Significance
Infection by HIV-1 requires fusion of viral and host cell membranes, a process mediated by viral protein gp41. Although extensive structural detail on both pre- and postfusion gp41 states is available from X-ray crystallography and cryo-EM studies, little is known about the actual transition. This NMR study of a trimeric gp41 ectodomain, which connects viral and host cell membranes in the prefusion state, suggests a fusion model, where this domain unzippers from opposite ends because of the affinity of its two α-helices for viral and host cell membranes. In this model, the change in orientation of the ectodomain helices, which is associated with membrane binding, provides the driving force that pulls the membranes into the close juxtaposition required for fusion.
Keywords: hemagglutinin, HIV-1 fusion inhibitor, RDC, 15N relaxation, chemical shift
Abstract
The envelope glycoprotein gp41 mediates the process of membrane fusion that enables entry of the HIV-1 virus into the host cell. The actual fusion process involves a switch from a homotrimeric prehairpin intermediate conformation, consisting of parallel coiled-coil helices, to a postfusion state where the ectodomains are arranged as a trimer of helical hairpins, adopting a six-helix bundle (6HB) state. Here, we show by solution NMR spectroscopy that a water-soluble 6HB gp41 ectodomain binds to zwitterionic detergents that contain phosphocholine or phosphatidylcholine head groups and phospholipid vesicles that mimic T-cell membrane composition. Binding results in the dissociation of the 6HB and the formation of a monomeric state, where its two α-helices, N-terminal heptad repeat (NHR) and C-terminal heptad repeat (CHR), become embedded in the lipid–water interface of the virus and host cell. The atomic structure of the gp41 ectodomain monomer, based on NOE distance restraints and residual dipolar couplings, shows that the NHR and CHR helices remain mostly intact, but they completely lose interhelical contacts. The high affinity of the ectodomain helices for phospholipid surfaces suggests that unzippering of the prehairpin intermediate leads to a state where the NHR and CHR helices become embedded in the host cell and viral membranes, respectively, thereby providing a physical force for bringing these membranes into close juxtaposition before actual fusion.
The first step of HIV infection involves fusion of the viral and target cell membranes, a process mediated by the viral envelope glycoprotein Env, consisting of subunits gp120 and gp41 (1). The envelope proteins form a noncovalent complex on the viral surface with the trimerized gp41 transmembrane subunit sequestered by three gp120 surface subunits (2–5). Binding of gp120 to the cell surface receptors CD4 and chemokine receptors CXCR4 or CCR5 triggers a cascade of conformational changes that disrupt the interactions between gp41 and gp120 and result in an extended gp41 conformation (1, 6). In this extended prefusion state, the highly hydrophobic N-terminal fusion peptide (FP) of gp41 anchors in the host cell membrane, while being spatially remote from its transmembrane domain (TM), which traverses the viral membrane (7, 8). After the host cell and viral membranes have fused, the gp41 ectodomain, which links the FP and TM domains, has transitioned into a C3-symmetric six-helix bundle (6HB), with the FP in physical proximity to the TM domain (9). The refolding of gp41 trimers into the highly stable 6HB arrangement is believed to overcome the large free-energy barrier of membrane fusion. Several atomic resolution structures of the 6HB postfusion state have been solved by X-ray crystallography, confirming that the C-terminal heptad repeat (CHR) helices pack in an antiparallel manner into the conserved hydrophobic grooves formed at the surface of the central trimer of N-terminal heptad repeat (NHR) helices (10–12).
Contrary to the postfusion state, structural features of the prehairpin intermediates of HIV-1 gp41 remain the subject of much debate. The functional requirement that gp41’s fusion peptide engages the membrane of spatially distant host cells dictates an extended conformation for the time point where FP engages the membrane of the host cell. Cartoon models commonly depict this prehairpin intermediate as an extended trimer of linear NHR and CHR helices (13–17). Recent cryo-EM studies provide more detailed insights into the relatively subtle rearrangement of the trimeric helical NHR core, which is associated with rearrangements of gp120 relative to gp41 on receptor activation of Env, that leads to the release of FP from its hydrophobic burial site at the gp41–gp120 interface (5, 18, 19). Subsequent dissociation of the gp120 subunits leaves the gp41 core in a state somewhat similar to the common cartoon models, lacking the trimer-stabilizing interactions supplied by gp120.
Although it seems clear that, initially, gp41 directly engages the viral and host cell membranes only by means of its TM and FP domains, there is evidence that, subsequently, the NHR region also interacts directly with the membranes and actively participates in the fusion process. In particular, the NHR-derived peptide, N36, binds to both zwitterionic and negatively charged phospholipid vesicles (20), whereas the N70 peptide, which encompasses the FP and NHR domains, is four times more fusogenic than FP alone for negatively charged membranes (21). The latter result suggests that the NHR segment takes an active role in destabilizing membranes and works synergistically with FP to increase the efficiency of lipid mixing. In another elegant set of experiments, Wexler-Cohen and Shai (14) showed that NHR-mimicking peptides, designed to interfere with formation of gp41’s 6HB state by competing with gp41 NHR insertion into the 6HB, have strongly increased inhibitory activity when they carry a membrane-anchoring alkyl chain. Increased inhibition is seen regardless of whether the alkyl chain is attached at the N or C terminus of the NHR peptide, suggesting that the gp41 NHR domain is embedded in the membrane surface. 6HB oligomers formed by NHR- and CHR-derived synthetic peptides dissociate in the presence of either zwitterionic or negatively charged phospholipid vesicles (20, 22). This lipid binding property has been postulated to facilitate membrane fusion by introducing an additional destabilization of the viral and target cell membranes, thereby lowering the free-energy barrier for fusion (23).
In the present study, we show that the 6HB complex formed by an ectodomain that contains large segments of the NHR and CHR helices, connected by a six-residue linker (CoreS), dissociates and forms stable monomers on binding to either dodecyl phosphocholine (DPC) micelles or phospholipid vesicles of a lipid composition that mimics the T-cell membrane. The transition from trimers to monomers is associated with a significant decrease in α-helicity and also observed for a longer ectodomain construct (CoreIL) that encompasses the native immunodominant loop (IL) connecting the NHR and CHR helices. The CoreS construct was chosen for detailed characterization of the structure and dynamics of the gp41 ectodomain monomer in the presence of DPC micelles. An atomic structure determination by NMR spectroscopy of the gp41 ectodomain monomer, based on residual dipolar coupling (RDC) and NOE restraints, reveals a monomeric, flexibly linked two-helical structure lying on the surface of the DPC micelle without any specific interaction between the stable and well-defined NHR and CHR helices. We propose that formation of this lipid-bound state, where CHR embeds in the viral membrane and NHR in the membrane of the host cell, provides the force for pulling the two membranes into close juxtaposition, thereby priming the system for membrane fusion. After fusion, close spatial proximity between the opposite ends of the ectodomain then permits their tight interaction, which is seen in 6HB crystal structures of the full-length gp41 ectodomain (9).
Results
Secondary Structure and Oligomeric State of gp41 Ectodomain.
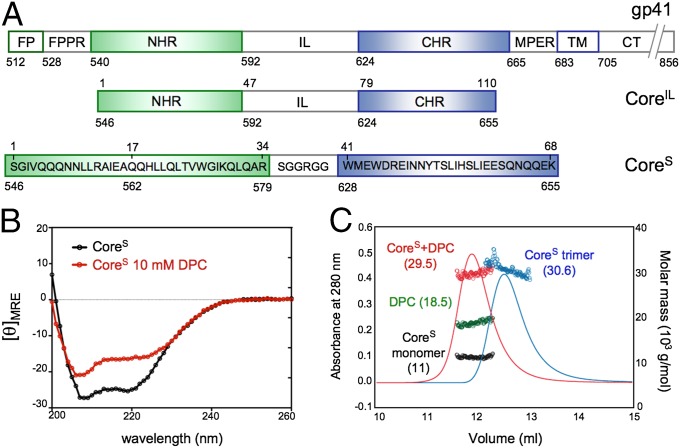
We expressed and purified a recombinant protein, CoreS, containing the NHR and CHR segments connected by a 6-residue linker (L6) (Fig. 1A), which is known to form a stable 6HB homotrimeric complex in aqueous solution (11). CD spectra of CoreS recorded at pH 4.0 show the characteristic signature of an α-helical protein with a deep minimum at 222 nm (Fig. 1B), corresponding to ca. 83% helical content. The addition of 10 mM DPC results in a 23% loss in helicity (Fig. 1B), indicating a substantial structural perturbation of CoreS on binding to the DPC micelle. The same change of the CD spectrum is observed when CoreS is mixed with dihexanoyl phosphatidylcholine (DHPC) micelles (Fig. S1A), which contrasts with virtually no change of the CD spectrum on addition of the detergents 3-([3-cholamidopropyl]dimethylammonio)-2-hydroxy-1-propanesulfonate (CHAPSO) (Fig. S1B) or lauryl maltose neopentyl glycol (MNG-3) (Fig. S1C), suggesting that the presence of phospholipid head groups is important for the binding of CoreS.
Fig. 1.
Sequence and properties of gp41. (A) Schematic representation of the gp41 sequence, including the FP, FPPR, NHR, IL, CHR, MPER, TM, and intraviral C-terminal domain (CT). The constructs used in the present study contain the NHR and CHR segments connected by either IL or L6. The numbering 512–704 refers to the Env precursor sequence, whereas the 1–68 numbering is used for CoreS. In addition, the CoreS sequence contains four extra residues (GSHM) at its N terminus, which correspond to an uncleaved fragment of the original tag (SI Materials and Methods). (B) CD spectra of CoreS, reported here as the mean residue ellipticity (103 degrees centimeter2 decimoles−1 residue−1), were recorded in the absence of detergent (black) and the presence of 10 mM DPC (red) at 310 K. (C) Molecular mass analysis of CoreS (including its N-terminal His-tag) (SI Materials and Methods) in the absence and presence of DPC as determined by SEC-MALS. The elution profiles monitored by the absorbance at 280 nm are shown for the CoreS trimer (blue, 30.6 ± 0.3 kDa) and a CoreS monomer bound to a DPC micelle (red, 29.5 ± 0.4 kDa), with the DPC micelle contribution in green (18.5 ± 0.4 kDa) and the CoreS monomer in black (11 ± 0.1 kDa).
CD spectra of CoreS were also recorded at pH 6.0, showing the same decrease in helicity on addition of DPC at pH 4.0 (Fig. S1E). Importantly, a very similar perturbation is observed when CoreS is mixed with vesicles known as LM3, which mimic the T-cell membrane lipid composition (24) (Fig. S1D).
Using size-exclusion chromatography coupled to multiangle light scattering, refractive index, and UV measurements (SEC-MALS), we find that the secondary structure perturbation described above is correlated with a change in the oligomeric state of CoreS. For a 5 μM protein solution in the absence of detergent, a single elution peak corresponding to the CoreS trimer (molecular mass = 30.6 kDa) is observed, which was expected for a stable 6HB trimer (Fig. 1C, blue trace). By contrast, in the presence of 10 mM DPC, the SEC-MALS data show an elution peak corresponding to monomeric CoreS (molecular mass = 11 kDa) bound to a DPC micelle (molecular mass = 18.5 kDa) (Fig. 1C, red trace). No elution peak corresponding to the trimeric form is observed under such conditions, indicating that an excess of DPC micelles completely shifts the equilibrium to the monomeric state of CoreS. SEC-MALS measurements were also performed for CoreS at pH 6.0, again showing a complete trimer-to-monomer transition in the presence of DPC and molecular masses for the trimer and the micelle-bound monomer very similar to the masses seen at pH 4.0 (Fig. S1F).
To exclude that the trimer-to-monomer transition observed for CoreS in the presence of DPC is a consequence of substituting the native immunodominant loop (IL) by L6, the same measurements were repeated for a construct that included IL instead of L6 (Fig. 1A). CD measurements on CoreIL show a similar decrease in helical content on addition of 10 mM DPC (Fig. S1 G and I), whereas the SEC-MALS data again indicate that the trimeric population of CoreIL undergoes a complete shift to a monomeric micelle-bound state in the presence of DPC (Fig. S1 H and J). These results, therefore, confirm that the trimer-to-monomer transition is a common property of both CoreIL and CoreS and not a simple consequence of the replacement of the IL region by a short linker.
Structure and Dynamics of the Trimeric and Monomeric States of CoreS.
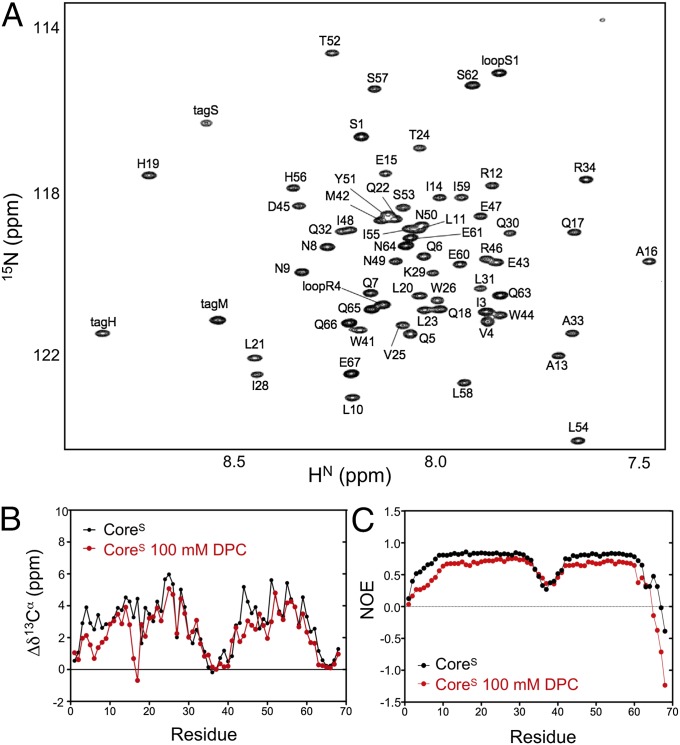
The structure and backbone dynamics of the trimeric and monomeric forms of CoreS were studied by solution NMR spectroscopy. In the absence of DPC, the 1H-15N TROSY-HSQC spectrum of CoreS at pH 4.0 presents all of the characteristics of a stably folded protein, with 68 well-dispersed amide chemical shifts and uniform resonance line widths, indicating that the trimer is C3-symmetric. The 1H-15N TROSY-HSQC spectrum recorded for the CoreS monomer in the presence of 100 mM DPC (Fig. 2A) also shows 68 well-dispersed amide resonances but with large chemical shift differences relative to the trimer (Table S1). 1H-15N TROSY-HSQC spectra, recorded at pH 6.0 in the absence and presence of 100 mM DPC, show the same characteristic chemical shift differences of the trimer-to-monomer transition (Fig. S2 A and B). 1H-15N TROSY-HSQC spectra were also recorded in the presence of LM3 vesicles at both pH 4.0 and 6.0 (Fig. S2 C and D), showing the disappearance of CoreS cross-peaks for all ordered residues in the slowly tumbling lipid-bound state; this observation confirmed that CoreS binds to these T cell-mimicking vesicles, while exhibiting resonances for the observable, dynamically disordered residues that fall close to the positions seen in the monomeric DPC-solubilized construct.
Fig. 2.
NMR characterization of CoreS at pH 4.0 and 310 K. (A) The most crowded region of the 1H-15N TROSY-HSQC spectrum in the presence of 100 mM DPC. Assignments are included in Table S1. (B) Comparison of the secondary 13Cα chemical shifts (ΔδCα) of CoreS in the absence (black) and presence (red) of 100 mM DPC (red). ΔδCα values represent the difference between the measured 13Cα chemical shifts and the temperature- and pH-corrected random coil values (25). (C) Steady state heteronuclear 15N-{1H} NOE values of CoreS (600 MHz 1H frequency) in the absence of detergent (black) and the presence of 100 mM DPC (red).
The secondary chemical shifts of the 13Cα, 13Cβ, and 13C′ nuclei, which refer to the difference between the observed chemical shifts and the corresponding residue-specific random coil values, are sensitive indicators of local secondary structure. With the exception of the two N-terminal and the six C-terminal residues, large positive 13Cα secondary chemical shifts are observed for the NHR and CHR regions in the absence of detergent, which is indicative of α-helical structure. The six residues composing the artificial linker between the NHR and CHR helices show chemical shifts close to random coil chemical shift values, indicating that this linker is dynamically disordered in solution (Fig. 2B). The 13Cα secondary chemical shifts measured for the CoreS monomer in the presence of 100 mM DPC also show large positive values for most of the CHR region, but significant differences relative to the shifts of the CoreS trimer are seen in the NHR segment, particularly for residues I3–N9, which exhibit decreased deviations from random coil values in the monomeric state (Fig. 2B). In addition, the very small and even negative values observed for A16 and Q17, respectively, point to the presence of a break in the NHR helix of the CoreS monomer.
Measurement of the 15N spin–lattice (R1) and spin–spin (R2) relaxation rates together with the heteronuclear 15N-{1H} NOE, which was carried out in both the absence and presence of 100 mM DPC, permits quantitative evaluation of the backbone dynamics of CoreS in the two states. With the exception of the eight N- and seven C-terminal residues, highly uniform NOE values (between 0.81 and 0.85) that fall close to their theoretical rigid limit are observed in the absence of DPC for both the NHR and CHR helices of CoreS (Fig. 2), indicating that they adopt a highly ordered conformation in the trimer. Uniform R2 and R1 relaxation rates of ca. 15 and 1 s−1, respectively (Fig. S3), correspond to a rotational correlation time of 9.9 ± 0.2 ns, close to the value expected for a 24.9 kDa globular protein. Addition of DPC resulted in a small but rather uniform decrease of the heteronuclear NOE (Fig. 2). Together with decreased R2 relaxation rates and increased R1 values (Fig. S3), these data indicate that both the NHR and CHR helices tumble more rapidly in the detergent-attached monomeric state. A model free analysis (26) of the 15N relaxation data additionally reveals increased internal motions for the NH vectors of the first nine N-terminal residues of the CoreS monomer, with S2 order parameters ranging between 0.4 and 0.7 (Fig. S3C and Table S2). Comparison of the R2 relaxation rates measured at 600 and 800 MHz (Fig. S3D) shows no evidence of slow (on the microsecond to millisecond timescale) conformational exchange. Therefore, any additional dynamics that may be present on a timescale slower than the overall molecular tumbling, which escapes the Lipari–Szabo relaxation analysis, must take place on a timescale faster than ca. 30 μs.
Comparison of the CoreS Trimer with 6HB Crystal Structures.
An X-ray structure of CoreS, crystallized in the absence of detergent (11), revealed the same 6HB structure seen for the ectodomain of other class I viral fusion proteins. RDCs are exquisitely sensitive probes for evaluating how close this structure resembles the structure present in aqueous solution. Therefore, we collected a nearly complete set of 1DNH, 1DNC′, 2DHNC′, and 1DCαC′ RDCs and fitted these couplings to the X-ray structure (Protein Data Bank ID code 1SZT) (11), yielding a Q factor of 0.274. Considering the limited crystallographic resolution at which the coordinates had been determined (2.4-Å resolution), this Q factor indicates good agreement with the crystal structure, with no obvious outliers or systematic differences (Fig. S4A). A higher-resolution crystal structure of CoreS, crystallized in the presence of n-octyl-β-glucopyranoside (Protein Data Bank ID code 1DF4; 1.45-Å resolution), had been interpreted as an alternative, micelle-bound conformation of the 6HB structure; however, the Cα coordinate rmsd between the 1SZT and 1DF4 structures is only modest (0.58 Å) (27). Interestingly, a fit of our experimental RDCs measured in the absence of detergent to the 1DF4 structure actually shows significantly better agreement, with a Q factor = 0.192 (Fig. S4B), than to the detergent-free structure. This result indicates that the fit of the experimental RDCs to 1SZT is principally limited by the accuracy of the atomic coordinates derived from a 2.4-Å map and that 1DF4 actually represents a more accurate coordinate representation of the detergent-free 6HB conformation. Importantly, these results confirm that CoreS in solution adopts the same trimeric 6HB structure seen in the crystalline state.
Structure of Monomeric CoreS.
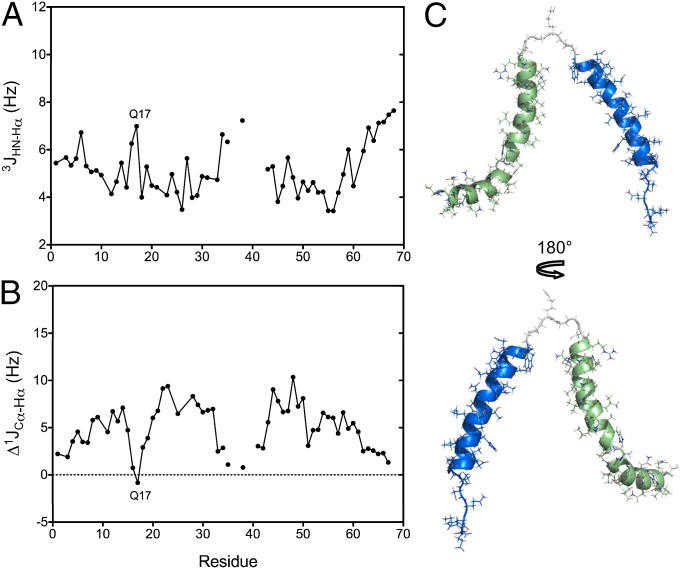
The structure of CoreS in the micelle-bound monomeric state was derived by simulated annealing calculations based on 465 sequential and short-range NOE distance restraints, 114 backbone dihedral angle restraints, and 157 backbone RDCs. The prediction of the backbone dihedral angles provided by the TalosN program (28) and based on the experimental 15N, 1HN, 1Hα, 13Cα, 13Cβ, and 13C′ chemical shifts was complemented by the measurements of 3JHN-Hα and 1JCα-Hα scalar couplings (Fig. 3). The two residues for which very small and even negative secondary 13Cα chemical shifts were measured, A16 and Q17 (Fig. 2B), also show nonhelical 3JHN-Hα couplings (6.3 and 7.0 Hz, respectively) (Fig. 3A) and reduced secondary 1JCα-Hα couplings (0.75 and −0.84 Hz, respectively) (Fig. 3B), confirming the nonhelical backbone torsion angles of these two residues. The somewhat extended conformations of A16 and Q17 result in a clear kink of the NHR helix (Fig. 3C), with the N-terminal segment at an angle of ca. 60° relative to the main axis of the helical segment, which is formed by residues 18–34. The CHR helix is largely preserved in the monomeric state, except for its eight most C-terminal residues. These C-terminal residues all have polar side chains, and their inability to engage the lipid surface in an α-helical conformation, therefore, is (not surprising) resulting in dynamic disorder. Analogously, five sequential polar residues near the N terminus, QQQNN, prevent lipid binding of this section of the NHR, also resulting in dynamic disorder and only transient helical character, which was judged by 13Cα secondary shifts and 3JHNHα and 1JCαHα couplings (Fig. 3).
Fig. 3.
CoreS in the presence of DPC. (A) 3JHN-Hα couplings and (B) secondary ∆1JCα−Hα values reporting on secondary structure. ∆1JCα−Hα is the difference between the measured 1JCα−Hα coupling and the residue-specific random coil value. Values measured for residues within the NHR and CHR segments are connected by solid lines for visual purposes. (C) Structure of CoreS in the micelle-bound monomeric state, with the NHR helix in green, the CHR in blue, and the flexible linker (residues 35–40) in gray. Although the fourfold degeneracy in the average relative orientations of NHR and CHR, intrinsic to RDC analysis (29), is broken by the requirement that both helices adhere with their lipophilic surface to the same micelle, their instantaneous relative orientation is subject to dynamic disorder. Without direct interhelical contacts, translationally, the relative position of the two helices also is ill-defined. For structural statistics, see Table S3.
Absence of NHR and CHR Interaction in the Monomeric State.
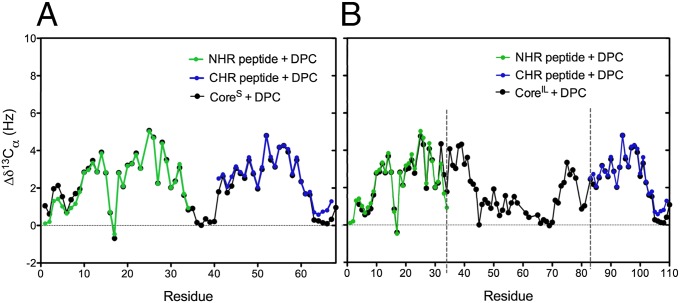
Although no interhelical interactions were observed in the 2D and 3D NOESY spectra of CoreS in the presence of DPC, weak transient interactions cannot be excluded a priori, because the corresponding NOEs would be notoriously difficult to detect. However, chemical shifts are exquisitely sensitive to even transient, weak interactions. To investigate the presence of potential weak interactions between NHR and CHR helices, we compared the chemical shifts observed for the 68-residue CoreS monomer with chemical shifts recorded for separately purified recombinant forms of the NHR and CHR peptides. CD spectra recorded in 50 mM sodium acetate (pH 4.0) at 310 K show that the NHR peptide is intrinsically disordered in the absence of detergent but adopts an α-helical conformation on addition of DPC (Fig. S5A). Comparison of the chemical shifts of CoreS in the presence of DPC with the shifts of the two separate peptide samples shows them to be essentially indistinguishable for not only 13Cα (Fig. 4A) but also, the amides 1HN (Fig. S5C) and 15N (Fig. S5D). The minor differences observed for the terminal regions of NHR and CHR reflect the presence of additional residues that extend the isolated NHR and CHR peptides, necessary for their isolation (SI Materials and Methods). Other than these minor differences, the very close correspondence between the chemical shifts measured for the isolated peptides and the micelle-associated CoreS indicates that the NHR–CHR interactions are completely disrupted in the monomeric state of CoreS. In addition, the very close correspondence between the 13Cα secondary chemical shifts measured for the two peptides and the shifts measured for CoreIL (Fig. 4B) suggests that the interhelical interactions are also disrupted in this longer construct. The absence of stabilizing interactions between the two helices together with the high flexibility of the interhelical linker suggest that the relative orientation and position on the lipid surface are subject to large dynamic disorder, with the structure depicted in Fig. 3C only representing an average view. Indeed, when immersed in anisotropically compressed acrylamide gel, the alignment strength of the larger N-terminal helix is found to be greater than for the shorter C-terminal helix, confirming their dynamic relative arrangement.
Fig. 4.
Comparison of the secondary 13Cα chemical shifts of the individual NHR (green) and CHR (blue) peptides with shifts of (A) CoreS (black) and (B) CoreIL [black; all in 50 mM sodium acetate (pH 4.0) in the presence of 100 mM DPC]. The individual NHR and CHR helix constructs contain additional residues at their N and C termini (SI Materials and Methods), respectively, that are not present in CoreS and presumed to be responsible for the small chemical shift differences near the termini, which seem dynamically disordered in the two peptides as well as in CoreS and CoreIL.
CoreS at the Phospholipid–Water Interface.
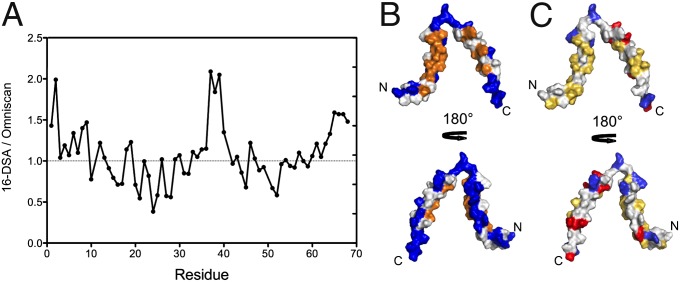
Paramagnetic relaxation enhancement has become a standard method to study the partitioning of peptides and proteins at the water–phospholipid interface. The solvent- and micelle-associated surfaces of CoreS in the monomeric state were identified by comparing the amide signal attenuation induced by two paramagnetic agents: 16-doxyl-stearic acid (16-DSA), which is confined to the hydrophobic interior of the DPC micelle, and gadodiamide (Omniscan), which remains free in solution. 1H-15N TROSY-HSQC spectra of CoreS with 100 mM DPC were recorded in the presence of either 2 mM 16-DSA or 2 mM Omniscan, and the attenuation profile of each amide group was calculated by comparing the cross-peak intensities in the presence and absence of paramagnetic agent (Table S2). The ratio between the attenuations induced by 16-DSA and Omniscan (16-DSA/Omniscan) (Fig. 5A) reveals which amide groups are more affected by Omniscan than 16-DSA (ratio > 1; therefore, closer to solvent) or micelle-exposed (ratio < 1). The largest and smallest 16-DSA/Omniscan ratios are marked on the structure of the CoreS monomer in blue and orange, respectively, in Fig. 5B, and they show a clear partitioning of the solvent- and micelle-exposed surfaces.
Fig. 5.
Probing of the interaction between monomeric CoreS and the phospholipid interface. (A) Ratios of attenuation induced by the paramagnetic agents, 16-DSA and Omniscan, as a function of residue number. The attenuation of each amide signal in the 1H-15N TROSY-HSQC spectra of CoreS with 100 mM DPC was independently determined on addition of either 2 mM 16-DSA or 2 mM Omniscan (Table S2). (B) Surface representation of the CoreS monomer structure, with the residues showing the largest 16-DSA/Omniscan ratios colored in blue (solvent-exposed) and the smallest ratios colored in orange (micelle-exposed). (C) Surface representation of the CoreS monomer colored on the basis of residue type (blue, positively charged; red, negatively charged; yellow, hydrophobic). The N and C termini are marked N and C, respectively.
Discussion
Before reaching the postfusion 6HB state, the NHR and CHR regions have the opportunity to interact with their adjacent membranes, and a growing body of evidence suggests that these heptad repeat regions may play an active role in destabilizing membranes by directly binding to the lipid bilayers (20–23). Our study shows that the 6HB trimeric structure of a recombinant ectodomain, lacking the membrane-interacting domains FP, FP proximal region (FPPR), membrane proximal external region (MPER), and TM, dissociates into stable monomers on binding to zwitterionic detergent micelles and behaves analogously in the presence of vesicles that mimic the T-cell membrane composition. Although the ability of gp41 constructs lacking the MPER and TM regions to induce lipid mixing and vesicle fusion has been shown to depend strongly on pH (30), we find that dissociation into monomers occurs both at pH 4.0 and 6.0. In the lipid-bound state, both the NHR and CHR helices are embedded at the water–lipid interface, thereby destabilizing their respective membranes and lowering the barrier for membrane fusion (31). The trimer-to-monomer transition introduces a significant kink in the NHR helix at residue Q562 (Q17 in CoreS numbering). The force needed to maintain the lipid-bound NHR in its kinked conformation can only be provided by its interaction with the lipids and therefore, must be accompanied by additional destabilization of the lipid interface, thereby also contributing to a lowering of the energy barrier associated with membrane fusion. In this respect, we note that, although the small degree of helical axis curvature observed for the NHR and CHR CoreS helices in our detergent-solubilized model system is likely to differ from any curvature present when these helices are embedded in the host cell and viral membranes, the kink in the micelle-bound NHR helix seems to be an essential attribute for its binding to a contiguous hydrophobic bilayer surface. This kink is necessary to avoid the polar side chain of residue Q562 from facing the bilayer interior, while allowing the hydrophobic side chains of L555, L556, and I559, which precede the kink, to engage the bilayer simultaneously with the side chains of L565, L566, V570, and I573.
Recently, we found that the CHR, MPER, and TM regions of a much longer gp41 construct (residues 512–705; see Fig. 1A) are subject to extensive conformational exchange processes in the presence of DPC (32). Although sedimentation equilibrium centrifugation and SEC-MALS data unambiguously showed that this gp41512–705 remains trimeric under such conditions, the chemical shifts of the NHR residues in gp41512–705 correlate much closer with the shifts of the CoreS monomer (R2 = 0.76 and 0.74 for the 1HN and secondary 13Cα chemical shifts, respectively) than with the CoreS trimer [R2 = 0.24 (δ1HN) and R2 = 0.55 (∆δ13Cα)] (Fig. S6). This paradox is resolved by recognizing that the membrane-associated TM region is responsible for retaining the trimeric state of gp41512–705, even in the absence of stable NHR–CHR interhelical interactions. The present CoreS data indicate that the affinity of the NHR and CHR segments for phospholipid surfaces is strong enough to break the thermodynamically very stable 6HB trimeric state and therefore, more than sufficient to disrupt the much weaker intermolecular NHR and potential CHR interactions in the initial extended prehairpin intermediate state (Fig. 6A), thereby rapidly transitioning to a collapsed state (Fig. 6B). This transition pulls the viral and host cell membranes closer to one another to a distance that is limited by the length of the IL (Fig. 1A). Considering that a significant segment of IL (residues I580–D589 after NHR and S618–T627 preceding CHR) also is lipophilic and α-helical (Fig. 4), the effective intermembrane distance likely is even shorter and also dependent on the oxidation state of the two Cys residues (C598 and C604) located in the nonlipophilic segment of IL.
Fig. 6.
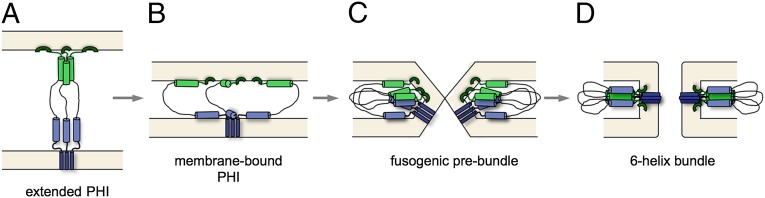
Model of the intermediate steps in gp41-driven fusion of the viral and target cell membranes showing the NHR (light green) and CHR (light blue) segments and the membrane-anchoring elements [FP (dark green) and TM (dark blue)] at four different stages of the fusion process. (A) The short-lived extended prehairpin intermediate (PHI) state, where both TM and NHR are presumed responsible for maintaining the trimeric nature. (B) The collapsed PHI state, where NHR and CHR have become embedded in the viral and host cell membranes, thereby pulling the membranes into juxtaposition. (C) Formation of fusogenic prebundles, which is possibly initiated by contacts between the short polar segments at opposing ends of the NHR and CHR. (D) Formation of mature, postfusion 6HB trimers, which are stabilized by FP–TM, FPPR–MPER, and 6HB NHR–CHR interactions.
Destabilization of the viral and target cell membranes introduced by the heptad helices and the FP in the collapsed prehairpin intermediate state (Fig. 6B) coupled with their spatial proximity then creates a state conducive to the formation of a hemifusion stalk, in which the outer leaflets of the viral and host cell membranes are fused (31), and which can progress to formation of a small fusion pore (Fig. 6C). Taking advantage of the temperature dependence of the fusion pore growth, Markosyan et al. (33) have shown that folding of the 6HB is not complete until the very late stage of pore formation. Based on this kinetic argument, we propose that the initial formation of fusion pores is driven by the association of gp41 trimers into prebundle complexes (Fig. 6C). Formation of these complexes then depends on a competition between intermolecular association of the NHR and CHR helices, including their FPPR and MPER extensions, and membrane binding of these lipophilic regions (32, 34). Bundling of MPER with FPPR residues stabilizes the 6HB state (9) but only becomes kinetically accessible after these regions have progressed to a state of close spatial proximity (Fig. 6C). We speculate that formation of this trimeric state may be initiated by interactions between the polar segments of the NHR (S546–N554) and CHR (E647–K655) regions, which lack high membrane affinity but make tight and specific interhelical contacts in the 6HB. Specific interactions between the FP and TM (35), which are only accessible after formation of the fusogenic prebundle, may further stabilize formation of the postfusion state. Competition between intermolecular and membrane association may also be impacted by a shift in lipid composition after outer leaflet lipids of the viral and host cells can mix with one another by translational diffusion through the hemifusion stalk, which would be a slow and strongly temperature-dependent process.
In our model, the fusogenic prebundle complexes, comprising both membrane- and self-associated heptad regions, represent the actual target for the peptides used for fusion inhibition (15, 16, 36). Such long-lived prebundle conformations would also represent an ideal target for the membrane-conjugated class of inhibitory NHR- and CHR-mimicking peptides (37–39) as well as neutralizing antibodies, which can tightly engage incomplete states of the 6HB core (40, 41).
Materials and Methods
CoreS, identical in sequence to the N34-L6-C28 construct described in ref. 11, was expressed with an N-terminal His-tag to aid its purification. The N-terminal nonnative residues were removed by thrombin cleavage, with the exception of four residues (GSHM) remaining at the N terminus of the CoreS sequence, which was followed by size exclusion chromatography under denaturing conditions and reverse-phase HPLC.
NMR measurements were carried out at 500, 600, 800, and 900 MHz on uniformly 2H/15N/13C-, 15N/13C-, and 2H/15N-enriched samples at protein concentrations of ca. 0.5 mM (monomer) in both the absence and presence of 100 mM DPC. The NMR structure of the CoreS monomer was calculated using NOE distance restraints, RDCs, and TalosN dihedral restraints (28) using X-PLOR-NIH v2.34 (42). Details are in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Annie Aniana for help with protein expression and purification and Robert Blumenthal and Leonid Chernomordik for helpful discussions, and we acknowledge support from the National Institute of Diabetes and Digestive and Kidney Diseases MS facility. This work was funded by the National Institutes of Health Intramural Research Programs of the National Institute of Diabetes and Digestive and Kidney Diseases and the Intramural AIDS-Targeted Antiviral Program of the Office of the Director, National Institutes of Health.
Footnotes
The authors declare no conflict of interest.
Data deposition: The NMR, atomic coordinates, chemical shifts, and restraints have been deposited in the Protein Data Bank, www.pdb.org (PDB ID code 2MK3).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1401397111/-/DCSupplemental.
References
- 1.Harrison SC. Viral membrane fusion. Nat Struct Mol Biol. 2008;15(7):690–698. doi: 10.1038/nsmb.1456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Roux KH, Taylor KA. AIDS virus envelope spike structure. Curr Opin Struct Biol. 2007;17(2):244–252. doi: 10.1016/j.sbi.2007.03.008. [DOI] [PubMed] [Google Scholar]
- 3.Merk A, Subramaniam S. HIV-1 envelope glycoprotein structure. Curr Opin Struct Biol. 2013;23(2):268–276. doi: 10.1016/j.sbi.2013.03.007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Julien JP, et al. Crystal structure of a soluble cleaved HIV-1 envelope trimer. Science. 2013;342(6165):1477–1483. doi: 10.1126/science.1245625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Lyumkis D, et al. Cryo-EM structure of a fully glycosylated soluble cleaved HIV-1 envelope trimer. Science. 2013;342(6165):1484–1490. doi: 10.1126/science.1245627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Furuta RA, Wild CT, Weng YK, Weiss CD. Capture of an early fusion-active conformation of HIV-1 gp41. Nat Struct Biol. 1998;5(4):276–279. doi: 10.1038/nsb0498-276. [DOI] [PubMed] [Google Scholar]
- 7.Gallo SA, et al. The HIV Env-mediated fusion reaction. Biochim Biophys Acta. 2003;1614(1):36–50. doi: 10.1016/s0005-2736(03)00161-5. [DOI] [PubMed] [Google Scholar]
- 8.Blumenthal R, Durell S, Viard M. HIV entry and envelope glycoprotein-mediated fusion. J Biol Chem. 2012;287(49):40841–40849. doi: 10.1074/jbc.R112.406272. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Buzon V, et al. Crystal structure of HIV-1 gp41 including both fusion peptide and membrane proximal external regions. PLoS Pathog. 2010;6(5):e1000880. doi: 10.1371/journal.ppat.1000880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Chan DC, Fass D, Berger JM, Kim PS. Core structure of gp41 from the HIV envelope glycoprotein. Cell. 1997;89(2):263–273. doi: 10.1016/s0092-8674(00)80205-6. [DOI] [PubMed] [Google Scholar]
- 11.Tan KM, Liu JH, Wang JH, Shen S, Lu M. Atomic structure of a thermostable subdomain of HIV-1 gp41. Proc Natl Acad Sci USA. 1997;94(23):12303–12308. doi: 10.1073/pnas.94.23.12303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weissenhorn W, Dessen A, Harrison SC, Skehel JJ, Wiley DC. Atomic structure of the ectodomain from HIV-1 gp41. Nature. 1997;387(6631):426–430. doi: 10.1038/387426a0. [DOI] [PubMed] [Google Scholar]
- 13.Weissenhorn W, et al. Structural basis for membrane fusion by enveloped viruses. Mol Membr Biol. 1999;16(1):3–9. doi: 10.1080/096876899294706. [DOI] [PubMed] [Google Scholar]
- 14.Wexler-Cohen Y, Shai Y. Membrane-anchored HIV-1 N-heptad repeat peptides are highly potent cell fusion inhibitors via an altered mode of action. PLoS Pathog. 2009;5(7):e1000509. doi: 10.1371/journal.ppat.1000509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Eggink D, Berkhout B, Sanders RW. Inhibition of HIV-1 by fusion inhibitors. Curr Pharm Des. 2010;16(33):3716–3728. doi: 10.2174/138161210794079218. [DOI] [PubMed] [Google Scholar]
- 16.Ashkenazi A, Shai Y. Insights into the mechanism of HIV-1 envelope induced membrane fusion as revealed by its inhibitory peptides. Eur Biophys J. 2011;40(4):349–357. doi: 10.1007/s00249-010-0666-z. [DOI] [PubMed] [Google Scholar]
- 17.Garg H, Viard M, Jacobs A, Blumenthal R. Targeting HIV-1 gp41-induced fusion and pathogenesis for anti-viral therapy. Curr Top Med Chem. 2011;11(24):2947–2958. doi: 10.2174/156802611798808479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bartesaghi A, Merk A, Borgnia MJ, Milne JLS, Subramaniam S. Prefusion structure of trimeric HIV-1 envelope glycoprotein determined by cryo-electron microscopy. Nat Struct Mol Biol. 2013;20(12):1352–1357. doi: 10.1038/nsmb.2711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tran EEH, et al. Structural mechanism of trimeric HIV-1 envelope glycoprotein activation. PLoS Pathog. 2012;8(7):e1002797. doi: 10.1371/journal.ppat.1002797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Korazim O, Sackett K, Shai Y. Functional and structural characterization of HIV-1 gp41 ectodomain regions in phospholipid membranes suggests that the fusion-active conformation is extended. J Mol Biol. 2006;364(5):1103–1117. doi: 10.1016/j.jmb.2006.08.091. [DOI] [PubMed] [Google Scholar]
- 21.Sackett K, Shai Y. The HIV-1 gp41 N-terminal heptad repeat plays an essential role in membrane fusion. Biochemistry. 2002;41(14):4678–4685. doi: 10.1021/bi0255322. [DOI] [PubMed] [Google Scholar]
- 22.Kliger Y, Peisajovich SG, Blumenthal R, Shai Y. Membrane-induced conformational change during the activation of HIV-1 gp41. J Mol Biol. 2000;301(4):905–914. doi: 10.1006/jmbi.2000.4004. [DOI] [PubMed] [Google Scholar]
- 23.Lev N, et al. Conformational stability and membrane interaction of the full-length ectodomain of HIV-1 gp41: Implication for mode of action. Biochemistry. 2009;48(14):3166–3175. doi: 10.1021/bi802243j. [DOI] [PubMed] [Google Scholar]
- 24.Aloia RC, Tian HR, Jensen FC. Lipid composition and fluidity of the human immunodeficiency virus envelope and host cell plasma membranes. Proc Natl Acad Sci USA. 1993;90(11):5181–5185. doi: 10.1073/pnas.90.11.5181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Kjaergaard M, Brander S, Poulsen FM. Random coil chemical shift for intrinsically disordered proteins: Effects of temperature and pH. J Biomol NMR. 2011;49(2):139–149. doi: 10.1007/s10858-011-9472-x. [DOI] [PubMed] [Google Scholar]
- 26.Lipari G, Szabo A. Model-free approach to the interpretation of nuclear magnetic resonance relaxation in macromolecules. 1. Theory and range of validity. J Am Chem Soc. 1982;104(17):4546–4559. [Google Scholar]
- 27.Shu W, Ji H, Lu M. Interactions between HIV-1 gp41 core and detergents and their implications for membrane fusion. J Biol Chem. 2000;275(3):1839–1845. doi: 10.1074/jbc.275.3.1839. [DOI] [PubMed] [Google Scholar]
- 28.Shen Y, Bax A. Protein backbone and sidechain torsion angles predicted from NMR chemical shifts using artificial neural networks. J Biomol NMR. 2013;56(3):227–241. doi: 10.1007/s10858-013-9741-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Al-Hashimi HM, et al. Variation of molecular alignment as a means of resolving orientational ambiguities in protein structures from dipolar couplings. J Magn Reson. 2000;143(2):402–406. doi: 10.1006/jmre.2000.2049. [DOI] [PubMed] [Google Scholar]
- 30.Sackett K, TerBush A, Weliky DP. HIV gp41 six-helix bundle constructs induce rapid vesicle fusion at pH 3.5 and little fusion at pH 7.0: Understanding pH dependence of protein aggregation, membrane binding, and electrostatics, and implications for HIV-host cell fusion. Eur Biophys J. 2011;40(4):489–502. doi: 10.1007/s00249-010-0662-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Chernomordik LV, Kozlov MM. Protein-lipid interplay in fusion and fission of biological membranes. Annu Rev Biochem. 2003;72(2003):175–207. doi: 10.1146/annurev.biochem.72.121801.161504. [DOI] [PubMed] [Google Scholar]
- 32.Lakomek NA, et al. Internal dynamics of the homotrimeric HIV-1 viral coat protein gp41 on multiple time scales. Angew Chem Int Ed Engl. 2013;52(14):3911–3915. doi: 10.1002/anie.201207266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Markosyan RM, Cohen FS, Melikyan GB. HIV-1 envelope proteins complete their folding into six-helix bundles immediately after fusion pore formation. Mol Biol Cell. 2003;14(3):926–938. doi: 10.1091/mbc.E02-09-0573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sun Z-YJ, et al. HIV-1 broadly neutralizing antibody extracts its epitope from a kinked gp41 ectodomain region on the viral membrane. Immunity. 2008;28(1):52–63. doi: 10.1016/j.immuni.2007.11.018. [DOI] [PubMed] [Google Scholar]
- 35.Reuven EM, et al. HIV-1 gp41 transmembrane domain interacts with the fusion peptide: Implication in lipid mixing and inhibition of virus-cell fusion. Biochemistry. 2012;51(13):2867–2878. doi: 10.1021/bi201721r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kilby JM, Eron JJ. Novel therapies based on mechanisms of HIV-1 cell entry. N Engl J Med. 2003;348(22):2228–2238. doi: 10.1056/NEJMra022812. [DOI] [PubMed] [Google Scholar]
- 37.Hildinger M, et al. Membrane-anchored peptide inhibits human immunodeficiency virus entry. J Virol. 2001;75(6):3038–3042. doi: 10.1128/JVI.75.6.3038-3042.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Melikyan GB, Egelhofer M, von Laer D. Membrane-anchored inhibitory peptides capture human immunodeficiency virus type 1 gp41 conformations that engage the target membrane prior to fusion. J Virol. 2006;80(7):3249–3258. doi: 10.1128/JVI.80.7.3249-3258.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Hollmann A, Matos PM, Augusto MT, Castanho MARB, Santos NC. Conjugation of cholesterol to HIV-1 fusion inhibitor C34 increases peptide-membrane interactions potentiating its action. PLoS One. 2013;8(4):e60302. doi: 10.1371/journal.pone.0060302. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Golding H, et al. Dissection of human immunodeficiency virus type 1 entry with neutralizing antibodies to gp41 fusion intermediates. J Virol. 2002;76(13):6780–6790. doi: 10.1128/JVI.76.13.6780-6790.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gustchina E, et al. Structural basis of HIV-1 neutralization by affinity matured Fabs directed against the internal trimeric coiled-coil of gp41. PLoS Pathog. 2010;6(11):e1001182. doi: 10.1371/journal.ppat.1001182. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schwieters CD, Kuszewski JJ, Tjandra N, Clore GM. The Xplor-NIH NMR molecular structure determination package. J Magn Reson. 2003;160(1):65–73. doi: 10.1016/s1090-7807(02)00014-9. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.