multipartite genomes, as it has been demonstrated in Vibrio cholerae that the replication initiation of the smaller chromosome occurs after that of the larger chromosome to ensure that replication of both chromosomes terminates at the same time (35).
Fig. 2.
Cell cycle gene expression of genes involved in various cellular processes. (A) Expression of genes involved in DNA processes (replication, repair, and segregation) in S. meliloti. Genes not included in our list of cell cycle-regulated transcripts are denoted by an asterisk (*). The color scale bar corresponding to log-fold expression is included at the bottom of the heatmaps. Values represent raw log fold change values. (B) Cell cycle expression patterns for genes involved in cell growth (ribosome and cell envelope) and division. (C) Cell cycle gene expression patterns of genes involved in motility and attachment including flagellar biosynthesis genes (and regulators), chemotaxis machinery, and genes required for pili biogenesis.
Another set of genes whose cell cycle-regulated transcription was not strongly conserved between S. meliloti and C. crescentus were those involved in nucleotide biosynthesis and DNA repair (18). Only the gene encoding lexA, a regulator of the SOS response (36), demonstrated strong cell cycle-regulated transcription in both C. crescentus and S. meliloti (Fig. 2A) (18). In S. meliloti, lexA expression peaks late in the cell cycle suggesting a function for LexA at the end of S phase. A possible function of LexA during the S. meliloti cell cycle would be to inhibit the SOS response as the replicating chromosomes and megaplasmids are resolved.
Components of the S. meliloti cell division machinery fell into two separate clusters suggesting the existence of early and late cell division genes (37). The homologs of E. coli division genes ftsA, ftsZ, and minC were in cluster 4, whereas ftsQ, ftsI, ftsK, minD, and minE were in cluster 5 (Fig. 1C). Consistent with their predicted function in S. meliloti, these genes were most highly expressed coincident with the timing of septum formation (Figs. 1A and 2B). The expression pattern of the minCDE operon, which is required to restrict the septum to midcell in S. meliloti, was surprising (37). It has been demonstrated that minCDE genes are coexpressed from a promoter directly upstream of minC (13), but our data indicate that minC expression greatly precedes that of minDE during the cell cycle. This uncoupling of expression may be due to a terminator sequence or the 43-bp partial rhizobium-specific intergenic mosaic element (RIME) in the 101-bp region between minC and minD (13). Repetitive elements similar to the rhizobia-specific RIME element have been shown to alter gene expression in polycistronic operons (38).
The set of genes demonstrating the most robust cell cycle-regulated transcription were motility genes. Flagellar biosynthesis genes dominated cluster 4, whereas chemotaxis genes were present in cluster 5 (Figs. 1C and 2C). In S. meliloti, swimming motility is restricted to the exponential phase of growth by a three-class hierarchy of flagellar and chemotaxis genes (39). Class I regulators VisNR and Rem are at the top of the hierarchy controlling the expression of class II genes (flg, flh, fli, and mot) and class III genes (che and fla) (39). Our analysis revealed that the motility genes are highly repressed in the beginning of the cell cycle and that expression of class I and II genes is activated between 100–120 min and expression of class III genes is activated at 140 min (Fig. 2C). This pattern of gene expression is consistent with the known regulatory hierarchy, but raises the intriguing question of how the expression of these genes during the cell cycle is regulated.
Transcription of Important Cell Cycle Regulatory Genes Occurs Late in the Cell Cycle.
Homologs of the many well-characterized genes involved in the C. crescentus cell cycle regulatory network have cell cycle-regulated transcripts in S. meliloti (Fig. 3). These conserved regulatory genes fell into three clusters: clusters 4 (pleC and podJ1), cluster 5 (ctrA, sciP, divK, divJ, ccrM, and chpT), and cluster 6 (cpdR1) (Fig. 1B). As indicated by the clustering and heatmap in Fig. 3, many of these genes are repressed until mid-S phase, which is consistent with their defined roles in establishing morphological and replicative asymmetry in the C. crescentus predivisional cell (17). The S. meliloti homologs of genes required for the cell type-specific phosphorylation of CtrA in C. crescentus, pleC, divJ, and divK, demonstrated robust cell cycle-regulated transcription (40). Transcription of pleC was activated at 120 min, whereas transcription of divJ and divK was activated at 140 min (Fig. 3). This pattern of transcription combined with recently published biochemical and genetic analyses, help to solidify the role of DivK, DivJ, and PleC in regulating CtrA activity in the S. meliloti predivisional cell (25, 40).
Fig. 3.
Expression profiles of conserved cell cycle regulators. Values depicted represent raw log fold change values. The color scale bar corresponds to log-fold expression change in comparison with unsynchronized culture. Genes not included in our list of cell cycle-regulated transcripts are denoted by an asterisk (*).
In addition, the gene encoding SciP is first up-regulated during G1 arrest and then again up-regulated in predivisional cells, which is consistent with its function in C. crescentus (Fig. 3). In C. crescentus, SciP accumulates specifically in swarmer cells to inhibit the activity of CtrA as a transcriptional activator, but not its ability to bind to the origin and repress the initiation of DNA replication (41). Surprisingly, the expression of ctrA during the S. meliloti cell cycle was less pronounced and occurred later than its counterpart in the C. crescentus cell cycle (Fig. 3 and Fig. S1). It is also interesting that the expression of the sensor histidine kinase CbrA is high in G1 cells, but is not significantly cell cycle regulated. Loss of CbrA activity in S. meliloti has been shown to reduce the polar localization of the response regulator DivK and to be detrimental to symbiosis with Medicago sativa (14, 16). Thus, a function for CbrA may be to mediate DivK activity in G1 cells and/or during the starvation response, which may be important during symbiosis.
A Small Core of Cell Cycle-Regulated Transcripts Is Conserved Between the Mostly Divergent S. meliloti and C. crescentus Cell Cycle Transcriptional Regulons.
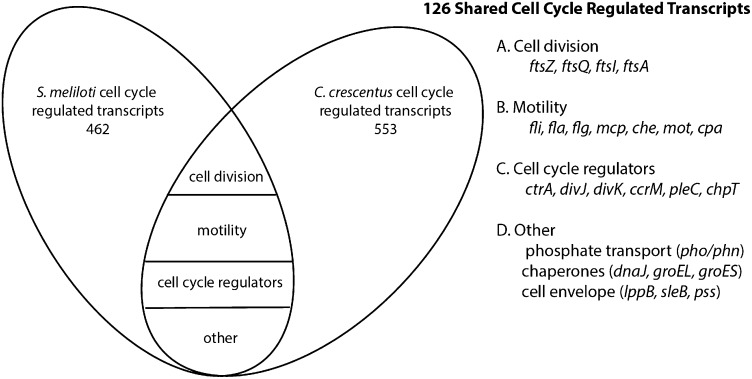
To understand the conservation of transcriptional regulation in cell cycle progression in α-proteobacteria, bidirectional BLAST and clusters of orthologous groups (COG) analysis was used to compare the S. meliloti set of 462 genes with cell cycle-regulated gene expression with the set of 553 genes previously identified in C. crescentus (18). Because different methods were used to synchronize the two bacteria, genes with cell cycle-regulated transcripts conserved between the two species not only represent a core set of α-proteobacterial cell cycle-regulated genes, but also represent genes demonstrating cell cycle-regulated transcription independent of synchronization method. Our analysis revealed 126 genes with cell cycle-regulated transcripts that are conserved between S. meliloti and C. crescentus (Fig. 4 and Dataset S2), which included cell division genes (ftsZ, ftsI, ftsA, and ftsQ), cell cycle regulators (ctrA, pleC, divK, divJ, cpdR, and chpT), and many flagellar and chemotaxis genes (Fig. 4). Genes encoding phosphate and phosphonate transporters (pho/phn), chaperones (dnaJ and groELS), and cell envelope proteins (lppB, sleB, and pss) were also transcriptionally regulated as a function of the cell cycle in both species. The processes controlled by these conserved genes are fundamental to the proliferation and architecture of α-proteobacteria and thus likely represent a core set of genes with cell cycle-regulated transcription conserved in many α-proteobacteria, despite their differing specific ecological niches.
Fig. 4.
Comparison of genes demonstrating cell cycle-regulated transcription in S. meliloti and C. crescentus. Diagram of genes conserved between the S. meliloti and C. crescentus cell cycle-regulated datasets (18). One hundred twenty-six cell cycle-regulated genes were conserved between the two species and fell into the functional groups as illustrated in the Venn diagram.
Intriguingly, the above analysis revealed that many of the 336 cell cycle-regulated genes specific to S. meliloti are crucial for its distinctive lifestyle. For example, megaplasmid replication and segregation genes (repABC) and a putative cyclic β-1,2 glucan ABC transporter (ndvA) do not have homologs in C. crescentus and are required for symbiosis (42). Also, several cell envelope genes (lpxB, olsB, opgC, ddl, and rlpA) and the genes coding for the Min system (minCDE) had cell cycle-regulated transcripts in S. meliloti, but either did not have homologs in C. crescentus or did not demonstrate cell cycle-regulated transcription in C. crescentus, demonstrating that the cell cycle regulon is highly specialized in each species.
Binding Site Analysis Reveals Conserved CtrA- and DnaA-Binding Sites Among Genes with Cell Cycle-Regulated Transcripts.
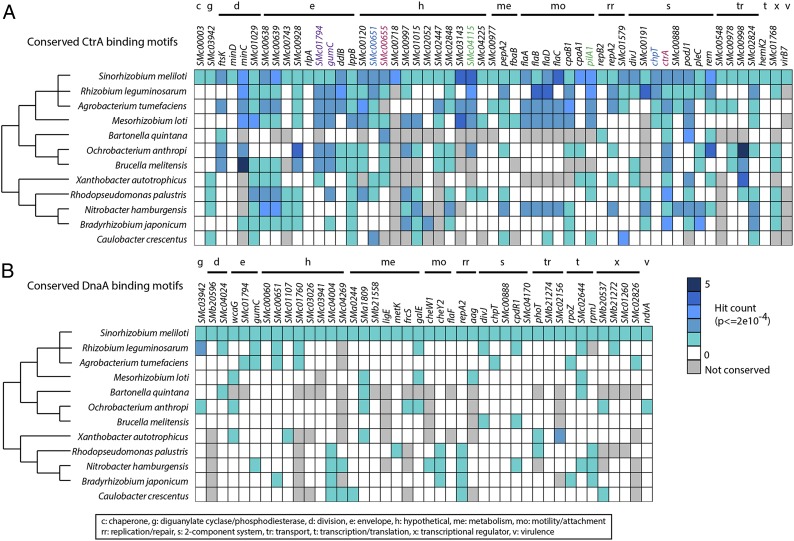
In C. crescentus, CtrA directly controls the transcription of 95 genes in 55 operons primarily involved in late cell cycle events and in establishing asymmetry, whereas DnaA controls the transcription of 40 genes primarily involved in early cell cycle events, including replication and early cell polarity (20, 21). To determine which of the genes with cell cycle-regulated expression in S. meliloti may be directly controlled by CtrA and DnaA, we searched for CtrA- and DnaA-binding motifs in the upstream regulatory regions of the 462 S. meliloti cell cycle-regulated genes using previously described position weight matrices describing the 16 nucleotide CtrA and 9 nucleotide DnaA-binding motifs in α-proteobacteria (22). This analysis revealed 64 genes with predicted CtrA-binding sites and 47 with predicted DnaA-binding sites (Fig. 5 A and B and Dataset S3 A and B). Several CtrA-binding motifs previously identified in S. meliloti were found, including motifs in the promoter regions of minC, chpT, pleC, ftsK, flaC, flaA, flaD, SMc00651, and podJ1 (22, 43). Surprisingly, there was very little overlap between previously defined C. crescentus CtrA and DnaA direct regulons and the corresponding predicted CtrA and DnaA cell cycle direct regulons of S. meliloti. Only eight genes were shared in the CtrA regulons and only four genes were shared in the DnaA regulons of the two species (20, 21).
Fig. 5.
Conservation of putative CtrA- and DnaA-binding sites in cell cycle-regulated genes. The upstream regulatory regions of genes with cell cycle-regulated transcripts were scanned for CtrA-and DnaA-binding motifs. “Hit count” signifies the number of significantly conserved full-length binding motifs that occur within the examined DNA segment. Homologs of genes containing hits from 11 related α-proteobacteria were also scanned for conserved CtrA and DnaA motifs. The tree to the left of the species’ names represents their evolutionary relationship as described in ref. 22. (A) S. meliloti cell cycle-regulated genes with the highest-scoring CtrA-binding motifs are displayed and the number of hits is denoted by different shades of blue as described in the legend. The presence of CtrA-binding sites in the homologous genes of related species is denoted in the same manner. Color-coded genes (i.e., pilA and SMc04115) share promoter regions. (B) S. meliloti cell cycle-regulated genes with the highest-scoring DnaA-binding motifs. The number of binding motifs and conservation of these motifs in other α-proteobacteria are denoted in the same manner as in A. A key explaining the putative function of the various genes displayed is provided at the bottom of the figure.
Because the predicted S. meliloti CtrA and DnaA cell cycle regulons were so strikingly diverged from the corresponding C. crescentus regulons, we decided to test the conservation of these regulons in a sampling of α-proteobacterial species more closely related to S. meliloti. Homologs of the S. meliloti cell cycle-regulated genes with strong CtrA- and DnaA-binding motifs were found in 11 such α-proteobacterial species and then scanned for conserved CtrA- and DnaA-binding motifs. The results for a selection of genes with the highest-scoring motifs are displayed in Fig. 5 (Dataset S3 A and B). Although the CtrA-binding motifs in S. meliloti genes with cell cycle-regulated transcripts identified by this analysis were not well conserved in C. crescentus, they were much more strongly conserved in α-proteobacterial species more closely related to S. meliloti (Fig. 5A) (20). The conservation of the CtrA regulon in these α-proteobacterial species strongly implies that the regulation of these genes by CtrA during the cell cycle is critical in these species. For example, in all organisms in which the septum formation regulator minC is conserved, the CtrA-binding motifs in its promoter region are also conserved (Fig. 5A). Therefore, not only is the transcription of minC cell cycle regulated in S. meliloti, but the role of CtrA in controlling the timing of minC expression is highly conserved and likely crucial in α-proteobacteria with the Min system as was previously suggested in ref. 23. This analysis also identified many genes of unknown function as predicted members of the CtrA regulon, such as SMc04115, which is an uncharacterized gene with homology to the N terminus of pili genes. SMc04115 shares a promoter region with pilin gene pilA, which also harbors CtrA-binding motifs. In addition, SMc04115 has a very strong co-occurrence in the genome with pilA1 and the pilus assembly genes of the cpa operon in α-proteobacteria suggesting a conserved function in pilus assembly.
A primary role of CtrA in C. crescentus is to establish morphological asymmetry in the predivisional cell, which CtrA accomplishes by directly regulating the expression genes involved in flagellar biogenesis and chemotaxis. Interestingly, the binding sites found in the regulatory regions of the motility genes of C. crescentus are not conserved in S. meliloti. Instead, there are conserved CtrA-binding motifs preceding the gene encoding the motility regulator Rem. This suggests that cell cycle regulation of flagellar and chemotaxis genes in S. meliloti may be achieved indirectly through regulation of Rem by CtrA, instead of through direct regulation of the flagellar and chemotaxis genes by CtrA. Rem has already been described as an important node for the regulation of S. meliloti flagellar gene expression in response to several environmental cues (44). Furthermore, the CtrA-binding motifs upstream of rem are conserved in many α-proteobacteria surveyed in our analysis, including the two organisms most closely related to S. meliloti, Rhizobium leguminosarum and Agrobacterium tumefaciens (Fig. 5A). Thus, in certain α-proteobacteria, including S. meliloti, CtrA likely regulates the expression of flagella and chemotaxis genes indirectly during the cell cycle through the intermediate regulator Rem.
Another interesting deviation from the C. crescentus cell cycle regulatory network is the absence of CtrA-binding motifs upstream of the gene encoding the essential cell cycle-regulated methylase, CcrM (12, 22). In C. crescentus, CcrM methylates newly replicated DNA at the adenine of GANTC site and the activation of ccrM transcription by CtrA at the end of S phase results in the remethylation of the chromosome (45). Our data indicate that the expression of ccrM in S. meliloti is indeed cell cycle regulated (Fig. 3), but conserved CtrA-binding motifs are absent from the upstream regulatory region of ccrM (Fig. 5A). Previous analysis identified GANTC sites upstream of ccrM, suggesting a method of autoregulation, but it is still unclear how transcription of ccrM is up-regulated after completion of S phase in S. meliloti (12).
The DnaA-binding motifs identified in S. meliloti genes with cell cycle-regulated transcripts were much less conserved in the α-proteobacterial species we surveyed (Fig. 5B). This suggests that the function of DnaA as a transcriptional regulator in α-proteobacteria might be much less constrained by evolution than that of CtrA. For example, the CtrA-binding motif upstream of the polarity factor podJ1 is conserved among α-proteobacterial species, but the DnaA-binding motif upstream of C. crescentus podJ is not (Fig. 5 A and B) (21). A previously verified DnaA-binding motif upstream of the repABC operon of pSymA was identified by this analysis, as well as a conserved CtrA-binding motif 90 bp downstream of the DnaA site (11). The presence of these binding motifs suggests a possible interplay between CtrA and DnaA in the regulation of expression of this repABC operon on pSymA during the cell cycle. It is surprising that neither DnaA nor CtrA motifs were found in the regulatory regions of repC1 or repAB3 on pSymB because the expression of these genes is also cell cycle regulated. How the expression of these genes is coordinated with the S. meliloti cell cycle remains unclear.
Discussion
This study describes the first published method for the robust synchronization of S. meliloti growth as well as detailed microarray analysis of global gene expression during the S. meliloti cell cycle, the only published analysis for an α-proteobacterium with multiple replicons. The microarray analysis revealed that the expression of 462 genes is modulated as a function of the cell cycle. Comparing this set of S. meliloti genes with the list of 553 previously identified C. crescentus genes with cell cycle-regulated transcripts (18), a core set of 126 genes exhibiting conserved cell cycle-dependent transcription in both bacteria was discovered. Despite this conservation, 72% of the S. meliloti cell cycle-regulated genes identified in our analysis either did not have counterparts or were not cell cycle-regulated in C. crescentus and there was very little overlap between the predicted CtrA and DnaA cell cycle-dependent regulons between the two species. These observations suggest that, despite the conservation of core elements of the cell cycle regulatory machinery, the cell cycle transcriptional regulons of α-proteobacteria are as diverse as their lifestyles and the environmental niches they occupy (22, 23).
Cell Cycle-Regulated Transcription in α-Proteobacteria.
Temporal regulation of transcription during the cell cycle was at one time thought to be a purely eukaryotic phenomenon (46). However, groundbreaking work by Laub et al. (18) demonstrated that temporally regulated transcription is also used by the α-proteobacterium C. crescentus to control cell cycle progression. Due to the conservation of the genes encoding the cell cycle regulatory circuit in a large percentage of α-proteobacteria, it was postulated that transcriptional control of cell cycle progression was conserved throughout the α-proteobacterial group (22, 23). However, the degree to which this paradigm is globally conserved had remained untested. Therefore, our discovery that cell cycle-regulated transcription is conserved in S. meliloti, which lacks the morphological asymmetry of C. crescentus and also thrives in an ecological niche completely different than C. crescentus, is highly significant. The conservation of cell cycle-regulated transcription in S. meliloti also strongly suggests that the function of temporally regulated gene expression in α-proteobacteria is not simply to produce dimorphism.
Genes with cell cycle-regulated transcripts conserved between C. crescentus and S. meliloti include many of the components of the complex core regulatory circuit that governs cell cycle progression in C. crescentus. The transcription of these factors (which include ctrA, pleC, divK, divJ, cpdR, chpT, and sciP) was mostly up-regulated coincident with the timing of their prescribed function in C. crescentus. This observation, along with previous molecular studies, strongly suggests that the basic functions of these cell cycle proteins are well conserved between S. meliloti and C. crescentus (16, 24, 25, 40). Interestingly, many of the S. meliloti genes with cell cycle-regulated transcripts that are either not conserved or not cell cycle regulated in C. crescentus, are required for the symbiotic relationship between S. meliloti and its legume host—including the repABC genes controlling megaplasmid replication and maintenance, as well as genes involved in cyclic glucan production.
CtrA as a Modulator of Asymmetry and Cellular Differentiation in α-Proteobacteria.
Because CtrA is such an important regulator of morphological and replicative asymmetry in C. crescentus, it is remarkable that CtrA-binding sites found in C. crescentus are only conserved in eight genes with cell cycle-regulated transcripts in S. meliloti. A striking illustration of this divergence is the difference in cell cycle regulatory schemes for the flagella and chemotaxis genes in the two organisms. In C. crescentus, CtrA directly regulates genes in all four tiers of the flagellar regulatory hierarchy including flg, fli, flm and flagellin genes (20). In S. meliloti, however, CtrA-binding motifs are only present in the promoters of the motility regulator rem and of the flagellin genes (flaABCD). It has been shown that S. meliloti is able to up- or down-regulate production of its flagella and chemotaxis machinery in response to different environmental cues through the intermediate regulator Rem (39, 44, 47). Thus, S. meliloti CtrA likely exerts a major influence on the transcription of these genes during the cell cycle by directly regulating rem transcription. This regulatory strategy could allow for environmental cues to be integrated with or even override the cell cycle-coordinated production of flagella and chemotaxis machinery, which might be crucial not only for S. meliloti to adapt to life in the soil, but also life within the plant during symbiosis. It is well known that flagellar proteins can activate innate immunity in eukaryotic hosts, so this indirect regulatory adaption may complement the rhizobial strategy of reducing flagella recognition by using Nod factors to suppress the plant innate immune system (48).
Interestingly, our analysis also revealed a high degree of conservation of the predicted S. meliloti-direct CtrA regulon in α-proteobacteria whose lifestyles are more similar to S. meliloti than to C. crescentus. One example is the Min system for regulation of septation, which is present in S. meliloti and a number of other α-proteobacteria, but not C. crescentus. CtrA-binding motifs are present upstream of the minCDE operon in every α-proteobacterium surveyed in this analysis that contains the operon. The conservation of these motifs lends support to the theory first presented in Hallez et al. (23) that CtrA mediates cell division through direct regulation of minC transcription in α-proteobacteria containing the Min system and through direct regulation of ftsZ in C. crescentus and other α-proteobacteria that lack the Min system. It is intriguing that a single regulatory factor can retain its role in a process so highly conserved as cell division but simultaneously demonstrates the flexibility to modulate diverse cellular differentiation events central to the lifestyles of α-proteobacteria, which can range from cyst cell formation in Rhodospirillum centenum to intracellular pathogenesis in Brucella (49, 50).
Further research will be required to gain a more complete understanding of the wiring of the cell cycle regulatory circuit in S. meliloti and how this regulatory circuit is modified by NCR peptides and perhaps other plant factors during symbiosis to achieve the specific cellular differentiation required during bacteroid development. In this issue of PNAS, Penterman et al. (9) found that a specific NCR peptide (NCR247) can alter S. meliloti cell cycle progression to cause a cell division block by at least in part manipulating the CtrA regulatory circuit. It will be important to continue to probe the role of the S. meliloti cell cycle regulation during symbiosis, including carrying out in vivo experiments to verify the predicted CtrA and DnaA regulons, as well as further elucidating the molecular mechanisms by which NCR247 and other NCR peptides modulate the S. meliloti cell cycle and prepare the organism for its important symbiotic role.
Methods
Growth Conditions and Cell Synchronization.
S. meliloti strain Sm1021 was cultured at 30 °C in LBMC media [LB with 2.5 mM magnesium sulfate (MgSO4) and 2.5 mM CaCl2] or Mops-GS media lacking mannitol (50 mM Mops, 1 mM MgSO4, 0.25 mM CaCl2, 19 mM glutamic acid, and 0.004 mM biotin, pH 7.4) on a platform shaker. For synchronization, 1 L of LBMC was inoculated with 100 μL of saturated Sm1021 liquid culture and grown to OD600 = 0.10–0.15. Cells were then collected by centrifugation at 6,500 rpm (Sorvall Slc-4000) for 5 min, washed twice with 0.85% saline, and resuspended in modified Mops-GS. Cells arrested in G1 after a 270-min incubation in Mops-GS. Cells arrested in G1 were then collected by centrifugation and cultured in LBMC.
Flow Cytometry.
To quantify DNA content per cell in S. meliloti cultures, 200 μL culture was fixed in 933 μL of 100% ethanol. Cells were collected by centrifugation and incubated for 2 h in 1 mL 50 mM sodium citrate with 3.3 μg/mL RNase A at 50° C to eliminate cellular RNA. To label DNA, 1 μL 8.3-μM SYTOX Green dye was added to each sample. Samples were analyzed with a BD FACScan flow cytometer (BD Biosciences) and data were analyzed with FlowJo 9.6.3 software (Tree star).
RNA Isolation and Gene Expression Profiling.
RNA was obtained from 10 mL time points taken from four separate synchronized cultures and from a control (OD600 = 0.15) unsynchronized cultures by first treating with Qiagen RNAprotect bacterial reagent to stabilize the RNA and then using the Qiagen RNeasy plus kit for RNA isolation. A total of 600 ng of RNA was used for cDNA synthesis (Ambion MessageAmp II-Bacterial kit) and cDNA labeling with Cy3 and Cy5 fluorophores was carried out using the Ambion Amino Allyl MessageAmp II cRNA Amplification kit. Labeled cDNA was hybridized to a custom Agilent gene expression array (Smexpr1:00 AMADID: 036667) of optimized 60-mer probes to 6,046 S. meliloti ORFs (51). The array data were processed using the limma package in R. Median red/green hybridization signals were background corrected and the red/green hybridization signal log-ratios, M = log2(R/G), were normalized for each array using locally weighted scatterplot smoothing. Normalization between arrays was also performed to increase consistency. The fourth biological replicate of time point 140 was removed from this analysis due to poor correlation with the other replicates.
Selection and Clustering of Genes with Cell Cycle-Regulated Transcription.
The replicate log ratio values for time points 40–160 were averaged and used to calculate the SD for each gene. The SDs were compared with the distribution of SDs generated from 10,000 randomly permuted gene expression profiles and genes with empirical P values <0.05 were denoted as cell cycle regulated and considered for clustering. Fuzzy c-means clustering was applied using R software package e1071 (http://cran.r-project.org/web/packages/e1071/index.html). The gene expression profiles were mean centered normalized and the fuzzy parameter was estimated as described previously (32). The optimal cluster number (six) was determined using methods described in ref. 52 and a bagged clustering wrapper, bclust (http://cran.r-project.org/web/packages/bclust/index.html), was used to reduce the variability in the clustering results.
S. meliloti and C. crescentus Cell Cycle Gene Comparisons.
The amino acid sequences for 6,201 Sm1021 genes were acquired and blasted against 3,737 CB15 protein sequences. The BLAST results were used to determine which of the 553 C. crescentus cell cycle-regulated genes identified in ref. 18 were similar to the 462 identified in S. meliloti. The BLAST analysis was complemented with COGs (53). The results were filtered using E-values <1 × 10−20, as well as manually filtered, to match genes and remove replicates.
CtrA- and DnaA-Binding Motif Discovery.
CtrA and DnaA motifs (22) were used in find individual motif occurrences (FIMO) (54) with the 400 bp upstream and 100 bp downstream of the S. meliloti cell cycle gene promoters. A P value cutoff of 2 × 10−4 was used to establish the S. meliloti genes containing the motifs. The protein sequences of these genes were reciprocal blasted against the proteins of 11 closely related species to identify homologous genes and those with protein BLAST E-values <0.05 were considered for FIMO analysis.
Quantitative PCR Analysis.
Primers were designed to amplify a 100-bp region of the ctrA gene (SMc00654) and control gene SMc00128, which demonstrated stable expression across the cell cycle (55). To quantify cell cycle gene expression, RNA was isolated from four representative time points in synchronized S. meliloti cultures (t = 40, 80, 120, and 160 min). RNA was isolated by the same method as it was for microarray analysis. cDNA was generated using the BioRad iScript cDNA Synthesis kit, and qPCR analysis was carried out on a Roche LightCycler 480 using Roche LightCycler 480 SYBR Green I Master Mix. The log10 expression ratios were calculated for each time point with respect to the expression of the SMc00128, such that log expression ratio = log[ctrA] − log[SMc00128].
Supplementary Material
Acknowledgments
We thank Stuart Levine and Manlin Luo [Massachusetts Institute of Technology (MIT) BioMicro Center] for their support and contributions in the microarray studies. We also thank all the members of the G.C.W. laboratory for their input and support, and Michael T. Laub for many thoughtful discussions and comments on this manuscript. This work was supported by National Institutes of Health (NIH) Grant GM31010 (to G.C.W.) and P30 ES002109 (to the MIT Center for Environmental Health Sciences). G.C.W. is an American Cancer Society professor. The microarray studies were also partially funded by the National Cancer Institute of the NIH under Award P30 CA14051.
Footnotes
The authors declare no conflict of interest.
Data deposition: The microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE54208).
See Profile on page 3201.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1400421111/-/DCSupplemental.
References
- 1.Ettema TJ, Andersson SG. The alpha-proteobacteria: The Darwin finches of the bacterial world. Biol Lett. 2009;5(3):429–432. doi: 10.1098/rsbl.2008.0793. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Jones KM, Kobayashi H, Davies BW, Taga ME, Walker GC. How rhizobial symbionts invade plants: the Sinorhizobium-Medicago model. Nat Rev Microbiol. 2007;5(8):619–633. doi: 10.1038/nrmicro1705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Perret X, Staehelin C, Broughton WJ. Molecular basis of symbiotic promiscuity. Microbiol Mol Biol Rev. 2000;64(1):180–201. doi: 10.1128/mmbr.64.1.180-201.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Oldroyd GE. Speak, friend, and enter: Signalling systems that promote beneficial symbiotic associations in plants. Nat Rev Microbiol. 2013;11(4):252–263. doi: 10.1038/nrmicro2990. [DOI] [PubMed] [Google Scholar]
- 5.Mergaert P, et al. Eukaryotic control on bacterial cell cycle and differentiation in the Rhizobium-legume symbiosis. Proc Natl Acad Sci USA. 2006;103(13):5230–5235. doi: 10.1073/pnas.0600912103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Galibert F, et al. The composite genome of the legume symbiont Sinorhizobium meliloti. Science. 2001;293(5530):668–672. doi: 10.1126/science.1060966. [DOI] [PubMed] [Google Scholar]
- 7.Van de Velde W, et al. Plant peptides govern terminal differentiation of bacteria in symbiosis. Science. 2010;327(5969):1122–1126. doi: 10.1126/science.1184057. [DOI] [PubMed] [Google Scholar]
- 8.Wang D, et al. A nodule-specific protein secretory pathway required for nitrogen-fixing symbiosis. Science. 2010;327(5969):1126–1129. doi: 10.1126/science.1184096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Penterman J, et al. 2014. Host plant peptides elicit a transcriptional response to control the Sinorhizobium meliloti cell cycle during symbiosis. Proc Natl Acad Sci USA 111:3560–3565.
- 10.Latch JN, Margolin W. Generation of buds, swellings, and branches instead of filaments after blocking the cell cycle of Rhizobium meliloti. J Bacteriol. 1997;179(7):2373–2381. doi: 10.1128/jb.179.7.2373-2381.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sibley CD, MacLellan SR, Finan T. The Sinorhizobium meliloti chromosomal origin of replication. Microbiology. 2006;152(Pt 2):443–455. doi: 10.1099/mic.0.28455-0. [DOI] [PubMed] [Google Scholar]
- 12.Wright R, Stephens C, Shapiro L. The CcrM DNA methyltransferase is widespread in the alpha subdivision of proteobacteria, and its essential functions are conserved in Rhizobium meliloti and Caulobacter crescentus. J Bacteriol. 1997;179(18):5869–5877. doi: 10.1128/jb.179.18.5869-5877.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cheng J, Sibley CD, Zaheer R, Finan TM. A Sinorhizobium meliloti minE mutant has an altered morphology and exhibits defects in legume symbiosis. Microbiology. 2007;153(Pt 2):375–387. doi: 10.1099/mic.0.2006/001362-0. [DOI] [PubMed] [Google Scholar]
- 14.Gibson KE, Campbell GR, Lloret J, Walker GC. CbrA is a stationary-phase regulator of cell surface physiology and legume symbiosis in Sinorhizobium meliloti. J Bacteriol. 2006;188(12):4508–4521. doi: 10.1128/JB.01923-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kobayashi H, De Nisco NJ, Chien P, Simmons LA, Walker GC. Sinorhizobium meliloti CpdR1 is critical for co-ordinating cell cycle progression and the symbiotic chronic infection. Mol Microbiol. 2009;73(4):586–600. doi: 10.1111/j.1365-2958.2009.06794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Sadowski CS, Wilson D, Schallies KB, Walker G, Gibson KE. The Sinorhizobium meliloti sensor histidine kinase CbrA contributes to free-living cell cycle regulation. Microbiology. 2013;159(Pt 8):1552–1563. doi: 10.1099/mic.0.067504-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Tsokos CG, Laub MT. Polarity and cell fate asymmetry in Caulobacter crescentus. Curr Opin Microbiol. 2012;15(6):744–750. doi: 10.1016/j.mib.2012.10.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Laub MT, McAdams HH, Feldblyum T, Fraser CM, Shapiro L. Global analysis of the genetic network controlling a bacterial cell cycle. Science. 2000;290(5499):2144–2148. doi: 10.1126/science.290.5499.2144. [DOI] [PubMed] [Google Scholar]
- 19.Jonas K, Chen YE, Laub MT. Modularity of the bacterial cell cycle enables independent spatial and temporal control of DNA replication. Curr Biol. 2011;21(13):1092–1101. doi: 10.1016/j.cub.2011.05.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Laub MT, Chen SL, Shapiro L, McAdams HH. Genes directly controlled by CtrA, a master regulator of the Caulobacter cell cycle. Proc Natl Acad Sci USA. 2002;99(7):4632–4637. doi: 10.1073/pnas.062065699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Hottes AK, Shapiro L, McAdams HH. DnaA coordinates replication initiation and cell cycle transcription in Caulobacter crescentus. Mol Microbiol. 2005;58(5):1340–1353. doi: 10.1111/j.1365-2958.2005.04912.x. [DOI] [PubMed] [Google Scholar]
- 22.Brilli M, et al. The diversity and evolution of cell cycle regulation in alpha-proteobacteria: A comparative genomic analysis. BMC Syst Biol. 2010;4:52. doi: 10.1186/1752-0509-4-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hallez R, Bellefontaine AF, Letesson JJ, De Bolle X. Morphological and functional asymmetry in alpha-proteobacteria. Trends Microbiol. 2004;12(8):361–365. doi: 10.1016/j.tim.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 24.Barnett MJ, Hung DY, Reisenauer A, Shapiro L, Long SR. A homolog of the CtrA cell cycle regulator is present and essential in Sinorhizobium meliloti. J Bacteriol. 2001;183(10):3204–3210. doi: 10.1128/JB.183.10.3204-3210.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pini F, et al. The DivJ, CbrA and PleC system controls DivK phosphorylation and symbiosis in Sinorhizobium meliloti. Mol Microbiol. 2013;90(1):54–71. doi: 10.1111/mmi.12347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Evinger M, Agabian N. Envelope-associated nucleoid from Caulobacter crescentus stalked and swarmer cells. J Bacteriol. 1977;132(1):294–301. doi: 10.1128/jb.132.1.294-301.1977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Ferullo DJ, Cooper DL, Moore HR, Lovett ST. Cell cycle synchronization of Escherichia coli using the stringent response, with fluorescence labeling assays for DNA content and replication. Methods. 2009;48(1):8–13. doi: 10.1016/j.ymeth.2009.02.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wells DH, Long SR. The Sinorhizobium meliloti stringent response affects multiple aspects of symbiosis. Mol Microbiol. 2002;43(5):1115–1127. doi: 10.1046/j.1365-2958.2002.02826.x. [DOI] [PubMed] [Google Scholar]
- 29.Krol E, Becker A. ppGpp in Sinorhizobium meliloti: Biosynthesis in response to sudden nutritional downshifts and modulation of the transcriptome. Mol Microbiol. 2011;81(5):1233–1254. doi: 10.1111/j.1365-2958.2011.07752.x. [DOI] [PubMed] [Google Scholar]
- 30.de Lichtenberg U, et al. Comparison of computational methods for the identification of cell cycle-regulated genes. Bioinformatics. 2005;21(7):1164–1171. doi: 10.1093/bioinformatics/bti093. [DOI] [PubMed] [Google Scholar]
- 31.Galardini M, Pini F, Bazzicalupo M, Biondi EG, Mengoni A. Replicon-dependent bacterial genome evolution: The case of Sinorhizobium meliloti. Genome Biol Evol. 2013;5(3):542–558. doi: 10.1093/gbe/evt027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Schwämmle V, Jensen ON. A simple and fast method to determine the parameters for fuzzy c-means cluster analysis. Bioinformatics. 2010;26(22):2841–2848. doi: 10.1093/bioinformatics/btq534. [DOI] [PubMed] [Google Scholar]
- 33.Eisen MB, Spellman PT, Brown PO, Botstein D. Cluster analysis and display of genome-wide expression patterns. Proc Natl Acad Sci USA. 1998;95(25):14863–14868. doi: 10.1073/pnas.95.25.14863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Ramírez-Romero MA, Soberón N, Pérez-Oseguera A, Téllez-Sosa J, Cevallos MA. Structural elements required for replication and incompatibility of the Rhizobium etli symbiotic plasmid. J Bacteriol. 2000;182(11):3117–3124. doi: 10.1128/jb.182.11.3117-3124.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rasmussen T, Jensen RB, Skovgaard O. The two chromosomes of Vibrio cholerae are initiated at different time points in the cell cycle. EMBO J. 2007;26(13):3124–3131. doi: 10.1038/sj.emboj.7601747. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Butala M, Zgur-Bertok D, Busby SJ. The bacterial LexA transcriptional repressor. Cell Mol Life Sci. 2009;66(1):82–93. doi: 10.1007/s00018-008-8378-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Errington J, Daniel RA, Scheffers DJ. Cytokinesis in bacteria. Microbiol Mol Biol Rev. 2003;67(1):52–65. doi: 10.1128/MMBR.67.1.52-65.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Newbury SF, Smith NH, Higgins CF. Differential mRNA stability controls relative gene expression within a polycistronic operon. Cell. 1987;51(6):1131–1143. doi: 10.1016/0092-8674(87)90599-x. [DOI] [PubMed] [Google Scholar]
- 39.Rotter C, Mühlbacher S, Salamon D, Schmitt R, Scharf B. Rem, a new transcriptional activator of motility and chemotaxis in Sinorhizobium meliloti. J Bacteriol. 2006;188(19):6932–6942. doi: 10.1128/JB.01902-05. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lam H, Matroule JY, Jacobs-Wagner C. The asymmetric spatial distribution of bacterial signal transduction proteins coordinates cell cycle events. Dev Cell. 2003;5(1):149–159. doi: 10.1016/s1534-5807(03)00191-6. [DOI] [PubMed] [Google Scholar]
- 41.Gora KG, et al. A cell-type-specific protein-protein interaction modulates transcriptional activity of a master regulator in Caulobacter crescentus. Mol Cell. 2010;39(3):455–467. doi: 10.1016/j.molcel.2010.06.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Breedveld MW, Hadley JA, Miller KJ. A novel cyclic beta-1,2-glucan mutant of Rhizobium meliloti. J Bacteriol. 1995;177(22):6346–6351. doi: 10.1128/jb.177.22.6346-6351.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schlüter JP, et al. Global mapping of transcription start sites and promoter motifs in the symbiotic α-proteobacterium Sinorhizobium meliloti 1021. BMC Genomics. 2013;14:156. doi: 10.1186/1471-2164-14-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bahlawane C, McIntosh M, Krol E, Becker A. Sinorhizobium meliloti regulator MucR couples exopolysaccharide synthesis and motility. Mol Plant Microbe Interact. 2008;21(11):1498–1509. doi: 10.1094/MPMI-21-11-1498. [DOI] [PubMed] [Google Scholar]
- 45.Collier J, McAdams HH, Shapiro L. A DNA methylation ratchet governs progression through a bacterial cell cycle. Proc Natl Acad Sci USA. 2007;104(43):17111–17116. doi: 10.1073/pnas.0708112104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Spellman PT, et al. Comprehensive identification of cell cycle-regulated genes of the yeast Saccharomyces cerevisiae by microarray hybridization. Mol Biol Cell. 1998;9(12):3273–3297. doi: 10.1091/mbc.9.12.3273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hoang HH, Gurich N, González JE. Regulation of motility by the ExpR/Sin quorum-sensing system in Sinorhizobium meliloti. J Bacteriol. 2008;190(3):861–871. doi: 10.1128/JB.01310-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Liang Y, et al. Nonlegumes respond to rhizobial Nod factors by suppressing the innate immune response. Science. 2013;341(6152):1384–1387. doi: 10.1126/science.1242736. [DOI] [PubMed] [Google Scholar]
- 49.Berleman JE, Bauer CE. Characterization of cyst cell formation in the purple photosynthetic bacterium Rhodospirillum centenum. Microbiology. 2004;150(Pt 2):383–390. doi: 10.1099/mic.0.26846-0. [DOI] [PubMed] [Google Scholar]
- 50.Roop RM, 2nd, Gaines JM, Anderson ES, Caswell CC, Martin DW. Survival of the fittest: How Brucella strains adapt to their intracellular niche in the host. Med Microbiol Immunol (Berl) 2009;198(4):221–238. doi: 10.1007/s00430-009-0123-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Rüberg S, et al. Construction and validation of a Sinorhizobium meliloti whole genome DNA microarray: Genome-wide profiling of osmoadaptive gene expression. J Biotechnol. 2003;106(2-3):255–268. doi: 10.1016/j.jbiotec.2003.08.005. [DOI] [PubMed] [Google Scholar]
- 52.Yeung KY, Haynor DR, Ruzzo WL. Validating clustering for gene expression data. Bioinformatics. 2001;17(4):309–318. doi: 10.1093/bioinformatics/17.4.309. [DOI] [PubMed] [Google Scholar]
- 53.Tatusov RL, et al. The COG database: New developments in phylogenetic classification of proteins from complete genomes. Nucleic Acids Res. 2001;29(1):22–28. doi: 10.1093/nar/29.1.22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Grant CE, Bailey TL, Noble WS. FIMO: Scanning for occurrences of a given motif. Bioinformatics. 2011;27(7):1017–1018. doi: 10.1093/bioinformatics/btr064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Krol E, Becker A. Global transcriptional analysis of the phosphate starvation response in Sinorhizobium meliloti strains 1021 and 2011. Mol Genet Genomics. 2004;272(1):1–17. doi: 10.1007/s00438-004-1030-8. [DOI] [PubMed] [Google Scholar]
- 56.Pleier E, Schmitt R. Expression of two Rhizobium meliloti flagellin genes and their contribution to the complex filament structure. J Bacteriol. 1991;173(6):2077–2085. doi: 10.1128/jb.173.6.2077-2085.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.