Significance
Noncovalent interactions between proteins modulate their functions and occur widely in biological regulation. A large proportion of such regulatory proteins are homo-oligomeric, with multiple copies of a single polypeptide assembled into higher-order quaternary structures. Understanding the regulatory interactions between homo-oligomeric proteins is difficult because their periodic structural configuration may allow different modes of interaction with differing functions. We apply a powerful combination of analytical techniques to study the interaction between TRAP (trp RNA-binding attenuation protein), an 11-mer that regulates tryptophan metabolism by binding RNA, and its trimeric inhibitor protein anti-TRAP. We show that anti-TRAP condenses multiple TRAP oligomers into heterocomplexes, thereby blocking TRAP’s RNA-binding sites. These findings and our approach may have broad implications for other oligomeric regulatory proteins.
Keywords: small-angle X-ray scattering, minimal ensembles, native mass spectrometry, NMR, reversible oligomerization
Abstract
The control of tryptophan production in Bacillus is a paradigmatic example of gene regulation involving the interplay of multiple protein and nucleic acid components. Central to this combinatorial mechanism are the homo-oligomeric proteins TRAP (trp RNA-binding attenuation protein) and anti-TRAP (AT). TRAP forms undecameric rings, and AT assembles into triskelion-shaped trimers. Upon activation by tryptophan, the outer circumference of the TRAP ring binds specifically to a series of tandem sequences present in the 5′ UTR of RNA transcripts encoding several tryptophan metabolism genes, leading to their silencing. AT, whose expression is up-regulated upon tryptophan depletion to concentrations not exceeding a ratio of one AT trimer per TRAP 11-mer, restores tryptophan production by binding activated TRAP and preventing RNA binding. How the smaller AT inhibitor prevents RNA binding at such low stoichiometries has remained a puzzle, in part because of the large RNA-binding surface on the tryptophan-activated TRAP ring and its high affinity for RNA. Using X-ray scattering, hydrodynamic, and mass spectrometric data, we show that the polydentate action of AT trimers can condense multiple intact TRAP rings into large heterocomplexes, effectively reducing the available contiguous RNA-binding surfaces. This finding reveals an unprecedented mechanism for substoichiometric inhibition of a gene-regulatory protein, which may be a widespread but underappreciated regulatory mechanism in pathways that involve homo-oligomeric or polyvalent components.
The tryptophan (Trp)-bound trp RNA-binding attenuation protein (TRAP) directly affects the transcription and translation of several genes responsible for Trp metabolism by binding with nanomolar affinity to tandem (G/U)AG triplet repeats (one per TRAP subunit) in the 5′ UTR of several RNA transcripts (Fig. 1A) (1–3). Transcriptional regulation of the trpECDFBA operon is achieved by the interaction of Trp–TRAP with the 5′ UTR of a nascent transcript, which results in the formation of an RNA terminator stem–loop structure and an abortive hypertranslocation of the RNA polymerase (4). In addition, the trpE gene and at least three other genes are subject to translational regulation by TRAP through sequestration of the transcript’s Shine–Dalgarno sequence upon TRAP binding (Fig. S1) (5).
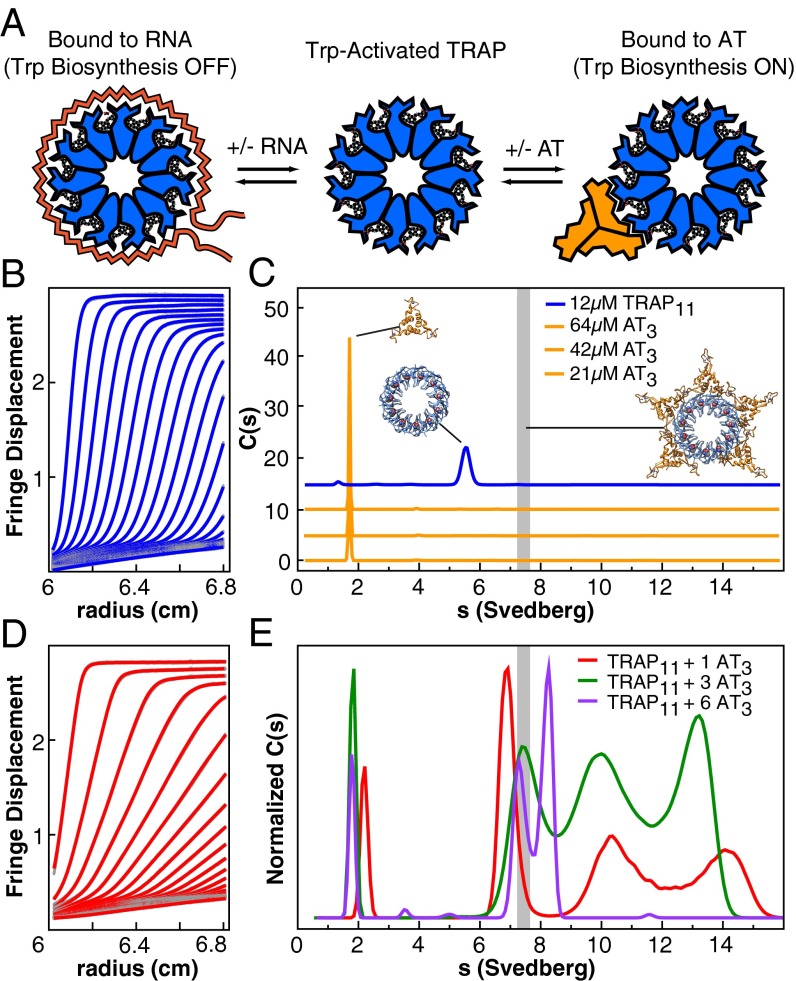
Fig. 1.
In B. subtilis the oligomeric proteins TRAP and AT integratively regulate gene expression through interactions with RNA, Trp, and each other; analytical ultracentrifugation helps describe these interactions. (A) Trp binding activates TRAP for binding to RNA, leading to transcriptional attenuation and translational repression. AT competes with RNA for Trp-activated TRAP, thereby relieving repression. (B) Fringe displacement traces from sedimentation velocity experiments of TRAP alone, illustrating a narrow boundary and homogeneous sedimentation. (C) Continuous sedimentation coefficient distributions for TRAP and AT. A range of concentrations is dominated by single species for each component. Cartoons reflect the AT3 NMR structure, the crystal structure of TRAP11, and a model of 5AT3 bound to TRAP11. The vertical gray bar represents the predicted sedimentation coefficient for the putative TRAP11–5AT3 complex. (D) Fringe displacement traces recorded at an AT3:TRAP11 ratio of 1 reveal the presence of heterogeneous, rapidly sedimenting species. (E) Sedimentation coefficient distributions for samples assembled at AT3:TRAP11 ratios of 1, 3, and 6. Multiple species and/or reaction boundaries are observed. Experiments were performed at pH 7; sedimentation experiments at pH 8 are shown in Fig. S3.
Anti-TRAP (AT) relieves TRAP-mediated repression of the trp operon expression by binding directly to Trp-bound activated TRAP and excluding RNA binding (Fig. 1A) (6, 7). AT expression is up-regulated in response to Trp depletion through sensing of elevated levels of uncharged tRNATrp via a T box anti-termination mechanism (6, 8). Quantitative Western blotting of cell lysates at conditions mimicking Trp starvation indicate that AT is maximally expressed to fewer than two AT3 per TRAP11 and that at these substoichiometric ratios AT is maximally able to restore trpECDFBA operon transcription to 70–80% that of TRAP-free levels (9). Order-of-addition experiments demonstrate that AT inhibition of the TRAP–RNA interaction is determined kinetically, because AT can block RNA binding if present before incubation with RNA but cannot compete effectively with RNA in a preformed complex (7).
Structural and oligomeric asymmetries have complicated the understanding of how AT inhibits the interaction of TRAP with RNA. AT comprises an N-terminal zinc-binding domain and a C-terminal helix, which serves as the assembly point for three protomers into a trimer, AT3, via a helical bundle (10). At physiological pH, AT3 further associates into a tetramer of trimers, (AT3)4, via ion pairs involving the protonated N terminus of the protein and conserved aspartate carboxylates (6, 11, 12). Because it is the trimeric form of AT that is competent for Trp–TRAP binding (13), this equilibrium underlies a means for oligomerization-mediated pH sensing (11). Complications arising from the oligomeric and stoichiometric asymmetry between TRAP11 and AT3 were overcome partially by crystallization of TRAP in a dodecameric form, TRAP12 (13). In this form, density was observed for six AT3 circumscribing the RNA-binding surface of TRAP, making extensive contacts to two adjacent Trp-bound TRAP protomers via one of the AT3 protomers (13), as is consistent with predictions from mutagenesis experiments (7, 14).
The available structural models of the TRAP–AT complex have remained insufficient to clarify crucial mechanistic matters such as the observation that substoichiometric ratios of AT3 are sufficient to inhibit the RNA-binding activity of TRAP11 in vivo (9, 15). The affinity of Trp–TRAP for the full-length trp leader RNA (11 G/UAG repeats) has been shown by filter-binding assays to be less than 1 nM (16, 17), whereas RNA binding to only six contiguous binding sites is sufficient to approach this affinity (16). However, the affinity of AT for TRAP appears to be orders of magnitude weaker (apparent Kd ∼1 µM) (Fig. S2) but is comparable to the cellular concentration of TRAP in Bacillus subtilis, which has been estimated at 0.5 µM based on 300 TRAP rings per cell (18) and a volume of 10−15 L (19). Analytical ultracentrifugation of mixtures of TRAP and AT has resulted in confusing and contradictory interpretations of their interaction (12, 13). In one case, sedimentation data suggested that AT and TRAP form large aggregates (12), with the largest occurring at low AT3:TRAP11 ratios, but a second study under alkaline conditions suggested much smaller complexes, consistent with one or more AT3s bound to a single TRAP oligomer (13). Interpretation of these results is complicated further by the effect of pH upon the 4AT3 ↔ (AT3)4 equilibrium.
Using small-angle X-ray scattering (SAXS), analytical ultracentrifugation, and native mass spectrometry, we show that the polydentate action of trimeric AT can reversibly condense multiple intact TRAP rings into large heterocomplexes, effectively reducing the available contiguous RNA-binding surfaces. This finding reveals an unprecedented mechanism for substoichiometric inhibition of a gene-regulatory protein, which may turn out to be a widespread regulatory mechanism in pathways that involve homo-oligomeric or polyvalent components.
Results
Heterocomplex Assembly and Disassembly.
To resolve these structural and mechanistic discrepancies, we used N-formyl AT (ƒAT), which strongly favors its trimeric form because of the loss of an ion pair involving the N terminus (11). This strategy allows us to examine the interaction of AT with TRAP in the absence of the complicating 4AT3 ↔ (AT3)4 equilibrium. Lamm equation modeling (20) of data obtained from sedimentation velocity experiments on ƒAT and TRAP revealed that, over a range of concentrations, the free components sediment as >95% single species with sedimentation coefficients of 1.7 S for AT and 5.4 S for TRAP (Fig. 1 B and C). These values are consistent with those observed previously for AT3 and TRAP11 (12) and are reasonably close to those predicted by hydrodynamic considerations (21) from their crystal structures (2.1 S and 6.3 S, respectively).
The sedimentation coefficient distributions, c(s), for Trp–TRAP with increasing ratios of ƒAT revealed a dramatic and discontinuous dependence on component concentrations at neutral pH, whereas controls performed at higher pH (Fig. S3 A–D) replicated previous results (11, 13). When mixed at a ratio of one AT3 per TRAP11 under conditions in large excess of the apparent Kd (i.e., 120 μM TRAP), four distinct sedimenting species are observed, with S values of 2.2, 6.9, 10.3, and 14.1 (Fig. 1 D and E). We interpret the broad peaks at 10.3 and 14.1 S as corresponding to large, heterogeneous TRAP-AT complexes, because they are not present in sedimentation data of the isolated components. The sharp peak at 2.2 S is assigned to the reaction boundary of AT trimers shifted to a larger apparent s resulting from the fast exchange with more rapidly sedimenting components; such a shift is diagnostic for rapidly interacting macromolecules (20, 22). Similarly, we interpret the species sedimenting at 6.9 S to be the reaction boundary of TRAP11, demonstrating a greater apparent sedimentation rate because of rapid exchange with the larger complexes. Although species with very large s are observed at low AT:TRAP ratios, increasing the ratio results in decreased overall s values for the TRAP–ƒAT complexes. Experiments performed with deformyl-AT and TRAP produced comparable but more convoluted results (Fig. S3E). When mixed at an AT3:TRAP11 ratio of 3, four sedimenting species also are resolved, but with reduced sedimentation coefficients. The proportion of the c(s) peak from the free AT reaction boundary increases and demonstrates a reduced shift toward greater sedimentation rates, indicating either less occupancy in larger complexes or reduced complex size. Because significant free AT is evident in the data, broad peaks at 7.5, 10, and 13 S likely represent complexes with varying numbers of TRAP11 rings with bound AT3. At an AT3:TRAP11 ratio of 6, only three predominant species remain, at 1.7, 7.3, and 8.2 S.
The sedimentation data are insufficiently precise to allow determination of exact stoichiometries of TRAP–AT complexes, but we compared the observed and predicted sedimentation coefficients for several modeled TRAP11–AT3 configurations. We used rigid-body transformations to model complexes of TRAP11 bound to varying numbers of AT3 using as templates the available structures of the components and the crystal structure of the Bacillus stearothermophilus TRAP12–6AT3 complex (13). The predicted sedimentation coefficients from hydrodynamic modeling (21) for fully saturated TRAP11–5AT3 and TRAP12–6AT3 complexes were 7.4 and 8.0S, respectively, far smaller than those observed experimentally. These analyses indicate that the rapidly sedimenting species cannot be explained by the binding of multiple AT3 to single TRAP rings and must be composed of higher-order TRAP-AT oligomers.
Heterocomplex Structural Comparisons.
To interrogate the oligomeric structure of the TRAP–AT complexes, we recorded SAXS (23) profiles from solutions of TRAP, ƒAT, and their mixtures. Guinier plots of scattering profiles obtained from solutions of the individual components were linear at low scattering angles, indicating monodisperse solutions and little or no aggregation, even at 10-mg/mL concentrations (Fig. 2A). Calculated ab initio scattering envelopes (24) for TRAP11 and AT3 on their own (Fig. 2B) accurately recapitulate the available high-resolution models of TRAP11 and AT3 obtained by conventional crystallographic and NMR methods (10, 25). The scattering envelopes generated for the free components then were used to deconvolute SAXS data recorded on their mixtures. Guinier plots of scattering profiles recorded from solutions at low AT3:TRAP11 ratios were nonlinear (Fig. 2C), revealing significant heterogeneity, although the feature-rich scattering profiles themselves are inconsistent with nonspecific aggregation (23). Pairwise electron distribution functions, P(r), computed from the scattering profiles demonstrated a peak near 50 Å, with the largest calculated radius of gyration (RG) and maximal scattering distance (Dmax) values observed at ratios of one AT3 per TRAP11. These curves exhibit the clearly discernable “ski slope” characteristic of elongated particles (Fig. 2D). Dmax was found to be relatively constant at 250 Å and to be independent of total protein concentration. Even at the lowest tested concentrations, the average RG at an AT3:TRAP11 ratio of 1 was significantly higher than predicted from hydrodynamic data for complexes containing a single TRAP. At AT3:TRAP11 ratios in excess of 1, Dmax and RG decreased steadily with increasing AT3, regardless of TRAP concentration (Fig. 2F). These results are consistent with large sedimenting species observed in area under the curve (AUC) experiments, indicating that large heterocomplexes are populated at low AT3:TRAP11 ratios and are titrated out as the AT:TRAP ratio increases.
Fig. 2.
Solution SAXS describes the size and shape of oligomers composed of TRAP, AT, and their complexes. (A) Experimental and fitted scattering profiles of 3 mg/mL AT (yellow) and TRAP (blue); lines are fits calculated from averaged envelopes shown in B. (Inset) Guinier plots with linear fits. (B) Ab initio GASBOR reconstructions of TRAP (Left) and AT (Right) determined assuming axially symmetric 11- and 3-mers, respectively. (C) Experimental SAXS profiles obtained from samples with a range of AT3:TRAP11 ratios; solid lines represent regularized GNOM fits. (Inset) Guinier plots demonstrate deviation from linearity at low AT:TRAP ratios. (D) Electron pairwise distribution plots of data in C; data show large (>200 Å) intraparticle distances at low AT3:TRAP11 ratios; structures become more compact at higher ratios. (E) Scattering profile obtained from a 1:1 mixture of AT3:TRAP11 (10 mg/mL TRAP) cannot be fit by a weighted sum of all possible single TRAP11–nAT3 models and free AT3 (blue). A much better fit generated from predicted scattering curves of the minimal ensemble complexes is depicted below the inset (red). (Inset) Fit deviations from experimental data, with goodness-of-fit χ2 values. (F) Rg and Dmax (Inset) as determined from GNOM fits to experimental data at various AT3:TRAP11 ratios and TRAP concentrations. The horizontal dashed line represents the Rg predicted from the model of a TRAP11–5AT3 complex. Data demonstrate that Dmax and Rg are largest at low ratios and decrease at high saturating concentrations of AT3. Vertical shading indicates range of AT3:TRAP11 ratios detected in cells.
To model the structures of the observed TRAP–AT complexes, we generated all sterically allowed configurations of multiple (one to five) AT3 molecules bound to a single TRAP11, guided by the structure of the TRAP12–6AT3 complex (13) and computed their theoretical scattering profiles using the program CRYSOL (Fig. S4C) (26). The scattering curves were used to reconstruct the experimental SAXS profiles by fitting their variably weighted sum to the experimental data using the program OLIGOMER (27). Fits to experimental data at AT3:TRAP11 ratios >3 using these models were quite good (Fig. S4B), suggesting that at these ratios single TRAP11 rings decorated with multiple AT3 are the primary species scattering X-rays in solution. However, fits to experimental data at low AT3:TRAP11 ratios (0.5–2.5) were quite poor (Fig. 2E and Fig. S4A), with the goodness-of-fit metric χ2 >100, as implied by the difference between the experimental Dmax of 250 Å and the Dmax of ∼150 Å for multiple AT3s around one TRAP11.
AT Flexibility Enables Polydentate Binding.
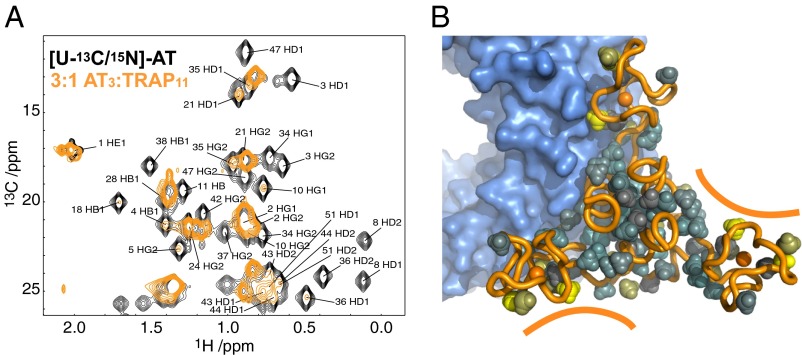
Because complexes comprised of single TRAP rings bound to variable numbers of AT3 could not explain the large apparent molecular weights implied by the AUC and SAXS data, we considered how larger oligomeric structures might be formed. Titrations of [U-13C/15N]-AT with unlabeled TRAP monitored by 2D 1H-13C correlation spectra revealed that at low AT:TRAP ratios the resonances from AT were severely broadened, consistent with large oligomeric structures. At high AT:TRAP ratios, resonance broadening was more selective (Fig. 3), with the resonances least broadened by TRAP binding corresponding to those at the extreme ends of each of the zinc-binding domains that otherwise are expected to interact with TRAP rings. These data suggested that when an AT3 interacts with a single TRAP ring, one AT protomer interacts directly with TRAP and the other two protomers in the trimer remain flexible and thus are able to present two additional TRAP-binding sites. This recognition led us to the remarkable hypothesis that AT could function by chaining together multiple TRAP rings.
Fig. 3.
A homo-trimer of AT, AT3, presents three binding sites for up to three TRAP rings. (A) 2D 1H-13C HMQC spectra recorded of [U-13C/15N]-AT in the absence (black) and presence of limiting TRAP (3:1 AT3:TRAP11) (orange). AT3 methyl resonance assignments are indicated. Because of the large size and asymmetry of the AT3–TRAP11 complex, methyl signals that persist through the titration are interpreted as being flexible and away from the TRAP–AT interface. (B) Model of one AT3 bound to a TRAP11; solid lines highlight two available TRAP-binding sites. Methyl groups in yellow are those whose resonances persist the longest as TRAP is titrated into AT.
Minimal Ensemble Analysis of Heterocomplex Configuration.
To test the hypotheses that AT3 molecules could chain together multiple TRAP rings, we generated a library of thousands of potential structures comprised of multiple TRAP and AT oligomers by systematic geometric transposition of possible TRAP–AT arrangements. Random combinations of these structures then were fit to the experimental SAXS data using summed scattering profiles calculated from these larger component complexes. This process successfully identified minimal ensembles with as few as three representatives that could produce high agreement with experimental data (Fig. 2E). Representative three-member minimum ensembles retained AT3:TRAP11 stoichiometries of 2:1, 3:2, and 3:4 and were weighted at 49%, 29%, and 22%, respectively, in the ensemble average. Notably, the larger oligomers adopted extended configurations, with AT present on opposing faces of the TRAP oligomer (Fig. 2E). These findings provide a structural explanation for the observations from bulk measurements that AT3 promotes oligomerization of TRAP rings and clarifies the type of TRAP–AT complexes formed at low ratios: long, extended complexes in which one or more AT3 triskelions effectively stitch together multiple TRAP11 rings. Although this minimal ensemble approach yielded a gratifying explanation for complex molecular behavior, the approach by design identifies a small subset of structures that is sufficient to explain an experimental observation but is not always able to comment on the accuracy of individual solutions or necessarily describe the breadth of structures that may be populated in a statistical fashion—i.e., the heterogeneity of the ensemble.
Composition and Variability of Heterocomplexes.
To validate the stoichiometric identity and heterogeneity of TRAP–AT complexes detected by this minimal ensemble approach, we used native mass spectrometry coupled to ion mobility (IM-MS) to separate TRAP–AT complexes by their collisional cross-section and mass-to-charge ratios. Mass spectra of TRAP and ƒAT alone displayed the expected ion series matching masses of homo-oligomers (Fig. S5 and Table S1). ƒAT spectra exhibited signals arising from the homo-trimer as the major species but also weak ions corresponding to the monomer and oligomers of trimers (AT3)n. TRAP spectra were dominated by signals from TRAP11 rings, although TRAP12 ions were present in the spectra, as were signals from stacked rings, as previously reported (28). Contrasting with this relative simplicity, the mass spectra obtained from solutions consisting of 0.5 equivalents of ƒAT3 per TRAP11 reveal highly heterogeneous mixtures with each species exhibiting several resolved features in the spectra corresponding to a continuous series of charge states. These signals could be assigned from their charge distributions to at least seven distinct species of complexes (Fig. 4A and Table S2). These complexes spanned a range of AT:TRAP compositions and include several of the complexes suggested in Fig. 3 as well as three TRAP rings assembled onto a single AT trimer. Additionally, the IM-MS experiments respond in a similar way as the solution methods to changes in component concentrations, with experiments performed at an AT3:TRAP11 ratio of 2 yielding ions whose calculated masses correspond mostly to multiple AT3 bound to single-TRAP rings (Fig. 4B and Table S2). Control experiments with deformyl-AT provided similar trends with elevated abundances of multiple AT3-bound single-TRAP rings at high AT3:TRAP1 ratios. Thus, although we expect some quantitative differences between the solution and gas-phase experiments in regard to the abundances of the species, the mass spectrometry data confirm the presence of multiple TRAP oligomers bound to multiple AT trimers and provide important qualitative insights into the heterogeneity of the populations.
Fig. 4.
Polydentate interactions between oligomeric AT and TRAP result in heterogeneous, polymeric assemblies. (A) IM-MS spectrum of TRAP–AT mixed at a ratio of 0.5:1 AT3:TRAP11. Color saturation is linearly proportional to ion intensity. Ions are separated by their differences in mass-to-charge (x-axis) and drift time (y-axis). Species with the same mass-to-charge ratio can be separated in drift time if they have different collisional cross-sections, allowing greater resolving power for the highly heterogeneous multicomponent mixture than achieved using the mass-to-charge dimension alone. Major species are enclosed in black boxes labeled with heterocomplex models possessing molecular weights consistent with the deconvoluted masses. The stacked double-TRAP ions have been previously reported as both TRAP11 and TRAP12 (28). (B) IM-MS at an AT3:TRAP11 ratio of 2:1. The presence of additional AT results in the formation of ions consisting primarily of single TRAPs decorated with varying numbers of ATs. (C) At low AT:TRAP ratios, polymeric chains of multiple TRAPs linked by AT trimers are formed, minimizing the area of TRAP available for RNA binding. At extraphysiological AT:TRAP ratios, smaller complexes consisting of multiple AT trimers bound to single TRAP rings are favored.
Discussion
The mechanism of TRAP inhibition by AT has remained poorly understood, in part because of uncertainty regarding their stoichiometry and because AT binding appears to be much weaker than RNA binding (10, 12–14). Armed with the results set forth in this report, we describe a mechanism in which the condensation of multiple activated TRAP molecules by AT trimers minimizes the availability of TRAP’s RNA-binding sites and thereby kinetically prevents the formation of the otherwise thermodynamically favored TRAP–RNA complex. Without this condensation mechanism, a substantially larger number of ATs per TRAP oligomer would plausibly be required to block TRAP’s RNA-binding ability. Such a model also is consistent with our understanding of the mechanism by which TRAP regulates transcription, facilitating dissociation of stalled transcription complexes (4). Large TRAP–AT heterocomplexes not only would diffuse more slowly in the crowded cellular environment but also would create a kinetic barrier by reducing available RNA-binding surfaces and retarding conformational search. Furthermore, because the sequestration of TRAP by AT into inactive clusters does not degrade TRAP irreversibly, such complexes also may act as a reservoir for activated TRAP, allowing an efficient response to changing metabolic cues.
The ability of TRAP and AT to form heteropolymers follows from their oligomeric structures and polydentate display of binding surfaces. Although in vivo formation of these heteropolymers has yet to be confirmed, the 0.5-μM intracellular concentration of TRAP is close to the apparent Kd in vitro for the AT–TRAP interaction of ∼1 μM. Together with the effects of local concentration gradients and molecular crowding (29), we believe these values are sufficiently close for the phenomenon to be plausible in cells. Condensation of multiple effector proteins by a polydentate ligand, although not previously described for a transcriptional regulator, is reminiscent of the crosslinking of cell-surface receptors by polyvalent ligands; the bivalent receptor system forms receptor aggregates in response to IgE (30) as merely one example of receptor aggregation processes (31). The condensation of a multimeric transcriptional attenuator by polyvalent sequestration by AT thus represents a powerful mechanism for relieving transcriptional attenuation and thereby up-regulating gene expression.
Materials and Methods
B. subtilis TRAP, AT, and fAT were prepared as previously described (11), and their proper mass was verified via MALDI mass spectrometry. The structural integrity of refolded fAT was verified by comparison of 1H-15N NMR spectra obtained from 15N-labeled native AT and refolded fAT.
Analytical Ultracentrifugation.
After purification, AT and TRAP stock solutions were split and dialyzed against sample buffers consisting of 50 mM sodium phosphate, 100 mM NaCl, 0.5 mM Trp, and 0.02% NaN3 at pH 7 and 8. Sedimentation samples were made subsequently from the concentrated stock solutions with the corresponding dialysis buffer and were allowed to equilibrate overnight. Sample concentrations were kept at 120 µM TRAP (monomer), with the concentration of AT varied to produce the desired AT:TRAP ratio. Sedimentation coefficient distributions were obtained from Lamm analysis using SEDFIT (20) with a fitted uniform frictional value f/f0, as previously described (12) and a sampling interval of 10 points per Svedberg. Sedimentation velocity data were obtained at 20 °C using a Beckman Coulter ProteomeLab XL-I ultracentrifuge equipped with an eight-position rotor and double-sector cells with sapphire windows, using a matched dialysis buffer in the reference sector. Data were recorded using interference optics with a scan interval of 1 min and 300 scans per cell at a rotor speed of 50,000 rpm. Data at pH 7 are shown in Fig. 1, and data for controls performed at pH 8 are shown in Fig. S3.
SAXS.
SAXS data were obtained on the SIBYLS beamline at Lawrence Berkeley National Laboratory Advanced Light Source (32, 33). Samples were prepared as for sedimentation experiments, and scattering profiles of TRAP, AT, and their mixtures were collected at 1, 5, and 10 mg/mL in sample buffer at pH 8. For solutions of TRAP containing AT, the concentration of TRAP was held constant, and the concentration of AT was varied to obtain the desired AT:TRAP ratio. For each sample, datasets for exposure times of 0.5, 1, and 6 s were recorded, and scattering from the buffer solution alone was subtracted. Selective merging of the 1- and 6-s profiles was performed when detector saturation occurred at low scattering angles.
Merged scattering profiles were regularized using GNOM (34), and Dmax was obtained by iteratively increasing its value until the χ2 fit of the regularized profile to experimental data was less than 1.4. The program GASBOR (24) was used to generate 100 chain-compatible bead models from the scattering profiles of AT and TRAP alone, enforcing p11 and p3 symmetry restraints for TRAP and AT, respectively. The 10 GASBOR models that best fit each experimental dataset were averaged using DAMAVER (35), using default cutoff parameters.
NMR.
Unlabeled B. stearothermophilus TRAP and uniformly 15N,13C-labeled AT were expressed and purified as previously described (11, 36). Samples were prepared at 2.04 mM AT3, and 2.04 mM AT3 + 0.614 mM TRAP11 and were dialyzed against 20 mM Tris (pH 8), 200 mM potassium glutamate, and 50 µM Trp; 1H-13C heteronuclear multiple-quantum correlation (HMQC) data were acquired at 55 °C on a Bruker DRX 800-MHz spectrometer equipped with a triple-resonance inverse TXI cryoprobe. All data were recorded using gradient coherence selection at a spectral width of 12,500 Hz sampled over 1,024 complex points in ω2(1H) and a spectra width of 8,048 Hz sampled over 128 points in ω1(13C). Data were processed in NMRPipe (37) and analyzed in NMRView (38).
Native Mass Spectrometry.
Mass spectrometry experiments were carried out on a Synapt G2 mass spectrometer (Waters Corporation). The details of the instrument setup can be found elsewhere (39). TRAP and AT samples were buffer exchanged into 100 mM ammonium acetate (pH 7, uncorrected) using size-exclusion spin columns (MicroBioSpin-6; Bio-Rad); the concentration of TRAP was 10 µM TRAP11. Trp was added to the protein at equal molar ratio to the TRAP monomer. The concentration of AT trimer was varied to be either 0.5 or two times TRAP11. Samples were introduced and ionized using nanoelectrospray, and the following instrument conditions were used: capillary voltage, 1.2 kV; cone voltage, 50 V; helium gas flow, 120 mL/min; ion mobility nitrogen gas flow, 60 mL/min; source at room temperature; source backing pressure, 5.6 mbar; time-of-flight analyzer pressure, 6.8 × 10−7 mbar.
Supplementary Material
Acknowledgments
We thank Kevin Dyer and Greg Hura of the Lawrence Berkeley National Laboratory and the staff at the Advanced Light Source for assistance in collecting the SAXS data. This work was supported by National Institutes of Health Grant GM077234 (to M.P.F. and P.G.) and National Science Foundation Division of Biological Infrastructure Grant 0923551 (to V.H.W.). The Advanced Light Source is supported by the Director, Office of Science, Office of Basic Energy Sciences, of the US Department of Energy under contract DE-AC02-05CH11231.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1315281111/-/DCSupplemental.
References
- 1.Antson AA, et al. Structure of the trp RNA-binding attenuation protein, TRAP, bound to RNA. Nature. 1999;401(6750):235–242. doi: 10.1038/45730. [DOI] [PubMed] [Google Scholar]
- 2.Hopcroft NH, et al. The interaction of RNA with TRAP: The role of triplet repeats and separating spacer nucleotides. J Mol Biol. 2004;338(1):43–53. doi: 10.1016/j.jmb.2004.02.038. [DOI] [PubMed] [Google Scholar]
- 3.Babitzke P, Stults JT, Shire SJ, Yanofsky C. TRAP, the trp RNA-binding attenuation protein of Bacillus subtilis, is a multisubunit complex that appears to recognize G/UAG repeats in the trpEDCFBA and trpG transcripts. J Biol Chem. 1994;269(24):16597–16604. [PubMed] [Google Scholar]
- 4.Potter KD, Merlino NM, Jacobs T, Gollnick P. TRAP binding to the Bacillus subtilis trp leader region RNA causes efficient transcription termination at a weak intrinsic terminator. Nucleic Acids Res. 2011;39(6):2092–2102. doi: 10.1093/nar/gkq965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Merino E, Babitzke P, Yanofsky C. trp RNA-binding attenuation protein (TRAP)-trp leader RNA interactions mediate translational as well as transcriptional regulation of the Bacillus subtilis trp operon. J Bacteriol. 1995;177(22):6362–6370. doi: 10.1128/jb.177.22.6362-6370.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Valbuzzi A, Yanofsky C. Inhibition of the B. subtilis regulatory protein TRAP by the TRAP-inhibitory protein, AT. Science. 2001;293(5537):2057–2059. doi: 10.1126/science.1062187. [DOI] [PubMed] [Google Scholar]
- 7.Valbuzzi A, Gollnick P, Babitzke P, Yanofsky C. The anti-trp RNA-binding attenuation protein (Anti-TRAP), AT, recognizes the tryptophan-activated RNA binding domain of the TRAP regulatory protein. J Biol Chem. 2002;277(12):10608–10613. doi: 10.1074/jbc.M111813200. [DOI] [PubMed] [Google Scholar]
- 8.Sarsero JP, Merino E, Yanofsky C. A Bacillus subtilis operon containing genes of unknown function senses tRNATrp charging and regulates expression of the genes of tryptophan biosynthesis. Proc Natl Acad Sci USA. 2000;97(6):2656–2661. doi: 10.1073/pnas.050578997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Yang WJ, Yanofsky C. Effects of tryptophan starvation on levels of the trp RNA-binding attenuation protein (TRAP) and anti-TRAP regulatory protein and their influence on trp operon expression in Bacillus subtilis. J Bacteriol. 2005;187(6):1884–1891. doi: 10.1128/JB.187.6.1884-1891.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Shevtsov MB, Chen Y, Gollnick P, Antson AA. Crystal structure of Bacillus subtilis anti-TRAP protein, an antagonist of TRAP/RNA interaction. Proc Natl Acad Sci USA. 2005;102(49):17600–17605. doi: 10.1073/pnas.0508728102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Sachleben JR, McElroy CA, Gollnick P, Foster MP. Mechanism for pH-dependent gene regulation by amino-terminus-mediated homooligomerization of Bacillus subtilis anti-trp RNA-binding attenuation protein. Proc Natl Acad Sci USA. 2010;107(35):15385–15390. doi: 10.1073/pnas.1004981107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Snyder D, Lary J, Chen Y, Gollnick P, Cole JL. Interaction of the trp RNA-binding attenuation protein (TRAP) with anti-TRAP. J Mol Biol. 2004;338(4):669–682. doi: 10.1016/j.jmb.2004.03.030. [DOI] [PubMed] [Google Scholar]
- 13.Watanabe M, et al. The nature of the TRAP-Anti-TRAP complex. Proc Natl Acad Sci USA. 2009;106(7):2176–2181. doi: 10.1073/pnas.0801032106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Chen Y, Gollnick P. Alanine scanning mutagenesis of anti-TRAP (AT) reveals residues involved in binding to TRAP. J Mol Biol. 2008;377(5):1529–1543. doi: 10.1016/j.jmb.2008.02.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cruz-Vera LR, Gong M, Yanofsky C. Physiological effects of anti-TRAP protein activity and tRNA(Trp) charging on trp operon expression in Bacillus subtilis. J Bacteriol. 2008;190(6):1937–1945. doi: 10.1128/JB.01820-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Elliott MB, Gottlieb PA, Gollnick P. The mechanism of RNA binding to TRAP: Initiation and cooperative interactions. RNA. 2001;7(1):85–93. doi: 10.1017/s135583820100173x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Baumann C, Otridge J, Gollnick P. Kinetic and thermodynamic analysis of the interaction between TRAP (trp RNA-binding attenuation protein) of Bacillus subtilis and trp leader RNA. J Biol Chem. 1996;271(21):12269–12274. doi: 10.1074/jbc.271.21.12269. [DOI] [PubMed] [Google Scholar]
- 18.McCabe BC, Gollnick P. Cellular levels of trp RNA-binding attenuation protein in Bacillus subtilis. J Bacteriol. 2004;186(15):5157–5159. doi: 10.1128/JB.186.15.5157-5159.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Whatmore AM, Reed RH. Determination of turgor pressure in Bacillus subtilis: A possible role for K+ in turgor regulation. J Gen Microbiol. 1990;136(12):2521–2526. doi: 10.1099/00221287-136-12-2521. [DOI] [PubMed] [Google Scholar]
- 20.Schuck P. Size-distribution analysis of macromolecules by sedimentation velocity ultracentrifugation and lamm equation modeling. Biophys J. 2000;78(3):1606–1619. doi: 10.1016/S0006-3495(00)76713-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ortega A, Amorós D, García de la Torre J. Prediction of hydrodynamic and other solution properties of rigid proteins from atomic- and residue-level models. Biophys J. 2011;101(4):892–898. doi: 10.1016/j.bpj.2011.06.046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Schuck P. On the analysis of protein self-association by sedimentation velocity analytical ultracentrifugation. Anal Biochem. 2003;320(1):104–124. doi: 10.1016/s0003-2697(03)00289-6. [DOI] [PubMed] [Google Scholar]
- 23.Putnam CD, Hammel M, Hura GL, Tainer JA. X-ray solution scattering (SAXS) combined with crystallography and computation: Defining accurate macromolecular structures, conformations and assemblies in solution. Q Rev Biophys. 2007;40(3):191–285. doi: 10.1017/S0033583507004635. [DOI] [PubMed] [Google Scholar]
- 24.Svergun DI, Petoukhov MV, Koch MHJ. Determination of domain structure of proteins from X-ray solution scattering. Biophys J. 2001;80(6):2946–2953. doi: 10.1016/S0006-3495(01)76260-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Antson AA, et al. The structure of trp RNA-binding attenuation protein. Nature. 1995;374(6524):693–700. doi: 10.1038/374693a0. [DOI] [PubMed] [Google Scholar]
- 26.Svergun D, Barberato C, Koch MHJ. CRYSOL - A program to evaluate x-ray solution scattering of biological macromolecules from atomic coordinates. J Appl Cryst. 1995;28:768–773. [Google Scholar]
- 27.Konarev PV, Volkov VV, Sokolova AV, Koch MHJ, Svergun DI. PRIMUS: A Windows PC-based system for small-angle scattering data analysis. J Appl Cryst. 2003;36:1277–1282. [Google Scholar]
- 28.McCammon MG, Hernández H, Sobott F, Robinson CV. Tandem mass spectrometry defines the stoichiometry and quaternary structural arrangement of tryptophan molecules in the multiprotein complex TRAP. J Am Chem Soc. 2004;126(19):5950–5951. doi: 10.1021/ja0317170. [DOI] [PubMed] [Google Scholar]
- 29.Zimmerman SB, Minton AP. Macromolecular crowding: Biochemical, biophysical, and physiological consequences. Annu Rev Biophys Biomol Struct. 1993;22:27–65. doi: 10.1146/annurev.bb.22.060193.000331. [DOI] [PubMed] [Google Scholar]
- 30.Goldstein B, Perelson AS. Equilibrium theory for the clustering of bivalent cell surface receptors by trivalent ligands. Application to histamine release from basophils. Biophys J. 1984;45(6):1109–1123. doi: 10.1016/S0006-3495(84)84259-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Kahn CR, Baird KL, Jarrett DB, Flier JS. Direct demonstration that receptor crosslinking or aggregation is important in insulin action. Proc Natl Acad Sci USA. 1978;75(9):4209–4213. doi: 10.1073/pnas.75.9.4209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Classen S, et al. Implementation and performance of SIBYLS: A dual endstation small-angle X-ray scattering and macromolecular crystallography beamline at the Advanced Light Source. J Appl Cryst. 2013;46(Pt 1):1–13. doi: 10.1107/S0021889812048698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hura GL, et al. Robust, high-throughput solution structural analyses by small angle X-ray scattering (SAXS) Nat Methods. 2009;6(8):606–612. doi: 10.1038/nmeth.1353. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Svergun DI. Determination of the Regularization Parameter in Indirect-Transform Methods Using Perceptual Criteria. J Appl Cryst. 1992;25:495–503. [Google Scholar]
- 35.Volkov VV, Svergun DI. Uniqueness of ab initio shape determination in small-angle scattering. J Appl Cryst. 2003;36(3):860–864. doi: 10.1107/S0021889809000338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Chen Xp, et al. Regulatory features of the trp operon and the crystal structure of the trp RNA-binding attenuation protein from Bacillus stearothermophilus. J Mol Biol. 1999;289(4):1003–1016. doi: 10.1006/jmbi.1999.2834. [DOI] [PubMed] [Google Scholar]
- 37.Delaglio F, et al. NMRPipe: A multidimensional spectral processing system based on UNIX pipes. J Biomol NMR. 1995;6(3):277–293. doi: 10.1007/BF00197809. [DOI] [PubMed] [Google Scholar]
- 38.Johnson BA, Blevins RA. NMR View: A computer program for the visualization and analysis of NMR data. J Biomol NMR. 1994;4(5):603–614. doi: 10.1007/BF00404272. [DOI] [PubMed] [Google Scholar]
- 39.Zhou MW, Huang CS, Wysocki VH. Surface-induced dissociation of ion mobility-separated noncovalent complexes in a quadrupole/time-of-flight mass spectrometer. Anal Chem. 2012;84(14):6016–6023. doi: 10.1021/ac300810u. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.