Significance
Using microfiltration as a liquid biopsy for the recovery of circulating tumor cells (CTCs) has revealed an accompanying macrophage subset that we use as a highly sensitive biomarker for solid tumors. We supply evidence that this circulating giant cell is a subset of disseminated tumor-associated macrophages capable of binding CTCs in peripheral blood of cancer patients. The presence of this cell expands the concept of using a liquid biopsy not only to indicate cancer presence but also to track cancer treatment effects sequentially using other circulating blood cells. Further, we supply observational evidence hypothesizing a metastasis pathway model in which CTCs migrate with pro-angiogenic macrophages, linking cancer cell intravasation, migration, and extravasation and the formation of metastatic microenvironments.
Keywords: liquid biopsy, blood cell biomarkers, cancer metastasis, cancer biomarker, cancer screening
Abstract
Tumor-associated macrophages (TAMs) derived from primary tumors are believed to facilitate circulating tumor cell (CTC) seeding of distant metastases, but the mechanisms of these processes are poorly understood. Although many studies have focused on the migration of CTCs, less attention has been given to TAMs that, like CTCs, derive from tumor sites. Using precision microfilters under low-flow conditions, we isolated circulating cancer-associated macrophage-like cells (CAMLs) from the peripheral blood of patients with breast, pancreatic, or prostate cancer. CAMLs, which are not found in healthy individuals, were found to express epithelial, monocytic, and endothelial protein markers and were observed bound to CTCs in circulation. These data support the hypothesis that disseminated TAMs can be used as a biomarker of advanced disease and suggest that they have a participatory role in tumor cell migration.
Tumor-associated macrophages (TAMs) are specialized differentiated macrophages found within most tumors, which can be used as prognostic indicators of either tumor invasiveness or tumor suppression (1–3). TAMs, recruited to the stroma from circulating monocytes, are required for tumor cell intravasation, migration, extravasation, and angiogenesis (2–7). Tumors attract monocytes via chemoattractants (e.g., MCP-1, CCL-2) (2–4). In turn TAMs secrete cytokines and growth factors (e.g., MMP-1, CXCL12) which stimulate tumor cells with the potential to become circulating tumor cells (CTCs) (2–4). TAMs and CTCs then migrate via the lymphatic system or intravasate across intratumor capillary barriers into peripheral circulation (4–9).
Pathological evidence detailing the dissemination of CTCs via a metastatic cascade remains inconclusive. Typically, cancer cell dissemination requires three steps: CTC separation from the tumor, movement away from the parent mass, and migration into the circulatory system (10). Although various theories have explained selected aspects of this dissemination and involved various cell types in this process, including endothelial progenitor cells (EPCs), cancer mesenchymal stem cells, and hybrid cancer cells (10–12), among others, none of these single components explains the entire metastatic process. Recent, in vivo studies have shown that circulating monocytic cells are intricately involved in tumor cell invasiveness, motility, and metastatic potential (1–6). Interactions between myeloid-lineage cells and tumor cells have been documented in patients and modeled in mice, suggesting that the pathway for cancer cell intravasation occurs in conjunction with macrophages via transendothelial migration (4–7).
Here we report evidence of the existence of highly differentiated giant circulating (macrophage-like) cells isolated from the peripheral blood of patients with breast, prostate, or pancreatic cancer, which we hypothesize to be disseminated TAMs (DTAMs). Although giant cells resembling these have been observed sporadically in the past, only now have their systematic isolation, identification, and characterization for proper in-depth study become technologically possible (13–15). We isolated this cell type by developing a low-pressure filtration system equipped with precision microfilters, allowing histological identification of cellular morphology (16). We term this giant cell a “circulating cancer-associated macrophage-like cell” (CAML), because it exhibits CD14+ expression and vacuoles of phagocytosed material and has been observed exclusively in cancer patients (Fig. 1 and Table S1). We propose that this cell population, which is not detected in healthy individuals, could serve as a robust cellular biomarker of a previously undefined innate immune response to cancer presence and of cancer aggressiveness and could be useful in monitoring chemotherapy-induced responses. Observations of these giant cells interacting with CTCs while in circulation support evidence that a patient’s immune cells have an observable effect on the migration or elimination of CTCs. Furthermore, angiopoietin-1 receptor (TIE-2) positive markers expressed by macrophages (4, 5) suggest that CAMLs have a possible a role as cellular initiators of neovascularization within tumor metastases. We have uncovered supporting in vivo evidence that CAMLs may play an associated role in the migration of CTCs in circulation.
Fig. 1.
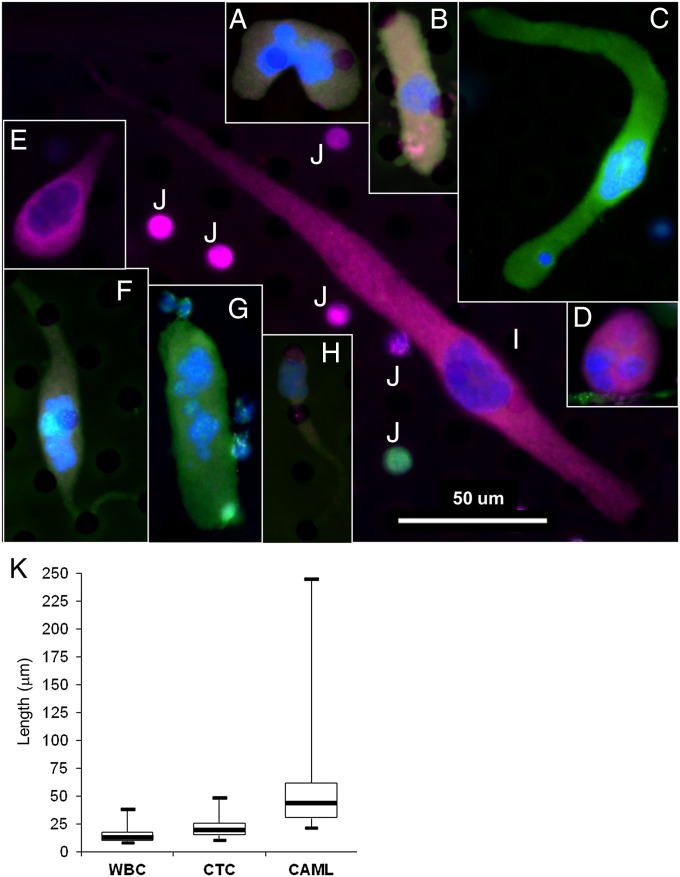
(Upper) Representative collage of the five morphologies, signal variation, and cytoplasmic diameters. (A, F, and G) Pancreatic cells. (B, C, and D) breast cells. (E, H, and I) Prostate cells. (J) Typical WBCs. Morphology variants are as follows: amorphous (A), oblong (B and G), spindle-shaped (C, F, and I), round (D), and tadpole-shaped (E and H). Color differences result from varying degrees of staining for DAPI (blue), cytokeratins (green), EpCAM (red), and CD45 (violet) (Fig. S1). (Scale bar, 50 μm.) (K, Lower) Whisker plot of cytoplasmic diameters showing diameters of WBCs, CTCs, and CAMLs (n = 75) from pooled prostate, breast, and pancreatic samples.
Results
Defining CAMLs.
Giant fused macrophages are a poorly understood type of immune cells found in a multitude of tissues. They are hybrids of multinucleated cells originating from myeloid lineage (17) and are involved in numerous physiological and pathological processes, including phagocytosis of foreign/necrotic tissue, tissue reabsorption, and inflammation (17). We show that CAMLs are giant cells of myeloid lineage (CD14+/CD11c+) presenting with enlarged nuclei, are CD45+, and exhibit cytoplasmic staining by cytokeratin 8, 18, and 19 and epithelial cell adhesion molecule (EpCAM) that is characteristic of epithelial cells (Fig. S1). Multiple individual nuclei can be found in CAMLs, although fused nucleoli (14–64 µm in diameter) are also common (Fig. S1). CAML cytoplasm, defined by a cytokeratin border (21–300 µm in length), can be observed on the filter with five morphological phenotypes (Fig. 1 A and B). CAMLs are defined by a large atypical nucleus and a cytokeratin-positive cytoplasmic signal that is diffuse in nature.
Although morphological identification of these cells by their extreme size, large nuclear profile, and cytoplasmic signature is straightforward, they have highly heterogeneous phenotypes. The degree of expression of cytokeratin, EpCAM, and CD45 can each vary from the absence of any expression to very intense expression (Fig. S1). This heterogeneity is further varied in the five cellular configurations, the size ranges, and the nuclear profiles (Fig. S1). Highly heterogeneous marker expression implies that CAMLs, like many myeloid-derived cells, either may represent different stages of differentiation or are the result of nonspecific engulfment of cells of epithelial origin. Notably, macrophages are highly plastic cells capable of differentiating into numerous phenotypes (8, 11, 12, 17, 18).
CAMLs Complement CTC Enumeration.
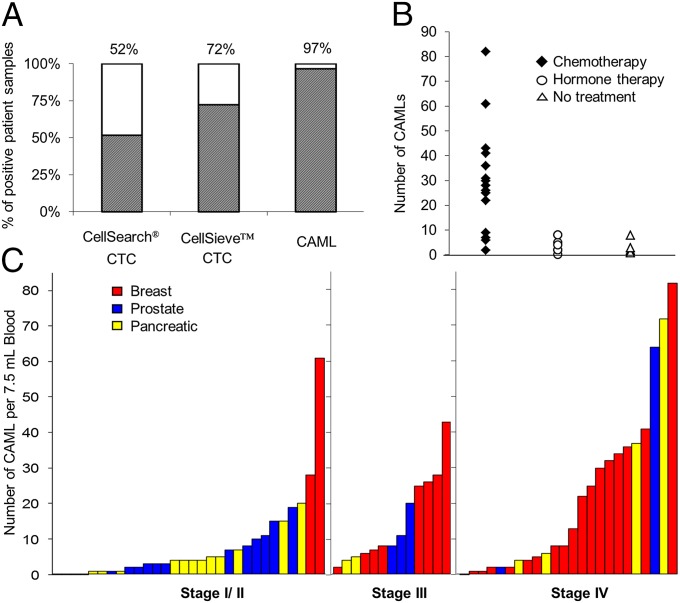
Given the potential biological role of CAMLs, we hypothesize that the detection of these cells in the peripheral blood of patients with advanced cancer may serve as an independent prognostic indicator of cancer progression and may complement CTC enumeration (19). We sought to compare the number of CAMLs and CTCs using the CellSieve low-pressure microfiltration assay (Creatv MicroTech, Inc.) and the CellSearch CTC test (Janssen Diagnostics, LLC). Isolating CTCs has been challenging because of their rarity and limited occurrence (in only 10–50% of cancer patients with metastatic disease) (19). TAM enumeration and phenotyping has prognostic utility but currently lacks sequential testing for tracking primary to metastatic progression, because this task requires numerous invasive tumor biopsies (20–22). Enumerations of CTCs enriched by the CellSearch and CellSieve systems were compared directly with the enumerations of CAMLs isolated by CellSieve using blood from 29 patients with cancer (Fig. 2A). CellSearch uses EpCAM antibodies to enrich CTCs, whereas CellSieve uses size exclusion to isolate both CAMLs and CTCs (16). Both technologies phenotypically identify CTCs using an antibody panel. The CellSieve system identified at least one CTC in 72% of the patient samples, whereas CellSearch identified at least one CTC in only 58% of the samples. In contrast, CellSieve identified CAMLs in 97% of the samples (Fig. 2A). Interestingly, the only CAML− sample was from a breast cancer patient being treated with systemic therapy combined with bisphosphonates, a class of drugs that inhibits the formation of osteoclasts, giant myeloid cells of the bone composed of fused cells. Thus, because the specificity of CAMLs is 100% for the samples tested, we hypothesize that the presence of CAMLs may provide a method for noninvasive sequential testing that could be used as an additional indicator of metastatic disease in a broad range of cancer patients.
Fig. 2.
Data comparing the presence of CAMLs versus CTCs. (A) CTCs were isolated in 15/29 samples by CellSearch and in 21/29 duplicate samples by CellSieve. CAMLs were isolated in 28/29 samples by CellSieve. (B) In 29 different breast cancer patients, CAML number was affected by treatment vs. no treatment. No treatment (open triangle), n = 5 patients; average CAML number = 3. Hormone-based treatment (open circle), n = 7 patients, average CAML number = 4.1. Chemotherapy (solid diamond), n = 17 patients, average CAML number= 29. (C) CAMLs are markers found in the blood of patients with breast (red bars), prostate (blue bars), and pancreatic (yellow bars) cancer in all cancer stages. Each bar represents a single blood sample from a patient with known staging (n = 67).
CAMLS Are Found in a Broad Patient Population.
To assess the sensitivity and specificity of CAMLs as an indication of malignant disease, we examined samples from early- to late-stage cancer patients and healthy subjects. Samples from 79 patients were run on CellSieve microfilters. The patient distribution compared stage I through stage IV and unknown stage, breast, pancreatic, and prostate cancers (Fig. 2 B and C and Tables S1 and S2). We included newly diagnosed/untreated patients as well as patients undergoing systemic therapies. The study included healthy subjects, including two with benign disease (one fibroadenoma and one basal cell carcinoma). No CAMLs were found in this control group. CAMLs were found in 97% patients with stage III/IV cancer, in 83% of patients with stage I/II cancer, and in 92% of patients overall, regardless of cancer type. CAMLs were found in 86% of patients with prostate cancer, 93% of patients with pancreatic cancer, and 97% of patients with breast cancer. CAML number were slightly lower in prostate samples, possibly because of the relatively high proportion of stage I patients ( n = 7/22). Analysis on CellSieve microfilters identified CTCs in 54% across all patients tested, and CAMLs were found in 92% across the same cohort, with no correlation between the two counts (Table S1). Although further study of additional patients with various conditions and illnesses must be assessed, these findings demonstrate that CAML detection, with an associated diagnosis of invasive disease, could provide a robust and widely applicable assessment of cancer status.
CAMLs Contain Engulfed Material from the Site of the Tumor.
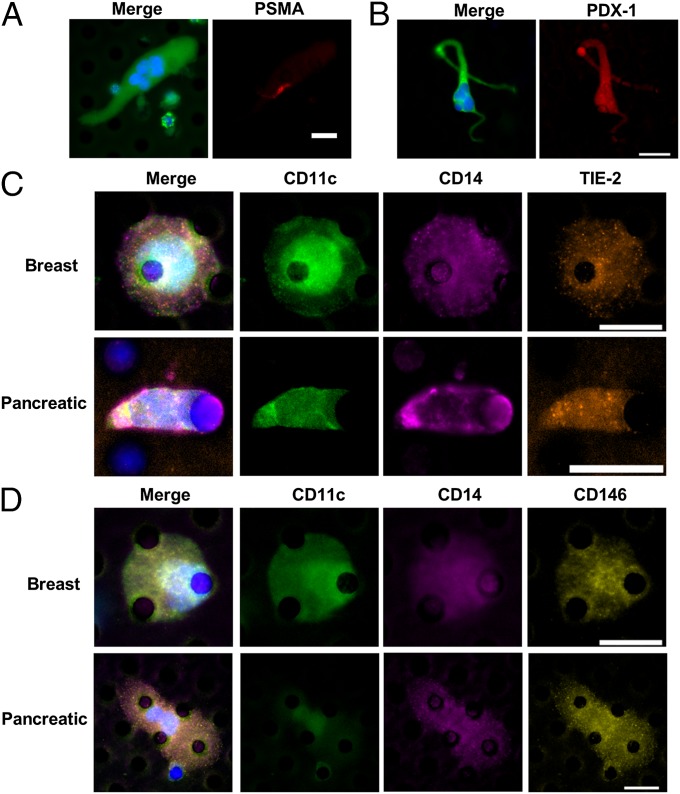
We propose that CAMLs represent a specialized DTAM likely originating at the site of the tumor and disseminating into circulation. This suggestion is consistent with previous publications characterizing TAMs at the primary tumor site (1–4) and with our observations that CAMLs appear to be CD14+, contain engulfed epithelial tissue, and occur exclusively in cancer patients. To understand the origin of CAMLs, we investigated whether they arise from circulating monocytes or directly from TAMs. Because TAMs and monocytes would present with similar protein markers, we assessed the presence of engulfed organ-specific markers, pancreatic duodenal homeobox-1 (PDX-1) for patients with pancreatic cancer or prostate-specific membrane antigen (PSMA) for patients with prostate cancer. PDX-1 is a differentiation and development marker detected in adult endocrine organs, namely pancreatic cells. PSMA is a membrane glycoprotein that is highly expressed in prostate cells. After isolating and enumerating CAMLs, we restained preidentified CAMLs for PSMA or PDX-1. PSMA+ debris was demonstrated in CAMLs from prostate samples (Fig. 3 and Fig. S2 A and B), and PDX-1 was found in all CAMLs from pancreatic samples (Fig. 3 and Fig. S2 C and D). Although it is possible that cellular fusion or ingestion of debris occurred away from the tumor site, the high concentration of markers coupled with the scarcity of tumor debris in circulation make this possibility less likely. Instead, these findings suggest that CAMLs participate in a process of phagocytosis of necrotic debris or engulfment of neoplastic cells, likely from a tumor site, as part of the innate immune recognition of and response to the tumor.
Fig. 3.
CAMLs stain for monocytic, endothelial, and specific tissue markers. (A) CAML from a prostate cancer patient stained with DAPI (blue) and anti–cytokeratins-FITC (green) (Left) and with anti–PSMA-Dylight594 (red) (Right). (B) CAML from a pancreatic cancer patient stained with DAPI (blue) and anti–cytokeratins-FITC (green) (Left) and anti–PDX-1-Dylight594 (red) (Right). (C) CAMLs from samples from patients with breast (Upper Row) or pancreatic (Lower Row) cancer. Images from left to right show a merged image with DAPI, anti-CD11c (monocyte marker), anti-CD14 (monocyte marker), and anti-TIE-2 (angiogenic marker) staining and images of staining for individual markers. (D) CAMLs from samples from patients with breast (Upper Row) or pancreatic (Lower Row) cancer. Images from left to right show a merged image with DAPI, anti-CD11c, anti-CD14, and anti-CD146 (endothelial marker) staining and images of staining for individual markers. (Scale bars, 20 µm.)
Chemotherapy Affects CAML Numbers.
To test further whether CAMLs are a DTAM subtype, we compared therapy regimes and changes over time in the number of CAMLs in relation to benefit of systemic therapy and outcome. We hypothesized that if CAMLs are associated with the phagocytosis of cancer cell debris (Fig. 3 and Fig. S2) resulting from cytotoxicity occurring at the tumor site, i.e., derived from TAMs, then patients who are untreated or nonresponsive to therapy would not produce additional cellular debris and thus would have low CAML numbers. Conversely, patients who are responsive to systemic therapy, either chemotherapy or endocrine therapy, would have high CAML numbers. Analysis of therapeutic regimes showed that chemotherapy, but not endocrine or nontherapy, was associated with high CAML levels (Fig. 2B). We find that nontreated and hormone-treated patients who should show little change in tumor size have low CAML numbers, fewer than 3–4.1 per 7.5-mL sample. Conversely, when a chemotherapeutic regime was in use, CAML numbers averaged 29 per 7.5-mL sample. The lack of changes seen with hormone therapy and the substantial increase upon implementation of chemotherapy indicate that the mechanism for CAML phagocytosis and/or release into circulation is affected specifically by therapy type. Additionally, these data suggest that CAMLs may provide a sensitive representation of phagocytosis at the tumor site that could quantify a cell-specific innate immune reaction to the extent of cellular debris caused by chemotherapy.
CAMLs Interact with CTCs in the Circulation.
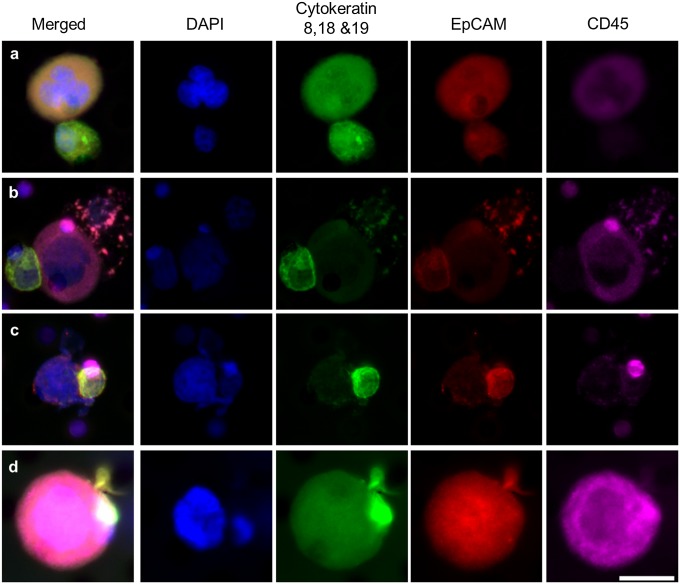
CTCs originate at a tumor site, circulate in peripheral blood, and have the ability to seed metastatic sites. However, the pathway for CTC detachment and invasion into the circulation is a complex process. We analyzed CAMLs isolated from patient samples for evidence of a CTC/CAML interaction. CTCs were found bound to CAMLs in 4 of 79 patients, all with metastatic disease (Fig. 4 A–C and Figs. S3A and S4). In addition, 4 of 79 patients had CAMLs with engulfed cells that appeared to have an epithelial phenotype (Fig. 4D) and an amplified mutation genotype matching the primary tumor (Fig. S3B). The observed interaction of a dual CTC/CAML pair in 10% of patients is indicative of two possibilities. The first is that these cells interact while in circulation, implying that CAMLs are an active immune response to cancer cells in circulation. Alternatively, these cells might bind at the tumor site and disseminate together into circulation, implying a similar pathway of intravasation. In either case, this pairing of cells is more common than expected, and although there is no evidence implying facilitation or hindrance of the bound CTCs, this observation does suggest that CAMLs may play some participatory role in the migration of cancer cells in the peripheral blood of cancer patients.
Fig. 4.
CAMLs interacting with CTCs in patient blood. Images from left to right show a merged image of DAPI, cytokeratins, EpCAM, and CD45 immunostains and images staining for individual markers. CTCs are filamentous cytokeratin-positive and CD45− cells (Fig. S7). CAMLs are the large CD45+ cells. CTCs and CAMLs were observed with varying levels of interactions. (A) Loose association of a CAML cell (above) and CTC (below). (B) CAML cell (center) bound to a CTC (far left) and an apoptotic cell (far right). (C) Attachment with membranes fusing (image includes a CD45+ blood cell behind the CTC, top right). (D) Engulfment of a putative CTC within the CAMLs cytoplasmic area. (Scale bars, 20 µm.)
Proangiogenic Surface Markers Can Be Found on CAMLs.
Circulating monocytes can extravasate into any tissue compartment of the body, including lymph nodes, bone marrow, most organs, and even across the blood–brain barrier (8). Angiogenic EPCs with neovascular potential are capable of being derived from macrophages, and TIE-2+ (CD202b) macrophages are intricately involved in tumor vascularization (4, 5, 12, 23–25). Recent in vitro and mouse in vivo experiments have shown that EPCs derived from CD14+/CD11c+ monocytes differentiate into CD146+/TIE-2+ endothelial cells capable of proangiogenic activity (5, 23–25). Because CAMLs presented with an EPC-like spindle phenotype, we analyzed CAMLs for evidence of this pathway. Preidentified CAML samples were stained with panels of monocytic markers (CD11c and CD14) and angiogenic endothelial markers (CD146 or TIE-2) (Fig. 3 C and D). We observed CAMLs that were positive for both monocytic and endothelial markers. The monocytic marker CD11c was the most reactive, found in all CAMLs stained, and CD14 was the least reactive, at times being absent entirely. The proangiogenic marker TIE-2 and endothelial marker CD146 stained positive in CAMLs, but staining intensity was variable. The findings of endothelial/myeloid overlap are not surprising, because circulating monocytes have high morphological and marker heterogeneity. Even current literature debates over the mononuclear phagocytic system for classification as endothelial cells and monocytic cell express overlapping markers and similar developmental pathways. Nevertheless, the presence of CD146 or TIE-2 on CD14+ cells indicates a specialized proangiogenic macrophage with neovascular potential (4, 5).
Discussion
These results support the hypothesis that CAMLs provide a robust indicator of invasive cancer that may substantially improve on the sensitivity and specificity of existing biomarkers. The ability to track DTAMs provides an opportunity to monitor tumor activity and therapeutic response to chemotherapy routinely by noninvasive sequential sampling of peripheral blood. Currently, long-term prospective studies are underway to track changes in CAMLs over time from surgical resection (Fig. S5) through therapy and to provide a more complete clinical perspective (Fig. S6).
Although many studies have focused on CTC dissemination, TAMs are involved in the seeding, proliferation, and neovascularization of metastases. Most relevant to our observations, recent studies involving 3D in vivo invasion assays with intravital imaging of solid tumors have solidified the theory that CTCs intravasate into the circulatory system in conjunction with TAMs via transendothelial migration (4, 7). Our findings now supply further clinical evidence of a continued interaction of macrophages with tumor cells found within the circulation of cancer patients. We demonstrate that macrophages originating at the tumor site disseminate into the circulation in large numbers. We show that CAMLs bind to and migrate through the circulation attached to CTCs in 10% of late-stage patients, a surprising observation given the rarity of both cell types and the probability that the cell interaction would be pulled apart by the shear stresses within the vascular circulation. Finally, we describe proangiogenic TIE-2/CD146 expression on CAMLs, suggesting an ability to neovascularize a metastatic microenvironment (4, 5). Although only observational, these data provide clinical evidence that proangiogenic cells migrate bound to CTCs. Using these clinical observations, we suggest a theory linking intravasation, migration, and extravasation of CTCs via a single macrophage interaction. Furthermore, we believe these data indicate the complexity of interactions between cancer cells and innate immune cells and suggest that the detection, characterization, and monitoring of the various cellular components present in the peripheral blood of cancer patients are essential elements in the comprehensive biological characterization of solid tumors. Establishing rigorous criteria for the evaluation in monitoring the “liquid phase” of solid tumors would represent an important future approach, potentially useful in the selection and monitoring of standard systemic therapies or combined with more innovative immunotherapies.
Materials and Methods
Blood Sample Collection.
In total, peripheral blood samples from 79 cancer patients were supplied through a collaboration agreement with Fox Chase Cancer Center, University of Maryland Baltimore, Northwestern University, and Medical College of Wisconsin, in accordance with the local institutional review board (IRB) approval and with patients’ informed consent. Anonymized blood samples were collected into CellSave preservative tubes (Veridex, LLC) or K2EDTA purple-top tubes (Becton Dickinson). In addition, blood samples from 30 healthy volunteers were collected in either CellSave preservative tubes or K2EDTA tubes, according to the local IRB approval and with informed consent.
To compare the CellSearch and CellSieve assays, peripheral blood samples (∼10 mL) from 29 cancer patients were drawn in tandem into two CellSave Preservative tubes. One tube was used to enumerate CTCs using the CellSearch system. The second tube was used to enumerate CTCs and CAMLs using the CellSieve microfiltration assay (Fig. 2A).
CellSieve Low-Flow Microfiltration Procedure.
Samples were run at Fox Chase Cancer Center, Northwestern University, University Maryland Baltimore, or Creatv Microtech, Inc. with a CellSieve Microfiltration Assay using a low-pressure vacuum system (16). The CellSieve Microfiltration assay isolates CTCs by size exclusion and identifies CTCs by the morphological features and the phenotypic expression of EpCAM, cytokeratins 8, 18, and 19, and DAPI (Fig. S7). The low-pressure vacuum system uses the bottom half of a filter holder assembly (Millipore), attached to 3 in of silicone tubing placed into the stopper of a 250-mL glass Erlenmeyer flask. The Erlenmeyer arm then was attached to a VacuGene Pump (GE Healthcare) with a regulated pressure gauge set at ∼15 mbar, removing the possibility of increasing pressure gradients. A CellSieve microfilter was washed with 5 mL PBS, placed onto the filter holder, and centered 5 mm beneath a 30-mL syringe. Peripheral blood (7.5 mL) collected in CellSave preservative tubes was prefixed for 15 min, placed into the 30-mL syringe, and drawn through the filter in ∼3 min. The filter was washed with 6 mL PBS, postfixed for 15 min, and permeabilized for 15 min. The filter and cells were stained with an antibody mixture and blocking buffer of FITC-conjugated anti-cytokeratin 8, 18, 19, phycoerythrin (PE)-conjugated EpCAM, and Cy5-conjugated CD45 (Creatv Microtech, Inc.). Filters were washed with PBS + 0.1%Tween-20 (PBST), placed onto a microscope slide, and mounted with Fluoromount-G with DAPI (Southern Biotech).
Identification and Enumeration of CAMLs.
For this study, CAMLs were morphologically identified using the following criteria: a single cell with an enlarged nuclear profile (14–64 μm in diameter) or separated polymorphic nuclei contained within the cell. In addition, the nuclei had to be surrounded by a larger cytokeratin cytoplasmic signal (21–300 μm in length). The cytokeratin signal was identified by a nonfilamented diffuse signal found throughout the cellular cytoplasmic area. CAMLs could be identified further by vacuoles containing DAPI+ and/or epithelial debris, EpCAM, or cytokeratin (Fig. 1 and Figs. S1 and S2).
CellSearch Analysis.
The enumeration of CTCs by the CellSearch system was run according to pre-established protocols at Fox Chase Cancer Center (26). Peripheral blood (7.5 mL) was collected in CellSave preservative tubes (Veridex, LLC) and maintained at ambient temperature until processed within 96 h of collection. CellSearch Epithelial Cell kits (Veridex, LLC) were used for the isolation of CTCs. All automated isolations were performed on the CellTracks AutoPrep System (Veridex, LLC). Data were collected and analyzed on the CellTracks Analyzer II (Veridex, LLC) (Fig. 2A).
Anti–pan cytokeratin (cytokeratin 8, 18, 19)-PE, anti–CD45-APC, and DAPI stain (CellSearch Epithelial Cell kit reagents) were used to label the CTCs. Immunomagnetic enrichment of CTCs using the CellTracks AutoPrep System has been described in detail (26). Briefly, ferrofluid particles conjugated with anti-EpCAM were used to capture CTCs from 7.5 mL of blood via a magnetic separation. Captured cells were washed, permeabilized, and labeled with fluorescent antibodies. After labeling, cells were washed, resuspended in cell fixative, and loaded into cartridges. Cartridges were placed in magnetic holders (MagNest) which align the ferrofluid-captured cells with the cartridge surface. The MagNests were placed into the CellTracks Analyzer II where the fluorescently labeled cells were scanned and images were captured. Images for analyses were sorted by computer-assisted software selecting events based on the parameters of CD45−, cytokeratin-positive, and DAPI+. Captured images were displayed in thumbnails and reviewed. Images depicting complete cells were selected as a CTC (26).
PSMA or PDX-1 Staining Assay.
PSMA (BioLegend) and PDX-1 (eBioscience) were purchased and conjugated to Dylight594, using a Dylight594 conjugation kit (Pierce Thermo). PSMA-Dylight594 or PDX-1-Dylight594 was added to the antibody mixture of cytokeratin 8, 18, 19-FITC, EpCAM-PE, and CD45-Cy5. The residual blood (2 mL) from patient samples (n = 4 pancreatic cancer and n = 4 prostate cancer) that had been preidentified as having CAMLs was run on the CellSieve microfilters, using the protocol described above for the filtration through permeabilization steps (Fig. 3 A and B). The filters then were stained using the additional antibody mixture with PSMA-Dylight594 for the four prostate cancer samples (Fig. 3A and Fig. S2 A and B) or with PDX-1-Dylight594 for the four pancreatic cancer samples (Fig. 3B and Fig. S2 C and D). Filters then were washed with PBST and mounted as above. Cross-reactivity of the antibodies was tested by applying the PDX-1-Dylight594 antibody to three sets of prostate CAML samples or applying the PSMA-Dylight594 mixture to three sets of pancreatic CAML samples. No reaction was observed.
CD11c, CD14, CD146, and CD202b Staining Assay.
FITC-labeled anti-CD11c (eBioscience), Cy5-labeled anti-CD14 (eBioscience), PE-labeled anti-CD146 (eBioscience), and PE-labeled CD202b (BioLegend) were purchased preconjugated. The residual blood (2 mL) from patient samples (n = 3 pancreatic cancer, n = 3 breast cancer, and n = 3 prostate cancer), which had been preidentified as having CAMLs, was run on the CellSieve Microfilters, using the protocol described above. After the permeabilization step, filters were blocked in 150 µL PBS with 10% (vol/vol) FBS and 0.1 mg/mL whole mouse IgG (Jackson ImmunoResearch) for 1 h at room temperature. Filters were washed with 5 mL PBS and stained using a mixture combining anti–CD11c-FITC, anti–CD14-Cy5, and anti–CD146-PE or anti–CD11c-FITC, anti–CD14-Cy5, and anti–CD202b-PE (Fig. 3 C and D and Table S3). Filters then were washed with PBST and mounted as above.
Interacting CAMLS and CTCs.
We defined CAMLs and CTCs as interacting if they met the definitions given above of a CAML or a CTC and both cells were in direct contact or were engulfed.
For an approximate calculation of the probability that a CTC would contact a CAML at random on a filter, we assumed CTCs and CAMLs are round and that the average diameter of a CAML is 51.3 µm (D) and the average diameter of a CTC is 20.9 µm (d) (Fig. 1l). The average number of CAMLs per sample is 13.3, and the average number of CTCs is 21.7 (Table S1). The diameter of the area around a single CAML in which a CTC could fall and be in contact is D+2(1/2d), or 72.2 µm. The total area of the microfilter is 13 mm. Therefore the probability that one of the 21.7 CTCs would randomly contact one of the 13.3 CAMLs is 0.89% per microfilter, or less than one patient sample in the 79-patient cohort.
HER-2 FISH Probe on CAMLs and Their Engulfed Nuclear Debris.
The PathVysion HER-2 DNA Probe Kit was supplied by Abbott Molecular Inc. CAML+ samples identified as having engulfed or engulfing nuclear structures using the CellSieve Microfiltration Assay were imaged as described above and then were probed. Identified cells were FISH probed directly on the filter or were transferred onto glass slides for FISH probing. In each case the x/y/z placement of cells was marked by etching the sample substrate, and placement was recorded using Zen2011 Blue software (Carl Zeiss). Samples were demounted in a 2× SSC solution for 10 min and dried by air. The protease solution was added to each sample for 20 min in a 37 °C incubator. Slides were washed twice in 2× SSC for 5 min and were dried on a 45 °C warmer. Slides were placed in the denaturing solution at 72 °C for 5 min and were washed with 70% ethanol for 1 min, 85% ethanol for 1 min, and 100% ethanol for 1 min and were dried on a 45 °C warmer. Then 10 µL of probe was added to the slides, a coverslip was added and sealed with rubber cement, and the slide was incubated for 22 h in a 37 °C hybridization chamber. The coverslip was removed in the posthybridization wash buffer at room temperature, washed in the posthybridization wash buffer at 72 °C for 2 min, rewashed in 2× SSC for 10 min, and dried at room temperature. Samples were mounted in Fluoromount-DAPI (Southern Biotech) and imaged on an Olympus BX54WI Fluorescent microscope with a Carl Zeiss AxioCam. Images were overlaid using Zen2011 Blue software (Carl Zeiss). Nuclear debris within CAMLs could be found with and without HER-2 amplification (Fig. S3).
Testing Blood Storage Conditions Using K2EDTA Tubes.
To verify that CAMLs were not an artifact of the preservation tubes, CAML presence in patient samples was tested using K2EDTA tubes (n = 16). Samples were run on CellSieve filters as above. All samples from patients with stage IV cancer (n = 6) and from patients with stage II cancer (n = 5) were positive for CAMLs. Two samples from patients with stage I cancer (n = 5) were positive for CAMLs.
Testing Temporal Changes of CAMLs in Patient Blood.
To verify that the number of CAMLs was a result of therapy change and not random events, we tracked five patients at two separate time points. CAMLs were enumerated in three patients (two with pancreatic cancer and on with prostate cancer) at the time of diagnosis and in two patients with breast cancer on a set therapy regime. A subsequent sample was run for CAMLs at follow-up visits (30, 50, 67, 90, or 180 d) and before any change or addition of treatment (Fig. S5A). The number of CAMLs found in cancer patients remained steady over various time periods.
Testing Temporal Changes of CAMLs in Patient Blood Before and After Cancer Resection.
To demonstrate further that the number of CAMLs was the result of an immune response to cancer presence, we tracked five patients with early-stage pancreatic (n = 3) or prostate (n = 2) cancer at two separate time points, before and after surgical resection. Blood was drawn from patients <30 d before surgery, and CAMLs were enumerated. Blood was redrawn from patients 30–45 d after surgical resection, and CAMLs were enumerated (Fig. S5B). In contrast to the aforementioned stability in CAML numbers over time in patients not undergoing treatment (Fig. S5A), the number of CAMLs was 65–100% lower after surgical resection than before surgical resection. This result suggests that the immune response responsible for CAML formation is still active directly after surgery and for weeks after tumor resection.
Microscope Imaging and Measurement of Cell Size.
After samples were filtered and processed according to the protocols described above, all slides were sent to the Creatv MicroTech, Inc. core facility for CAML enumeration. An Olympus BX54WI Fluorescent microscope with a Carl Zeiss AxioCam (0.16 µm per pixel) was used to image all the filters for both CTCs and CAMLs. Exposures were preset at 5 s for Cy5, 2 s for PE, 750 ms for Dylight594, 750 ms for FITC, and 10–50 ms for DAPI. A Zen2011 Blue (Carl Zeiss) was used to process the images. Minimum/maximum (Min/Max) ranges for Fig. 1 and Fig. S1 were between 400/2,000 for Cy5, 300/1,000 for PE, 350/2,000 for FITC, and 50/2,000 for DAPI. Min/Max ranges for Fig. 3 A and B were set at 200/2,000 for Dylight594, 250/2,000 for FITC, and 50/2,000 for DAPI. Min/Max ranges for Fig. 3C were set at 150/700 for Cy5, 200/460 for PE, 250/882 for FITC, and 81/1,100 for DAPI. Min/Max ranges for Fig. 3D were set at 225/700 for Cy5, 275/750 for PE, 375/1,000 for FITC, and 50/2,500 for DAPI. Min/Max ranges for Fig. 4 A–D were set at 200/2,000 for Cy5, 300/2,000 for PE, 150/2,000 for FITC, and 50/2,500 for DAPI.
To measure cell size, Zen2011 Blue software (Carl Zeiss) was precalibrated by the manufacturer. The automatic measurement software was used to find the length of CAMLs, CTCs, and WBCs. In total, 75 WBCs, CAMLs, and CTCs were taken at random and measured by the Zen2011 Blue. The median values were 12.4 µm for WBCs, 18.8 µm for CTCs, and 43.5 µm for CAMLs (Fig. 1).
Interaction Between a CAML and a CTC.
After a CAML+ sample with an attached CTC was preidentified on a filter, the cells were stained for the cytoskeletal markers vimentin and α-tubulin. Rabbit polyclonal anti-vimentin (Cell Signaling Technology) and mouse monoclonal anti–α-tubulin (Sigma) were purchased unconjugated. The preidentified filter was stained using a mixture combining anti-vimentin and anti–α-tubulin for 1 h. Filters were washed with 5 mL PBST and blocked in 150 µL PBS with 10% FBS and 0.1 mg/mL whole mouse IgG (Jackson ImmunoResearch) for 1 h at room temperature. Filters then were stained with goat anti-mouse Alexa Fluor-555 (Invitrogen) and goat anti-rabbit Alexa Fluor-647 (Invitrogen) for 30 min. Filters then were washed with PBST and mounted as described above. Fig. S4 shows an example of a vimentin-positive and α-tubulin–positive CTC bound to a cytokeratin-positive CAML, which appears negative for the markers. This cell then was imaged using an Olympus FV-1000 confocal microscope at the University Maryland Baltimore to show the size of the CAML and CTC relative to other WBCs and the intact protrusions from the CTC (Fig. S4 and Movie S1).
Statistical Calculations.
We calculated and plotted the Kaplan–Meier curve for the overall survival of patients in Fig. S6 using a cumulative distribution function in MATLAB R2013A. We calculated the Pearson’s correlation between the number of CTCs and number of CAMLs in Table S1 using a Student t test distribution, again using MATLAB R2013A.
Supplementary Material
Acknowledgments
We thank all the patients and healthy volunteers who contributed to this study; C. Haudenschild for pathology expertise and for editing this manuscript; and S. Stefansson, D. Adams, A. Sethi, S. Li, P. Zhu, P. Amstutz, and O. Makarova for editing this manuscript. This work was supported by an award from the Maryland Technology Development Corporation/Maryland Technology Transfer Commercialization Fund; Grant R01-CA154624 from the National Cancer Institute; Grant KG100240 from the Susan G. Komen Foundation; Grant BC100675 from an Era of Hope Scholar Award from the Department of Defense; Advancing a Healthier Wisconsin; Grants CA90386 and CA122985 from the National Institutes of Health (to R.C.B.); and Grant CA90386 from the Prostate Specialized Program of Research Excellence.
Footnotes
Conflict of interest statement: D.L.A. and C.-M.T. have filed a patent application regarding the clinical utility of cancer-associated macrophage-like cells and are employees at Creatv Microtech, Inc.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1320198111/-/DCSupplemental.
References
- 1.Heusinkveld M, van der Burg SH. Identification and manipulation of tumor associated macrophages in human cancers. J Transl Med. 2011;9:216. doi: 10.1186/1479-5876-9-216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Shih JY, Yuan A, Chen JJW, Yang PC. Tumor-associated macrophage: Its role in cancer invasion and metastasis. J Cancer Mol. 2006;2(3):101–106. [Google Scholar]
- 3.Solinas G, Germano G, Mantovani A, Allavena P. Tumor-associated macrophages (TAM) as major players of the cancer-related inflammation. J Leukoc Biol. 2009;86(5):1065–1073. doi: 10.1189/jlb.0609385. [DOI] [PubMed] [Google Scholar]
- 4.Condeelis J, Pollard JW. Macrophages: Obligate partners for tumor cell migration, invasion, and metastasis. Cell. 2006;124(2):263–266. doi: 10.1016/j.cell.2006.01.007. [DOI] [PubMed] [Google Scholar]
- 5.De Palma M, et al. Tie2 identifies a hematopoietic lineage of proangiogenic monocytes required for tumor vessel formation and a mesenchymal population of pericyte progenitors. Cancer Cell. 2005;8(3):211–226. doi: 10.1016/j.ccr.2005.08.002. [DOI] [PubMed] [Google Scholar]
- 6.Wyckoff JB, et al. Direct visualization of macrophage-assisted tumor cell intravasation in mammary tumors. Cancer Res. 2007;67(6):2649–2656. doi: 10.1158/0008-5472.CAN-06-1823. [DOI] [PubMed] [Google Scholar]
- 7.Roussos ET, et al. Mena invasive (MenaINV) promotes multicellular streaming motility and transendothelial migration in a mouse model of breast cancer. J Cell Sci. 2011;124(Pt 13):2120–2131. doi: 10.1242/jcs.086231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Ovchinnikov DA. Macrophages in the embryo and beyond: Much more than just giant phagocytes. Genesis. 2008;46(9):447–462. doi: 10.1002/dvg.20417. [DOI] [PubMed] [Google Scholar]
- 9.Dovas A, et al. Visualization of actin polymerization in invasive structures of macrophages and carcinoma cells using photoconvertible β-actin-Dendra2 fusion proteins. PLoS ONE. 2011;6(2):e16485. doi: 10.1371/journal.pone.0016485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Mego M, Mani SA, Cristofanilli M. Molecular mechanisms of metastasis in breast cancer—clinical applications. Nat Rev Clin Oncol. 2010;7(12):693–701. doi: 10.1038/nrclinonc.2010.171. [DOI] [PubMed] [Google Scholar]
- 11.Pawelek JM, Chakraborty AK. Fusion of tumour cells with bone marrow-derived cells: A unifying explanation for metastasis. Nat Rev Cancer. 2008;8(5):377–386. doi: 10.1038/nrc2371. [DOI] [PubMed] [Google Scholar]
- 12.Coffelt SB, et al. Elusive identities and overlapping phenotypes of proangiogenic myeloid cells in tumors. Am J Pathol. 2010;176(4):1564–1576. doi: 10.2353/ajpath.2010.090786. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Raker JW, Taft PD, Edmonds EE. Significance of megakaryocytes in the search for tumor cells in the peripheral blood. N Engl J Med. 1960;263:993–996. doi: 10.1056/NEJM196011172632003. [DOI] [PubMed] [Google Scholar]
- 14.Hume R, West JT, Malmgren RA, Chu EA. Quantitative Observations of Circulating Megakaryocytes in the Blood of Patients with Cancer. N Engl J Med. 1964;270:111–117. doi: 10.1056/NEJM196401162700301. [DOI] [PubMed] [Google Scholar]
- 15.Bobek V, Hoffman RM, Kolostova K. Site-specific cytomorphology of disseminated PC-3 prostate cancer cells visualized in vivo with fluorescent proteins. Diagn Cytopathol. 2013;41(5):413–417. doi: 10.1002/dc.22843. [DOI] [PubMed] [Google Scholar]
- 16.Adams DL, et al. The Systematic Study of Circulating Tumor Cell Isolation using Lithographic Microfilters. RSC Advances. 2014;4(9):4334–4342. doi: 10.1039/C3RA46839A. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Quinn MT, Schepetkin IA. Role of NADPH oxidase in formation and function of multinucleated giant cells. J Innate Immun. 2009;1(6):509–526. doi: 10.1159/000228158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Horrevoets AJ. Angiogenic monocytes: Another colorful blow to endothelial progenitors. Am J Pathol. 2009;174(5):1594–1596. doi: 10.2353/ajpath.2009.090198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Allard WJ, et al. Tumor cells circulate in the peripheral blood of all major carcinomas but not in healthy subjects or patients with nonmalignant diseases. Clin Cancer Res. 2004;10(20):6897–6904. doi: 10.1158/1078-0432.CCR-04-0378. [DOI] [PubMed] [Google Scholar]
- 20.Chen JJ, et al. Tumor-associated macrophages: The double-edged sword in cancer progression. J Clin Oncol. 2005;23(5):953–964. doi: 10.1200/JCO.2005.12.172. [DOI] [PubMed] [Google Scholar]
- 21.Fiumara A, et al. In situ evidence of neoplastic cell phagocytosis by macrophages in papillary thyroid cancer. J Clin Endocrinol Metab. 1997;82(5):1615–1620. doi: 10.1210/jcem.82.5.3909. [DOI] [PubMed] [Google Scholar]
- 22.Medrek C, Pontén F, Jirström K, Leandersson K. The presence of tumor associated macrophages in tumor stroma as a prognostic marker for breast cancer patients. BMC Cancer. 2012;12:306. doi: 10.1186/1471-2407-12-306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Venneri MA, et al. Identification of proangiogenic TIE2-expressing monocytes (TEMs) in human peripheral blood and cancer. Blood. 2007;109(12):5276–5285. doi: 10.1182/blood-2006-10-053504. [DOI] [PubMed] [Google Scholar]
- 24.Rehman J, Li J, Orschell CM, March KL. Peripheral blood “endothelial progenitor cells” are derived from monocyte/macrophages and secrete angiogenic growth factors. Circulation. 2003;107(8):1164–1169. doi: 10.1161/01.cir.0000058702.69484.a0. [DOI] [PubMed] [Google Scholar]
- 25.Schmeisser A, et al. Monocytes coexpress endothelial and macrophagocytic lineage markers and form cord-like structures in Matrigel under angiogenic conditions. Cardiovasc Res. 2001;49(3):671–680. doi: 10.1016/s0008-6363(00)00270-4. [DOI] [PubMed] [Google Scholar]
- 26.Kagan M, et al. A sample preparation and analysis system for identification of circulating tumor cells. J Clin Ligand Assay. 2002;25(1):104–110. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.