Significance
Mutations within the BCR-ABL1 kinase domain lead to resistance to tyrosine kinase inhibitor (TKI) therapy in chronic myeloid leukemia. We show a high prevalence of compound BCR-ABL1 mutations (polymutants) in patients failing TKIs. Molecular dynamics analyses of the most frequent polymutants in complex with TKIs unveiled the basis of TKI resistance. Modeling of BCR-ABL1 in complex with ponatinib, a potent pan-BCR-ABL1 TKI, highlighted the presence of complex BCR-ABL1 mutant proteins capable of escaping all available TKI therapy.
Keywords: compound mutation, ponatinib resistance
Abstract
The acquisition of mutations within the BCR-ABL1 kinase domain is frequently associated with tyrosine kinase inhibitor (TKI) failure in chronic myeloid leukemia. Sensitive sequencing techniques have revealed a high prevalence of compound BCR-ABL1 mutations (polymutants) in patients failing TKI therapy. To investigate the molecular consequences of such complex mutant proteins with regards to TKI resistance, we determined by cloning techniques the presence of polymutants in a cohort of chronic-phase patients receiving imatinib followed by dasatinib therapy. The analysis revealed a high frequency of polymutant BCR-ABL1 alleles even after failure of frontline imatinib, and also the progressive exhaustion of the pool of unmutated BCR-ABL1 alleles over the course of sequential TKI therapy. Molecular dynamics analyses of the most frequent polymutants in complex with TKIs revealed the basis of TKI resistance. Modeling of BCR-ABL1 in complex with the potent pan-BCR-ABL1 TKI ponatinib highlighted potentially effective therapeutic strategies for patients carrying these recalcitrant and complex BCR-ABL1 mutant proteins while unveiling unique mechanisms of escape to ponatinib therapy.
The BCR-ABL1 tyrosine kinase inhibitor (TKI) imatinib mesylate is highly effective in chronic myeloid leukemia in chronic phase (CML-CP) (1), being associated with complete cytogenetic (CCyR) and major molecular response rates of 83% and 86%, and progression-free and overall survival rates of 92% and 85%, respectively (2). However, after 8 y of follow-up, 45% of the patients failed imatinib therapy (2), frequently due to the acquisition of mutations within the kinase domain of BCR-ABL1 (3–9). More than 100 distinct BCR-ABL1 point mutations have been reported in patients (4, 9–15), and many others have been generated in vitro by random mutagenesis of BCR-ABL1 (13, 16). Most patients exhibiting imatinib resistance receive a second-generation TKI, such as nilotinib or dasatinib (17, 18), which inhibit most clinically relevant BCR-ABL1 mutations, except for T315I (19, 20). Sequential TKI therapy has been associated with the acquisition of more than one mutation in the same BCR-ABL1 protein (i.e., compound mutant or polymutant) (21–23). In transformation assays, the accumulation of more than one mutation within the same allele has been associated with increased oncogenic potential compared with each individual mutation (21). As such, the emergence of polymutants might represent a powerful mechanism of resistance, perhaps as deleterious as that of developing single point mutations at gatekeeper residues (e.g., T315I). Inadequate selection of subsequent TKI therapy may result in a recrudescence of this phenomenon via selection pressure of complex polymutants highly resistant to available TKIs. We sought to investigate the structural, thermodynamic, and biochemical impact of polymutant BCR-ABL1 proteins detected in patients with CML on TKI binding and activity. As multiple additional genetic lesions other than BCR-ABL1 mutations may contribute to TKI resistance in patients with accelerated or blast phase, we intentionally limited our study to patients with CML-CP, in whom response or resistance to TKI therapy is largely determined by BCR-ABL1 mutational status. Molecular dynamics analyses of the most frequently detected polymutants were validated by biochemical assays, which demonstrated profound resistance to imatinib and dasatinib. In silico modeling of polymutant BCR-ABL1 kinases in complex with ponatinib (24–26) further revealed that the presence of polymutant BCR-ABL1 proteins might represent a critical mechanism of escape of CML cells against this pan-BCR-ABL1 inhibitor.
Results
Generation of Polymutant BCR-ABL1 Proteins During Sequential TKI Therapy.
Seventy patients with CML-CP receiving imatinib followed by dasatinib were evaluated by DNA expansion of specific clones (SI Appendix, Table S1). Upon imatinib failure, BCR-ABL1 kinase domain mutations were detected in 61/70 patients (87%), including 38 (54%) with mutations detected in ≥20% of sequenced clones. Overall, 125 mutations at 113 amino acid positions were detected (SI Appendix, Table S2). Polymutant BCR-ABL1 proteins were detected in 41% of patients, with individual clones frequently expressing two (but up to five) single point mutations (SI Appendix, Fig. S1 and Table S1). At variance with prior data (21), polymutants were found not only after failure of a second TKI (e.g., dasatinib), but very frequently upon failure of frontline imatinib therapy. DNA expansion of specific clones was performed in 32 (47%) patients during dasatinib therapy and revealed 20 additional mutations not present at dasatinib start (SI Appendix, Table S2).
Dynamics of Unmutated BCR-ABL1 Alleles During Sequential TKI Therapy.
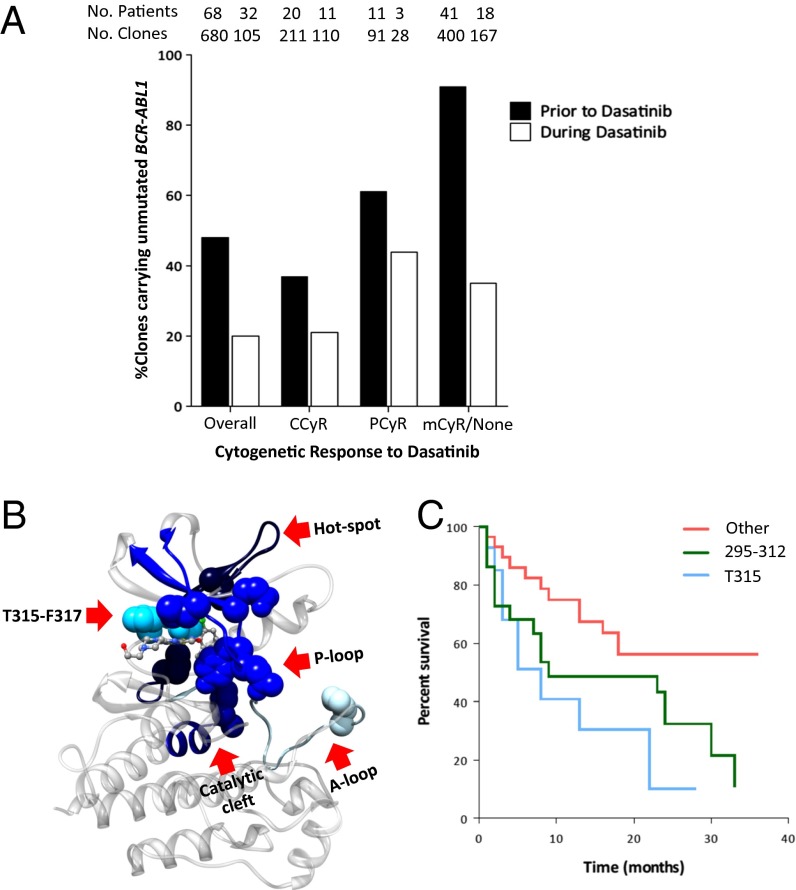
Next, we examined the dynamics of unmutated BCR-ABL1 alleles after imatinib failure and during second-line therapy with dasatinib according to the cytogenetic response achieved on this TKI (Fig. 1A). The percentage of clones expressing unmutated BCR-ABL1 decreased significantly during dasatinib therapy (P = 0.001), particularly in patients carrying highly dasatinib-resistant mutants. The percentage of mutated BCR-ABL1–bearing clones increased significantly upon exposure to dasatinib in nonresponders. This increase occurred rapidly after dasatinib start because the median time elapsed until a follow-up mutational analysis by DNA expansion of specific clones was only 16 wk (range: 4–84 wk). Though differences were observed for each level of cytogenetic response during dasatinib therapy, the proportion of clones carrying unmutated BCR-ABL1 was remarkably lower among patients who either did not achieve a cytogenetic response or had only a minor cytogenetic response compared with those who achieved a major cytogenetic response [MCyR, i.e., ≤35% Ph-positive cells in metaphase (P = 0.0001)]. These data suggest that BCR-ABL1 genetic instability evolves during second-line TKI therapy. The selection pressure exerted by dasatinib on clones carrying unmutated or mutated but sensitive BCR-ABL1 proteins promotes the expansion of clones carrying highly resistant proteins and rapidly shifts the balance between mutated and unmutated clones, resulting in exhaustion of unmutated clones and expansion of mutant (and polymutant) clones associated with clinical resistance to dasatinib. Our results suggest a correlation between achieving clinically meaningful cytogenetic responses (i.e., at least MCyR) and the ratio of mutated/unmutated BCR-ABL1–positive clones. Notably, we had previously shown the critical importance of achieving at least MCyR within the first 12 mo of second-line TKI therapy to improve long-term survival (27).
Fig. 1.
Mutated and unmutated BCR-ABL1 in CML clones during sequential tyrosine kinase inhibitor therapy. (A) Dynamics of unmutated BCR-ABL1–bearing clones. Patients without cytogenetic response during dasatinib therapy after imatinib failure bore a lower proportion of unmutated clones compared with those who achieved at least a partial cytogenetic response (P = 0.01), suggesting exhaustion of unmutated BCR-ABL1–bearing clones in patients not responding to sequential TKI therapy. (B) Mapping of the clinically relevant mutations found onto the BCR-ABL1 protein in complex with dasatinib. The different colors reflect the density of mutations found in each region of the kinase: the darker the color the higher the density of mutations in that specific region. Color code: light blue (9%): A-loop (residues 379–398); sky blue (13%): T315-F317 region; blue (16%): P-loop (residues 242–261); navy blue (17%): catalytic cleft (residues 351–359); dark blue (18%): hotspot region (residues 295–312). (C) Survival of patients carrying mutations in the hotspot 295–312 region. Patients carrying mutations at T315I or at the 295–312 region exhibit a shorter overall survival compared with those carrying other BCR-ABL1 mutations (P = 0.03).
The Hotspot 295–312 Region.
In accord with previous reports, most mutations mapped to four areas of the BCR-ABL1 kinase domain: the P-loop (16%), the catalytic cleft (17%), the 315–317 region (13%), and the activation loop (9%) (Fig. 1B and SI Appendix, Fig. S1B). However, a significant number of mutations clustered at high frequency to other regions of the ABL1 kinase domain. One of these hotspot regions spans the residues flanked by amino acid positions 295 and 312 and contained 18% of all mutations detected in the treated cohort (Fig. 1B and SI Appendix, Fig. S1B). In addition to five patients who acquired the highly dasatinib-resistant V299L mutation, 16 of 61 patients (25%) harbored mutations mapping to the 295–312 region before the start of dasatinib therapy; 11 (69%) never achieved any cytogenetic response on dasatinib, and this was associated with significantly worse overall survival than that of patients expressing any other BCR-ABL1 mutation (P = 0.02), except for T315I (Fig. 1C). Notably, 5 of 16 patients (32%) with mutations within the 295–312 region carried clones expressing more than one mutation within that same region. Mutations in this region are clinically relevant because none of the latter achieved a cytogenetic response to TKI therapy, and all died after a median of 9 mo (Fig. 1C).
In Silico Modeling of Polymutants in Complex with Imatinib and Dasatinib.
Exposure to specific TKIs is associated with the acquisition of distinct ABL1 kinase mutations (SI Appendix, Fig. S2). To gain mechanistic insight regarding the TKI resistance posed by polymutant BCR-ABL1 proteins and potential ways to therapeutically overcome it, we next performed a thorough in silico analysis of clinically relevant BCR-ABL1 mutant alleles. First, we modeled the resistance imposed by single point mutations frequently acquired by patients with CML failing imatinib or dasatinib using molecular dynamics simulations (SI Appendix, Table S3 and SI Results). Notably, our in silico methodology (28) ranked the affinity of the different BCR-ABL1 isoforms similarly to available experimental data obtained in biochemical assays with imatinib, dasatinib, and nilotinib (SI Appendix, Figs. S3–S5) (6, 19, 20, 29–31), thus validating the adopted computational approach and supporting its use to discern differences in the mechanisms of resistance to specific TKIs among distinct mutants (SI Appendix, Tables S4–S8).
Next, we analyzed the impact of compound mutants (polymutants) identified from our clinical samples in the same BCR-ABL1 protein with regards to TKI binding affinity (Table 1). All double BCR-ABL1 mutants analyzed were highly resistant to imatinib. In most cases, both mutations were synergistic in enhancing imatinib resistance, as seen from the calculated IC50 values and the fold change (FC) values vs. those obtained from each single mutation (Table 1, far right column). Similarly, all these double mutants were resistant to dasatinib, with both single point mutations within any given BCR-ABL1 polymutant protein acting synergistically to enhance dasatinib resistance (Table 1 and SI Appendix, Table S3). For example, the synergistic effect of the T315I/V299L and F317L/V299L polymutants on the destabilization of the binding site is apparent from the corresponding per-residue energy deconvolution shown in SI Appendix, Tables S9 and S10. The concomitant presence of two point mutations dramatically diminished the affinity of the ATP-binding site for imatinib as well as for the second-generation TKIs dasatinib and nilotinib (Table 1 and SI Appendix, Fig. S6). Interestingly, for both TKIs, the double mutant T315I/V299L was predicted to be extremely deleterious, followed by the double mutant T315I/V304D.
Table 1.
Predicted ΔGbind, IC50, and FC values for BCR-ABL1 double mutants in complex with imatinib, dasatinib, or ponatinib
| BCR-ABL1 | ΔGbind, kcal/mol | IC50, nM | FC |
| Imatinib | |||
| Unmutated | −10.47 ± 0.03 | 21 | — |
| T315I/Y253H | −5.04 ± 0.01 | 203,000 | 9,667 |
| T315I/V299L | −6.01 ± 0.02 | 39,000 | 1,857 |
| T315I/V304D | −6.12 ± 0.02 | 32,500 | 1,548 |
| Y253H/V299L | −6.49 ± 0.01 | 17,600 | 838 |
| V304D/Y253H | −7.11 ± 0.03 | 6,100 | 290 |
| V304D/V299L | −9.18 ± 0.02 | 185 | 8.8 |
| Dasatinib | |||
| Unmutated | −12.39 ± 0.01 | 0.8 | — |
| T315I/F317L | −5.47 ± 0.02 | 970,000 | 1,212,500 |
| T315I/V299L | −4.83 ± 0.02 | 290,000 | 3,625 |
| T315I/V304D | −6.02 ± 0.03 | 38,000 | 47,500 |
| V299L/F317L | −7.42 ± 0.02 | 3,650 | 4,563 |
| V304D/V299L | −8.05 ± 0.01 | 1,250 | 1,563 |
| F317L/V304D | −9.01 ± 0.02 | 250 | 313 |
| Ponatinib | |||
| Unmutated | −12.67 ± 0.02 | 0.52 | — |
| T315I/F359V | −10.23 ± 0.03 | 32 | 61 |
| T315I/F317L | −10.48 ± 0.02 | 21 | 40 |
| V299L/F317L | −10.56 ± 0.01 | 18 | 34 |
| V304D/F317L | −10.68 ± 0.02 | 15 | 29 |
| T315I/V299L | −10.92 ± 0.02 | 10 | 19 |
| T315I/V304D | −11.01 ± 0.01 | 8.8 | 16 |
| T315I/Y253H | −11.05 ± 0.03 | 8 | 15 |
| Y253H/V299L | −11.58 ± 0.02 | 3.3 | 6.3 |
| V304D/V299L | −11.84 ± 0.02 | 2.1 | 4 |
| V304D/Y253H | −12.31 ± 0.03 | 0.96 | 1.8 |
Activity of the Third-Generation TKI Ponatinib Against Polymutant BCR-ABL1 Proteins.
Given the high frequency and variety of highly resistant polymutants observed in patients failing sequential imatinib/dasatinib therapy, we next performed 3D structural analyses to model the activity of the pan–BCR-ABL1 inhibitor ponatinib against unmutated BCR-ABL1, single mutant, and some of the most frequently encountered polymutant BCR-ABL1 proteins (Fig. 2 A–C). Ponatinib, which is highly active against all BCR-ABL1 single point mutants tested to date (24), was rationally designed following a structure-guided drug-design strategy with the double intent of targeting the inactive conformation of the ABL1 kinase and avoiding interaction with the side chain of the mutated 315 gatekeeper residue. Ponatinib received accelerated approval by the Food and Drug Administration in December 2012 for the treatment of patients with CML after failure of prior TKI therapy, although it was temporarily withdrawn from the US market in October 2013 due to the risk of life-threatening vascular events. We tested 21 BCR-ABL1 mutants (11 single and 10 double mutants) in complex with ponatinib, including both active site (i.e., drug contact) and nonactive site residues, nonpolar-to-nonpolar residue mutants (e.g., V299L), nonpolar-to-polar residue mutants (e.g., G250E), and polar-to-polar residue mutants (e.g., E255K). The estimated binding free energy for the unmutated and the 21 mutant BCR-ABL1/ponatinib complexes are listed in Table 1 and SI Appendix, Table S3. As expected from biochemical data (24), ponatinib was predicted to potently inhibit both native (IC50 = 0.52 nM) and T315I BCR-ABL1 (IC50 = 7.4 nM) in addition to multiple imatinib-, nilotinib-, and dasatinib-resistant BCR-ABL1 point mutant isoforms. Most single mutants should not result in high levels of resistance to ponatinib, because the calculated fold change values ranged from 1.1 for Y253H (IC50 = 0.55 nM) to 17 for E255K (IC50 = 8.8 nM), the latter being the most resistant single point mutant (Table 1, Fig. 2C, and SI Appendix, Table S3).
Fig. 2.
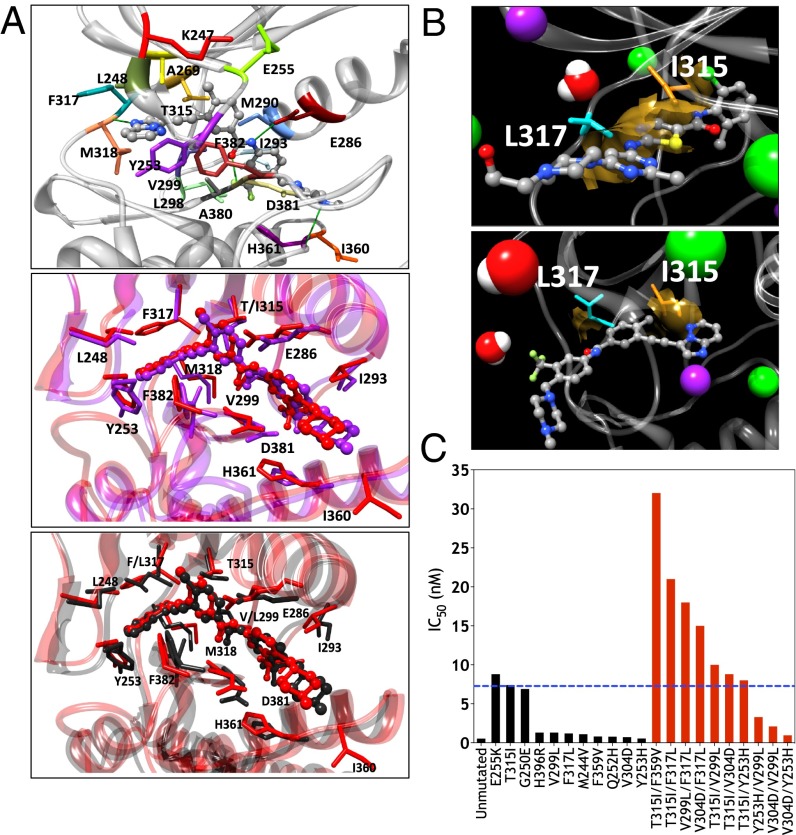
In silico modeling of ponatinib binding to BCR-ABL1 polymutants. (A) (Top) Details of the binding site of unmutated BCR-ABL1 in complex with ponatinib as obtained from equilibrated MD simulation snapshots. The protein backbone is portrayed as a transparent gray ribbon; the main residues involved in drug interactions are shown as labeled colored sticks. Hydrogen bonds are shown as green lines. (Middle) Superposition of the binding site of unmutated BCR-ABL1 (red) and the BCR-ABL1T315I (purple) in complex with ponatinib. In the two complexes, the drug is depicted in red (unmutated) and purple (T315I) sticks and balls, respectively. (Bottom) Superposition of the binding site of unmutated BCR-ABL1 (red) and the BCR-ABL1V299L/F317L double mutant (black) in complex with ponatinib. In the two complexes, the drug is depicted in red (unmutated) and black (F317L/V299L) sticks and balls, respectively. (B) MD snapshots of dasatinib (Upper) and ponatinib (Lower) bound BCR-ABL1T315I/F317L. The mutant I315 and L317 are depicted as orange and cyan sticks, respectively. Note the marked difference in steric clash (golden areas in both panels), accounting for the higher affinity of ponatinib for the double-mutant protein. Hydrogen atoms, water molecules, and ions are omitted for clarity. (C) Sensitivity of a series of BCR-ABL1 proteins with single point mutations (black bars) or dual mutants (red bars) against ponatinib, based on calculated IC50. Dotted blue line represents the half-maximal inhibitory concentration (IC50) of ponatinib against BCR-ABL1T315.
In sharp contrast with imatinib and dasatinib, the predicted high sensitivity of the BCR-ABL1 mutants to ponatinib binding probably reflects an intrinsic spatial adaptability of the inhibitor to bind different conformational states of BCR-ABL1 kinase (Fig. 2A, Table 1, and SI Appendix, Tables S3 and S11) (25), producing the substantially smaller area of steric clash between ponatinib and the mutant catalytic site relative to other TKIs such as dasatinib (Fig. 2B). Indeed, structurally, different conformations of the activation loop and the α-helix C do not result in dramatic changes in the binding profiles of ponatinib (Table 1 and SI Appendix, Table S3). P-loop mutations (usually associated with high TKI resistance and poor clinical outcomes with imatinib and nilotinib) are involved in shifting the conformational equilibrium of the kinase toward the active conformation and are characterized by computed binding affinities for ponatinib (Table 1 and SI Appendix, Table S3) similar to those calculated for the unmutated BCR-ABL1 kinase (e.g., IC50 = 0.52 nM for the unmutated protein, and 0.79 nM and 0.55 nM for the Q252H and Y253H mutants, respectively). The effect of deleterious drug contact mutations (F317L, V299L, and particularly T315I) is marginally different from the binding free energies associated with P-loop mutants, with the exception of G250E and E255K. In our analysis, the latter are several-fold more resistant to ponatinib than most other studied mutations, in keeping with biochemical data (24).
Our modeling confirms that the contribution of residue T315 to ponatinib binding is very small (SI Appendix, Table S11). However, though both T315I and the contact mutant residues F317L and V299L showed high sensitivity to ponatinib, the association of any two of these three mutations in a dual polymutant synergized to produce mutant BCR-ABL1 proteins that exhibited markedly high resistance, with fold-change values ranging from 19 to 40 that of the unmutated protein (Table 1 and Fig. 2C). Additionally, the double mutant T315I/F359V displayed the greatest resistance to ponatinib, with a 61-fold change and calculated IC50 of 32 (Table 1 and Fig. 2C). Modeling of ponatinib binding to the BCR-ABL1T315I/F359V polymutant revealed a cavity that breaks the drug–protein surface complementarity, producing a difference in ΔGbind of 2.4 kcal/mol vs. the unmutated BCR-ABL1 protein (Fig. 3A and SI Appendix, Fig. S7). These results suggest a potential mechanism of escape to ponatinib therapy through the acquisition and/or selection of complex mutant BCR-ABL1 proteins.
Fig. 3.
Effect of single and dual BCR-ABL1 kinase mutations on Ba/F3 cellular sensitivity to tyrosine kinase inhibitors. (A) The dual mutant T315I/F359V induces a rearrangement of the BCR-ABL1 binding pocket, with the addition of the F359V mutation resulting in loss of drug–protein surface complementarity and formation of a cavity (Lower, in blue), that promotes a weaker protein/inhibitor interaction with respect to the unmutated BCR-ABL1 protein (Upper, in red). (B) Activity of imatinib, dasatinib, and ponatinib against unmutated BCR-ABL1 and BCR-ABL1 carrying T315I, T315I/V299L, T315I/F317L, and T315I/F359V mutations in Ba/F3-based cellular assays. (C) Stat5 and CrkL phosphorylation was examined in lysates obtained from Ba/F3 cells carrying unmutated or mutated BCR-ABL1 proteins upon exposure to increasing concentrations of ponatinib for 4 h. Samples were analyzed by immunoblot analysis with antibodies against phospho-Stat5, phospho-CrkL, and actin (loading control).
Overall, our data suggest that ponatinib favorably binds the majority of assessed BCR-ABL1 single mutants irrespective of the conformational state of the enzyme and of the potential switching between structurally different inactive states that these mutations may cause. However, some polymutants, particularly those including one or more mutant contact residues, exhibit very high levels of resistance to ponatinib, thus anticipating a potential mechanism of resistance to this TKI.
Ponatinib Inhibits BCR-ABL1–Mediated Signaling and Growth in Cells Expressing Polymutant BCR-ABL1 Alleles.
To validate our in silico analyses regarding ponatinib resistance, we exposed Ba/F3 cells expressing BCR-ABL1T315I or a variety of engineered polymutant BCR-ABL1 alleles predicted to produce ponatinib-resistant BCR-ABL1 proteins to imatinib, dasatinib, or ponatinib (Fig. 3 B and C). Although all agents effectively suppressed the growth of Ba/F3 cells expressing unmutated BCR-ABL1, only ponatinib exhibited activity against Ba/F3 cells carrying BCR-ABL1T315I or any of the polymutants tested. However, the polymutants tested exhibited very high resistance to ponatinib, which was ∼10-fold higher than that exhibited by cells carrying BCR-ABL1T315I. As predicted by our in silico modeling, BCR-ABL1T315I/F359V exhibited the highest resistance of all of the polymutants tested (Fig. 3 and SI Appendix, Fig. S7). The growth inhibitory activity of these BCR-ABL1 kinase inhibitors was coupled with inhibition of phosphorylation of CrkL and Stat5, both surrogate markers of BCR-ABL1 kinase activity (Fig. 3C) (32). The mutant BCR-ABL1 proteins demonstrated progressively greater resistance to inhibition of Stat5 and CrkL phosphorylation by ponatinib than wild type. Ponatinib consistently suppressed the phosphorylation of Stat5 and CrkL at concentrations of 30 nM or greater, which are clinically achievable in patients with CML. However, ponatinib, while suppressing Stat5 phosphorylation, could not suppress CrkL phosphorylation in cells expressing the BCR-ABL1T315I/F359V polymutant kinase, even at a concentration of 100 nM, which is 50-fold the IC50 required to inhibit BCR-ABL1T315I in kinase-based assays and 10-fold the IC50 required in Ba/F3 cells forced to express BCR-ABL1T315I. These experimental results confirm that our in silico modeling methodology accurately predicts response to TKIs in the presence of complex mutations. Importantly, they alert us of potential molecular and cellular mechanisms of resistance to ponatinib through the acquisition of resistant BCR-BL1 polymutants.
Discussion
Our work demonstrates that most patients failing imatinib therapy carry BCR-ABL1 mutations and that polymutants (22, 23) are readily detected not only after sequential therapy but immediately after failure of imatinib therapy. We also mapped mutations to a hotspot region (residues 295–312) within the BCR-ABL1 kinase domain, which were associated with high resistance and poor clinical outcomes. Because our analysis only included patients with CML-CP, where TKI resistance is largely dependent on the BCR-ABL1 genotype, our results indicate a progressive malignant behavior of BCR-ABL1–bearing clones, even in incipient stages of the disease. This heightened genetic instability even in CML-CP has been proposed to be mediated by BCR-ABL1 kinase-induced reactive oxygen species, enhanced spontaneous DNA damage, and compromised fidelity of DNA repair mechanisms in CML cells (33, 34). TKI-induced selection pressure perpetuates this “mutator phenotype,” inducing attrition of the pool of CML clones carrying unmutated BCR-ABL1 proteins, and ultimately hampering TKI activity.
Importantly, our work provides a detailed analysis of the structural and thermodynamic impact that clinically relevant polymutants have on TKI binding. Similar to our prior work using this methodology to analyze clinical TKI resistance in gastrointestinal stromal tumors (35), the affinity data obtained in silico for BCR-ABL1 single point mutants in complex with TKIs was extremely close to those previously reported experimentally. In addition, molecular dynamics simulation-based study of native and fourteen single point mutant BCR-ABL kinases (M244V, G250E, Q252H, Y253F, Y253H, E255K, E255V, T315A, T315I, F317L, F317V, M351T, F359V, and H396P) in complex with ponatinib has been recently published (32). Though using a different computational approach and limited to single mutant BCR-ABL1 proteins, it is worth noting that the reported results are in agreement with those herein presented, thus further supporting our in silico approach.
Although the computational methods used in this work provide a quantitative approach to the structural and energetic consequences of BCR-ABL1 mutations on TKI binding by evaluating the contributions of electrostatic and van der Waals interactions and changes in solvation and entropy, such an approach is not exempt of potential shortcomings. Computer-based simulation results rely on and may be affected by the availability of validated and reliable starting structures, the choice of a force field that incompletely or incorrectly describes the protein, the ligand and/or their interactions, a limited sampling of time and phase space, and the underlying challenge in accurately accounting for solvation and entropic effects. In our experience, the computational approached we used (35) may occasionally perform poorly, suggesting that its success may be system- and/or protocol-dependent. Therefore, caution must be exercised when formulating/performing the computational ansatz (36–38) (see SI Appendix, Fig. S9 and relevant discussion) and, most importantly, as presented here, in silico results must be validated experimentally.
We showed that the accumulation of more than one BCR-ABL1 mutation within the same allele promotes a higher than expected level of TKI resistance compared with that of each single mutation alone. In fact, some of the polymutants exhibited levels of resistance to imatinib and dasatinib remarkably higher than those of the highly resistant T315I gatekeeper mutation, suggesting that the generation of polymutant BCR-ABL1 alleles represents a powerful mechanism of escape for resistant CML clones exposed to TKI-induced selection pressure. However, the complexity of the polymutants is limited, which suggests that, in addition to increasing TKI resistance, the successive accumulation of single mutations within the same clone may also compromise kinase activity and cellular fitness, the latter being a function of the number and type of single mutants involved in any given polymutant. Poorly fit clones are detected only transiently, and their clinically relevance is likely to be negligible.
Preventing the generation of resistant polymutants and the emergence of clinical resistance may require the use of frontline therapy with TKIs, which, like ponatinib, are active against all clinically relevant single point BCR-ABL1 mutations, including the gatekeeper mutation T315I (24–26). In keeping with experimental data, molecular dynamics analyses predicted ponatinib to maintain a notable affinity for most clinically relevant BCR-ABL1 single mutants but also for some of the polymutants detected in our patient cohort. Ponatinib accommodated the structural changes induced by some of these complex mutants, suggesting polymutant kinase inhibition at doses frequently within the clinically achievable range. These findings were validated in cell-based biochemical assays, thus validating the clinical applicability of in silico modeling as a means to predict the TKI approach with the highest probability of clinical success. Importantly, some polymutants required ponatinib concentrations unlikely to be reached in humans without unacceptable toxicity, thus anticipating potential mechanisms of escape to ponatinib through selection and expansion of clones carrying highly TKI-resistant complex BCR-ABL1 mutant proteins.
Materials and Methods
Patient Cohort.
Seventy patients with CML-CP resistant to imatinib therapy signed informed consent and received dasatinib salvage therapy in phase I or II clinical trials, approved by the MD Anderson Cancer Center Institutional Review Board. Initial dasatinib dose ranged from 15 mg/d to 180 mg/d, administered as a single or a divided dose.
Analysis of BCR-ABL1 Mutations.
Mutations in BCR-ABL1 were analyzed in CML patient specimens as previously described (3, 20). In brief, total RNA was extracted (RNeasy Protect Mini Kit; Qiagen) and used to prime a one-step RT-PCR (Invitrogen) using the following primers: CM10 (5′-GAAGCTTCTCCCTGACATCCGT-3′, BCR: 2609–2630) and 3′ ABL1 KD (5′GCCAGGCTCTCGGGTGCAGTCC-3′, ABL1: 1292–1271), resulting in a 1.3-kb fragment. The 1.3-kb gel-purified fragment was used as a template to prime a second PCR [5′ ABL1 KD (5′GCGCAACAAGCCCACTGTCTATGG-3′) and 3′ ABL1 KD], resulting in amplification of the complete ABL1 kinase domain. In all patient samples available, the product remaining from the primary PCR was used to prime a seminested second PCR followed by direct sequencing of the resultant product (10). Second PCR reactions were performed using PCR SuperMix High Fidelity (Invitrogen; catalog no. 10790-020). The resultant 0.6-kb fragment was isolated by QIAquick Gel Extraction Kit (catalog no. 28704) and subcloned into the pGEM-T vector (Promega; catalog no. A3600). At least 10 clones containing the ABL1 kinase domain were directly sequenced using an ABI377 automated sequencer. All mutations are reported in the c-ABL1 gene 1a orientation.
Response Criteria.
Response criteria were as previously described (39). Cytogenetic analysis was performed by standard G-banding techniques, and at least 20 metaphases were analyzed. BCR-ABL1 transcripts were detected by RT-PCR analysis on peripheral blood and bone marrow aspirate (40).
In Silico Modeling of Mutant BCR-ABL1.
The free energies of binding ΔGbind for the unmutated and all mutant BCR-ABL1 kinase/inhibitor complexes were obtained using an extensively validated procedure based on the molecular mechanics/Poisson–Boltzmann surface area methodology (see SI Appendix for full computational details) (28, 35, 41–43). According to this computational recipe, ΔGbind values are calculated for equilibrated structures extracted from the corresponding molecular dynamics (MD) trajectories. The average value of ΔGbind is estimated as the sum of different energetic contributions, corresponding to the average MD energies (ΔEMM = ΔEele + ΔEvdW), the average solvation free energy (ΔGsolv = ΔGsolv,pol + ΔGsolv,nonpol), and the entropic contribution (−TΔS). The MD energies (ΔEMM) were evaluated from a single 50-ns MD trajectory of each kinase/inhibitor complex. The solvation energies were obtained solving the Poisson–Boltzmann equation for the polar part (ΔGsolv,pol) plus a nonpolar contribution (ΔGsolv,nonpol), proportional to the solvent-accessible surface area and including the entropy cost of creating a solute-size cavity in the solvent. Finally, the variation of entropy upon binding was evaluated by using the quasi-harmonic approach (44), according to which the atomic fluctuation matrix is calculated as the mass-weighted covariance matrix obtained from the snapshots of the MD trajectories of each complex. Calculations were carried out using the AMBER 9/11 platform (45, 46) with Amber ff03 (47) and GAFF force field (48) on the SP6 supercomputer (CINECA, Bologna, Italy).
Construction of Single and Double BCR-ABL1 Point Mutants.
The construction of eGFP C-terminally tagged wild-type and T315I mutant Bcr/Abl was previously described (49). The pMX/eGFP-T315I-BcrAbl template was used to generate double mutants by site-directed mutagenesis (Stratagene). Following mutagenesis, the 4-kb XhoI/SgrAI sequence carrying the double mutant was sequenced and subcloned into the pMX/eGFP-BcrAbl template (at XhoI/SgrAI sites) to ensure that no other mutations were introduced during mutagenesis.
Transformation of Ba/F3 Cells with pMX/eGFP-BCR-ABL1.
Parental Ba/F3 was maintained in RPMI medium with 10% FBS and 1 ng/mL mouse IL-3. Ba/F3 cells was verified to be IL-3 dependent by depriving cells of IL-3 before transfection. The parental Ba/F3 cells at 2.5 × 106 were electroporated with 2.5 μg plasmid using the Amaxa Nucleofector system. After electroporation, Ba/F3 cells were rested overnight, and puromycin was then added to the medium at a final concentration of 1 μg/mL. Ba/F3 cells were cultured in the RPMI medium/IL-3/puromycin media for more than a week. To ensure equivalent expression of BCR-ABL1 between wild-type and mutant cells, Ba/F3 cells were sorted by flow cytometry for eGFP positivity. Sorted eGFP-positive Ba/F3 cells were further selected in medium without IL-3. Cell growth rates were equivalent in all BCR-ABL1–transformed cells. Cells maintained in the absence of IL-3 for more than 1 wk were used to examine inhibitor activity.
ABL1 Kinase Inhibitor Studies.
Dasatinib was provided by Bristol–Myers Squibb. Imatinib was synthesized and purified by William Bornmann (MD Anderson Cancer Center). Ponatinib was obtained from SelleckChem. All reagents were made up as 10-mM stocks in DMSO and diluted into culture media just before use. Cells were treated with the indicated concentration of inhibitor for 4 h before examining expression of phosphoproteins in cell lysates as previously described (49, 50). Phosphospecific antibodies against Stat5 and CrkL were obtained from Cell Signaling. Actin antibody was purchased from Santa Cruz Biologicals. Cells were incubated for 72 h with inhibitors to determine their effect on growth and survival of BaF3 cells as previously described (51, 52).
Statistical Considerations.
Statistical significance of the differences was analyzed by using unpaired Student t test for comparisons of two groups or by one-way ANOVA for comparisons of more than two groups. Survival curves were estimated by the Kaplan–Meier method and compared by the log rank (Mantel–Cox) test. All P values were two-sided (statistical significance was set at less than 0.05).
Supplementary Material
Acknowledgments
Support for this work was provided by National Cancer Institute Grant K08 CA151651 (to D.L.G.), the Division of Cancer Medicine Advanced Scholars Program at MD Anderson Cancer Center, Start-Up Funds from the MD Anderson Cancer Center, grant support from Dr. Frederick Hagemeister at MD Anderson Cancer Center (to A.Q.-C.), a Michigan Institute for Clinical and Health Research grant (to N.D.), and the Leukemia Lymphoma Society (N.D.). Part of this work was carried out in the framework of the High Performance Computing (HPC)-Europa2 Projects MONALISA, NANOHEALTH, and INSIDER (CINECA Supercomputing Center, Bologna, Italy), funded by European Commission Directorate General Research in the Seventh Framework Program Grant 228398 (to S.P. and E.L.).
Footnotes
Conflict of interest statement: M.T. received grant support from Ariad, Novartis, and Bristol–Myers Squibb.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1321173111/-/DCSupplemental.
References
- 1.Druker BJ, et al. Effects of a selective inhibitor of the Abl tyrosine kinase on the growth of Bcr-Abl positive cells. Nat Med. 1996;2(5):561–566. doi: 10.1038/nm0596-561. [DOI] [PubMed] [Google Scholar]
- 2. Deininger M, et al. (2009) International randomized study of interferon vs STI571 (IRIS) 8-year follow up: Sustained survival and low risk for progression or events in patients with newly diagnosed chronic myeloid leukemia in chronic phase (CML-CP) treated with imatinib. Blood 114(22):1126 (abstr)
- 3.Hochhaus A, La Rosée P. Imatinib therapy in chronic myelogenous leukemia: Strategies to avoid and overcome resistance. Leukemia. 2004;18(8):1321–1331. doi: 10.1038/sj.leu.2403426. [DOI] [PubMed] [Google Scholar]
- 4.Shah NP, et al. Multiple BCR-ABL kinase domain mutations confer polyclonal resistance to the tyrosine kinase inhibitor imatinib (STI571) in chronic phase and blast crisis chronic myeloid leukemia. Cancer Cell. 2002;2(2):117–125. doi: 10.1016/s1535-6108(02)00096-x. [DOI] [PubMed] [Google Scholar]
- 5.Löwenberg B. Minimal residual disease in chronic myeloid leukemia. N Engl J Med. 2003;349(15):1399–1401. doi: 10.1056/NEJMp038130. [DOI] [PubMed] [Google Scholar]
- 6.Corbin AS, La Rosée P, Stoffregen EP, Druker BJ, Deininger MW. Several Bcr-Abl kinase domain mutants associated with imatinib mesylate resistance remain sensitive to imatinib. Blood. 2003;101(11):4611–4614. doi: 10.1182/blood-2002-12-3659. [DOI] [PubMed] [Google Scholar]
- 7.Gambacorti-Passerini CB, et al. Molecular mechanisms of resistance to imatinib in Philadelphia-chromosome-positive leukaemias. Lancet Oncol. 2003;4(2):75–85. doi: 10.1016/s1470-2045(03)00979-3. [DOI] [PubMed] [Google Scholar]
- 8.Apperley JF. Part I: Mechanisms of resistance to imatinib in chronic myeloid leukaemia. Lancet Oncol. 2007;8(11):1018–1029. doi: 10.1016/S1470-2045(07)70342-X. [DOI] [PubMed] [Google Scholar]
- 9.Soverini S, et al. BCR-ABL kinase domain mutation analysis in chronic myeloid leukemia patients treated with tyrosine kinase inhibitors: Recommendations from an expert panel on behalf of European LeukemiaNet. Blood. 2011;118(5):1208–1215. doi: 10.1182/blood-2010-12-326405. [DOI] [PubMed] [Google Scholar]
- 10.Schindler T, et al. Structural mechanism for STI-571 inhibition of abelson tyrosine kinase. Science. 2000;289(5486):1938–1942. doi: 10.1126/science.289.5486.1938. [DOI] [PubMed] [Google Scholar]
- 11.Nagar B, et al. Crystal structures of the kinase domain of c-Abl in complex with the small molecule inhibitors PD173955 and imatinib (STI-571) Cancer Res. 2002;62(15):4236–4243. [PubMed] [Google Scholar]
- 12.Hantschel O, et al. A myristoyl/phosphotyrosine switch regulates c-Abl. Cell. 2003;112(6):845–857. doi: 10.1016/s0092-8674(03)00191-0. [DOI] [PubMed] [Google Scholar]
- 13.Azam M, Latek RR, Daley GQ. Mechanisms of autoinhibition and STI-571/imatinib resistance revealed by mutagenesis of BCR-ABL. Cell. 2003;112(6):831–843. doi: 10.1016/s0092-8674(03)00190-9. [DOI] [PubMed] [Google Scholar]
- 14. Quintas-Cardama AG, Gibbons DL, Kantarjian H, et al. (2007) Mutational analysis of chronic myeloid leukemia (CML) clones reveals heightened BCR-ABL1 genetic instability and wild-type BCR-ABL1 exhaustion in patients failing sequential imatinib and dasatinib therapy. Blood 110:1938 (abstr)
- 15.Hughes T, et al. Monitoring CML patients responding to treatment with tyrosine kinase inhibitors: Review and recommendations for harmonizing current methodology for detecting BCR-ABL transcripts and kinase domain mutations and for expressing results. Blood. 2006;108(1):28–37. doi: 10.1182/blood-2006-01-0092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Hochhaus A, et al. Molecular and chromosomal mechanisms of resistance to imatinib (STI571) therapy. Leukemia. 2002;16(11):2190–2196. doi: 10.1038/sj.leu.2402741. [DOI] [PubMed] [Google Scholar]
- 17.Talpaz M, et al. Dasatinib in imatinib-resistant Philadelphia chromosome-positive leukemias. N Engl J Med. 2006;354(24):2531–2541. doi: 10.1056/NEJMoa055229. [DOI] [PubMed] [Google Scholar]
- 18.Kantarjian H, et al. Nilotinib in imatinib-resistant CML and Philadelphia chromosome-positive ALL. N Engl J Med. 2006;354(24):2542–2551. doi: 10.1056/NEJMoa055104. [DOI] [PubMed] [Google Scholar]
- 19.Shah NP, et al. Overriding imatinib resistance with a novel ABL kinase inhibitor. Science. 2004;305(5682):399–401. doi: 10.1126/science.1099480. [DOI] [PubMed] [Google Scholar]
- 20.O’Hare T, et al. In vitro activity of Bcr-Abl inhibitors AMN107 and BMS-354825 against clinically relevant imatinib-resistant Abl kinase domain mutants. Cancer Res. 2005;65(11):4500–4505. doi: 10.1158/0008-5472.CAN-05-0259. [DOI] [PubMed] [Google Scholar]
- 21.Shah NP, et al. Sequential ABL kinase inhibitor therapy selects for compound drug-resistant BCR-ABL mutations with altered oncogenic potency. J Clin Invest. 2007;117(9):2562–2569. doi: 10.1172/JCI30890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Khorashad JS, et al. BCR-ABL1 compound mutations in tyrosine kinase inhibitor-resistant CML: Frequency and clonal relationships. Blood. 2013;121(3):489–498. doi: 10.1182/blood-2012-05-431379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Soverini S, et al. Unraveling the complexity of tyrosine kinase inhibitor-resistant populations by ultra-deep sequencing of the BCR-ABL kinase domain. Blood. 2013;122(9):1634–1648. doi: 10.1182/blood-2013-03-487728. [DOI] [PubMed] [Google Scholar]
- 24.O’Hare T, et al. AP24534, a pan-BCR-ABL inhibitor for chronic myeloid leukemia, potently inhibits the T315I mutant and overcomes mutation-based resistance. Cancer Cell. 2009;16(5):401–412. doi: 10.1016/j.ccr.2009.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Gibbons DL, Pricl S, Kantarjian H, Cortes J, Quintás-Cardama A. The rise and fall of gatekeeper mutations? The BCR-ABL1 T315I paradigm. Cancer. 2012;118(2):293–299. doi: 10.1002/cncr.26225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Cortes JE, et al. Ponatinib in refractory Philadelphia chromosome-positive leukemias. N Engl J Med. 2012;367(22):2075–2088. doi: 10.1056/NEJMoa1205127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tam CS, et al. Failure to achieve a major cytogenetic response by 12 months defines inadequate response in patients receiving nilotinib or dasatinib as second or subsequent line therapy for chronic myeloid leukemia. Blood. 2008;112(3):516–518. doi: 10.1182/blood-2008-02-141580. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Laurini E, et al. Through the open door: Preferential binding of dasatinib to the active form of BCR-ABL unveiled by in silico experiments. Mol Oncol. 2013;7(5):968–975. doi: 10.1016/j.molonc.2013.06.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Fabbro D, et al. Inhibitors of the Abl kinase directed at either the ATP- or myristate-binding site. Biochim Biophys Acta. 2010;1804(3):454–462. doi: 10.1016/j.bbapap.2009.12.009. [DOI] [PubMed] [Google Scholar]
- 30.O’Hare T, et al. SGX393 inhibits the CML mutant Bcr-AblT315I and preempts in vitro resistance when combined with nilotinib or dasatinib. Proc Natl Acad Sci USA. 2008;105(14):5507–5512. doi: 10.1073/pnas.0800587105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Weisberg E, et al. Characterization of AMN107, a selective inhibitor of native and mutant Bcr-Abl. Cancer Cell. 2005;7(2):129–141. doi: 10.1016/j.ccr.2005.01.007. [DOI] [PubMed] [Google Scholar]
- 32.Tanneeru K, Guruprasad L. Ponatinib is a pan-BCR-ABL kinase inhibitor: MD simulations and SIE Study. PLoS ONE. 2013;8(11):e78556. doi: 10.1371/journal.pone.0078556. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Skorski T. Genomic instability: The cause and effect of BCR/ABL tyrosine kinase. Curr Hematol Malig Rep. 2007;2(2):69–74. doi: 10.1007/s11899-007-0010-6. [DOI] [PubMed] [Google Scholar]
- 34.Penserga ET, Skorski T. Fusion tyrosine kinases: A result and cause of genomic instability. Oncogene. 2007;26(1):11–20. doi: 10.1038/sj.onc.1209756. [DOI] [PubMed] [Google Scholar]
- 35.Pierotti MA, Tamborini E, Negri T, Pricl S, Pilotti S. Targeted therapy in GIST: In silico modeling for prediction of resistance. Nat Rev Clin Oncol. 8(3):161–170. doi: 10.1038/nrclinonc.2011.3. [DOI] [PubMed] [Google Scholar]
- 36.Deng Y, Roux B. Computations of standard binding free energies with molecular dynamics simulations. J Phys Chem B. 2009;113(8):2234–2246. doi: 10.1021/jp807701h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hou T, Wang J, Li Y, Wang W. Assessing the performance of the MM/PBSA and MM/GBSA methods. 1. The accuracy of binding free energy calculations based on molecular dynamics simulations. J Chem Inf Model. 2011;51(1):69–82. doi: 10.1021/ci100275a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Wang W, Donini O, Reyes CM, Kollman PA. Biomolecular simulations: Recent developments in force fields, simulations of enzyme catalysis, protein-ligand, protein–protein, and protein–nucleic acid noncovalent interactions. Annu Rev Biophys Biomol Struct. 2001;30:211–243. doi: 10.1146/annurev.biophys.30.1.211. [DOI] [PubMed] [Google Scholar]
- 39.Kantarjian HM, et al. Prolonged survival in chronic myelogenous leukemia after cytogenetic response to interferon-α therapy. Ann Intern Med. 1995;122(4):254–261. doi: 10.7326/0003-4819-122-4-199502150-00003. [DOI] [PubMed] [Google Scholar]
- 40.Cortes J, et al. Molecular responses in patients with chronic myelogenous leukemia in chronic phase treated with imatinib mesylate. Clin Cancer Res. 2005;11(9):3425–3432. doi: 10.1158/1078-0432.CCR-04-2139. [DOI] [PubMed] [Google Scholar]
- 41.Cheatham TE, 3rd, Kollman PA. Molecular dynamics simulation of nucleic acids. Annu Rev Phys Chem. 2000;51:435–471. doi: 10.1146/annurev.physchem.51.1.435. [DOI] [PubMed] [Google Scholar]
- 42.Lee MR, Duan Y, Kollman PA. Use of MM-PB/SA in estimating the free energies of proteins: Application to native, intermediates, and unfolded villin headpiece. Proteins. 2000;39(4):309–316. [PubMed] [Google Scholar]
- 43.Wang W, Kollman PA. Computational study of protein specificity: the molecular basis of HIV-1 protease drug resistance. Proc Natl Acad Sci USA. 2001;98(26):14937–14942. doi: 10.1073/pnas.251265598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Andricioaei I, Karplus M. On the calculation of entropy from covariance matrices of the atomic fluctuations. J Chem Phys. 2001;115(14):6289–6292. [Google Scholar]
- 45. Case D, et al. (2006) AMBER 9 (University of California, San Francisco)
- 46. Case D, et al. (2010) AMBER 11 (University of California, San Francisco)
- 47.Duan Y, et al. A point-charge force field for molecular mechanics simulations of proteins based on condensed-phase quantum mechanical calculations. J Comput Chem. 2003;24(16):1999–2012. doi: 10.1002/jcc.10349. [DOI] [PubMed] [Google Scholar]
- 48.Wang J, Wolf RM, Caldwell JW, Kollman PA, Case DA. Development and testing of a general amber force field. J Comput Chem. 2004;25(9):1157–1174. doi: 10.1002/jcc.20035. [DOI] [PubMed] [Google Scholar]
- 49.Donato NJ, et al. Imatinib mesylate resistance through BCR-ABL independence in chronic myelogenous leukemia. Cancer Res. 2004;64(2):672–677. doi: 10.1158/0008-5472.can-03-1484. [DOI] [PubMed] [Google Scholar]
- 50.Donato NJ, et al. BCR-ABL independence and LYN kinase overexpression in chronic myelogenous leukemia cells selected for resistance to STI571. Blood. 2003;101(2):690–698. doi: 10.1182/blood.V101.2.690. [DOI] [PubMed] [Google Scholar]
- 51.Wu J, et al. ON012380, a putative BCR-ABL kinase inhibitor with a unique mechanism of action in imatinib-resistant cells. Leukemia. 2010;24(4):869–872. doi: 10.1038/leu.2009.300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Wu J, et al. Association between imatinib-resistant BCR-ABL mutation-negative leukemia and persistent activation of LYN kinase. J Natl Cancer Inst. 2008;100(13):926–939. doi: 10.1093/jnci/djn188. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.