Abstract
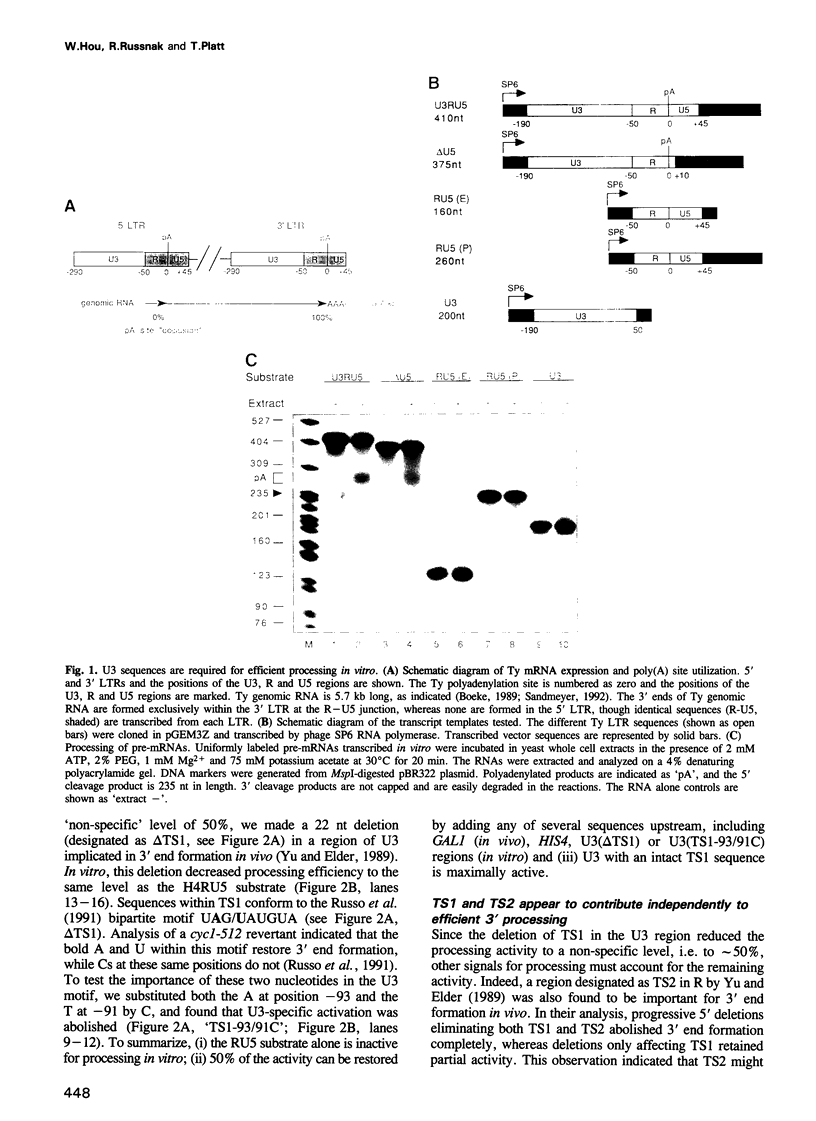
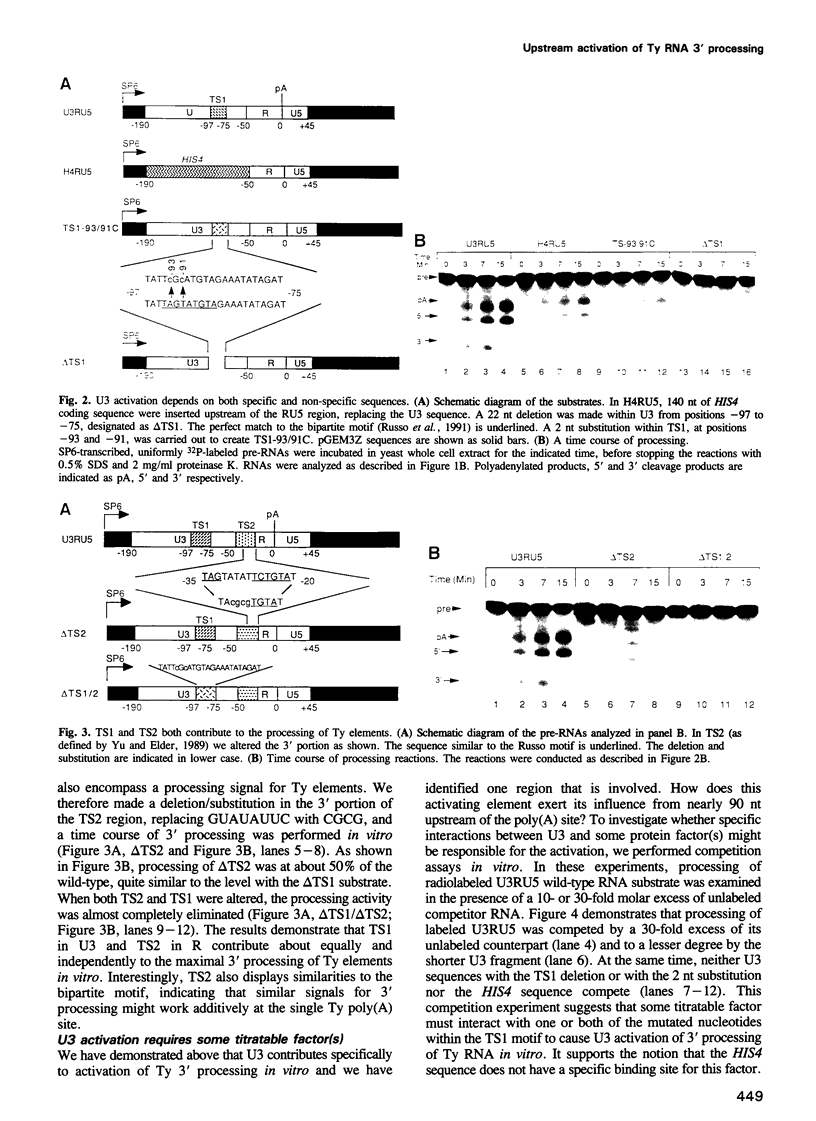
In the Ty retrotransposon of Saccharomyces cerevisiae, as in most retroelements, the polyadenylation site of the 5' long terminal repeat (LTR) is ignored and the one in the 3' LTR is efficiently used. We examine here the contribution to this poly(A) site selection of the region termed 'U3', corresponding to the upstream non-transcribed portion of the 5' LTR. Using an established assay in vitro, we find that 3' processing is accurate and efficient with an RNA substrate corresponding to most of the LTR, whereas none is detectable with a shorter transcript lacking the U3 region, thus explaining why the 5' poly(A) site is ignored in genomic Ty mRNA. When HIS4 coding RNA, representing 'non-specific' sequence, replaces the U3 region, the Ty polyadenylation site is activated to 50% of the wild-type level. Within one specific region (TS1) in U3, 90-95 nt upstream of the poly(A) site, the change of UAGUAU to UCGCAU reduces processing efficiency by half, to the non-specific level provided by other sequences or by a deletion of the TS1 region. Another region (TS2) near the poly(A) site appears to be independently responsible for the remaining half of the processing activity. Alteration of both TS1 and TS2 eliminates processing entirely. In competition assays, excess unlabeled U3, but not its mutated counterparts, reduces the processing of radiolabeled Ty mRNA, suggesting the involvement of some sequence-specific titratable factor(s) in the whole cell extract for U3-specific activation.(ABSTRACT TRUNCATED AT 250 WORDS)
Full text
PDF




Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Ahmed Y. F., Gilmartin G. M., Hanly S. M., Nevins J. R., Greene W. C. The HTLV-I Rex response element mediates a novel form of mRNA polyadenylation. Cell. 1991 Feb 22;64(4):727–737. doi: 10.1016/0092-8674(91)90502-p. [DOI] [PubMed] [Google Scholar]
- Brown P. H., Tiley L. S., Cullen B. R. Efficient polyadenylation within the human immunodeficiency virus type 1 long terminal repeat requires flanking U3-specific sequences. J Virol. 1991 Jun;65(6):3340–3343. doi: 10.1128/jvi.65.6.3340-3343.1991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Butler J. S., Platt T. RNA processing generates the mature 3' end of yeast CYC1 messenger RNA in vitro. Science. 1988 Dec 2;242(4883):1270–1274. doi: 10.1126/science.2848317. [DOI] [PubMed] [Google Scholar]
- Butler J. S., Sadhale P. P., Platt T. RNA processing in vitro produces mature 3' ends of a variety of Saccharomyces cerevisiae mRNAs. Mol Cell Biol. 1990 Jun;10(6):2599–2605. doi: 10.1128/mcb.10.6.2599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carswell S., Alwine J. C. Efficiency of utilization of the simian virus 40 late polyadenylation site: effects of upstream sequences. Mol Cell Biol. 1989 Oct;9(10):4248–4258. doi: 10.1128/mcb.9.10.4248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen J., Moore C. Separation of factors required for cleavage and polyadenylation of yeast pre-mRNA. Mol Cell Biol. 1992 Aug;12(8):3470–3481. doi: 10.1128/mcb.12.8.3470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherrington J., Ganem D. Regulation of polyadenylation in human immunodeficiency virus (HIV): contributions of promoter proximity and upstream sequences. EMBO J. 1992 Apr;11(4):1513–1524. doi: 10.1002/j.1460-2075.1992.tb05196.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherrington J., Russnak R., Ganem D. Upstream sequences and cap proximity in the regulation of polyadenylation in ground squirrel hepatitis virus. J Virol. 1992 Dec;66(12):7589–7596. doi: 10.1128/jvi.66.12.7589-7596.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeZazzo J. D., Imperiale M. J. Sequences upstream of AAUAAA influence poly(A) site selection in a complex transcription unit. Mol Cell Biol. 1989 Nov;9(11):4951–4961. doi: 10.1128/mcb.9.11.4951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeZazzo J. D., Kilpatrick J. E., Imperiale M. J. Involvement of long terminal repeat U3 sequences overlapping the transcription control region in human immunodeficiency virus type 1 mRNA 3' end formation. Mol Cell Biol. 1991 Mar;11(3):1624–1630. doi: 10.1128/mcb.11.3.1624. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilmartin G. M., Fleming E. S., Oetjen J. Activation of HIV-1 pre-mRNA 3' processing in vitro requires both an upstream element and TAR. EMBO J. 1992 Dec;11(12):4419–4428. doi: 10.1002/j.1460-2075.1992.tb05542.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heidmann S., Obermaier B., Vogel K., Domdey H. Identification of pre-mRNA polyadenylation sites in Saccharomyces cerevisiae. Mol Cell Biol. 1992 Sep;12(9):4215–4229. doi: 10.1128/mcb.12.9.4215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hyman L. E., Seiler S. H., Whoriskey J., Moore C. L. Point mutations upstream of the yeast ADH2 poly(A) site significantly reduce the efficiency of 3'-end formation. Mol Cell Biol. 1991 Apr;11(4):2004–2012. doi: 10.1128/mcb.11.4.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Imperiale M. J., DeZazzo J. D. Poly(A) site choice in retroelements: deja vu all over again? New Biol. 1991 Jun;3(6):531–537. [PubMed] [Google Scholar]
- Irniger S., Sanfaçon H., Egli C. M., Braus G. H. Different sequence elements are required for function of the cauliflower mosaic virus polyadenylation site in Saccharomyces cerevisiae compared with in plants. Mol Cell Biol. 1992 May;12(5):2322–2330. doi: 10.1128/mcb.12.5.2322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Iwasaki K., Temin H. M. The efficiency of RNA 3'-end formation is determined by the distance between the cap site and the poly(A) site in spleen necrosis virus. Genes Dev. 1990 Dec;4(12B):2299–2307. doi: 10.1101/gad.4.12b.2299. [DOI] [PubMed] [Google Scholar]
- Ju G., Cullen B. R. The role of avian retroviral LTRs in the regulation of gene expression and viral replication. Adv Virus Res. 1985;30:179–223. doi: 10.1016/s0065-3527(08)60451-0. [DOI] [PubMed] [Google Scholar]
- Kunkel T. A., Roberts J. D., Zakour R. A. Rapid and efficient site-specific mutagenesis without phenotypic selection. Methods Enzymol. 1987;154:367–382. doi: 10.1016/0076-6879(87)54085-x. [DOI] [PubMed] [Google Scholar]
- Mann K. P., Weiss E. A., Nevins J. R. Alternative poly(A) site utilization during adenovirus infection coincides with a decrease in the activity of a poly(A) site processing factor. Mol Cell Biol. 1993 Apr;13(4):2411–2419. doi: 10.1128/mcb.13.4.2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mogen B. D., MacDonald M. H., Leggewie G., Hunt A. G. Several distinct types of sequence elements are required for efficient mRNA 3' end formation in a pea rbcS gene. Mol Cell Biol. 1992 Dec;12(12):5406–5414. doi: 10.1128/mcb.12.12.5406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Proudfoot N. Poly(A) signals. Cell. 1991 Feb 22;64(4):671–674. doi: 10.1016/0092-8674(91)90495-k. [DOI] [PubMed] [Google Scholar]
- Russnak R. H. Regulation of polyadenylation in hepatitis B viruses: stimulation by the upstream activating signal PS1 is orientation-dependent, distance-independent, and additive. Nucleic Acids Res. 1991 Dec 11;19(23):6449–6456. doi: 10.1093/nar/19.23.6449. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russnak R., Ganem D. Sequences 5' to the polyadenylation signal mediate differential poly(A) site use in hepatitis B viruses. Genes Dev. 1990 May;4(5):764–776. doi: 10.1101/gad.4.5.764. [DOI] [PubMed] [Google Scholar]
- Russo P., Li W. Z., Hampsey D. M., Zaret K. S., Sherman F. Distinct cis-acting signals enhance 3' endpoint formation of CYC1 mRNA in the yeast Saccharomyces cerevisiae. EMBO J. 1991 Mar;10(3):563–571. doi: 10.1002/j.1460-2075.1991.tb07983.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sadhale P. P., Sapolsky R., Davis R. W., Butler J. S., Platt T. Polymerase chain reaction mapping of yeast GAL7 mRNA polyadenylation sites demonstrates that 3' end processing in vitro faithfully reproduces the 3' ends observed in vivo. Nucleic Acids Res. 1991 Jul 11;19(13):3683–3688. doi: 10.1093/nar/19.13.3683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sanfaçon H., Brodmann P., Hohn T. A dissection of the cauliflower mosaic virus polyadenylation signal. Genes Dev. 1991 Jan;5(1):141–149. doi: 10.1101/gad.5.1.141. [DOI] [PubMed] [Google Scholar]
- Sanfaçon H., Hohn T. Proximity to the promoter inhibits recognition of cauliflower mosaic virus polyadenylation signal. Nature. 1990 Jul 5;346(6279):81–84. doi: 10.1038/346081a0. [DOI] [PubMed] [Google Scholar]
- Schek N., Cooke C., Alwine J. C. Definition of the upstream efficiency element of the simian virus 40 late polyadenylation signal by using in vitro analyses. Mol Cell Biol. 1992 Dec;12(12):5386–5393. doi: 10.1128/mcb.12.12.5386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valsamakis A., Schek N., Alwine J. C. Elements upstream of the AAUAAA within the human immunodeficiency virus polyadenylation signal are required for efficient polyadenylation in vitro. Mol Cell Biol. 1992 Sep;12(9):3699–3705. doi: 10.1128/mcb.12.9.3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valsamakis A., Schek N., Alwine J. C. Elements upstream of the AAUAAA within the human immunodeficiency virus polyadenylation signal are required for efficient polyadenylation in vitro. Mol Cell Biol. 1992 Sep;12(9):3699–3705. doi: 10.1128/mcb.12.9.3699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Valsamakis A., Zeichner S., Carswell S., Alwine J. C. The human immunodeficiency virus type 1 polyadenylylation signal: a 3' long terminal repeat element upstream of the AAUAAA necessary for efficient polyadenylylation. Proc Natl Acad Sci U S A. 1991 Mar 15;88(6):2108–2112. doi: 10.1073/pnas.88.6.2108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wahle E., Keller W. The biochemistry of 3'-end cleavage and polyadenylation of messenger RNA precursors. Annu Rev Biochem. 1992;61:419–440. doi: 10.1146/annurev.bi.61.070192.002223. [DOI] [PubMed] [Google Scholar]
- Weichs an der Glon C., Ashe M., Eggermont J., Proudfoot N. J. Tat-dependent occlusion of the HIV poly(A) site. EMBO J. 1993 May;12(5):2119–2128. doi: 10.1002/j.1460-2075.1993.tb05860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weichs an der Glon C., Monks J., Proudfoot N. J. Occlusion of the HIV poly(A) site. Genes Dev. 1991 Feb;5(2):244–253. doi: 10.1101/gad.5.2.244. [DOI] [PubMed] [Google Scholar]
- Weiss E. A., Gilmartin G. M., Nevins J. R. Poly(A) site efficiency reflects the stability of complex formation involving the downstream element. EMBO J. 1991 Jan;10(1):215–219. doi: 10.1002/j.1460-2075.1991.tb07938.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilson-Gunn S. I., Kilpatrick J. E., Imperiale M. J. Regulated adenovirus mRNA 3'-end formation in a coupled in vitro transcription-processing system. J Virol. 1992 Sep;66(9):5418–5424. doi: 10.1128/jvi.66.9.5418-5424.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu K., Elder R. T. Some of the signals for 3'-end formation in transcription of the Saccharomyces cerevisiae Ty-D15 element are immediately downstream of the initiation site. Mol Cell Biol. 1989 Jun;9(6):2431–2444. doi: 10.1128/mcb.9.6.2431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaret K. S., Sherman F. DNA sequence required for efficient transcription termination in yeast. Cell. 1982 Mar;28(3):563–573. doi: 10.1016/0092-8674(82)90211-2. [DOI] [PubMed] [Google Scholar]