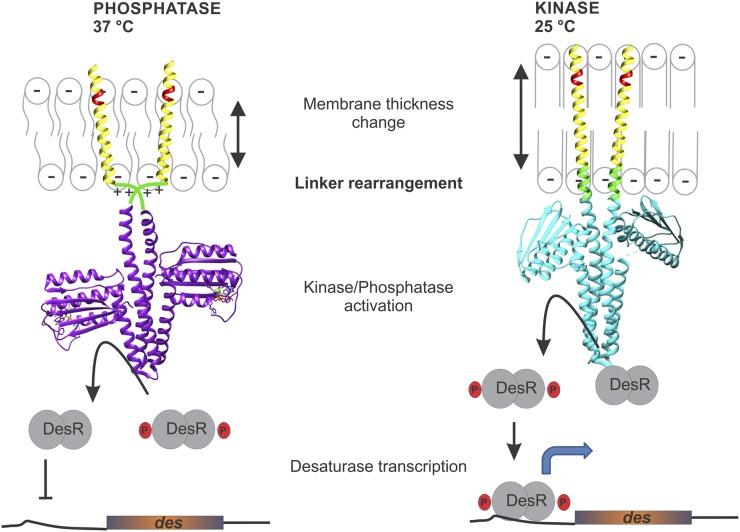
Significance
Environmental temperature variations affect most biological processes and reactions, and cells must adapt accordingly. One such example is the fine-tuned regulation of membrane fluidity and its impact on a large array of cell physiological processes. How do cells “sense” membrane fluidity? Here, we discovered that the bacterial histidine-kinase, DesK, transmits temperature information through lipid-mediated conformational changes of a region of the protein that links its membrane sensor domain with its catalytic domain and activates the expression of a desaturase, which allows membrane fluidity to be recovered at this lower temperature. Because many thermosensors and other types of sensors contain similar linker domains, the mechanism we describe here could prove a general theme.
Keywords: signal transduction, membrane–protein interaction, two-component system, sensor biophysics
Abstract
The thermosensor DesK is a multipass transmembrane histidine-kinase that allows the bacterium Bacillus subtilis to adjust the levels of unsaturated fatty acids required to optimize membrane lipid fluidity. The cytoplasmic catalytic domain of DesK behaves like a kinase at low temperature and like a phosphatase at high temperature. Temperature sensing involves a built-in instability caused by a group of hydrophilic residues located near the N terminus of the first transmembrane (TM) segment. These residues are buried in the lipid phase at low temperature and partially “buoy” to the aqueous phase at higher temperature with the thinning of the membrane, promoting the required conformational change. Nevertheless, the core question remains poorly understood: How is the information sensed by the transmembrane region converted into a rearrangement in the cytoplasmic catalytic domain to control DesK activity? Here, we identify a “linker region” (KSRKERERLEEK) that connects the TM sensor domain with the cytoplasmic catalytic domain involved in signal transmission. The linker adopts two conformational states in response to temperature-dependent membrane thickness changes: (i) random coiled and bound to the phospholipid head groups at the water-membrane interface, promoting the phosphatase state or (ii) unbound and forming a continuous helix spanning a region from the membrane to the cytoplasm, promoting the kinase state. Our results uphold the view that the linker is endowed with a helix/random coil conformational duality that enables it to behave like a transmission switch, with helix disruption decreasing the kinase/phosphatase activity ratio, as required to modulate the DesK output response.
DesK is the histidine kinase of the DesKR two-component system. It is a membrane protein with five transmembrane segments and a cytoplasmic catalytic domain containing the dimerization and histidine phosphotransferase (DHp) domain and the ATP-binding (ABD) domain. DesK is a bifunctional enzyme such that when stimulated acts as a kinase and when not stimulated acts as a phosphatase for its cognate response regulator, DesR (1–3). Conditions that decrease the order of membrane lipids (cold-shock, growth in media devoid of branched-chain amino acids) activate the kinase conformation and result in phosphorylation of the response regulator DesR (4, 5). Once phosphorylated, DesR activates transcription of the target gene Δ5-des. The Δ5-desaturase enzyme inserts into the membrane and catalyzes introduction of double bonds into the lipids to restore membrane fluidity (6). On the contrary, conditions that increase membrane fluidity (growth at warm temperatures or in the presence of branched-chain amino acids) favor DesK phosphatase activity, removing the phosphoryl group from the response regulator to terminate Δ5-des transcription (4, 5, 7).
Remarkably, the multimembrane-spanning domain of DesK can be simplified into a chimerical single membrane-spanning segment, which results from linking the N terminus region of the first transmembrane segment with the C terminus of the fifth transmembrane segment. This chimerical segment connected to the cytoplasmic catalytic domain is still able to respond to changes in lipid fluidity like full-length DesK. It has been called minimal sensor-DesK (MS-DesK) (Fig. 1) (3) and can be used as a tool to decipher the mechanism of DesK thermosensing.
Fig. 1.
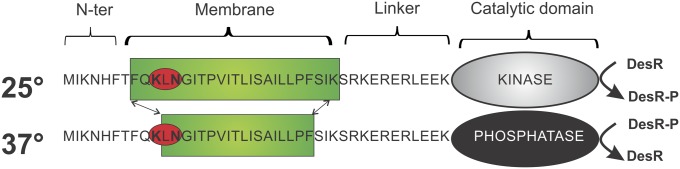
Schematic representation of MS-DesK at 25 °C and 37 °C. The sequence of the transmembrane region and the connecting linker is indicated with a one-letter code. The SB motif is highlighted with a red circle, the catalytic cytoplasmic domain is represented with an oval, and the lipid bilayer is represented with a green rectangle. At 25 °C, the membrane is thicker, and the hydrophilic SB motif is buried in the hydrophobic core of the membrane. The catalytic domain has kinase activity (gray oval) and phosphorylates DesR. At 37 °C the membrane is thinner, and the SB floats at the water–lipid interface. The catalytic domain has phosphatase activity (black oval) and dephosphorylates DesR-P.
An important contribution that has highlighted DesK functioning is the finding that a region located at the N-terminal transmembrane segment (Lys10-Asn12), which has been called “the sunken buoy” (SB), is involved in the detection of changes in membrane thickness that occur as a consequence of temperature variations (3). It has been proposed that, at lower temperatures, the membrane is thicker due to a more ordered packing of lipids (8, 9) and forces the SB motif to be buried in the hydrophobic region of the lipid bilayer, favoring the kinase activity of DesK. At higher temperatures, the lipids are in a more disordered state, and the membrane becomes thinner. The motif may now “buoy” to the membrane interface, stabilizing the protein in a “kinase-off” state (3) (Fig.1). Another key contribution to understand DesK functioning is a crystallographic study of DesKC (DesK catalytic core), which has revealed that DesK dimers are stabilized through hydrophobic interactions between two helical hairpins that form a dimerization motif, DHp (10). The structural comparison of DesK in the phosphatase or kinase functional states suggested that interhelical rearrangements that change the twisting in the DHp domain could favor one or the other signaling state (10).
In this scenario, the emerging question is: How is the information sensed by the transmembrane region transmitted to the cytoplasm and converted into a rearrangement in the catalytic domain that allows switching between kinase and phosphatase states? In this paper, we assessed the role of a linker region (KSRKERERLEEK) (Fig. 1) that connects the transmembrane sensing domain with the cytoplasmic catalytic domain. This region, although present in different constructs, was not solved in the crystal structure of the phosphatase state whereas it was nearly completely solved in the kinase state (10).
Here we used genetic, spectroscopic, and biochemical techniques to decipher the role of the linker in DesK signal transduction. We propose and validate the view that the linker is endowed with a helix/random coil conformational duality that enables its role as a transmission switch. Thus, the disruption of the helical structure between the transmembrane segment and the intracellular domain is mechanistically exploited to transmit temperature-dependent conformational changes from the transmembrane to the intracellular region.
Results
Charged Residues of the Linker Are Critical for DesK Signaling.
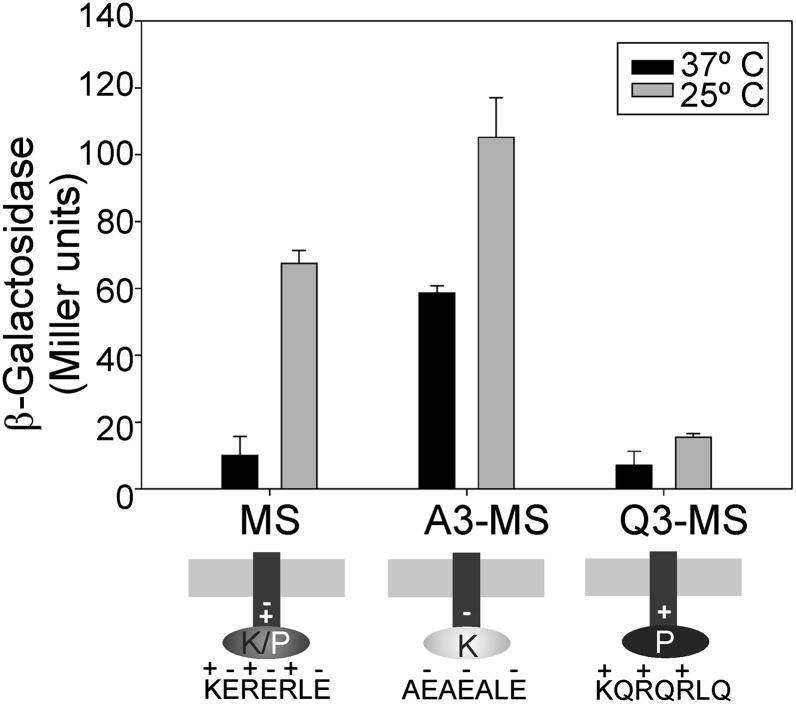
The linker region connects the transmembrane region with the cytoplasmic domain of DesK and is located in close proximity to the inner leaflet of the lipid bilayer. We took advantage of the minimized version of DesK, MS-DesK, to introduce point mutations in the linker (KERERLEEK) and replaced three positively or three negatively charged residues with neutral ones, shown in bold: AEAEALEEK (A3) and KQRQRLQEK (Q3) (Fig. 2, Lower). To evaluate the capacity of these MS-DesK variants to sense membrane-thickness variations, they were expressed in a desK− strain, and the activity of the reporter gene β-galactosidase fused to the desaturase promoter (Pdes-lacZ) was measured in conditions of high or low lipid membrane fluidity (cells grown at 37 °C or grown at 37 °C and then cold-shocked to 25 °C, respectively). As shown in Fig. 2, MS-DesK stimulates transcription of the reporter when cells are shifted from 37 °C to 25 °C. Surprisingly, the negatively charged linker variant A3 shows a constitutive kinase activity regardless of membrane fluidity variations. On the contrary, the positively charged linker variant Q3 locks the sensor in a state unable to stimulate des transcription at 25 °C or 37 °C (Fig. 2) although this variant displays high phosphatase activity (Fig. S1).
Fig. 2.
Linker charges are critical to determine the signaling state of DesK. Cells expressing MS-DesK linker variants were grown at 37 °C or grown at 37 °C and transferred to 25 °C at an OD525 = 0.3. β-galactosidase activity was assayed every hour in independent triplicates. The results shown are expressed as the average of three independent experiments and correspond to 2 h after the cold shock.
The Linker Binds to Lipid Membranes.
To analyze whether DesK linker variants interact with the phospholipid membrane and to characterize parameters that are required for such interaction, we designed the three corresponding peptides: WT (KSRKERERLEEK), A3 (KSRAEAEALEEK), and Q3 (KSRKQRQRLQEK) and prepared liposomes made of Escherichia coli lipids (mixture of anionic and zwitterionic phospholipids), which have a similar composition to that of Bacillus subtilis and are widely used for in vitro assays (3, 10) (Materials and Methods). Addition of the Q3 peptide to liposomes significantly increased turbidity, as measured by right-angle light scattering. Slow decrease of the scattering over time was due to visible flocculation observed after a few seconds. In contrast, no change in turbidity was observed after addition of A3 peptide, and, finally, a very moderate increase was observed for the WT peptide (Fig. S2A). A turbidity increase could be explained by liposome charge shielding due to the binding of positively charged peptides (WT and specially Q3) to the liposome surface, reducing electrostatic repulsion by liposomes and favoring flocculation.
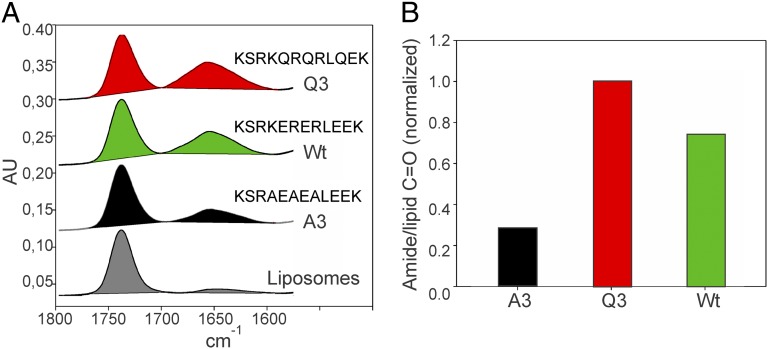
At increasing salt concentrations, the WT and Q3 peptides lost their capacity to increase liposome turbidity, suggesting an essentially electrostatic interaction between the WT or Q3 peptides and liposomes (Fig. S2B). Because turbidity increase does not necessarily mean that the peptides quantitatively interact with lipids, liposome–peptide mixtures were ultracentrifuged and analyzed for the simultaneous presence of lipids and peptides by attenuated total reflection (ATR)-FTIR spectroscopy in terms of lipid and peptide content. Lipids were detected at 1,735 cm−1 assigned to the C = O signal of lipid ester bonds and the peptides around 1,650 cm−1 assigned to the C = O of the amide bond (11). The intensity of the band at 1,650 cm−1 recorded for the three peptides follows the propensity of the linker variants to interact with charged lipid polar head groups (Fig. 3A). This experiment was validated by dialyzing the peptide–liposome complexes previous to ATR-FTIR analysis (Fig. S2C). The basic amino acid content of the linker seems essential for the interaction with lipids because increasing positive charges (Q3 mutant peptide) increases binding, whereas decreasing them (A3 mutant peptide) has the opposite effect. To confirm the ionic interaction, we performed microdialysis peptide-release assays and ultracentrifugation of the peptide–liposome mixture in the presence of salts and analyzed the complexes by ATR-FTIR. These expermiments show that peptides are released from liposomes in the presence of salts (Fig. S2 D and E).
Fig. 3.
Linker variants interact differentially with lipid membranes. (A) ATR-FTIR spectra of peptide–liposome complexes isolated by centrifugation. The 1,800–1,480 cm−1 region is displayed, with the absorption band of the C = 0 group of the lipid acyl chains (1,735 cm−1) and the amide I band of the peptide (1,600–1,700 cm−1); the absorbtion is expressed in arbitrary units (AU). (B) Quantification of bound peptide expressed as the ratio of the integrated amide I band of the peptide between 1,600 cm−1 and 1,700 cm−1 over the integrated C = O band at 1,735 cm−1 of the lipids shown in A. Results are corrected from the small-lipid contribution in the amide I region.
Does the Linker Change Its Conformation During DesK Activation?
ATR-FTIR measurements were performed to determine the secondary structure of the peptides and to detect possible structural changes when incubated in the presence of liposomes. Table 1 shows a tentative secondary structure content prediction according to Goormaghtigh et al. (12). The secondary structure of the three peptides is mainly a mixture of helical and random structures. The A3 mutant peptide is more helical-structured than the other peptides (Table 1).
Table 1.
Secondary-structure prediction for DesK-linker variants
| Peptide | Alpha | Beta | Turns | 3–10 helix | Random | Increase in random |
| Wt | 29.1 | 5.8 | 14.5 | 0 | 32.1 | |
| Wt + liposomes | 29.5 | 1.8 | 19.3 | 2 | 41.8 | 9.7 |
| A3 | 41.3 | 1.5 | 13.5 | 0.1 | 28 | |
| A3 + liposomes | 42.6 | 1.6 | 15.8 | 1.5 | 32.4 | 4.3 |
| Q3 | 17.1 | 5.8 | 20.5 | 2.4 | 45.2 | |
| Q3 + liposomes | 14.3 | 2.3 | 23.1 | 3 | 50.7 | 5.6 |
All numbers are expressed as percentages.
Nevertheless, the presence of liposomes stabilized the random-coil state of the WT even though a slight increase was also observed for the Q3 and A3 peptides, respectively (Table 1).
Analysis of the amide I and II bands in the 1,750–1,500 cm−1 region revealed a sharp maximum around 1,656 cm−1 in the amide I band and at 1,545 cm−1 in the amide II region characterizing an alpha-helical structure. Nevertheless, the hydrogen bonds are highly exchangeable because, when submitted to a D2O-saturated vapor flux, the amide I peak shifted quickly and the amide II band, sensitive to the amide bond proton–deuterium exchange, showed its maximal exchange after 5 min. This observation suggests that the flexibility of the helix makes H-bonds accessible very quickly and might explain the propensity of the linker peptide to adopt a random structure and to interact with the lipids at the membrane interface (Fig. S3).
Membrane–Linker Electrostatic Interactions Play a Role in DesK Activity.
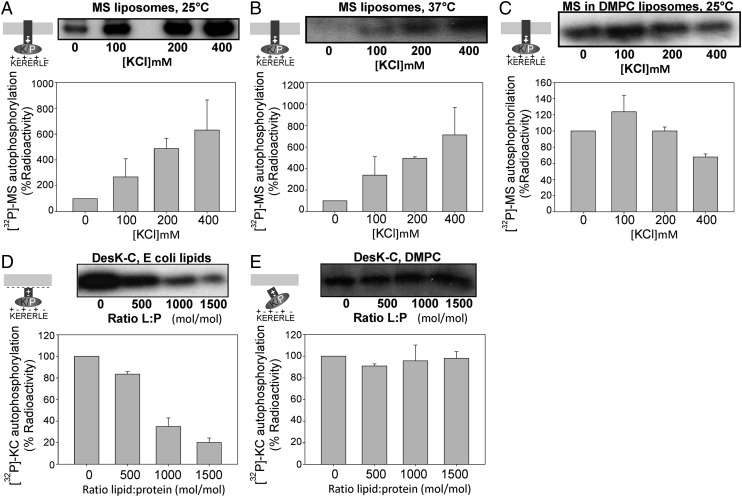
MS-DesK was reconstituted in liposomes of commercial E. coli lipids.The autokinase activity was measured at 25 °C, a temperature that activates the kinase conformation, in the presence of γ32-ATP at different salt concentrations. Consistent with the idea that electrostatic interactions affect the interaction of the linker with the membrane and modify the signaling state of the linker domain, we observed that the autophosphorylation of MS-DesKC was markedly dependent on salt concentration, with its highest activity at 400 mM KCl (Fig. 4A). The same effect was observed when NaCl was added to the reaction mixture (Fig. S4A).
Fig. 4.
Disruption of electrostatic interactions favor DesK autokinase activity. MS-DesK was reconstituted in liposomes of E. coli polar lipids (A and B) or DMPC lipids (C). Autokinase activity, shown in Upper, was assayed at 25 °C (A and C) or at 37 °C (B) in the presence of γ-32 ATP and increasing concentrations of KCl . The soluble truncated DesKC protein was incubated with increasing amounts of liposomes made of E. coli (D) or DMPC (E), and autokinase activity was measured at 25 °C. The total amounts of phosphorylated protein present in each well were determined by densitometry and plotted as the percentage of activity, considering 100% for the activity in the absence of salts (lane 1). The results correspond to the average of at least three independent experiments.
At 37 °C, a temperature that activates the phosphatase conformation, the autokinase activity is negligible in the absence of salts (lane 1, Fig. 4B) but is recovered at increasing salt concentrations (lanes 2–4, Fig. 4B). A feasible interpretation is that salts disrupt the interaction of the linker with the lipid membrane favoring the kinase state. On the contrary, if the protein is not inserted into liposomes (the soluble truncated cytoplasmic catalytic domain DesKC or the detergent-treated MS-DesK), increasing salt concentrations do not raise autokinase activity (Fig. S4B). These findings support the hypothesis that protein–membrane electrostatic interactions are involved in signal transmission.
To test the idea that the electrostatic interaction in mainly due to the negatively charged membrane surface and positively charged residues of the linker, we reconstituted the MS-DesK in 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC), which has a zwitterionic (neutral) polar headgroup, and tested the effect of salts in the kinase activity. As expected, in these neutral lipids, salts do not potentiate kinase activity (Fig. 4C).
To test our hypothesis that the kinase state of DesK is favored when the linker is detached from the membrane, liposomes made of E. coli lipids were incubated with the truncated cytoplasmic domain of DesK (DesKC). DesKC includes the linker region and in vivo shows constitutive kinase activity because it lacks the sensor transmembrane (TM) domain (2). Fig. 4D shows that the in vitro autokinase constitutive activity of truncated DesKC decreases when it is incubated with increasing amounts of liposomes containing anionic lipids. This experiment suggests that, in the presence of a lipid bilayer, a fraction of DesKC interacts via the linker with the negatively charged membrane surface, decreasing the kinase activity. This hypothesis is supported by the experiment showing that the activity of truncated DesKC is not inhibited by increasing the DMPC:protein ratio (Fig. 4E).
Can the Linker Conformation Reroute Transmission of the Signal?
The critical region involved in membrane-thickness sensing is the sunken buoy (SB) motif located at the N terminus of the transmembrane segment of MS-DesK. Mutations that increase the hydrophilicity of this motif increase kinase activity whereas mutations that decrease the SB hydrophilicity abolish this activity (3). At this stage, we wondered which one, the SB motif or the linker, was dictating the final DesK-signaling state.
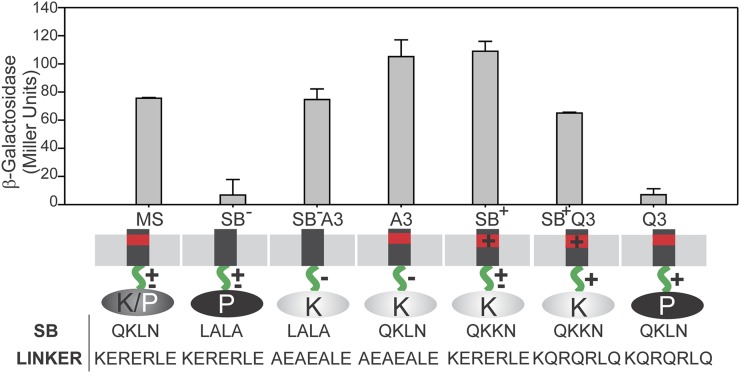
To analyze whether the linker is involved in signal transmission from the membrane to the cytoplasm and to determine whether the linker conformation contributes to define DesK signaling states, we combined point mutations in the transmembrane region that have been shown to lock DesK in the kinase or phosphatase state, respectively, with mutations in the linker. The inactive sensing mutant SB− (Q9L,K10A,N12A), which shows a constitutive phosphatase state regardless of the temperature due to elimination of the sensor sunken buoy motif in the transmembrane region (3), was combined with the linker mutant A3 (constitutive kinase), giving rise to SB−/A3 MS-DesK. On the other hand, the hyperactive mutant SB+ (L11K), which shows constitutive kinase activity due to the presence of an extra charge in the sunken buoy (3), was combined with the linker mutant Q3 (constitutive phosphatase), giving rise to SB+/Q3. Because the linker would be located downstream in the mechanism of signaling from the membrane to the cytoplasmic catalytic core, we expected that mutant SB−/A3 would have a predominant kinase activity and SB+/Q3 a predominant phosphatase activity, regardless of the signaling state dictated by the transmembrane region. These DesK variants were expressed in a desK− strain and tested for activity. We found that linker mutations that favor the kinase conformation can rescue SB− transmembrane mutants locked in the phosphatase conformation (Fig. 5). Similarly, the behavior of the double mutant SB+/ Q3 suggests that the Q3 linker mutations, which favor the phosphatase conformation, can rescue transmembrane mutants locked in the kinase conformation, decreasing kinase activity to wild-type levels (Fig. 5). Together, these results indicate that the linker is critical to transduce the signal and that charged residues play a crucial role in redirecting the signaling sensed by the transmembrane domain to the catalytic cytoplasmic domain.
Fig. 5.
Linker–membrane interaction dictates DesK signaling. B. subtilis desK− cells were complemented with either wild-type MS-DesK or MS-DesK variants with single mutations at the level of the membrane SB (SB−, SB+) or in the linker region (A3, Q3) or MS-DesK variants with combined mutations at both the transmembrane or linker region (SB− A3/ SB+ Q3). β-Galactosidase–specific activities were determined every hour as described for Fig. 2. The results shown are the average of three independent experiments made 2 h after the cold shock.
Discussion
Rerouting Transmembrane Information.
We identified a highly charged cluster of twelve residues (K32–K43) (Fig. 1) connecting the transmembrane domain with the cytoplasmic catalytic domain of MS-DesK. We propose that this motif, highly conserved in most species of the genus, is critical for signal transduction. We characterized two linker mutants, A3 and Q3, conferring, respectively, high and low kinase activity not only to MS-DesK but also to full-length DesK (Fig. S5). Structural infrared analysis suggests that the A3 linker has a higher tendency to form alpha-helix compared with the wild-type or Q3 linker variants. In contrast, the Q3 linker has a higher tendency to adopt a random-coil structure, and its higher conformational flexibility enables interactions between charged side-chain and the polar head groups of the membrane lipids. The role of the interaction with the lipids is supported by light scattering and infrared assays, which clearly demonstrate that the peptide Q3 binds stronger to liposomes than the peptide A3. The analysis of double mutants suggests that the linker is downstream from the stimulus perception in DesK signal transduction because mutations in the linker are dominant over constitutive mutations in the TM region. The transmission of the signal can thus be rerouted to one or the other state by modulating the linker–membrane interaction.
Dual Nature of the Linker–Membrane Interaction.
We propose that the DesK linker has the ability to adopt a dual conformation: helix and random coil. The random-coil conformation is stabilized by electrostatic interaction with the lipid membrane interface whereas the helix is stabilized by the formation of intrahelical salt bridges between paired opposite charges. This conformational duality becomes the essence of a molecular transmission switch that will be at the same time sensitive to subtle conformational changes perceived by the sensing SB motif and able to transmit the signal.
A Working Model for Thermosensor DesK.
We propose that, at high temperatures (thin membranes), the N-terminal SB motif is located at the water–membrane interface. The hydrophobic region of the TM is long enough to allow the linker to adopt the unstructured hydrophilic state (Fig. 6, Left), interacting with the charged lipids on the cytosolic side. Distribution of positively and negatively charged residues does not allow efficient clustering of positively charged residues on a helical structure that would favor a strong interaction with the negatively charged lipid, and a more random distribution is the way to optimize such an interaction.
Fig. 6.
Model for DesK activity upon temperature changes. The increase of the bilayer thickness prevents the interaction of the linker region (green) with the lipid bilayer and contributes to stabilize and favor the helical continuity between the transmembrane segment and the intracellular domain. Reversible interaction of the linker with the lipid bilayer surface would allow DesK to alternate between the kinase and phosphatase states.
The interaction of the linker with the lipid bilayer interface causes helix disruption, which favors the phosphatase conformation. This experimental observation is compatible with the crystallographic data reported for the cytosolic domain of the phosphatase-competent conformation (H188V), in which the linker region was not solved, a possible consequence of its high conformational flexibility (10).
At lower temperature, membrane thickness increases; the hydrophobic region of the TM is not long enough to match the hydrophobic core of the membrane and tends to bury the SB motif and its opposite counterpart, the N-terminal part of the KSRKERERLEEK linker motif, imposing a stress along the whole transmembrane region, which would force the linker to line up with the transmembrane part and adopt a more helical structure, protecting the otherwise exposed linker backbone from the hydrophobic membrane core (Fig. 6, Right). Burial of unpaired charged groups would be thermodynamically unfavorable; therefore, the charged residues are expected to increase their mutual coulombic interactions in the apolar environment of the membrane, making opposite charge pairs and triggering the formation of a continuous helix from the transmembrane region to the cytoplasmic part. The linker is thus removed from the membrane interface, adopting a helical structure. The helix is stabilized by a peculiar distribution of positive and negative residues that contribute to the formation of salt bridges in the helical register: n with n + 3 or n with n + 4 (n = the contour variable or residue number along the peptide chain). These salt bridges become stabilized and enhanced in the lipid phase because of the great decrease in charge shielding in the low dielectric medium (13).
This structural reorganization of the linker might constitute the basis of transmission and amplification of a signal sensed by the TM domain and transmitted to the catalytic domain and would result in modulation of the DHp interhelical twisting required to regulate DesK activity. Consequently, a continuous helix from the membrane to the cytoplasm would favor the kinase conformation. This idea is compatible with structural studies that show that, in the kinase conformation, the helical structure of the DHp extends further toward the membrane-proximal side of the protein (10). It is tempting to speculate that changes in tilting or rotation of the transmembrane segments resulting from a change in the membrane thickness could also play a role in the signaling process (14), stabilizing the linker in the bound or unbound conformation to finally determine the DesK signaling state. Interestingly, a cluster of positive and negative residues located at the water–lipid interface is also present in the tandem-pore K channel (TREK-1), the potassium sensor KdpD, the transient receptor potential proteins (TRPs), the OpuA transporter, and the mechanosensitive MscL channels (Table 2). For some of them, it has been suggested that the charged residues could interact with the negative surface of the membrane, modulating activity (15–18). We propose that highly charged residue clusters involved in signaling proteins and channels (Table 2) are able to adopt alternative states (unbound or bound to the membrane) in response to changes in mechanical properties of the lipid membrane, allowing this region to behave as a molecular switch for downstream protein activity.
Table 2.
Cluster of charged residues at the junction between transmembrane and cytosolic domains in signaling proteins, channels, and transporters
| Protein | Protein organization | Organism | Source |
| DesK | N-TM 5-KSRKERERLEEK—C | Bacillus | This work |
| KdpD | N-TM 4-EYLHRKSME—C | E. coli | (15) |
| OpuA | N-TM 7-EKEEENK—C | L. lactis | (16, 17) |
| TREK-1 | N-TM 4-KKTKEE—C | Mammals | (18) |
| TREK-2 | N-TM 4-KKTKEE—C | Mammals | (19) |
| MscL | N-TM 2-RKKEE—C | E. coli | (20) |
| Osm-9 | N-TM 6-EERSESK—C | C. elegans | (21) |
| TRPA1 | N-TM 6-DRFKKE—C | Mammals | (22) |
| TRPV4 | N-TM 6-RLRRDR—C | Mammals | (23) |
C, cytosolic domain; C. elegans, Caenorhabditis elegans; L. lactis, Lactococcus lactis; N, N-terminal domain; TM, transmembrane domain.
Materials and Methods
Synthetic Peptides.
Synthetic peptides were purchased from GL Biochem Ltd. Peptides corresponding to the linker sequence KSRKERERLEEK (WT) or KSRAEAEALEEK (A3 mutant) and KSRKQRQRLQEK (Q3 mutant) were all N-terminal–acetylated, C-terminal–amidated, and HPLC-purified. Their mass was confirmed by mass spectrometry. For FTIR experiments, samples were diluted in 10 mM HCl and lyophilized (three times) to remove traces of TFA.
Liposome–Peptide Complexes.
Ten milligrams of E. coli polar lipid extract dissolved in chloroform (50 mg/mL) were dried under a nitrogen flux and then maintained overnight under vacuum in a lyophilisator. The dried film was rehydrated in a 10 mM Hepes-Na buffer at pH 7.3 and extensively vortexed to form liposomes. The resulting turbid suspension was transferred to a 1.5-mL polycarbonate tube and sonicated in a water bath sonicator (UCD-200 Bioruptor; Diagenode) to convert multilameller liposomes into unilamellar. Liposomes were mixed with peptides (10 mg/mL dissolved in water) in a 5:1 lipid/peptide ratio (wt/wt) in a final volume of 0.1 mL. After incubation at room temperature for 30 min, the liposomes were centrifuged at 120,000 rpm in a 42.2 Ti Beckman rotor at 20 °C for 1 h. When microdialysis was used, the peptide–liposome complexes were dialyzed by suspending a sample drop on a floating-disk filter with a 20-nm cutoff, which allows permeation of the free peptide, whereas the liposomes were retained on the filter. The sample recovered from the filter was used for ATR-FTIR measurements.
ATR-FTIR Measurement.
Peptide–liposome complexes were spread on a 1-mm-square diamond ATR element (Goldengate Bridge Harrick Scientific) fitted to a Brucker IFS-55 FTIR spectrometer. The lipid samples (1 μL) were partially dried under nitrogen. Serial measurements were recorded at a 2 cm−1 resolution and averaged in the 4,000–900 cm−1 spectral region with a subtracted background. Further processing of the spectra (scaling, integration, and vapor subtraction) was made with in-house-made software (Kinetics) running under Matlab.
In Vitro Autophosphorylation Assay.
These reactions were performed essentially as previously described (3). Briefly, for the autokinase assay, proteoliposomes containing 0.3 mg of MS-DesK protein were incubated at 25 °C or 37 °C in P buffer [50 mM Tris⋅HCl (pH 8), 200 mM NaCl, 1 mM DTT, 20% (vol/vol) glycerol, 50 mM KCl, 1 mM MgCl2, 25 mM ATP, and 0.25 mCi/mL [γ-32P]ATP]; at different time points, aliquots were taken and were run on SDS/PAGE 12%. The radioactivity of phosphorylated proteins in gels was visualized using a Typhoon 9200 PhosphorImager screen (STORM840; GE Healthcare) and quantified using ImageQuant software (version 5.2). The values obtained were expressed as the percentage of phosphorylated protein. Western blots were also performed to confirm the amount of protein loaded in each lane. All results shown are representative of at least three independent experiments.
Supplementary Material
Acknowledgments
This work was supported by grants from Consejo Nacional de Investigaciones Científicas y Técnicas (CONICET), Agencia Nacional de Promoción Científica y Tecnológica (FONCYT), and the following international cooperations: CONICET-Fonds National de la Recherche Scientifique (FNRS) and Ministerio de Ciencia, Tecnología e Innovación Productiva-FNRS. M.V. thanks the Fonds National de la Recherche Scientifique (FNRS, Belgium) for continuous support. M.E.I. is a Fellow of CONICET, and L.E.C., D.d.M., and A.F. are Career Investigators of CONICET.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1317147111/-/DCSupplemental.
References
- 1.Aguilar PS, Hernandez-Arriaga AM, Cybulski LE, Erazo AC, de Mendoza D. Molecular basis of thermosensing: A two-component signal transduction thermometer in Bacillus subtilis. EMBO J. 2001;20(7):1681–1691. doi: 10.1093/emboj/20.7.1681. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Albanesi D, Mansilla MC, de Mendoza D. The membrane fluidity sensor DesK of Bacillus subtilis controls the signal decay of its cognate response regulator. J Bacteriol. 2004;186(9):2655–2663. doi: 10.1128/JB.186.9.2655-2663.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Cybulski LE, Martín M, Mansilla MC, Fernández A, de Mendoza D. Membrane thickness cue for cold sensing in a bacterium. Curr Biol. 2010;20(17):1539–1544. doi: 10.1016/j.cub.2010.06.074. [DOI] [PubMed] [Google Scholar]
- 4.Cybulski LE, del Solar G, Craig PO, Espinosa M, de Mendoza D. Bacillus subtilis DesR functions as a phosphorylation-activated switch to control membrane lipid fluidity. J Biol Chem. 2004;279(38):39340–39347. doi: 10.1074/jbc.M405150200. [DOI] [PubMed] [Google Scholar]
- 5.Cybulski LE, et al. Mechanism of membrane fluidity optimization: Isothermal control of the Bacillus subtilis acyl-lipid desaturase. Mol Microbiol. 2002;45(5):1379–1388. doi: 10.1046/j.1365-2958.2002.03103.x. [DOI] [PubMed] [Google Scholar]
- 6.Altabe SG, Aguilar P, Caballero GM, de Mendoza D. The Bacillus subtilis acyl lipid desaturase is a delta5 desaturase. J Bacteriol. 2003;185(10):3228–3231. doi: 10.1128/JB.185.10.3228-3231.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Cybulski LE, de Mendoza D. Bilayer hydrophobic thickness and integral membrane protein function. Curr Protein Pept Sci. 2011;12(8):760–766. doi: 10.2174/138920311798841681. [DOI] [PubMed] [Google Scholar]
- 8.Nagle JF, Tristram-Nagle S. Structure of lipid bilayers. Biochim Biophys Acta. 2000;1469(3):159–195. doi: 10.1016/s0304-4157(00)00016-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Pan J, Tristram-Nagle S, Kucerka N, Nagle JF. Temperature dependence of structure, bending rigidity, and bilayer interactions of dioleoylphosphatidylcholine bilayers. Biophys J. 2008;94(1):117–124. doi: 10.1529/biophysj.107.115691. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Albanesi D, et al. Structural plasticity and catalysis regulation of a thermosensor histidine kinase. Proc Natl Acad Sci USA. 2009;106(38):16185–16190. doi: 10.1073/pnas.0906699106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Goormaghtigh E, Raussens V, Ruysschaert JM. Attenuated total reflection infrared spectroscopy of proteins and lipids in biological membranes. Biochim Biophys Acta. 1999;1422(2):105–185. doi: 10.1016/s0304-4157(99)00004-0. [DOI] [PubMed] [Google Scholar]
- 12.Goormaghtigh E, Ruysschaert J-M, Raussens V. Evaluation of the information content in infrared spectra for protein secondary structure determination. Biophys J. 2006;90(8):2946–2957. doi: 10.1529/biophysj.105.072017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Fernández A, Crespo A, Maddipati S, Scott R. Bottom-up engineering of peptide cell translocators based on environmentally modulated quadrupole switches. ACS Nano. 2008;2(1):61–68. doi: 10.1021/nn700239j. [DOI] [PubMed] [Google Scholar]
- 14.Matthews EE, Zoonens M, Engelman DM. Dynamic helix interactions in transmembrane signaling. Cell. 2006;127(3):447–450. doi: 10.1016/j.cell.2006.10.016. [DOI] [PubMed] [Google Scholar]
- 15.Zimmann P, Steinbrügge A, Schniederberend M, Jung K, Altendorf K. The extension of the fourth transmembrane helix of the sensor kinase KdpD of Escherichia coli is involved in sensing. J Bacteriol. 2007;189(20):7326–7334. doi: 10.1128/JB.00976-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.van der Heide T, Stuart MC, Poolman B. On the osmotic signal and osmosensing mechanism of an ABC transport system for glycine betaine. EMBO J. 2001;20(24):7022–7032. doi: 10.1093/emboj/20.24.7022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Biemans-Oldehinkel E, Mahmood NABN, Poolman B. A sensor for intracellular ionic strength. Proc Natl Acad Sci USA. 2006;103(28):10624–10629. doi: 10.1073/pnas.0603871103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Chemin J, et al. A phospholipid sensor controls mechanogating of the K+ channel TREK-1. EMBO J. 2005;24(1):44–53. doi: 10.1038/sj.emboj.7600494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lesage F, Terrenoire C, Romey G, Lazdunski M. Human TREK2, a 2P domain mechano-sensitive K+ channel with multiple regulations by polyunsaturated fatty acids, lysophospholipids, and Gs, Gi, and Gq protein-coupled receptors. J Biol Chem. 2000;275(37):28398–28405. doi: 10.1074/jbc.M002822200. [DOI] [PubMed] [Google Scholar]
- 20.Kloda A, Ghazi A, Martinac B. C-terminal charged cluster of MscL, RKKEE, functions as a pH sensor. Biophys J. 2006;90(6):1992–1998. doi: 10.1529/biophysj.105.075481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Colbert HA, Smith TL, Bargmann CI. OSM-9, a novel protein with structural similarity to channels, is required for olfaction, mechanosensation, and olfactory adaptation in Caenorhabditis elegans. J Neurosci. 1997;17(21):8259–8269. doi: 10.1523/JNEUROSCI.17-21-08259.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Jaquemar D, Schenker T, Trueb B. An ankyrin-like protein with transmembrane domains is specifically lost after oncogenic transformation of human fibroblasts. J Biol Chem. 1999;274(11):7325–7333. doi: 10.1074/jbc.274.11.7325. [DOI] [PubMed] [Google Scholar]
- 23.Liedtke W, et al. Vanilloid receptor-related osmotically activated channel (VR-OAC), a candidate vertebrate osmoreceptor. Cell. 2000;103(3):525–535. doi: 10.1016/s0092-8674(00)00143-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.