Significance
Classification of cancer provides crucial guidance for clinical treatment and mechanistic studies. Our work extends previous glioma classification studies in that we established EGFR module (EM)/PDGFRA module (PM) glioma classification scheme based on gene coexpression modules around key signaling pathways conserved in neural development and gliomagenesis. We identified coexpressed EM and PM genes as classifiers. Based on the EM and PM signatures, our classification scheme robustly assigns adult low-grade and high-grade diffuse gliomas into three major subtypes that are distinct in patient survival, and in transcriptomic and genomic patterns. Our work suggests that EM and PM genes may play currently unrecognized roles in gliomagenesis. EM/PM glioma classification scheme forms a framework toward establishing molecular diagnostic tools and identifying new therapeutic targets to combat gliomas.
Abstract
We hypothesized that key signaling pathways of glioma genesis might enable the molecular classification of gliomas. Gene coexpression modules around epidermal growth factor receptor (EGFR) (EM, 29 genes) or platelet derived growth factor receptor A (PDGFRA) (PM, 40 genes) in gliomas were identified. Based on EM and PM expression signatures, nonnegative matrix factorization reproducibly clustered 1,369 adult diffuse gliomas WHO grades II-IV from four independent databases generated in three continents, into the subtypes (EM, PM and EMlowPMlow gliomas) in a morphology-independent manner. Besides their distinct patterns of genomic alterations, EM gliomas were associated with higher age at diagnosis, poorer prognosis, and stronger expression of neural stem cell and astrogenesis genes. Both PM and EMlowPMlow gliomas were associated with younger age at diagnosis and better prognosis. PM gliomas were enriched in the expression of oligodendrogenesis genes, whereas EMlowPMlow gliomas were enriched in the signatures of mature neurons and oligodendrocytes. The EM/PM-based molecular classification scheme is applicable to adult low-grade and high-grade diffuse gliomas, and outperforms existing classification schemes in assigning diffuse gliomas to subtypes with distinct transcriptomic and genomic profiles. The majority of the EM/PM classifiers, including regulators of glial fate decisions, have not been extensively studied in glioma biology. Subsets of these classifiers were coexpressed in mouse glial precursor cells, and frequently amplified or lost in an EM/PM glioma subtype-specific manner, resulting in somatic copy number alteration-dependent gene expression that contributes to EM/PM signatures in glioma samples. EM/PM-based molecular classification provides a molecular diagnostic framework to expedite the search for new glioma therapeutic targets.
Gliomas, the most common primary tumors in the adult CNS, are currently diagnosed as astrocytomas or oligodendrogliomas based on their morphological resemblance to astrocytes or oligodendrocytes (1). The severity of gliomas is further distinguished by malignant grades (I to IV) according to features of cellular atypia, cell proliferation, angiogenesis, and necrosis. However, this classification scheme is highly subjective and inconsistent in its designation of glioma subtypes and grades (2). Many gliomas cannot be clearly diagnosed due to their nontypical morphology. The prognosis of glioma patients is poor; most of the grade IV glioma (also called glioblastoma multiforme, GBM) patients die within 1–2 y of diagnosis (1). It is as yet unknown why certain low-grade gliomas progress rapidly to higher grades whereas others progress slowly. Inaccurate diagnoses cause clinical confusion, create artificial heterogeneity and complexities in glioma investigations, and hinder the development of targeted therapies.
Molecular classification has the potential to overcome the limitations of morphological diagnoses. Based on prognosis-related gene expression signatures (3–5), or stable unbiased gene expression clusters among glioma samples (6), previous studies classified high-grade gliomas into proneural (PN), proliferative (Prolif), and mesenchymal (Mes) (4), or PN, neural (NL), classical (CL), and Mes molecular subgroups (6). PN gliomas are distinguished by their frequent occurrence in adults younger than 40 y and with longer survival (3), enriched expression of the platelet derived growth factor receptor A (PDGFRA) and frequent mutation in the isocitrate dehydrogenase 1 (IDH1); CL gliomas, on the other hand, are associated with frequent amplification of the epidermal growth factor receptor (EGFR) and deletions in or loss of chromosome 10. NL and Mes gliomas are enriched in neuronal and mesenchymal markers, respectively (6). Other studies identified characteristic gene expression signatures among high-grade gliomas (7–10) or all major glioma subtypes (11, 12), and found correlations between expression signatures and the prognoses of patients and the cellular and genomic abnormalities of gliomas. A DNA methylome-based approach has identified a glioma-CpG island methylator phenotype (G-CIMP) in 30% of the PN GBMs and in a large proportion of low-grade gliomas (13, 14). These studies have made important steps forward in establishing an objective molecular classification for gliomas. However, questions of the relatedness of classifiers to glioma pathogenesis, the applicability of the classification scheme to both low- and high-grade gliomas, the correlation to glial development and the genomic distinction of glioma molecular subtypes encourage further investigation.
Here, we extend previous studies (4, 6, 11, 12) by taking a conceptually different approach, exploring whether gene coexpression modules around key signaling pathways conserved between neural development and glioma genesis might enable molecular classification of gliomas. EGFR and PDGFRA are two receptor tyrosine kinases (RTKs) that govern cell fate specification, cell proliferation, migration in the neural stem cell (NSC) compartment and glial development (15, 16). These two RTKs are frequently amplified, mutated, and overexpressed in gliomas (6), and their enforced signaling is essential in the generation of mouse glioma models (17). We describe the identification and characterization of genes consistently coexpressed with EGFR (EGFR module, EM) or with PDGFRA (PDGFRA module, PM) in gliomas, and their application in classifying adult diffuse gliomas into three major subtypes that are distinct in prognosis, genetic abnormalities and correlation to the cell lineages and differentiation stages of glial genesis.
Results
Identification of EM/PM Classifiers.
Using Pearson correlation coefficient analysis, we defined the EM and the PM by identifying the top 37 and 44 known genes with similar expression patterns to EGFR and PDGFRA, respectively, in the glioma transcriptome data set GSE4290 (18) (SI Appendix, Table S1). This data set includes 157 adult diffuse gliomas, with WHO grades II–IV, and 23 epileptic control samples. We found that the gene sets so defined were not overlapping. Furthermore, except for a small subset in EM (EGFR, SEC61G, VAV3, CDKN2C, TNFRSF19, NES, PDGFA, and SOX9) and PM (PDGFRA, OLIG1, OLIG2, SOX8, SOX6, and ZEB1), the majority of these EM and PM genes have not been extensively studied in glioma biology (SI Appendix, Table S2). Twenty-nine of the EM and 40 of the PM genes were found to be differentially expressed between the glioma samples and enabled clustering of glioma samples in unsupervised hierarchical clustering analysis (SI Appendix, Fig. S1). These 69 genes were used as classifiers throughout this study.
Clinical Relevance of EM/PM Classification.
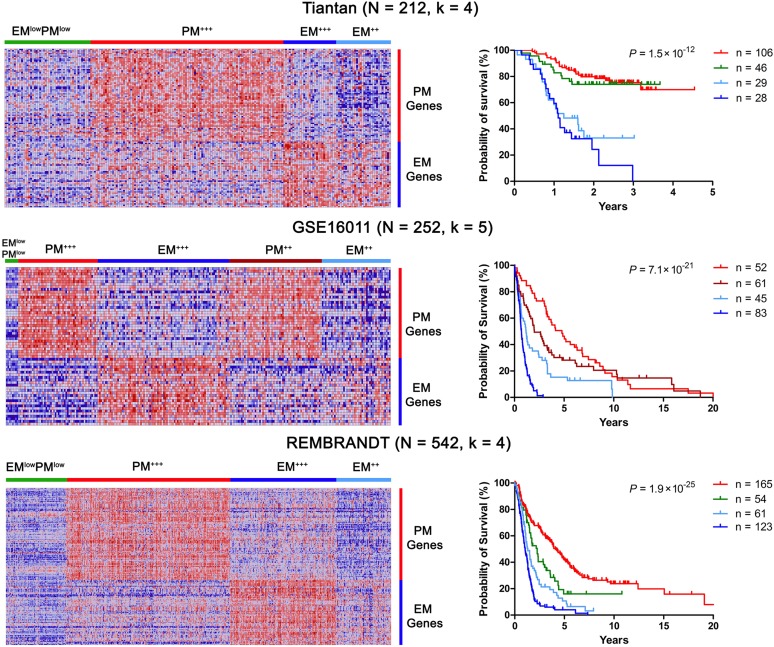
Using nonnegative matrix factorization (NMF), an unsupervised learning algorithm for identification of molecular patterns in gene expression data (19), we reproduced glioma clusters based on EM and PM signatures in three microarray-based data sets [Tiantan, the REMBRANDT (20) and GSE16011 (11)], and further validated the glioma clusters in a newly assembled, mRNA sequencing-based data set from TCGA. These data sets included 1,369 adult diffuse gliomas including diffuse astrocytomas WHO grade II, anaplastic astrocytomas WHO grade III, GBMs, oligodendrogliomas WHO grade II, anaplastic oligodendrogliomas WHO grade III, oligoastrocytomas WHO grade II, anaplastic oligoastrocytomas WHO grade III, and morphologically not clearly defined adult gliomas from patients in China, the Netherlands, and the US (Fig. 1 and SI Appendix, Fig. S2 and Tables S1 and S3), they were heterogeneous regarding the composition of patients and control, the platforms used to assess the gene expression data, and likely also in the details of sample processing and patient treatment. We reasoned that a fixed cluster number could not be applied to all data sets. The number of clusters was therefore independently determined in each data set based on large k numbers with stable cophenetic coefficients. Based on the extent of EM and PM expression, these analyses clustered gliomas into three major subtypes: EM (high EM but low PM expression), PM (high PM but low EM expression), and EMlowPMlow. EM gliomas were further subclustered into EM++ and EM+++ subtypes and PM gliomas into PM++ and PM+++ subtypes (Fig. 1 and SI Appendix, Figs. S1–S3). At the complete transcriptome level, gliomas within each of the EM/PM subtypes showed a high degree of subtype-specific similarity, the vast majority of the gliomas were clustered into one of the subtypes EM, PM, or EMlowPMlow (SI Appendix, Fig. S4A); the EMlowPMlow gliomas were distinct from the control brain samples that also showed an EMlowPMlow phenotype (SI Appendix, Fig. S4 B and C). The genes differentially enriched in each subtype are summarized in SI Appendix, Table S4.
Fig. 1.
EM/PM signatures define glioma subtypes with distinct prognosis. Glioma subtypes were classified by NMF based on the expression signatures of the EM/PM classifiers into the indicated number of clusters (k). Heat maps of EM/PM signatures in glioma samples are shown on the left. Tiantan (Top), GSE16011 (Middle), and REMBRANDT (Bottom) data sets are shown. (Right) Kaplan–Meier plots of the overall survival for the patients from each molecular subtype are shown. The overall survival data were analyzed using log-rank tests. The same color codes were used in the heat maps and the Kaplan–Meier plots. In all data sets, EMlowPMlow and PM gliomas were associated with better survival outcome than EM gliomas.
Nearly all adult diffuse glioma subgroups were present in each of the EM/PM subtypes. EM gliomas were more frequent in GBM; whereas PM gliomas were more frequent in grade II and III astrocytomas, oligodendrogliomas, and secondary GBMs (P < 0.0001, Pearson χ2 test, SI Appendix, Table S3). Notably, all of the morphologically undefined gliomas (SI Appendix, Table S3, “unknown” cases in the REMBRANDT data set) were clearly designated into EM/PM subtypes.
The median age at diagnosis for PM glioma patients in the GSE16011 and Tiantan data sets was 41.9 ± 12.3 y (n = 219), contrasting with that of EM glioma patients from the same data sets (53.3 ± 14.4 y, n = 188; P = 8.4 × 10−9, unpaired two-tailed t test). Patients with PM gliomas survived significantly longer than those with EM gliomas (log-rank test; Fig. 1 and SI Appendix, Fig. S2 and Table S3). The age at diagnosis and survival outcomes for EMlowPMlow gliomas were comparable to those of PM gliomas. EM/PM classification outperformed the morphological diagnosis in stratifying patients’ prognosis (SI Appendix, Fig. S5). The survival differences between patients with gliomas of PM or EMlowPMlow patterns and those with gliomas of EM patterns were shorter in high-grade than in low-grade gliomas. This effect could be either treatment-related or progression stage-related and probably also reflects the current lack of specific therapies for each glioma subtype. In multivariate Cox regression analysis, the prognostic value of EM/PM signatures was assessed after adjusting for the known prognostic factors, including IDH1 mutation, 1p19q codeletion, age, and Karnofsky performance score (KPS). Across all of these data sets, a more significant trend of EM score (the average of normalized expression levels for all EM classifier genes in each glioma sample) as a marker of poor prognosis was observed; the similarly calculated PM score was associated with a trend as a marker of favorable prognosis (SI Appendix, Table S5).
The classification schemes of Verhaak et al. (6) and Phillips et al. (4) were based on high-grade gliomas. The classification scheme of Verhaak et al. (6) did not predict overall patient survival in high-grade gliomas (14, 21). Using NMF clustering, we found that, in the Tiantan, GSE16011, and REMBRANDT data sets, patients with PN and NL gliomas defined by the classifiers of Verhaak et al. (6) were associated with better survival than patients with Mes and CL gliomas. PN gliomas predominantly contained PM gliomas and CL predominantly contained EM+++ gliomas (SI Appendix, Fig. S6). Notably, previous reports (22, 23) and our EM/PM clustering demonstrated that gliomas with EGFR amplification and PTEN deletion (present in >90% of EM+++ gliomas, SI Appendix, Table S3) or with codeletion of 1p36/19q13 (in ∼40% of PM+++ gliomas) represent two mutually exclusive glioma subtypes regarding survival profile, transcriptomic and genomic abnormalities, and association to neural lineages. These two types of gliomas were found in both NL and Mes subtypes in our analysis (SI Appendix, Table S6). Our NMF clustering of the Tiantan, GSE16011 and REMBRANDT data sets using the classifiers of Phillips et al. (4) showed that Mes and Prolif signatures overlapped with each other, or with PN signatures in glioma samples. The resulting glioma clusters were inconsistent in their patient survival profiles (SI Appendix, Fig. S7). Comparison with the G-CIMP classification scheme (14) showed that signatures of G-CIMP+ and G-CIMP− gliomas (24) were enriched in PM and EM gliomas, respectively; subsets of the G-CIMP+ and G-CIMP− signatures were concomitantly enriched in EMlowPMlow gliomas and control samples (SI Appendix, Fig. S8).
Relevance of EM/PM Classifiers to Glioma Pathogenesis.
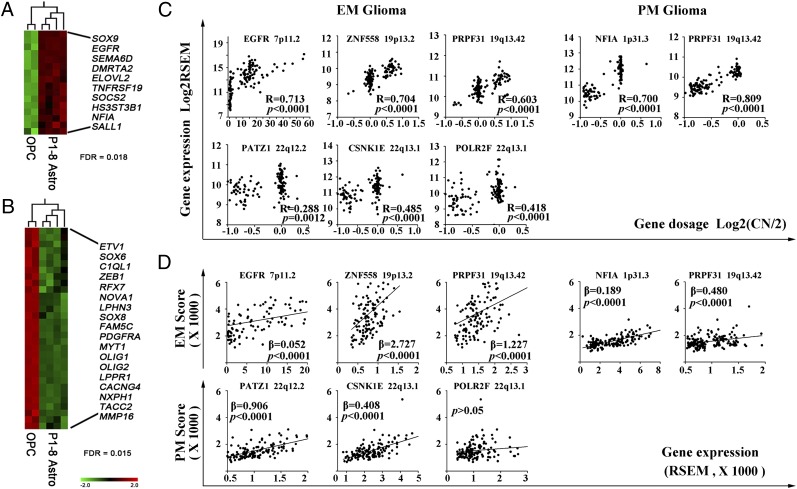
The EM/PM classifiers span diverse functional classes, including glial fate decision, Notch, TGF-β, and Jak-STAT signaling, cytoskeleton remodeling, protein transport, metabolism of lipid or the extracellular matrix, tumor suppression, chromatin modification, and regulation of transcription and pre-mRNA splicing (SI Appendix, Table S2). The activities of EM/PM classifiers may represent two coexpressed gene networks that provide multiple dependencies to glioma. In the mouse CNS developmental data set GSE9566 (25), subsets of EM and PM genes were found coexpressed in developing mouse astrocytes (Fig. 2A) and oligodendrocyte progenitor cells (OPCs) (Fig. 2B), respectively. In the glioma genome, analyses of the SNP data in both TCGA mRNA-seq and REMBRANDT data set concordantly show that other subsets of EM/PM classifiers, mostly not coexpressed in developing astrocytes or OPCs, were associated with frequent somatic copy number alterations (SCNA) in EM/PM glioma subtype-specific manner (SI Appendix, Tables S7 and S8 and Fig. S9). In agreement with previous reports on the low frequencies of mosaic coamplification of EGFR and PDGFRA in gliomas (26, 27), coamplification of EGFR and PDGFRA was observed in 10 of 123 EM+++ gliomas and 2 of 36 PM++(Chr7 Amp) gliomas of the TCGA mRNA-seq data set, but not in the other EM/PM subgroups (SI Appendix, Fig. S2). We focused our analyses on the consequence of subtype-specific SCNAs on the expression of EM/PM classifiers.
Fig. 2.
Coexpression in glial precursor cells and EM/PM glioma subtype-specific SCNAs of EM/PM classifiers. Heat maps of enriched coexpression of EM (A) and PM (B) genes in developing astrocytes and OPCs of mice. (C) EM/PM glioma subtype-specific SCNAs resulting in gene dosage-dependent expression of EM/PM classifiers was identified using Spearman’s rank correlation analysis; correlation coefficients (R) and their P values are indicated. (D) Linear regression analyses between the expression of individual classifiers and EM/PM signature in glioma samples show variations in the EM signature following the expression of ZNF558, PRPF31, and NFIA, and PM signature following the expression of PATZ1 and CSNK1E. Regression coefficients (β) and their P values are indicated.
Based on the SNP6.0 and the mRNA expression data in the TCGA mRNA-seq cohort, we first studied gene dosage-dependent expression of EM/PM classifiers. In EM gliomas, gene dosage-dependent expression was found for EM classifiers including EGFR, SEC61G, ITGB8, LFNG, PDGFA, DENND2A, CDKN2C, ZNF558, PRPF31, SNX5, NCOA3, and TRIOBP as well as PM classifiers including ETV1, NOVA1, PATZ1, CSNK1E, and POLR2F (Fig. 2C and SI Appendix, Fig. S10 and Table S7). In PM gliomas, gene dosage-dependent expression was observed for EM classifiers including NFIA and PRPF31 (located at Chr 1p or 19q and codeleted in 51 of 133 PM gliomas) and PM classifiers including ZEB1, EIF4EBP2, TACC2, MYT1, and NOVA1 (Fig. 2C and SI Appendix, Fig. S10 and Table S7). In EM gliomas, statistically significant cooccurrence of SCNAs among EM classifiers at Chr 7 (EGFR, SEC61G, ITGB8, LFNG, PDGFA, DENND2A) or at Chr 19 and Chr 20 (ZNF558, PRPF31, SNX5, NCOA3), and PM classifiers at Chr14q or Chr22q (NOVA1, PATZ1, CSNK1E, POLR2F) was observed (SI Appendix, Table S9). In PM gliomas, cooccurrence of SCNAs between EM classifiers NFIA and PRPF31, and among PM classifiers ZEB1, EIF4EBP2, TACC2, and MYT1 (at Chr 10 or 20q), was observed (SI Appendix, Table S9).
We next analyzed the contribution of the gene dosage-dependent expression of EM/PM classifiers to EM/PM signatures. Regression analyses show that in EM gliomas, the variable expression of frequently amplified EM classifier ZNF558 or PRPF31 strongly contributed to the variation of EM signature, whereas the expression of EGFR and SEC61G contributed to the EM signature predominantly due to its high expression level; the expression of the frequently lost PM classifiers PATZ1 and CSNK1E also strongly contributed to the variation of PM signature in EM gliomas. In PM gliomas, the expression of the frequently lost EM classifiers PRPF31 and NFIA strongly modulated the EM signature (Fig. 2D and SI Appendix, Table S7). The capacities of these classifiers in modulating the EM or PM signatures were validated in the REMBRANDT and GSE16011 data sets (SI Appendix, Table S10). These findings together suggest that common subtype-specific SCNAs lead to gene dosage-dependent altered expression of subsets of EM/PM classifiers resulting in modulation of the EM/PM signatures. These classifiers may play currently unrecognized roles in glioma pathogenesis.
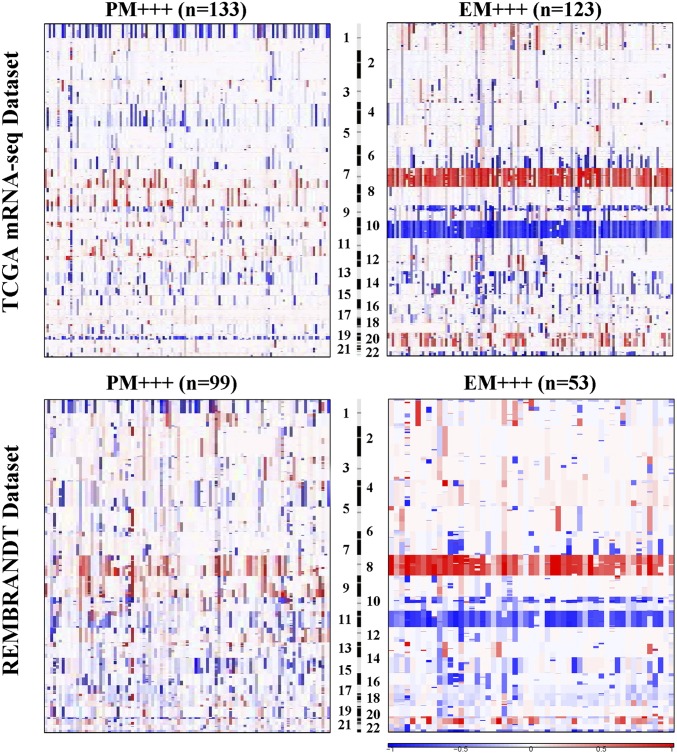
Distinct Genomic Alterations in EM/PM Glioma Subtypes.
Previous studies demonstrated frequent IDH1 mutation in the subset of the high-grade gliomas with better prognosis (28), and losses of heterozygosity (LOH) at Chr 1p/19q as a genomic hallmark in oligodendrogliomas (22); whereas other genomic alterations were not unambiguously specific to glioma subtypes (29). We found that IDH1 mutation was enriched in both PM and EMlowPMlow gliomas, LOH at 1p/19q in PM gliomas, whereas EGFR amplifications were enriched in EM gliomas (P < 0.0001, two-sided Fisher’s exact test, Tiantan and GSE16011 data sets; SI Appendix, Table S3). To assess SCNAs in EM/PM glioma subtypes, we used GISTIC2.0 (30) to analyze the SNP data from 349 gliomas in the TCGA mRNA-seq cohort and 205 gliomas in the REMBRANDT data set (20). EM and PM gliomas were associated with distinct chromosomal alterations (Fig. 3). Concomitant loss of Chr 1p/19q was found in ∼40% of the PM gliomas. Gain of Chr 7, loss of Chr 10, and Chr 22q were enriched in EM gliomas. According to the residual q value of the regional alterations, we identified the top 20 most significant amplification or deletion peaks in EM and PM gliomas. About 50% of these regional alterations were EM/PM glioma subtype-specific (SI Appendix, Figs. S11 and S12 and Tables S11 and S12). Furthermore, tumor specific SCNAs were also observed in the majority of EMlowPMlow gliomas, 18 of 25 EMlowPMlow gliomas in the REMBRANDAT data set showed tumor-specific SCNAs (SI Appendix, Fig. S12A), reinforcing that the transcriptomic signature of the EMlowPMlow gliomas indeed is derived from the tumor cells. Gene level SNP data of the TCGA mRNA-seq data set showed that focal amplification of EGFR and PDGFRA was more frequently observed in the EM+++ and PM++(Chr7 Amp) gliomas than in other EM/PM glioma subtypes (SI Appendix, Fig. S2 and Table S7). These results together suggest that EM/PM glioma subtypes are associated with distinct patterns of genomic alterations.
Fig. 3.
The raw genomic copy number of EM and PM gliomas. The SNP6.0 data from 349 gliomas in the TCGA mRNA-seq data set and the 50K HindIII SNP array data from 205 gliomas in the REMBRANDT data set were analyzed by GISTIC2.0 at an amplitude threshold of ± 0.2. The arm-level chromosomal gain or loss in PM+++ and EM+++ gliomas are depicted. The data for the raw genomic copy number of other subtypes are shown in SI Appendix, Figs. S11 and S12.
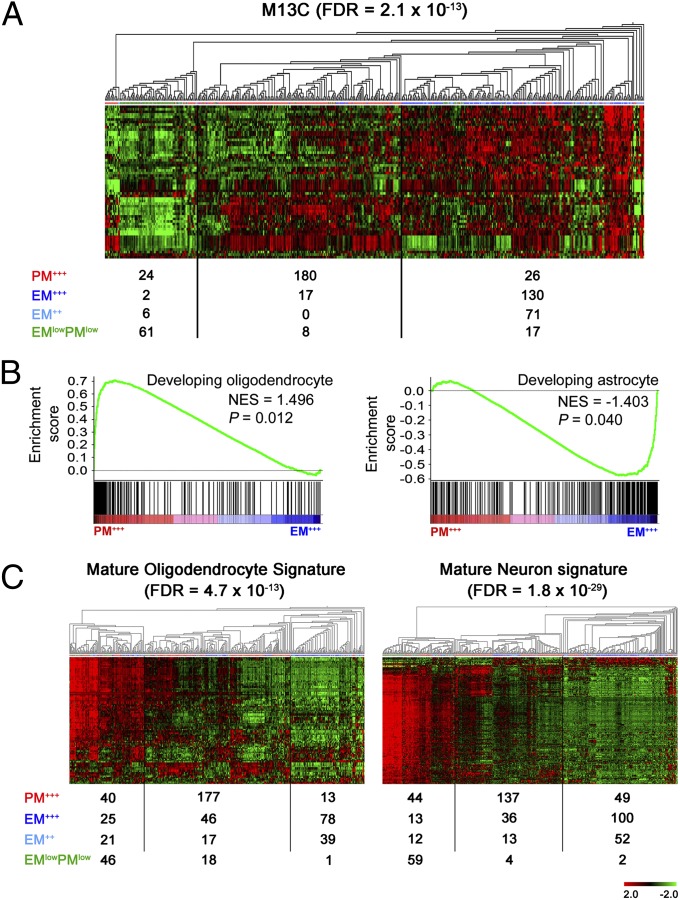
Differential Enrichment of Glial Genesis Activity in EM/PM Glioma Subtypes.
Gliomas have been suggested to arise from various cell types including NSCs (17), OPCs (31), and mature astrocytes (32). We found that the signature of NSCs, represented by the genes characteristic of the adult human neurogenic subventricular zone astrocytes (module M13C in Oldham et al.; ref. 33), was highly enriched in EM gliomas compared with PM and EMlowPMlow gliomas (Fig. 4 and SI Appendix, Fig. S13). Markers of NSC compartment (NES, EGFR) and key transcription factors regulating astrogenesis (POU3F2, NFIA, and SOX9; ref. 34) were significantly enriched in EM gliomas (Fig. 1 and SI Appendix, Figs. S1 and S2). Genes regulating oligodendrogenesis (PDGFRA, OLIG1, OLIG2, MYT1, SOX8, and SOX4; ref. 15) were highly enriched in PM gliomas (Fig. 1 and SI Appendix, Figs. S1 and S2). To further correlate EM/PM glioma subtypes with the immature or mature CNS cell types, we generated classifiers for developing astrocytes and oligodendrocytes using GSE9566 data set (25). Gene set enrichment analysis (GSEA) (35) identified significant enrichment of developing oligodendrocyte and astrocyte signatures in PM and EM glioma subtypes, respectively (Fig. 4 and SI Appendix, Figs. S13 and S14 and Table S13). We also generated classifiers for mature neurons, oligodendrocytes and astrocytes using GSE9566 (SI Appendix, Fig. S14). Compared with EM and PM gliomas, EMlowPMlow gliomas were enriched in the signatures of mature neurons and oligodendrocytes (Fig. 4 and SI Appendix, Fig. S13). The signature of mature astrocytes was dispersed over all EM/PM glioma subtypes.
Fig. 4.
Differential involvement of NSC compartment and glial lineage in EM/PM glioma subtypes. (A) The M13C coexpression module characteristic of subventricular zone astrocytes was enriched in EM gliomas. (B) Genes highly enriched in immature oligodendrocytes and immature astrocytes were enriched in PM+++ and EM+++ gliomas, respectively. (C) Genes highly enriched in mature oligodendrocytes and mature neurons were enriched in EMlowPMlow gliomas. Data are derived from the REMBRANDT data set.
Discussion
Using 69 genes that are coexpressed with EGFR or PDGFRA, we show that adult diffuse gliomas WHO grades II–IV from disparate institutions and ethnic backgrounds can be classified into EM, PM, and EMlowPMlow subtypes. This classification scheme is not guided by the morphological diagnosis of glioma or the survival outcome of the patients. EM gliomas are associated with enriched signatures of NSC compartment and astrogenesis, and they occur predominantly in patients older than 50 y and with an overall survival period of less than 2 y Conversely, PM gliomas are associated with enriched expression of genes regulating oligodendrogenesis and occur predominantly in patients younger than 50 y and with significantly longer survival time. EMlowPMlow gliomas are enriched in mature neuron and oligodendrocyte signatures with ages at diagnosis and survival outcomes similar to the PM gliomas. The three glioma subtypes show unique patterns of genomic alterations, including subtype-specific SCNAs in key regulators of glial fate decision. Our findings suggest that EM, PM, and EMlowPMlow glioma subtypes might represent biologically separate entities with distinct cellular origins, genetic alterations, and prognoses.
Our EM/PM classification scheme differs from previous glioma classification schemes in the following crucial aspects. Previous glioma molecular classification schemes depend on unbiased analyses of transcriptome (6) or DNA methylome (14). In contrast, EM/PM classification is based on the fundamental role of EGFR and PDGFRA in neural development and glioma pathogenesis (16, 17) and the gene coexpression modules around EGFR and PDGFRA. Whereas a previous classification scheme focused on classifiable core samples of GBMs and was unable to predict the survival of glioma patients (6, 14, 21), the EM/PM classification scheme is applicable to all diffuse gliomas WHO grades II–IV in adult, and robustly assigns gliomas into subtypes with distinct survival probabilities for patients. EM/PM classifiers are coexpressed in mouse immature glial cells and frequently amplified or lost in glioma genomes in EM/PM glioma subtype-specific manner. The expression of EM/PM classifiers with frequent SCNAs strongly contributes to the signature of the entire gene module around EGFR or PDGFRA. This analysis suggests that such EM/PM classifiers include currently unrecognized or poorly studied factors contributing to glioma pathogenesis.
The capacity of EM/PM classification scheme in assigning gliomas into subtypes with distinct genomic abnormalities and specific correlation to the known cell lineage and differentiation stages in neural development can provide important support to therapy development. Most of the genomic alterations in gliomas were previously found to be enriched in, but not specifically associated with, glioma subtypes. This inability to identify gliomas with distinct genomic abnormalities may have contributed to the failure of small molecule kinase inhibitors in the treatment of GBM patients, because it was not possible to identify gliomas that could respond to kinase inhibitors. Our analysis of SCNAs indicates that EM/PM glioma subtypes are associated with distinct genomic abnormalities, which may result in aberrant signaling activities in subtype-specific manner, or alternatively drive glioma transcriptome signatures toward a differentiation state which may be different from that of the cells in which the glioma originated. We envisage that signaling pathways specific for CNS cell lineages and differentiation stages are differentially involved in, and may account for, the etiology of EM/PM glioma subtypes. EM, PM, and EMlowPMlow gliomas may require different therapeutic strategies. Furthermore, we envisage that instead of individual signaling molecules or pathways, glioma therapy should ideally be designed to concomitantly target multiple pathways embedded in EM/PM coexpression modules.
In summary, the EM/PM classification scheme described here is applicable to diffuse gliomas WHO grades II–IV in adult and can predict the prognosis of glioma patients. This classification scheme creates a framework toward establishing molecular diagnostic tools and identifying new therapeutic targets to combat gliomas.
Materials and Methods
The Tiantan data set was generated by the Chinese Glioma Genome Atlas with materials from patients treated between 2006 and 2009 at Beijing Tiantan Hospital. Other data sets used in this study were retrieved from the TCGA, the Repository for Molecular Brain Neoplasia Data, and the Gene Expression Omnibus. Details regarding identification of EM/PM classifiers, clustering of gliomas, survival analysis, single nucleotide polymorphism analysis and lineage analysis are provided in SI Appendix.
Supplementary Material
Acknowledgments
We thank Håkan Olsson, Edgar Pera, Jiayi Li, Yuanji Chen, Claudia Jack, Robert Smail Jack, Andre Lieber, Srinivas Veerla, Ruiqiang Li, Xiaodong Su, Dan Csontos, and Clark Chen for helpful discussions. This study was supported by international collaborative program of the Ministry of Science and Technology of China (Grant 2012DFA30470), Beijing Normal University, National Natural Science Foundation of China (Grant 81072080), National High Technology Research and Development Program 863 (Grant 2012AA02A508), the Hans and Märit Rausing Charitable Foundation, and the Chinese Glioma Genome Atlas.
Footnotes
Conflict of interest statement: Beijing Normal University has applied a patent based on the EM/PM classification scheme.
*This Direct Submission article had a prearranged editor.
Data deposition: The gene expression data and the clinical and pathological parameters of all gliomas in the Tiantan data set can be retrieved from the Chinese Glioma Genome Atlas, www.cgga.org.cn under data portal, EMPM online.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1313814111/-/DCSupplemental.
References
- 1.Louis DN, et al. The 2007 WHO classification of tumours of the central nervous system. Acta Neuropathol. 2007;114(2):97–109. doi: 10.1007/s00401-007-0243-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Coons SW, Johnson PC, Scheithauer BW, Yates AJ, Pearl DK. Improving diagnostic accuracy and interobserver concordance in the classification and grading of primary gliomas. Cancer. 1997;79(7):1381–1393. doi: 10.1002/(sici)1097-0142(19970401)79:7<1381::aid-cncr16>3.0.co;2-w. [DOI] [PubMed] [Google Scholar]
- 3.Lee Y, et al. Gene expression analysis of glioblastomas identifies the major molecular basis for the prognostic benefit of younger age. BMC Med Genomics. 2008;1:52. doi: 10.1186/1755-8794-1-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Phillips HS, et al. Molecular subclasses of high-grade glioma predict prognosis, delineate a pattern of disease progression, and resemble stages in neurogenesis. Cancer Cell. 2006;9(3):157–173. doi: 10.1016/j.ccr.2006.02.019. [DOI] [PubMed] [Google Scholar]
- 5.Freije WA, et al. Gene expression profiling of gliomas strongly predicts survival. Cancer Res. 2004;64(18):6503–6510. doi: 10.1158/0008-5472.CAN-04-0452. [DOI] [PubMed] [Google Scholar]
- 6.Verhaak RG, et al. Cancer Genome Atlas Research Network Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Horvath S, et al. Analysis of oncogenic signaling networks in glioblastoma identifies ASPM as a molecular target. Proc Natl Acad Sci USA. 2006;103(46):17402–17407. doi: 10.1073/pnas.0608396103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liang Y, et al. Gene expression profiling reveals molecularly and clinically distinct subtypes of glioblastoma multiforme. Proc Natl Acad Sci USA. 2005;102(16):5814–5819. doi: 10.1073/pnas.0402870102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Murat A, et al. Stem cell-related “self-renewal” signature and high epidermal growth factor receptor expression associated with resistance to concomitant chemoradiotherapy in glioblastoma. J Clin Oncol. 2008;26(18):3015–3024. doi: 10.1200/JCO.2007.15.7164. [DOI] [PubMed] [Google Scholar]
- 10.Nutt CL, et al. Gene expression-based classification of malignant gliomas correlates better with survival than histological classification. Cancer Res. 2003;63(7):1602–1607. [PubMed] [Google Scholar]
- 11.Gravendeel LA, et al. Intrinsic gene expression profiles of gliomas are a better predictor of survival than histology. Cancer Res. 2009;69(23):9065–9072. doi: 10.1158/0008-5472.CAN-09-2307. [DOI] [PubMed] [Google Scholar]
- 12.Li A, et al. Unsupervised analysis of transcriptomic profiles reveals six glioma subtypes. Cancer Res. 2009;69(5):2091–2099. doi: 10.1158/0008-5472.CAN-08-2100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Turcan S, et al. IDH1 mutation is sufficient to establish the glioma hypermethylator phenotype. Nature. 2012;483(7390):479–483. doi: 10.1038/nature10866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Noushmehr H, et al. Cancer Genome Atlas Research Network Identification of a CpG island methylator phenotype that defines a distinct subgroup of glioma. Cancer Cell. 2010;17(5):510–522. doi: 10.1016/j.ccr.2010.03.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nicolay DJ, Doucette JR, Nazarali AJ. Transcriptional control of oligodendrogenesis. Glia. 2007;55(13):1287–1299. doi: 10.1002/glia.20540. [DOI] [PubMed] [Google Scholar]
- 16.Pastrana E, Cheng LC, Doetsch F. Simultaneous prospective purification of adult subventricular zone neural stem cells and their progeny. Proc Natl Acad Sci USA. 2009;106(15):6387–6392. doi: 10.1073/pnas.0810407106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huse JT, Holland EC. Targeting brain cancer: Advances in the molecular pathology of malignant glioma and medulloblastoma. Nat Rev Cancer. 2010;10(5):319–331. doi: 10.1038/nrc2818. [DOI] [PubMed] [Google Scholar]
- 18.Sun L, et al. Neuronal and glioma-derived stem cell factor induces angiogenesis within the brain. Cancer Cell. 2006;9(4):287–300. doi: 10.1016/j.ccr.2006.03.003. [DOI] [PubMed] [Google Scholar]
- 19.Brunet JP, Tamayo P, Golub TR, Mesirov JP. Metagenes and molecular pattern discovery using matrix factorization. Proc Natl Acad Sci USA. 2004;101(12):4164–4169. doi: 10.1073/pnas.0308531101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Madhavan S, et al. Rembrandt: Helping personalized medicine become a reality through integrative translational research. Mol Cancer Res. 2009;7(2):157–167. doi: 10.1158/1541-7786.MCR-08-0435. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Kim TM, Huang W, Park R, Park PJ, Johnson MD. A developmental taxonomy of glioblastoma defined and maintained by MicroRNAs. Cancer Res. 2011;71(9):3387–3399. doi: 10.1158/0008-5472.CAN-10-4117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Idbaih A, et al. BAC array CGH distinguishes mutually exclusive alterations that define clinicogenetic subtypes of gliomas. Int J Cancer. 2008;122(8):1778–1786. doi: 10.1002/ijc.23270. [DOI] [PubMed] [Google Scholar]
- 23.Labussière M, et al. All the 1p19q codeleted gliomas are mutated on IDH1 or IDH2. Neurology. 2010;74(23):1886–1890. doi: 10.1212/WNL.0b013e3181e1cf3a. [DOI] [PubMed] [Google Scholar]
- 24.Baysan M, et al. G-cimp status prediction of glioblastoma samples using mRNA expression data. PLoS ONE. 2012;7(11):e47839. doi: 10.1371/journal.pone.0047839. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cahoy JD, et al. A transcriptome database for astrocytes, neurons, and oligodendrocytes: A new resource for understanding brain development and function. J Neurosci. 2008;28(1):264–278. doi: 10.1523/JNEUROSCI.4178-07.2008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Szerlip NJ, et al. Intratumoral heterogeneity of receptor tyrosine kinases EGFR and PDGFRA amplification in glioblastoma defines subpopulations with distinct growth factor response. Proc Natl Acad Sci USA. 2012;109(8):3041–3046. doi: 10.1073/pnas.1114033109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Snuderl M, et al. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell. 2011;20(6):810–817. doi: 10.1016/j.ccr.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 28.Yan H, et al. IDH1 and IDH2 mutations in gliomas. N Engl J Med. 2009;360(8):765–773. doi: 10.1056/NEJMoa0808710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dunn GP, et al. Emerging insights into the molecular and cellular basis of glioblastoma. Genes Dev. 2012;26(8):756–784. doi: 10.1101/gad.187922.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Mermel CH, et al. GISTIC2.0 facilitates sensitive and confident localization of the targets of focal somatic copy-number alteration in human cancers. Genome Biol. 2011;12(4):R41. doi: 10.1186/gb-2011-12-4-r41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Persson AI, et al. Non-stem cell origin for oligodendroglioma. Cancer Cell. 2010;18(6):669–682. doi: 10.1016/j.ccr.2010.10.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bachoo RM, et al. Epidermal growth factor receptor and Ink4a/Arf: Convergent mechanisms governing terminal differentiation and transformation along the neural stem cell to astrocyte axis. Cancer Cell. 2002;1(3):269–277. doi: 10.1016/s1535-6108(02)00046-6. [DOI] [PubMed] [Google Scholar]
- 33.Oldham MC, et al. Functional organization of the transcriptome in human brain. Nat Neurosci. 2008;11(11):1271–1282. doi: 10.1038/nn.2207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kang P, et al. Sox9 and NFIA coordinate a transcriptional regulatory cascade during the initiation of gliogenesis. Neuron. 2012;74(1):79–94. doi: 10.1016/j.neuron.2012.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Subramanian A, et al. Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc Natl Acad Sci USA. 2005;102(43):15545–15550. doi: 10.1073/pnas.0506580102. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.