Significance
Our study shows that the clotting protein tissue factor (TF) controls the state of tumor dormancy and does so in conjunction with recruitment of inflammatory cells and blood vessels. We show that indolent glioma cells remain harmless in mice unless rendered TF positive. Our work also demonstrates the ability of TF to indirectly influence the DNA of cancer cells by facilitating gene mutations and silencing. This ability is important because injury, cardiovascular disease, or other conditions may activate the clotting system and contribute to the awakening of occult cancer cells. This understanding also may suggest a prophylactic use of blood thinners in cases where dormant cancer cells and clotting are suspected to coexist (e.g., after surgery).
Keywords: angiogenesis, oncogenes, macrophages, clotting, brain tumor
Abstract
The coagulation system links immediate (hemostatic) and late (inflammatory, angiogenic) tissue responses to injury, a continuum that often is subverted in cancer. Here we provide evidence that tumor dormancy is influenced by tissue factor (TF), the cancer cell-associated initiator of the coagulation system and a signaling receptor. Thus, indolent human glioma cells deficient for TF remain viable but permanently dormant at the injection site for nearly a year, whereas the expression of TF leads to a step-wise transition to latent and overt tumor growth phases, a process that is preceded by recruitment of vascular (CD105+) and myeloid (CD11b+ and F4/80+) cells. Importantly, the microenvironment orchestrated by TF expression drives permanent changes in the phenotype, gene-expression profile, DNA copy number, and DNA methylation state of the tumor cells that escape from dormancy. We postulate that procoagulant events in the tissue microenvironment (niche) may affect the fate of occult tumor cells, including their biological and genetic progression to initiate a full-blown malignancy.
The interplay between intrinsic (oncogenic) and external (microenvironmental) influences defines many aspects of the natural history of human cancers, including the process of tumor dormancy (1). This aspect is important because it remains unclear what controls the dormant state of solitary transformed cells that are detectable in various organs with prevalence that often far exceeds the site-specific incidence of the corresponding cancers (2–9).
Dormancy may entail a stable equilibrium between cell growth and death within the microscopic tumor nodule (3, 10, 11), a state which should be distinguished from slow, subclinical, but otherwise progressive (latent) disease (12). In addition to cell-intrinsic growth mechanisms, the dormant state also may be controlled by immune surveillance (13), interactions with bone marrow-derived inflammatory cells (14), or the angiogenic activity of the vasculature (3).
The coagulation system rarely is considered in this context, even though it constitutes the key element of the continuum of hemostasis, inflammation, angiogenesis, and tissue responses to injury (15–17). Activation of the coagulation system is relatively common in late-stage cancers (18), but emerging data suggest that cancer incidence also may increase in certain (but not all) thrombophilic syndromes (19) and may be affected by regulators of the coagulation system (20).
Tissue factor (TF) is the key inducer of the coagulation cascade and functions as the receptor for coagulation factor VII/VIIa, a zymogen that normally circulates in blood. The formation of TF/VIIa complexes on cellular surfaces triggers the generation of active coagulation proteases (especially factor Xa and thrombin), followed by deposition of activated platelets and fibrin. Protracted or unscheduled onset of these events in cancer may result in vascular occlusion and hypoxia, and in signals that influence inflammation, angiogenesis, and other cellular responses (21). These events are attributed, in part, to the ability of clotting factors (VIIa, Xa, thrombin) to induce changes in the gene-expression profile (22), largely through the activation of the protease-activated receptors PAR-1 and -2, integrins, and other TF pathway effectors (17).
The coagulation system has a special role in the pathogenesis of glioblastoma multiforme (GBM) (23), in which hypervascularity and thromboembolic vascular occlusions result in pseudopalisading necrosis, which is characteristic of this aggressive brain tumor (21). Moreover, GBM is associated with systemic deregulation of hemostasis (21, 24) and with the increased expression of TF (25) resulting from a combination of hypoxia (26) and genetic aberrations, including oncogenic mutations of EGF receptor (EGFR)/EGFRvIII (27–31). Although the temporal relevance of these events is presently unknown, studies have linked the incidence of GBM with head injury (32, 33), for reasons that remain poorly understood and are scarcely studied.
Here we show that TF expression may impact early stages of gliomagenesis by influencing the dormancy of transformed but indolent tumor cells. Thus, U373 glioma cells that express low levels of TF remain viable but permanently nontumorigenic in mice, and this dormant state can be disrupted by exogenous expression of TF. These events are preceded by the formation of a microenvironment containing angiogenic and inflammatory cells, triggering genetic tumor progression manifested by changes in gene copy number and DNA methylation.
Results
TF-Deficient Glioma Cells Exhibit Dormant Phenotype.
The fate of glioma cells before the onset of clinically apparent lesions is presently unknown, especially in the case of primary GBM (without the preceding low-grade disease) (23). To explore this process experimentally, we used an indolent subline of U373 cells that is completely nontumorigenic, as documented by several rounds of s.c. or intracranial injection of large numbers of cancer cells that yielded no growth even after extended observation time (31).
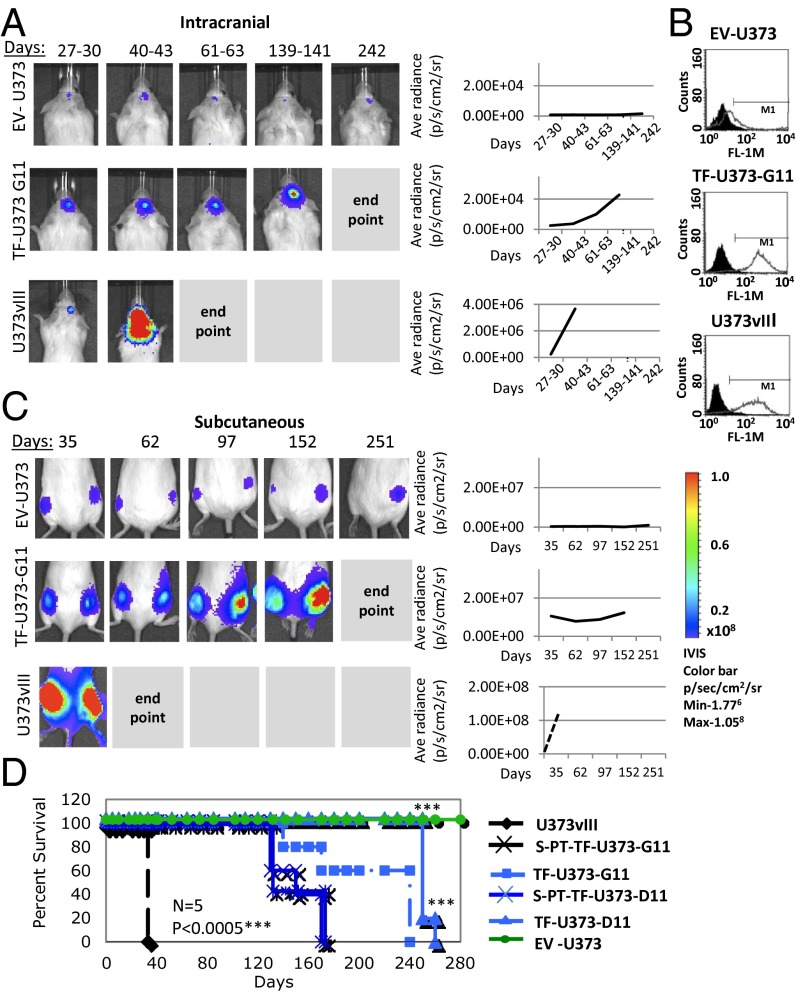
Interestingly, using Luciferase-tagged U373 cells (EV-U373), we observed that these cells are not eliminated upon inoculation into immune-deficient mice. Rather, they remain present permanently (i.e., for more than 1 y) at the injection site, viable and metabolically active, but without evidence of palpable tumor growth. In contrast, the EGFRvIII-expressing counterparts of these cells (U373vIII) form aggressive tumors in 100% of mice, reaching the clinical end point within 50–70 d (Fig. 1 A, C, and D). This pattern was observed regardless of whether glioma cells were inoculated s.c. or orthotopically (intracranially). Moreover, such differences could not be inferred from in vitro properties of these respective cell lines, both of which grow readily in culture, albeit at somewhat different rates (SI Appendix, Fig. S1 A and B). Indeed, the in vivo behavior of EV-U373 cells is one of the most striking examples of stable tumor dormancy described in the literature (3).
Fig. 1.
TF expression drives the escape of tumors from dormancy. (A) Dormant phenotype of EV-U373 cells is interrupted by the expression of TF. (Left) Bioluminescent imaging of Luciferase-expressing cell lines upon orthotopic injection into SCID mice. TF-U373-G11 cells injected intracranially remain latent but eventually emerge as detectable tumors. (Right) Quantitative plots of all tumors included in these experiments. (B) Flow cytometry shows immunostaining for the cell-surface expression of TF in parental/control cells (EV-U373), their derivatives expressing EGFRvIII (U373vIII), and an example of the TF transfectant clone (TF-U373-G11); compare with data in SI Appendix, Table S1. (C) Dormant phenotype of EV-U373 cells inoculated s.c. is interrupted by the enforced expression of TF. (Left) Bioluminescent images of s.c. tumors that were generated by EV-373, TF-U373-G11, and U373vIII cells. (Right) Cumulative growth plots combining all mice in the respective groups. (D) Impact of TF expression on survival of mice harboring dormant and aggressive glioma xenografts. Representative Kaplan–Meier plots depict survival of mice; n = 5 per group. EV-U373 cells remain dormant; TF-U373-G11 and TF-U373-D11 cell lines engineered to express TF form tumors after a long latency and with less than 100% tumor take rate (SI Appendix). PT, primary tumor cells derived by culturing cells isolated from TF-U373 tumors. Significance of differences was assessed by log-rank test; ***P < 0.0005.
Expression of TF Is Sufficient to Interrupt Tumor Cell Dormancy.
The contrasting biological properties of EV-U373 and U373vIII cells in vivo are paralleled by dramatic differences in their TF expression. Thus, U373 (or EV-U373) cells are largely TF negative, but their U373vIII derivatives express large amounts of TF protein and procoagulant activity, as described previously (27, 30). To assess whether the absence of TF activity contributes to the dormant phenotype, U373 cells were engineered to express full-length human TF, and several independent clones were characterized for procoagulant and tumorigenic properties (Fig. 1 and SI Appendix, Table S1 and Fig. S3A). Notably, all empty-vector transfectants (EV-U373) and TF-low expressors (TF-U373–low) retained their permanently dormant characteristics. However, TF-U373 cells expressing higher TF levels, comparable to those achievable endogenously in the presence of the EGFRvIII oncogene, exited the dormant state readily.
Of note, TF-expressing cells did not exhibit palpable tumors until 60–70 d postinjection; only after this latent period did they begin to form overt lesions rapidly (SI Appendix, Table S1). These characteristics were retained even when tumor cells were coinjected with Matrigel, which often facilitates experimental tumor formation. In this setting the TF-deficient parental population remained permanently dormant, but TF-expressing clones exhibited a somewhat increased tumor take rate and shortened time to progression. In all cases, after the initial tumor onset, full-blown disease developed within 5–6 mo postinjection and rapidly reached the clinical end point (Fig. 1 A, C, and D).
It is noteworthy that in only one of many series of such cell injections, one of 14 mice inoculated s.c. with the same preparation of control U373 cells developed an overt tumor (SI Appendix) after several weeks of latency. In this case the cell line reestablished in culture from the tumor mass (PT-EV-U373) exhibited high and stable levels of TF expression. A similar spontaneous increase in TF expression also had been observed earlier in several tumorigenic variants generated by in vivo passage of their indolent predecessors originating from colorectal cancer (Hhk-2), or melanoma (WM35; WM1341b) (SI Appendix). Thus in many cases the onset of TF expression is linked closely (causally) to a loss of indolent phenotype in vivo.
TF Provokes Early Recruitment of Blood Vessels and Myeloid Cells to Sites of Latent Tumor Growth.
Despite strikingly different behavior in vivo, TF-expressing U373 glioma sublines are indistinguishable from their control counterparts with respect to morphology and in vitro growth potential (SI Appendix). This similarity suggests that their differences are the result of interactions with the host cell compartment. In this regard, we were especially puzzled by the ability of TF to trigger tumor formation, not immediately but only after a relatively lengthy latency.
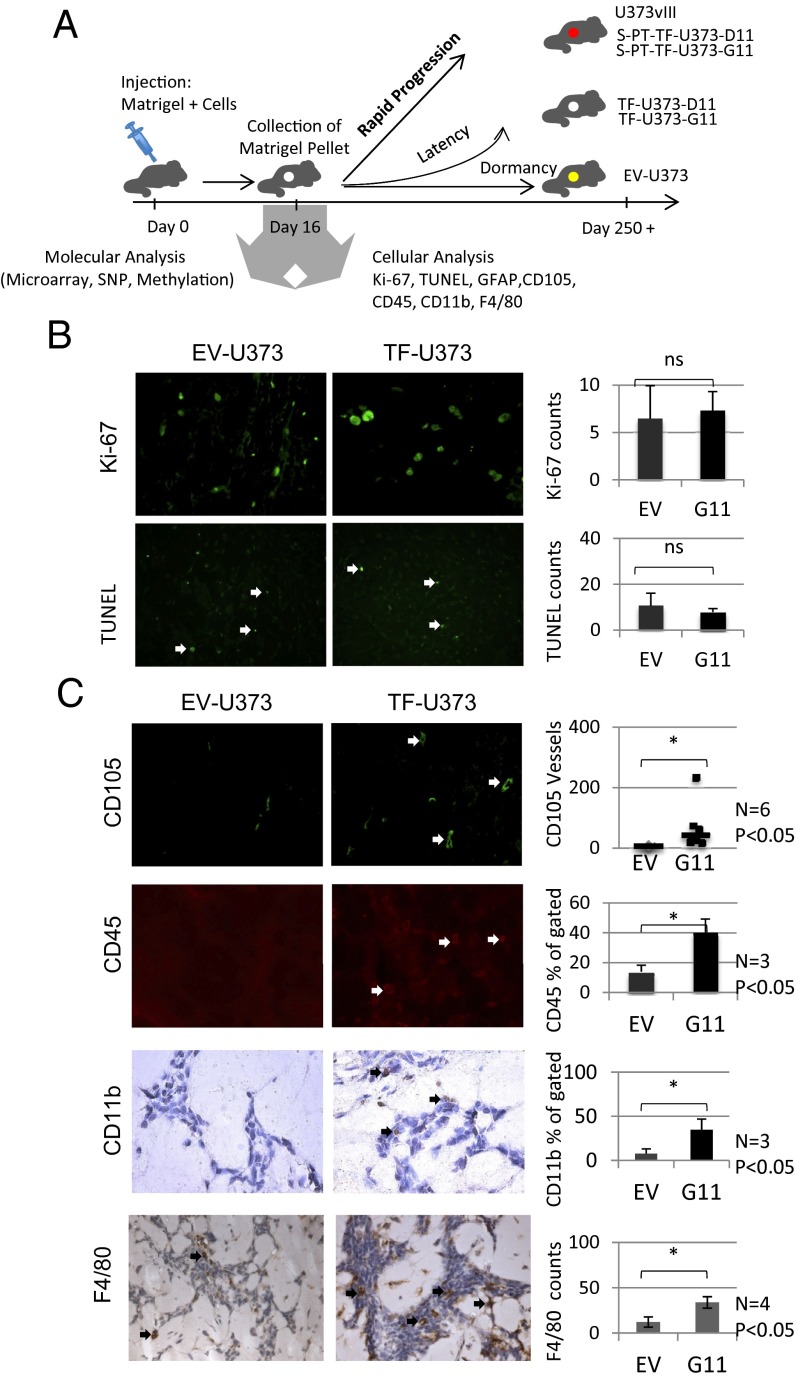
To capture the events occurring between the time of cancer cell inoculation and the onset of tumor growth, we chose to inject observable numbers of tumor cells embedded within s.c. Matrigel pellets. Unlike small cranial inocula, which often are difficult to locate, Matrigel pellets are readily extracted and analyzed histologically, molecularly, and by flow cytometry. Therefore, we chose to exploit these properties to observe the dormant behavior of U373 (EV-U373) cells and the latency of their TF-U373 counterparts (Fig. 2A and SI Appendix).
Fig. 2.
Angiogenesis and inflammation precede TF-driven tumor escape from dormancy. (A) Diagram showing the analysis of molecular and cellular changes preceding the escape from tumor dormancy. Tumor cells embedded in Matrigel were injected on day 0, and the pellets were retrieved on day 16. The cellular content of pellets was analyzed for cellular and molecular parameters as indicated (see text). (B) Staining of tumor inoculates for markers of proliferation (KI67) and apoptosis (TUNEL). EV-U373 and TF-U373-G11 pellets contain comparable numbers of dividing and apoptotic cells. (Upper) Quantification of KI67 at 40× magnification; n = 4; P > 0.05. (Lower) TUNEL staining; n = 4; P > 0.05. (C) Changes in the content of endothelial and leukocytic cells as a function of TF expression by glioma cells. (Top Row) Immunohistochemistry for CD105 reveals a differential influx of endothelial cells and blood vessels into Matrigel pellets containing EV-U373 and TF-U373-G11 cells. (Right) Quantification of CD105-positive vascular structures, n = 6; *P < 0.05. (Second Row) (Left) Immunohistochemistry for CD45+ cells (arrows). (Right) Flow cytometry quantification of CD45+ leukocytic cells within Matrigel pellets. n = 3; *P < 0.05. (Third Row) (Left) Immunostaining for CD11b+ myeloid cells in tumor cell-containing Matrigel pellets (black arrows). (Right) Flow cytometry quantification of CD11b+ cells in Matrigel pellets. n = 3; *P < 0.05. (Bottom Row) (Left) Immunostaining for the F4/80 marker of macrophages. (Right) Quantification of the F4/80 signal in pellets n = 4; *P < 0.05.
Using this approach, we noted that distinct cellular patterns began to emerge within the first 2–3 wk postinjection, that is, several weeks before the observable tumor onset. In this regard, day 16 postinoculation was chosen for consistency (Fig. 2B) and used to assess TF-dependent events such as rates of tumor cell proliferation (Ki67) and apoptosis (TUNEL), both of which were unremarkable.
In contrast, striking differences were observed in host cell recruitment. By day 16 after injection (1–2 mo before tumor onset), the TF-U373-containing Matrigel pellets became intensely vascularized with CD105+ capillary structures. This infiltration reached a median of 44.5 vessels per pellet (n = 6) as enumerated at high-power (40×) magnification, with at least 10 fields per pellet counted. At the same time postinoculation a median of only 7.3 vessels was observed in pellets containing control EV-U373 cells (P < 0.05; Fig. 2C, Top).
Moreover, pellets containing TF-U373 cells, unlike controls, were visibly infiltrated with CD45+ leukocytes, a difference that could be visualized by immunofluorescence and quantified by flow cytometry upon enzymatic dispersion of Matrigel and recovery of embedded cells (Fig. 2C, Middle). Interestingly, an even greater difference was observed in the content of cells with characteristics of CD11b+ myeloid bone marrow derived cells (BMDCs) (34) and F4/80+ macrophages (Fig. 2C, Bottom). Collectively, these observations suggest that the expression of TF in indolent U373 glioma results in transition from dormancy to latency, which is preceded (by 3–4 wk or more) by macrophage/myeloid infiltration and the onset of angiogenesis.
Molecular Responses of Glioma Cells to the TF-Modulated Microenvironment.
We reasoned that changes in gene expression would be expected to precede the profound biological transition accompanying the onset of tumor growth by TF-U373 cells. To address this question, Matrigel pellets containing TF-U373 (clone G11) cells or their isogenic EV-373 counterparts were extracted from several mice at 16 d after inoculation, and the expression of human transcripts was analyzed using the Affymetrix Human Genome Array U133 plus 2.0 platform (SI Appendix, Fig. S2). Indeed, we observed that multiple human genes were expressed differently in the inocula of TF-expressing and -nonexpressing cells. Of those genes, 319 were up-regulated at least twofold in TF-U373 cells relative to their EV-U373 controls, and 365 genes were down-regulated by at least twofold. The list in SI Appendix, Fig. S2A shows the most dramatic differences (the top 10 overexpressed and top 10 down-regulated genes), several of which were validated by RT-PCR (SI Appendix, Fig. S2B). Perhaps not surprisingly, these genes are reflective of developmental (KLHL4, SOX11), mitogenic (MYCN), or biosynthetic (RPS4Y1) functions, and their general pattern places the changes mostly within Cellular Movement, Cancer and Cellular Development regulatory pathways [Ingenuity Pathways Analysis (IPA)] (SI Appendix). Interestingly the connectivity mapping of molecular changes imparted by TF expression during tumor latency points to similarities with inflammatory and cell movement pathways, mostly converging on the respective cytokine networks (SI Appendix). This result is not unanticipated, because the procoagulant microenvironment is linked in several complex ways to pathways of inflammation (35). Thus, the expression of TF by tumor cells may cause changes in their pericellular microenvironment, which in turn could provoke a reciprocal alteration in cellular transcriptome.
TF-Imposed Exit from Dormancy Is Associated with Permanent Alterations in Tumor Cell Phenotype, Genome, and Epigenome.
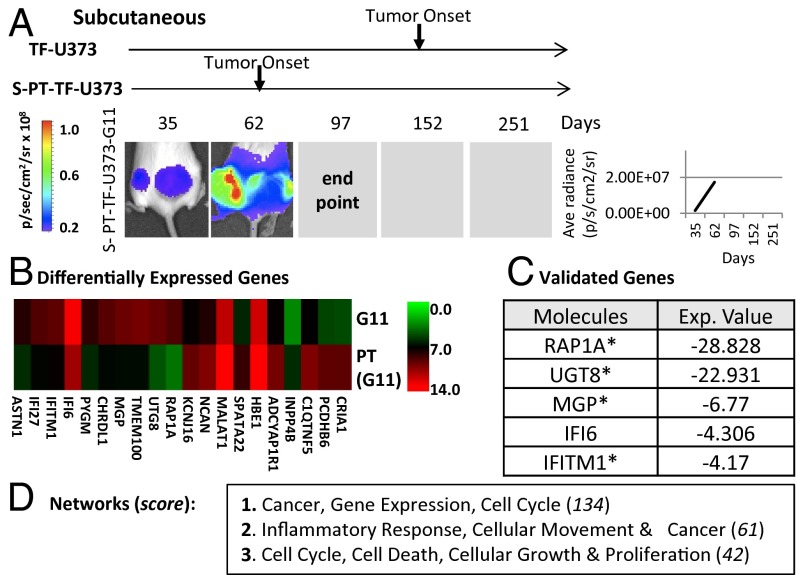
It is reasonable to ask whether passage through the latency period and the subsequent onset of aggressive growth in vivo, as happens with TF-U373 cells, can be recapitulated upon their reinjection into secondary recipients (Fig. 1D). To investigate this question, we established cell lines from several selected TF-U373 clones (G11, D11, B2, G2) after they had been allowed to form tumors either s.c. (S-PT-U373 series) or in the mouse brain (B-PT-U373 series). Upon short culture, these cells (S/B-PT-TF-U373) were injected into SCID mice either s.c. (in Matrigel), or intracranially and were followed over time using bioluminescence imaging (Fig. 3A). Surprisingly, S-PT-TF-U373 cells did not recapitulate the latent growth pattern of their TF-U373 predecessors but instead formed aggressive tumors shortly after inoculation. This observation suggests that TF-related cessation of dormancy results in a durable change in the cellular phenotype, an event that likely occurs during latent periods of growth within the procoagulant, inflammatory, and vascular microenvironment provoked by TF expression.
Fig. 3.
Growth in the TF-controlled microenvironment provokes permanent changes in cellular phenotype. (A) Expression of a highly tumorigenic phenotype in TF-expressing glioma cells after their passage as tumors in SCID mice. (Left) Bioluminescent images of s.c. primary tumors (S-PT-TF-U373-G11); arrows indicate the time of tumor onset in various experiments (SI Appendix). (Right) Line graph summarizing of tumor growth for all mice in the group (n = 5). (B) Changes in the gene-expression profile between TF-U373-G11 cells and their tumor-derived variant (S-PT-U373-G11). The heatmap generated from the output of the human Affymetrix U133 array compares the levels of 10 most different mRNA species between TF-U373-G11 and S-PT-TF-U373-G11 cells embedded in Matrigel pellets in vivo (day 16 postinjection). (C) List of genes validated by semiquantitative RT-PCR. (D) Table showing the top three networks according to the IPA software, including genes with a greater than twofold change.
To capture the molecular hallmarks of this transformation, Matrigel pellets containing TF-U373 G11 cells and their isogenic S-PT-TF-U373-G11 counterparts were recovered from the injection site at day 16 (before any overt tumor formation), and the human gene-expression profile was analyzed using the Affymetrix Human Genome Array U133 plus 2.0 platform (Fig. 3 B–D). We observed altered expression of several genes, including 1,186 genes up-regulated by twofold or more and 133 genes down-regulated to the same extent in S-PT-TF-U373-G11 inocula versus controls (TF-U373-G11). A number of these changes also were validated using semiquantitative RT-PCR. Once again, IPA analysis revealed a consistent involvement of transcripts linked to pathways such as Cancer, Inflammatory Response, and Cell Cycle (Fig. 3 C and D and SI Appendix).
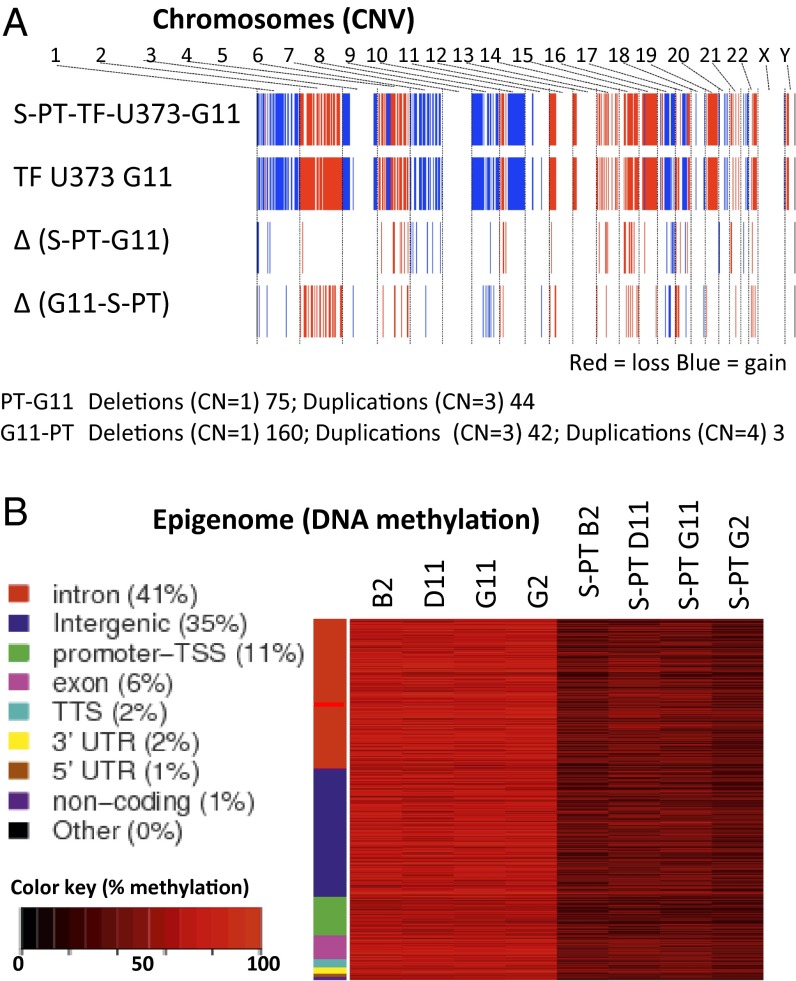
Notably, the S-PT-TF-U373 series of cell lines exhibited more elongated morphology in vitro, along with a somewhat enhanced growth potential and permanently altered expression of several genes (RAP1A, UGT8, MGP, IFI6, and IFITM1). In conjunction with the cells’ constitutively higher in vivo aggressiveness, these observations suggest that, these cells may have undergone not only a durable but, in fact, a permanent, genetic change during the course of their initial latency. To examine this possibility, we analyzed DNA copy number variation (CNV) using the Affymetrix SNP 6.0 platform to detect the possible genomic gains and losses that would separate S-PT-TF-U373-G11 cells from their isogenic TF-U373-G11 counterparts (Fig. 4A). Indeed, although the genome of both cell lines was a priori highly abnormal (as might be expected in the case of cancer cells), we did observe that the respective cell lines differed in several genomic regions, especially in chromosomes 2, 3, 5, 7, 12, and 22.
Fig. 4.
Growth of glioma cells in the TF-controlled microenvironment drives permanent changes in cellular genome and epigenome. (A) CNV differences between TF-U373-G11 cells and their derived isogenic variant S-PT-U373-G11 generated in vivo (see text). SNP analysis (Affymetrix 6.0) documents altered regions of genomic DNA. (B) Cellular methylome profiling using the RRBS strategy between indicated cell lines cultured in vitro. Hyper- and hypomethylated tiles with a greater than 20% difference in methylation between conditions and Fisher’s exact test (q-value <0.01) were used to create the heatmap. There is a clear separation between the GpC methylation profiles of several independent TF-expressing clonal cell lines and their tumor-derived counterparts (the PT series of cell lines).
Lasting changes in cellular properties also may be driven by the epigenome. An important hallmark of such changes is associated with aberrant DNA methylation, especially on cytosine in CpG dinucleotides. To explore this possibility, we used the reduced representation bisulfite sequencing (RRBS) method to detect genome-wide differences in DNA methylation at CG-dense regions that could separate TF transfectants and their tumor-derived (S-PT) counterparts. This analysis was performed on serially cultured cell lines in vitro and thus was outside the immediate influence of the host-tissue microenvironment and mouse cell contamination. Several independent, isogenic, TF-transfected clones exhibited a consistent and similar methylation pattern that was markedly different from that of their corresponding S-PT variants (Fig. 4B). A range of differentially methylated sequences was found between the TF and S-PT cells. Although loss of methylation was pronounced and widespread in S-PT variants, a portion of the affected sites were hypermethylated. This discrepancy in methylation is consistent and commonly observed in GBM and other tumor cells (36). The genomic regions found to be altered in this setting included mainly introns (41%), intergenic sequences (35%), and promoters (11%). The majority of the differentially methylated sequences did not correspond to regions where CNVs were found between the S-PT-TF-U373-G11 and TF-U373-G11 cells.
Overall, these observations suggest that microenvironmental changes orchestrated by tumor cell-associated TF (procoagulant/vascular responses) are capable of inducing permanent genetic and epigenetic alterations in these cells as they progress to an overtly tumorigenic phenotype.
Discussion
Dormancy is infrequently discussed in the context of GBM, in part because long periods of remission are rare in this devastating disease; instead, clinical demise is preceded by relatively short median survival of ∼12–15 mo postdiagnosis despite standard care, including surgery and radio/chemotherapy (23). Some variations may occur between clinically distinct molecular subsets of the disease (37) and between patients with primary and secondary pathways of GBM progression (38). Nonetheless, in the great majority of adult GBM cases tumors are discovered de novo (as primary GBM) and present as rapidly progressing, highly invasive, intractable lesions, seemingly without evidence of prior alterations or prolonged dormancy posttherapy.
However, we suggest that dormant behavior could be relevant to the biology of GBM cells before overt disease onset and might be subject to targetable changes in the tissue microenvironment. For example, puzzling observations in the literature suggest that clinically evident primary GBMs may emerge within 4–10 mo of ostensibly negative brain imaging results (32). These tumors are known to contain cells already endowed with a highly complex repertoire of chromosomal losses, gains, amplifications, mutations, and epigenetic changes involving several genes (e.g., EGFR, PDGFR, PTEN, MDM2, Rb, p16/Ink4a, PI3K) (23). It is unlikely that such aberrations could emerge during such a short period (i.e., within months) and suggest that a more lengthy and cryptic natural history could precede the diagnosis (39).
GBM also is reportedly more frequent in individuals who have sustained head injury or experienced unexplained seizures (33, 40). These considerations raise the possibility, which warrants study, of interactions between tissue repair processes in the brain and premalignant or dormant tumor cells before clinical detection. Such dormant disseminated cells also could exist at remote cranial sites, where microenvironmental perturbations could affect recurrent growth. In this regard our results suggest the possibility that activation of the coagulation system by changes in cellular phenotype, hypoxia, or microinjury could set off a cascade of inflammatory and vascular events promoting the onset of malignant lesion. Indeed, Karpatkin and colleagues previously suggested a possible role of thrombin in cancer dormancy (41).
Tumor progression increasingly is linked to the presence of local and bone marrow–derived CD11b+ cells and macrophages, features that we observed before TF-driven “awakening” of U373 glioma (42–45). Interestingly, cancers known to express high levels of TF often possess proinflammatory stroma, as do pancreatic (46) and colorectal cancers in which TF also may regulate the onset of tumor progression (20).
Whether the recruitment of macrophages by TF-U373 cells represents a true inflammatory response and the functional polarization of these infiltrating cells (M1/M2) remains to be determined. Nonetheless, these TF-dependent microenvironmental changes occur several weeks before the clinical manifestation of tumor growth and are followed by secondary events including phenotypic, genetic, and epigenetic alterations in cancer cells.
The mechanisms linking TF-driven changes in the microenvironment and alterations in the cancer cell genome and epigenome remain presently unknown, as are their key target genes. It is possible that inflammatory cells recruited to the site of chronic coagulation produce cellular stimulants (cytokines) and mutagens (e.g., reactive oxygen species) (47) with genome-altering consequences. Although carcinogenic effects of inflammation have been widely discussed in the context of chronic infections (48), our study suggests that the coagulation pathway also may be a part of similar processes in cancer. Indeed, the incidence of certain cancers is markedly elevated in patients with specific types of coagulopathy, as exemplified by the increased frequency of colon cancer in individuals with homozygous Factor V Leiden mutation (19). How the coagulation system interacts with cancer and dormant cells in different thrombophilias warrants further study.
The role of TF in the progression of various cancers is hardly uniform. Thus, in several experimental studies TF obliteration was shown to impede tumor growth (49–52), but other studies have shown that TF-negative cells may readily form aggressive tumors in mice (53–56). Similar differences could be expected also among patients with brain tumors. GBM represents a molecularly heterogeneous group of diseases, including subtypes with distinctively different profiles of tumor coagulome, only some of which include high levels of TF (29). We posit that the control of tumor dormancy by the coagulation/inflammation pathway also may be heterogeneous and potentially subtype specific. Indeed, high TF levels tend to segregate with high levels of EGFR, especially in the classical subtype of GBM. However, other GBM subtypes exhibit other prominent alterations in the expression of coagulation-related genes. It is possible that TF may play a different or lesser role in tumor–vascular interactions in these diverse settings (29).
Although many questions still remain unanswered, our study highlights the importance of the procoagulant microenvironment in the control of tumor dormancy and suggests that the clotting system may exert a hitherto unsuspected impact on genetic and epigenetic alterations, tumor dormancy, and progression. We propose that a better control of hemostatic perturbations may offer new means of therapy, control, and prevention in cancers, including, but possibly not restricted to, glioma.
Materials and Methods
Cell Lines, Culture Conditions, and Tumor Formation in Matrigel.
All cell lines were maintained under standard conditions unless otherwise indicated. Matrigel (BD Biosciences) was mixed with 5 × 106 cells (of the cell line of interest) and injected s.c. into SCID mice. All procedures involving animals were performed in accordance with the guidelines of the Canadian Council of Animal Care and the Animal Utilization Protocols approved by the Institutional Animal Care Committee at McGill University Health Centre Research Institute and McGill University. Tissues were collected for immunostaining as detailed in SI Appendix.
Molecular Profiling.
The platforms used for molecular profiling were Affymetrix 6.0 SNP Array, Affymetrix Human Genome U113 2.0 Array, and IPA (Ingenuity Systems; www.ingenuity.com). Heatmaps were generated using MeV software (57, 58). This profiling and reduced representation bisulfite sequencing (RRBS) methylation assays are delineated in SI Appendix (59) and were analyzed (60) as described earlier (61). Differentially methylated sequences were annotated using HOMER version 3.51 (62).
Statistical Analysis.
All experiments were reproduced at least twice with similar results, or independently validated, and are presented as number of replicates (n) and mean value of replicates ± SD. Statistical analyses were performed using JMP 10.0 (SAS Institute, Inc.) as indicated. Differences were considered statistically significant when P < 0.05. Wilcoxon and log-rank testing were performed for all mouse experiments. For all other experiments, statistical analysis was performed using one-tailed, unpaired t test.
Supplementary Material
Acknowledgments
We thank Alexandre Montpetit and the staff of the sequencing platform at the McGill University and Genome Quebec Innovation Centre for their expertise with sequencing required for the RRBS experiments and our colleagues for helpful discussions and inspiration, especially Dr. Ab Guha, who is sorely missed. This work was supported by Canadian Institutes of Health Research (CIHR) Grants MOP 102736 and MOP 111119 (to J.R.) and a CIHR grant (to J.T.). Fonds de la Recherche en Santé du Québec provided infrastructure support and fellowship support (N. Magnus). J.R. is the Jack Cole Chair in Pediatric Oncology. J.T. is a James McGill Professor of McGill University. The McGill Epigenomic Data Coordinating Centre is funded by CIHR Grant EP2-120609 through the Canadian Epigenetics, Environment and Health Research Consortium.
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1314118111/-/DCSupplemental.
References
- 1.Folkman J, Kalluri R. Cancer without disease. Nature. 2004;427(6977):787. doi: 10.1038/427787a. [DOI] [PubMed] [Google Scholar]
- 2.Aguirre-Ghiso JA. Models, mechanisms and clinical evidence for cancer dormancy. Nat Rev Cancer. 2007;7(11):834–846. doi: 10.1038/nrc2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Almog N. Molecular mechanisms underlying tumor dormancy. Cancer Lett. 2010;294(2):139–146. doi: 10.1016/j.canlet.2010.03.004. [DOI] [PubMed] [Google Scholar]
- 4.Black WC, Welch HG. Advances in diagnostic imaging and overestimations of disease prevalence and the benefits of therapy. N Engl J Med. 1993;328(17):1237–1243. doi: 10.1056/NEJM199304293281706. [DOI] [PubMed] [Google Scholar]
- 5.Goss PE, Chambers AF. Does tumour dormancy offer a therapeutic target? Nat Rev Cancer. 2010;10(12):871–877. doi: 10.1038/nrc2933. [DOI] [PubMed] [Google Scholar]
- 6.Heyn C, et al. In vivo MRI of cancer cell fate at the single-cell level in a mouse model of breast cancer metastasis to the brain. Magn Reson Med. 2006;56(5):1001–1010. doi: 10.1002/mrm.21029. [DOI] [PubMed] [Google Scholar]
- 7.Naumov GN, Folkman J, Straume O. Tumor dormancy due to failure of angiogenesis: Role of the microenvironment. Clin Exp Metastasis. 2009;26(1):51–60. doi: 10.1007/s10585-008-9176-0. [DOI] [PubMed] [Google Scholar]
- 8.Páez D, et al. Cancer dormancy: A model of early dissemination and late cancer recurrence. Clin Cancer Res. 2012;18(3):645–653. doi: 10.1158/1078-0432.CCR-11-2186. [DOI] [PubMed] [Google Scholar]
- 9.Uhr JW, Pantel K. Controversies in clinical cancer dormancy. Proc Natl Acad Sci USA. 2011;108(30):12396–12400. doi: 10.1073/pnas.1106613108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Arbiser JL, et al. Oncogenic H-ras stimulates tumor angiogenesis by two distinct pathways. Proc Natl Acad Sci USA. 1997;94(3):861–866. doi: 10.1073/pnas.94.3.861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Brabletz T. To differentiate or not—routes towards metastasis. Nat Rev Cancer. 2012;12(6):425–436. doi: 10.1038/nrc3265. [DOI] [PubMed] [Google Scholar]
- 12.Holmgren L, O’Reilly MS, Folkman J. Dormancy of micrometastases: Balanced proliferation and apoptosis in the presence of angiogenesis suppression. Nat Med. 1995;1(2):149–153. doi: 10.1038/nm0295-149. [DOI] [PubMed] [Google Scholar]
- 13.Quesnel B. Tumor dormancy and immunoescape. APMIS. 2008;116(7-8):685–694. doi: 10.1111/j.1600-0463.2008.01163.x. [DOI] [PubMed] [Google Scholar]
- 14.McAllister SS, et al. Systemic endocrine instigation of indolent tumor growth requires osteopontin. Cell. 2008;133(6):994–1005. doi: 10.1016/j.cell.2008.04.045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Nierodzik ML, Karpatkin S. Thrombin induces tumor growth, metastasis, and angiogenesis: Evidence for a thrombin-regulated dormant tumor phenotype. Cancer Cell. 2006;10(5):355–362. doi: 10.1016/j.ccr.2006.10.002. [DOI] [PubMed] [Google Scholar]
- 16.Rickles FR, Patierno SR, Fernandez PM. Targeting the endothelium in cancer—the importance of the interaction of hemostatic mechanisms and the vascular wall for tumor growth and angiogenesis. Pathophysiol Haemost Thromb. 2003;33(Suppl 1):1–4. doi: 10.1159/000073276. [DOI] [PubMed] [Google Scholar]
- 17.Ruf W. Redundant signaling of tissue factor and thrombin in cancer progression? J Thromb Haemost. 2007;5(8):1584–1587. doi: 10.1111/j.1538-7836.2007.02622.x. [DOI] [PubMed] [Google Scholar]
- 18.Rickles FR. Mechanisms of cancer-induced thrombosis in cancer. Pathophysiol Haemost Thromb. 2006;35(1-2):103–110. doi: 10.1159/000093551. [DOI] [PubMed] [Google Scholar]
- 19.Vossen CY, Hoffmeister M, Chang-Claude JC, Rosendaal FR, Brenner H. Clotting factor gene polymorphisms and colorectal cancer risk. J Clin Oncol. 2011;29(13):1722–1727. doi: 10.1200/JCO.2010.31.8873. [DOI] [PubMed] [Google Scholar]
- 20.Steinbrecher KA, et al. Colitis-associated cancer is dependent on the interplay between the hemostatic and inflammatory systems and supported by integrin alpha(M)beta(2) engagement of fibrinogen. Cancer Res. 2010;70(7):2634–2643. doi: 10.1158/0008-5472.CAN-09-3465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Brat DJ, Van Meir EG. Vaso-occlusive and prothrombotic mechanisms associated with tumor hypoxia, necrosis, and accelerated growth in glioblastoma. Lab Invest. 2004;84(4):397–405. doi: 10.1038/labinvest.3700070. [DOI] [PubMed] [Google Scholar]
- 22.Albrektsen T, et al. Transcriptional program induced by factor VIIa-tissue factor, PAR1 and PAR2 in MDA-MB-231 cells. J Thromb Haemost. 2007;5(8):1588–1597. doi: 10.1111/j.1538-7836.2007.02603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wen PY, Kesari S. Malignant gliomas in adults. N Engl J Med. 2008;359(5):492–507. doi: 10.1056/NEJMra0708126. [DOI] [PubMed] [Google Scholar]
- 24.Jenkins EO, Schiff D, Mackman N, Key NS. Venous thromboembolism in malignant gliomas. J Thromb Haemost. 2010;8(2):221–227. doi: 10.1111/j.1538-7836.2009.03690.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Takano S, Tsuboi K, Tomono Y, Mitsui Y, Nose T. Tissue factor, osteopontin, alphavbeta3 integrin expression in microvasculature of gliomas associated with vascular endothelial growth factor expression. Br J Cancer. 2000;82(12):1967–1973. doi: 10.1054/bjoc.2000.1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Rong Y, et al. PTEN and hypoxia regulate tissue factor expression and plasma coagulation by glioblastoma. Cancer Res. 2005;65(4):1406–1413. doi: 10.1158/0008-5472.CAN-04-3376. [DOI] [PubMed] [Google Scholar]
- 27.Milsom CCYJ, et al. Tissue factor regulation by epidermal growth factor receptor and epithelial-to-mesenchymal transitions: Effect on tumor initiation and angiogenesis. Cancer Res. 2008;68(24):10068–10076. doi: 10.1158/0008-5472.CAN-08-2067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Rong Y, et al. Epidermal growth factor receptor and PTEN modulate tissue factor expression in glioblastoma through JunD/activator protein-1 transcriptional activity. Cancer Res. 2009;69(6):2540–2549. doi: 10.1158/0008-5472.CAN-08-1547. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Magnus N, Gerges N, Jabado N, Rak J. Coagulation-related gene expression profile in glioblastoma is defined by molecular disease subtype. J Thromb Haemost. 2013;11(6):1197–1200. doi: 10.1111/jth.12242. [DOI] [PubMed] [Google Scholar]
- 30.Magnus N, Garnier D, Rak J. Oncogenic epidermal growth factor receptor up-regulates multiple elements of the tissue factor signaling pathway in human glioma cells. Blood. 2010;116(5):815–818. doi: 10.1182/blood-2009-10-250639. [DOI] [PubMed] [Google Scholar]
- 31.Al-Nedawi K, et al. Intercellular transfer of the oncogenic receptor EGFRvIII by microvesicles derived from tumour cells. Nat Cell Biol. 2008;10(5):619–624. doi: 10.1038/ncb1725. [DOI] [PubMed] [Google Scholar]
- 32.Chittiboina P, et al. Occult tumors presenting with negative imaging: Analysis of the literature. J Neurosurg. 2012;116(6):1195–1203. doi: 10.3171/2012.3.JNS112098. [DOI] [PubMed] [Google Scholar]
- 33.Hochberg F, Toniolo P, Cole P. Head trauma and seizures as risk factors of glioblastoma. Neurology. 1984;34(11):1511–1514. doi: 10.1212/wnl.34.11.1511. [DOI] [PubMed] [Google Scholar]
- 34.Mazzieri R, et al. Targeting the ANG2/TIE2 axis inhibits tumor growth and metastasis by impairing angiogenesis and disabling rebounds of proangiogenic myeloid cells. Cancer Cell. 2011;19(4):512–526. doi: 10.1016/j.ccr.2011.02.005. [DOI] [PubMed] [Google Scholar]
- 35.Gil-Bernabé AM, et al. Recruitment of monocytes/macrophages by tissue factor-mediated coagulation is essential for metastatic cell survival and premetastatic niche establishment in mice. Blood. 2012;119(13):3164–3175. doi: 10.1182/blood-2011-08-376426. [DOI] [PubMed] [Google Scholar]
- 36.Nagarajan RP, Costello JF. Epigenetic mechanisms in glioblastoma multiforme. Semin Cancer Biol. 2009;19(3):188–197. doi: 10.1016/j.semcancer.2009.02.005. [DOI] [PubMed] [Google Scholar]
- 37.Verhaak RG, et al. Cancer Genome Atlas Research Network Integrated genomic analysis identifies clinically relevant subtypes of glioblastoma characterized by abnormalities in PDGFRA, IDH1, EGFR, and NF1. Cancer Cell. 2010;17(1):98–110. doi: 10.1016/j.ccr.2009.12.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ohgaki H, Kleihues P. Genetic alterations and signaling pathways in the evolution of gliomas. Cancer Sci. 2009;100(12):2235–2241. doi: 10.1111/j.1349-7006.2009.01308.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Snuderl M, et al. Mosaic amplification of multiple receptor tyrosine kinase genes in glioblastoma. Cancer Cell. 2011;20(6):810–817. doi: 10.1016/j.ccr.2011.11.005. [DOI] [PubMed] [Google Scholar]
- 40.Moorthy RK, Rajshekhar V. Development of glioblastoma multiforme following traumatic cerebral contusion: Case report and review of literature. Surg Neurol. 2004;61(2):180–184, discussion 184. doi: 10.1016/s0090-3019(03)00423-3. [DOI] [PubMed] [Google Scholar]
- 41.Karpatkin S. Does hypercoagulability awaken dormant tumor cells in the host? J Thromb Haemost. 2004;2(12):2103–2106. doi: 10.1111/j.1538-7836.2004.01003.x. [DOI] [PubMed] [Google Scholar]
- 42.Kioi M, et al. Inhibition of vasculogenesis, but not angiogenesis, prevents the recurrence of glioblastoma after irradiation in mice. J Clin Invest. 2010;120(3):694–705. doi: 10.1172/JCI40283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Guo KT, et al. Isolation and characterization of bone marrow-derived progenitor cells from malignant gliomas. Anticancer Res. 2012;32(11):4971–4982. [PubMed] [Google Scholar]
- 44.Piao Y, et al. Glioblastoma resistance to anti-VEGF therapy is associated with myeloid cell infiltration, stem cell accumulation, and a mesenchymal phenotype. Neuro-oncol. 2012;14(11):1379–1392. doi: 10.1093/neuonc/nos158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Pyonteck SM, et al. CSF-1R inhibition alters macrophage polarization and blocks glioma progression. Nat Med. 2013;19(10):1264–1272. doi: 10.1038/nm.3337. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Guerra C, et al. Pancreatitis-induced inflammation contributes to pancreatic cancer by inhibiting oncogene-induced senescence. Cancer Cell. 2011;19(6):728–739. doi: 10.1016/j.ccr.2011.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Hasselbalch HC. Chronic inflammation as a promotor of mutagenesis in essential thrombocythemia, polycythemia vera and myelofibrosis. A human inflammation model for cancer development? Leuk Res. 2013;37(2):214–220. doi: 10.1016/j.leukres.2012.10.020. [DOI] [PubMed] [Google Scholar]
- 48.Ding SZ, Goldberg JB, Hatakeyama M. Helicobacter pylori infection, oncogenic pathways and epigenetic mechanisms in gastric carcinogenesis. Future Oncol. 2010;6(5):851–862. doi: 10.2217/fon.10.37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Hembrough TA, et al. Tissue factor/factor VIIa inhibitors block angiogenesis and tumor growth through a nonhemostatic mechanism. Cancer Res. 2003;63(11):2997–3000. [PubMed] [Google Scholar]
- 50.Ngo CV, et al. CNTO 859, a humanized anti-tissue factor monoclonal antibody, is a potent inhibitor of breast cancer metastasis and tumor growth in xenograft models. Int J Cancer. 2007;120(6):1261–1267. doi: 10.1002/ijc.22426. [DOI] [PubMed] [Google Scholar]
- 51.Yu JL, et al. Oncogenic events regulate tissue factor expression in colorectal cancer cells: Implications for tumor progression and angiogenesis. Blood. 2005;105(4):1734–1741. doi: 10.1182/blood-2004-05-2042. [DOI] [PubMed] [Google Scholar]
- 52.Zhao J, Aguilar G, Palencia S, Newton E, Abo A. rNAPc2 inhibits colorectal cancer in mice through tissue factor. Clin Cancer Res. 2009;15(1):208–216. doi: 10.1158/1078-0432.CCR-08-0407. [DOI] [PubMed] [Google Scholar]
- 53.Palumbo JS, et al. Tumor cell-associated tissue factor and circulating hemostatic factors cooperate to increase metastatic potential through natural killer cell-dependent and-independent mechanisms. Blood. 2007;110(1):133–141. doi: 10.1182/blood-2007-01-065995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Toomey JR, Kratzer KE, Lasky NM, Broze GJ., Jr Effect of tissue factor deficiency on mouse and tumor development. Proc Natl Acad Sci USA. 1997;94(13):6922–6926. doi: 10.1073/pnas.94.13.6922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yu J, et al. Contribution of host-derived tissue factor to tumor neovascularization. Arterioscler Thromb Vasc Biol. 2008;28(11):1975–1981. doi: 10.1161/ATVBAHA.108.175083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Liu Y, et al. Tissue factor-activated coagulation cascade in the tumor microenvironment is critical for tumor progression and an effective target for therapy. Cancer Res. 2011;71(20):6492–6502. doi: 10.1158/0008-5472.CAN-11-1145. [DOI] [PubMed] [Google Scholar]
- 57.Saeed AI, et al. 2006. TM4 Microarray Software Suite. Methods in Enzymology, eds Kimmel A, Oliver B (Academic, New York), Vol 411, pp 134–193.
- 58.Saeed AI, et al. TM4: A free, open-source system for microarray data management and analysis. Biotechniques. 2003;34(2):374–378. doi: 10.2144/03342mt01. [DOI] [PubMed] [Google Scholar]
- 59.Boyle P, et al. Gel-free multiplexed reduced representation bisulfite sequencing for large-scale DNA methylation profiling. Genome Biol. 2012;13(10):R92. doi: 10.1186/gb-2012-13-10-r92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Xi Y, Li W. BSMAP: Whole genome bisulfite sequence MAPping program. BMC Bioinformatics. 2009;10:232. doi: 10.1186/1471-2105-10-232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Akalin A, et al. methylKit: A comprehensive R package for the analysis of genome-wide DNA methylation profiles. Genome Biol. 2012;13(10):R87. doi: 10.1186/gb-2012-13-10-r87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Heinz S, et al. Simple combinations of lineage-determining transcription factors prime cis-regulatory elements required for macrophage and B cell identities. Mol Cell. 2010;38(4):576–589. doi: 10.1016/j.molcel.2010.05.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.