Significance
A highly conserved region of the HIV-1 RNA genome is responsible for regulating numerous steps of the retroviral life cycle, including initiation of reverse transcription. A complete understanding of the mechanisms controlling HIV-1 replication requires structural characterization of this RNA; unfortunately, however, its large size and conformational flexibility makes common methods of solving structures, such as X-ray crystallography and NMR, exceedingly difficult. The present study uses a solution technique, small-angle X-ray scattering coupled with computational molecular modeling, to characterize three ∼100-nucleotide RNAs that play central roles in HIV-1 replication. One of these domains mimics the L-shaped fold of tRNA, providing a structural basis for understanding how this genomic RNA coordinates interactions with a tRNA-binding host factor to facilitate initiation of reverse transcription.
Keywords: HIV-1 RNA structure, tRNA-like element, molecular modeling
Abstract
The most conserved region of the HIV type 1 (HIV-1) genome, the ∼335-nt 5′ UTR, is characterized by functional stem loop domains responsible for regulating the viral life cycle. Despite the indispensable nature of this region of the genome in HIV-1 replication, 3D structures of multihairpin domains of the 5′ UTR remain unknown. Using small-angle X-ray scattering and molecular dynamics simulations, we generated structural models of the transactivation (TAR)/polyadenylation (polyA), primer-binding site (PBS), and Psi-packaging domains. TAR and polyA form extended, coaxially stacked hairpins, consistent with their high stability and contribution to the pausing of reverse transcription. The Psi domain is extended, with each stem loop exposed for interactions with binding partners. The PBS domain adopts a bent conformation resembling the shape of a tRNA in apo and primer-annealed states. These results provide a structural basis for understanding several key molecular mechanisms underlying HIV-1 replication.
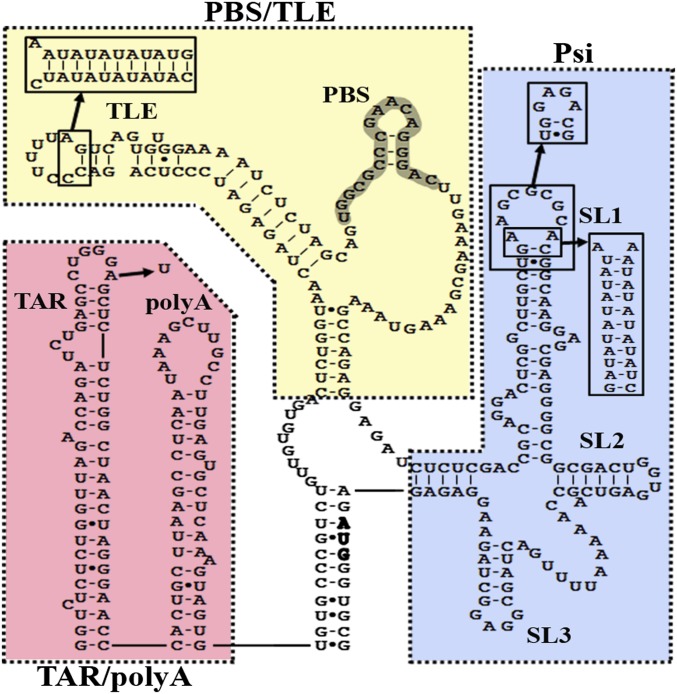
HIV type 1 (HIV-1) has a single-stranded RNA genome (vRNA) that is reverse-transcribed into double-stranded DNA and inserted into the host cell genome. Along with serving as genetic material, the vRNA plays a critical role in regulating the viral life cycle. The highly structured 5′ UTR dictates much of this control through interactions with viral and cellular factors (1, 2). The 5′ UTR consists of the transactivation response (TAR; nucleotides 1–57) element, polyadenylation (polyA; nucleotides 58–104) stem loop, primer-binding site (PBS; nucleotides 125–223) domain, and packaging domain (Psi; nucleotides 228–334) (Fig. 1). Mutations that disrupt the structures of these domains severely inhibit infectivity and replication (2–6), underscoring the importance of vRNA structure in the HIV-1 life cycle.
Fig. 1.
The HIV-1 5′ UTR secondary structure consists of the TAR/polyA, PBS/TLE, and Psi domains. Dotted lines encompass the individual 5′ UTR regions described herein. A single A34U point mutation and a GAGA tetraloop were introduced into the TAR loop of TAR/polyA and SL1 of Psi, respectively, to alleviate multimerization of the RNA constructs. The PBS/TLE domain is closed with three G:C pairs introduced at the 5′/3′ termini. Extended PBS/TLE and SL1 constructs containing 10 alternating A-U base pairs were prepared to aid the assignment of TLE and SL1 within the SAXS-derived envelopes.
The first 104-nt region of vRNA is sequentially composed of the TAR and polyA stem loops. Along with their function of stimulating transcription (7), these elements play roles in genome circularization and strand transfer during reverse transcription (8, 9), functions essential for promoting drug resistance through genetic diversity. There is also evidence that TAR plays a role in other processes, such as vRNA dimerization (10), although TAR is not strictly required for packaging (11). Results of secondary structure probing studies are consistent with TAR forming a stable hairpin (12–14). The 3D structure of the upper region of the HIV-1 TAR stem loop has been solved by NMR in the presence (15) and absence (16) of a bound Tat peptide. The polyA hairpin located downstream of TAR contains the polyadenylation signal. Although the function of polyA is not fully understood, its structural integrity is required for efficient vRNA packaging (3, 17). The 3D structure of polyA is unknown, but secondary structure probing suggests that the region adopts a helical stem with a large, unstructured loop, which may be involved in a long-range pseudoknot with the coding region of vRNA (13, 18).
Psi encompasses ∼120 nucleotides of the 5′ UTR upstream of the Gag start codon and is necessary, but not sufficient, for directing genome packaging via interactions with Gag (2). Psi also plays a critical role in vRNA dimerization via a dimerization initiation site (DIS) in SL1 (19, 20) and harbors the major splice donor site (21). The tertiary structure of the Psi region has been studied previously, providing two all-atom models. One model produced using chemical cross-linking and mass spectrometry suggested a globular structure stabilized by a loop–loop interaction between SL2 and SL3 and by a loop–stem interaction between SL1 and SL4 (22). Using chemical probing, Förster resonance energy transfer and molecular dynamics (MD), a more elongated model of the Psi region has been proposed in which the loops are solvent-exposed (23).
HIV-1 initiates reverse transcription from the 3′ end of tRNALys3 after annealing of the 3′ 18 nucleotides of tRNALys3 to the complementary PBS (24). Only a single copy of tRNALys3 is required to initiate reverse transcription, but all three human tRNALys isoacceptors (∼20–25 molecules) are selectively packaged into virions (25, 26) along with stoichiometric amounts of the major tRNALys binding protein, human lysyl-tRNA synthetase (LysRS) (27). LysRS is specifically packaged via its interaction with the viral Gag protein, thus leading to the enrichment of tRNALys in virions (28). We recently showed that LysRS binds with high affinity to the HIV-1 vRNA, owing in part to the presence of a tRNA-like element (TLE) in the U-rich stem loop proximal to the PBS that mimics the anticodon of tRNALys (29). Whether the 5′ UTR has more extensive tRNA structure mimicry is unknown, however. It remains unclear precisely how tRNALys3 annealing affects the conformation of the PBS domain; however, a recent study found that tRNA annealing promotes genome dimerization by inducing a conformational switch (30).
Whereas high-resolution structures of small portions of the HIV-1 5′ UTR have been solved by NMR spectroscopy (2, 15, 31–33), how these structures are organized in the context of the larger functional domains remains unclear. Small-angle X-ray scattering (SAXS) has proven to be a powerful and effective means of understanding the structures of large RNAs (34). Unlike X-ray crystallography and cryoelectron microscopy, SAXS can be performed in solution under physiological ionic conditions and does not require the introduction of labels or chemical probes. Thus, we used SAXS in combination with molecular modeling to examine the structural basis for the diverse functions regulated by the 5′ UTR. Our results reveal that the TAR, polyA, PBS, and Psi domains (NL4-3 isolate) fold into extended structures with solvent-exposed stem loops that allow for interactions with viral and host factors. In addition, we provide evidence that the PBS/TLE domain mimics the global tRNA tertiary fold. These results, along with our findings of a conformational change in the PBS/TLE domain on anti-PBS annealing and a dynamic SL2 in the Psi domain, underscore the importance of future efforts to elucidate how such conformational changes are propagated to regulate vRNA packaging.
Results
TAR and PolyA Adopt a Stable Coaxially Stacked Conformation.
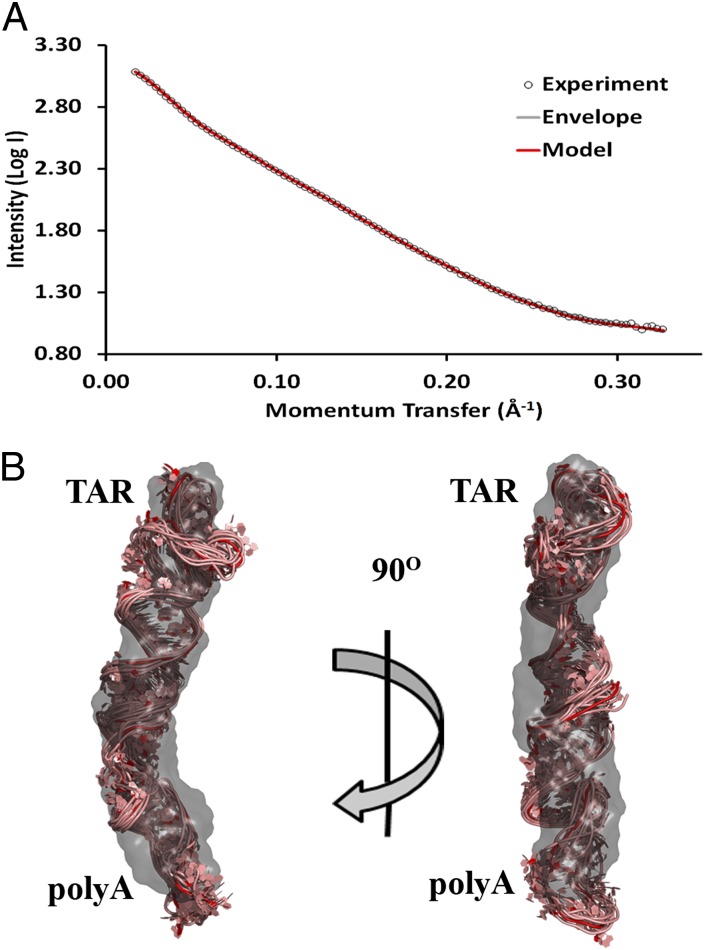
The predicted secondary structure of the HIV-1 5′ UTR (13) is shown in Fig. 1, highlighting the three domains investigated in this work. Size-exclusion chromatography (SEC) purification of a WT 104-nt RNA construct encompassing the TAR and polyA hairpins initially showed multimerization (Fig. S1A). This was alleviated using a previously described single point mutation (A34U) in the TAR loop (35), as confirmed by SEC. This A34U TAR/polyA variant was used for all SAXS experiments. Inspection of the SAXS data (Fig. 2A) and linearity of the Guinier region (Fig. S1B) confirmed the absence of aggregation. The radius of gyration (Rg), the mass distribution about a particle’s center of gravity, was calculated from the slope of the Guinier plot and, independently, from the pairwise distribution [P(r)] function, a histogram of distances between electron pairs within a particle. The Rg for TAR/polyA was determined to be 39 Å by the Guinier slope and 41 Å by the P(r) function. The maximum electron pair distance (Dmax) of TAR/polyA was 140 Å by the P(r) function (Table S1). Ab initio envelope reconstructions resulted in an elongated envelope (Fig. 2B), suggesting that the hairpins are coaxially stacked with a kink near the middle. A back-calculated scattering curve based on the ab initio envelope fits well with the SAXS data (χ2 = 1.15). The reproducibility of the ab initio calculations was determined using normalized spatial discrepancy (NSD), a measure of the degree to which each of the 20 envelopes that were averaged and filtered to produce the final envelope differs from one another. Values <1 are considered to indicate no systematic differences. The envelope for TAR/polyA yielded an NSD of 0.67.
Fig. 2.
SAXS-derived ab initio envelope and structural model of TAR/polyA. (A) Every fifth data point from SAXS experiments (open circles) was plotted along with the corresponding back-calculated scattering curves from the ab initio envelope (gray) and averaged intensity values from the 10 members of the modeled structural ensemble of A34U TAR/polyA (red). (B) The lowest-energy model was superimposed onto the SAXS-derived ab initio envelope using SUPCOMB. The remaining ensemble models were then aligned using the superimposed lowest-energy model as a register. Envelopes are shown in a gray 40% transparent surface representation, and all-atom models are represented in pink cartoon with the lowest-energy structure in red.
An ensemble of all-atom models was generated using the three-step approach involving de novo structure prediction, explicit solvent molecular dynamics (ESMD), and simulated annealing (SA) (SI Methods). The ensemble for TAR/polyA yielded a χ2 of 1.22 and an Rg of 41 Å, very close to the experimentally determined values of 39 Å and 41 Å (Table S1). The reproducibility of the structures was very good as well (all-atom rmsd from the average, 2.7 Å). Superimposing the all-atom models onto the ab initio envelope clearly shows that both methods converge on a structure in which the domain adopts two coaxially stacked hairpins with a kink at the interhelical junction of the two stems (Fig. 2B).
The Psi Domain Adopts a Nonglobular Conformation.
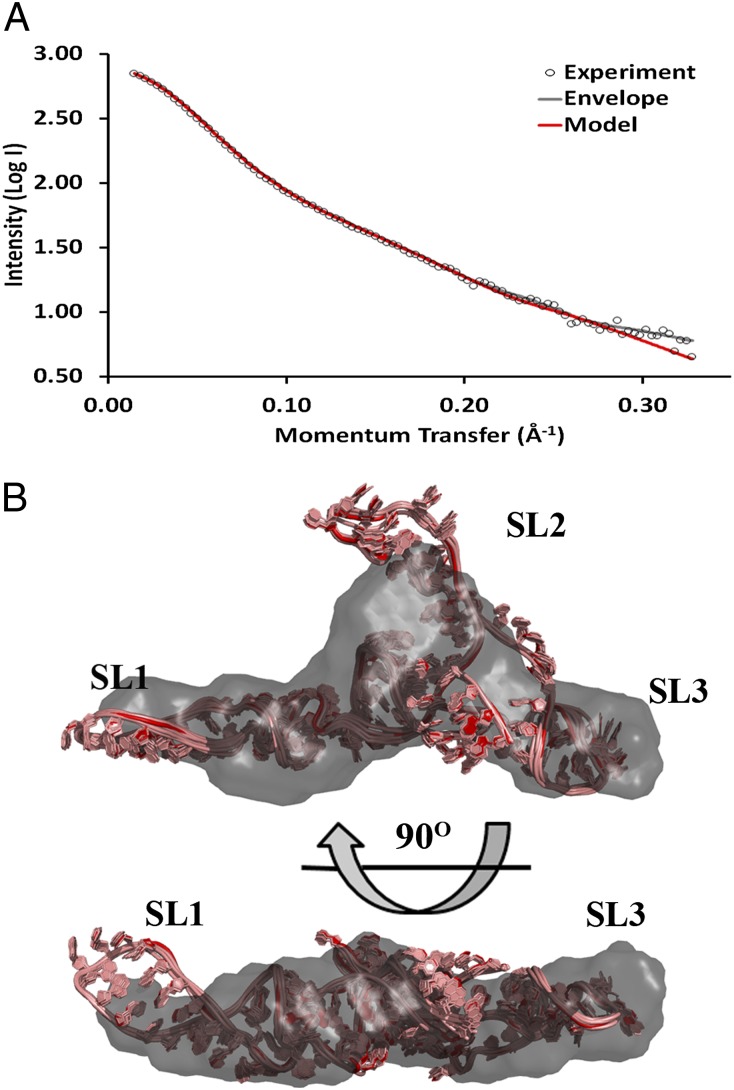
Psi contains the DIS in the loop of SL1 that promotes dimerization. To avoid conformational heterogeneity, we replaced the SL1 loop with a stable GNRA tetraloop (Fig. 1). The resulting construct yielded a single species in solution, as confirmed by SEC (Fig. S2A). The SAXS scattering curve (Fig. 3A) and Guinier plot (Fig. S2B) showed no evidence of aggregation or concentration-dependent effects. Analysis of the scattering curve revealed Guinier- and P(r)-derived Rg values of ∼34 Å and a Dmax of 121 Å (Table S1). The ab initio envelope reconstruction revealed a global conformation that was extended in solution. The high quality and reproducibility of the resulting envelope were confirmed based on the calculated χ2 and NSD values of 1.14 and 0.86, respectively (Table S1).
Fig. 3.
SAXS-derived ab initio envelope and structural model of Psi. (A) Every fifth data point from SAXS experiments (open circles) was plotted along with the corresponding back-calculated scattering curves from the ab initio envelope (gray) and averaged intensity values from the 10 members of the modeled structural ensemble of Psi with the SL1 loop mutated to a GNRA tetraloop (black). (B) The lowest-energy model was superimposed onto the SAXS-derived ab initio envelope using SUPCOMB. The remaining ensemble models were then aligned using the superimposed lowest-energy model as a register. Color-coding is as in Fig. 2.
Before MD simulations, potential secondary structures of the Psi construct were predicted using Mfold (36), revealing three likely conformations in addition to the structure predicted from chemical probing (13) (Fig. S3). All predicted secondary structures were modeled using RNAComposer (37) and screened for fit to our SAXS data. One secondary structure (Psi Mfold 1) yielded the best χ2 value (χ2 = 3.2), and the predicted Rg value of the chosen model (35 Å) also matched the experimentally determined value (Fig. S3). The chosen secondary structure is similar to one previously predicted based on phylogenetic analysis and chemical/enzymatic probing (38). An all-atom ensemble of models was produced with a χ2 of 0.99, Rg of 34 Å, and all-atom rmsd of 0.94 Å. Whereas SL1 and SL3 fit well within the filtered envelope on superposition (Fig. 3B), SL2 appears to be more flexible and is not as well defined by the envelope despite the low rmsd within the ensemble of models. The ab initio envelopes are intended to define a general molecular architecture, and so the convergence between the global shape of the envelope and the all-atom ensemble provides further confidence in the overall conformation of the models, but the specific orientation of SL2 remains ambiguous. Our assignment of individual stem loops within the envelope was confirmed using a construct in which SL1 was extended by 10 A-U base pairs (Fig. 1 and Fig. S4B).
Structure of the PBS/TLE Domain Reveals tRNA Mimicry.
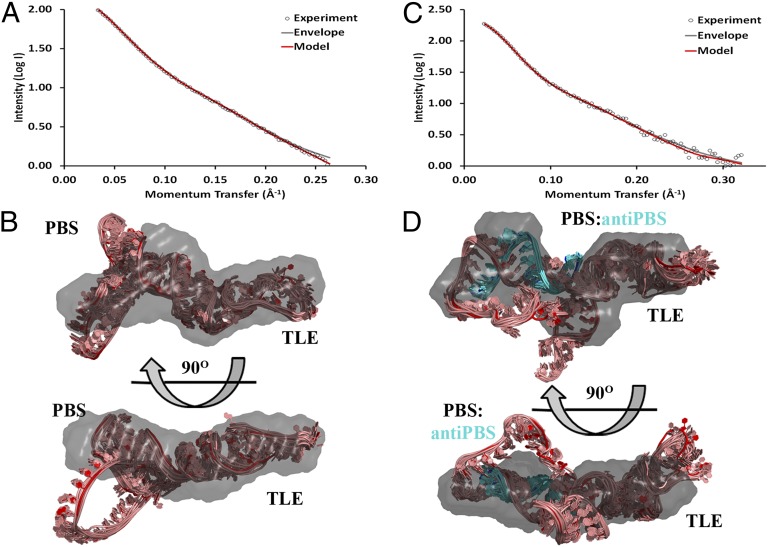
We examined a 99-nt PBS-containing construct (PBS/TLE) stabilized by the addition of three G-C base pairs (105 nt total) to the 5′/3′ ends (Fig. 1). SEC of the PBS/TLE construct demonstrated a single major peak, which was purified (Fig. 4A and Fig. S5A). Scattering was characterized by an Rg of 34 Å by both Guinier and P(r) analysis and a Dmax of 123 Å (Fig. S5B and Table S1). The ab intio envelope (χ2 = 0.90 and NSD = 0.76; Table S1) revealed a globular region and an extended arm corresponding to the largely single-stranded PBS loop and TLE helix, respectively (Fig. 4B). We confirmed these assignments by examining a PBS/TLE construct in which the TLE helix was extended by 10 A-U base pairs (Fig. 1 and Fig. S4A).
Fig. 4.
SAXS-derived ab initio envelopes and structural models of apo and annealed PBS/TLE. (A and C) Every fifth data point from SAXS experiments (open circles) was plotted along with the corresponding back-calculated scattering curves from the ab initio envelope (gray) and averaged intensity values from the 10 members of the modeled structural ensemble of PBS/TLE (red) (A) and PBS/TLE–anti-PBS (red) (C). (B and D) For both PBS/TLE (B) and PBS/TLE–anti-PBS (D), the lowest-energy model was superimposed onto the SAXS-derived ab initio envelope using SUPCOMB. The remaining ensemble models were then aligned using the superimposed lowest-energy model as a register. Envelopes are shown in a gray 40% transparent surface representation, and all-atom models are represented in pink (cyan for anti-PBS) cartoons, with the lowest-energy structure in red (blue for anti-PBS).
As with Psi, the secondary structure of PBS/TLE has not been unambiguously defined. Therefore, we screened the commonly predicted structures (Fig. S6), of which two gave reasonable fits with the experimental data (χ2 = 2.1 and 3.3). These two structures differed by the presence of a small hairpin within the PBS sequence, but both yielded an Rg similar to that determined experimentally (∼33 Å). Based on the lower χ2 value, the structure containing the PBS hairpin was used for all-atom modeling. The final refined ensemble was characterized by an Rg value of 34 Å, a χ2 value of 0.61, and an rmsd of 1.81 Å (Fig. 4 A and B and Table S1).
To investigate the effects of primer annealing to the PBS sequence, an 18-nt anti-PBS DNA was heat-annealed to the 105-nt PBS/TLE RNA construct. The resulting complex yielded multiple peaks via SEC (Fig. S7A). The major annealed fraction was purified, and the homogeneity of the sample was confirmed by native polyacrylamide electrophoresis before SAXS analysis. Analysis of the SAXS scattering curve (Fig. 4C) indicated that the Rg was not significantly increased relative to the apo PBS/TLE construct, and that the Dmax was increased only slightly (from 118 Å to 123 Å; Table S1). A high-quality ab initio envelope was generated, as evidenced by low χ2 and NSD values of 1.30 and 0.84, respectively (Fig. 4D). Modeling was then performed by taking the lowest-energy refined PBS/TLE structure and inserting an ideal A-form RNA:DNA duplex at the PBS region. This initial structure was then refined using our ESMD/SA strategy. The final ensemble was characterized by an Rg value of ∼35 Å, a χ2 value of 1.14, and an rmsd of 1.68 Å (Table S1). As with the SL2 region of Psi, the loop regions of both the apo and primer-annealed PBS/TLE do not fit well within the filtered ab intio envelopes. Thus, despite the high convergence in the ensemble of all-atom models, we are less confident in the exact conformation of these flexible regions.
Our recent finding that the TLE region of the PBS/TLE domain can specifically bind to LysRS (29) prompted us to investigate whether the tertiary structure of this domain resembles a tRNA in either the apo or primer-annealed state. We performed SAXS analysis on tRNALys3 (39), generated ab initio envelopes, and superimposed them onto the TLE regions of the PBS/TLE and PBS/TLE:anti-PBS envelopes (Fig. 5). The tRNA structure fits well into the envelopes for both constructs, with the anticodon stem loop positioned within the TLE arm, where the homologous anticodon-like sequence is located (29). The alignment suggests that both the unannealed and annealed PBS/TLE mimic the structure of tRNALys3 in solution.
Fig. 5.
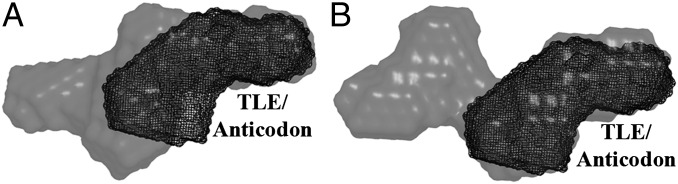
Assessment of tRNA-like structural characteristics of PBS/TLE and PBS/TLE–anti-PBS. The SAXS-derived ab initio envelope of human tRNALys3 was superimposed onto the envelopes for PBS/TLE (A) and PBS/TLE–anti-PBS (B) by aligning the anticodon region of the tRNALys3 envelope to the corresponding TLE region. In both panels, the viral RNA construct is shown in a gray 40% transparent surface representation, and the tRNALys3 envelope is represented as a black mesh.
Discussion
RNA structure and function are intimately related, and structural characterization of large functional RNAs contributes to a complete understanding of biological processes. The HIV-1 life cycle depends on the proper folding of domains present in the 5′ UTR. The feasibility of determining structural models of large, highly structured RNAs has recently improved with the development of techniques combining SAXS and molecular modeling (34, 40). This general approach has been used to characterize the structures of the hepatitis C virus internal ribosome entry site RNA (41), the cap-independent translation element/ribosome-binding structural element of turnip crinkle virus (42), RNase P RNA (43), and the U2/U6 snRNA (44). Herein we describe models derived using SAXS and molecular modeling of domains covering >92% of the 5′ UTR. Importantly, these models highlight mechanistic insights into the diverse functions of the 5′ UTR and provide a basis for further studies of the entire 5′ UTR and relevant RNA:protein complexes.
The SAXS-derived TAR/polyA structure reveals that it folds into extended, coaxially stacked helices (Fig. 2B). Coaxial stacking is well known to be a stabilizing structural feature of RNAs. The orientation observed for TAR/polyA hairpins is consistent with their distinct functional roles, which require binding of various factors to the apical loop and bulge region of TAR (7, 45). Although to our knowledge, no specific binding partners of the polyA domain have been reported, the observed structure would reinforce sequestration of the polyadenylation signal (46). The high stability of the coaxially stacked helices likely also contributes to the pausing of reverse transcription observed in this domain during minus-strand strong-stop DNA synthesis (47, 48), which results in increased efficiency of minus-strand transfer (47). Our structure also supports the requirement for HIV-1 nucleocapsid (NC) protein chaperone activity to partially destabilize these hairpins, thereby also facilitating minus-strand transfer (49, 50). To further evaluate the validity of our model, we compared it with a high-resolution NMR structure of the top end of the TAR hairpin [Protein Data Bank (PDB) ID code 1ANR] (15). The structures aligned with a heavy atom rmsd of 5.2 Å (Fig. S8); however, it should be noted that NMR experiments suggest a highly dynamic TAR loop (ranging from 4.0 to 9.7 Å rmsd from the lowest-energy structure) (15, 51). Nevertheless, as a qualitative comparison of the model, the overall kink in the helix at the base of the loop is recapitulated.
Two all-atom tertiary structure models of Psi (SL1–SL4) have been proposed. One model is globular with SL2–SL3 and SL1–SL4 interactions (22), whereas the other model is extended with all loops exposed (23). Our probe-free study supports the latter model in which SL1, SL2, and SL3 are solvent-exposed (Fig. 3B). It should be noted, however, that all three models have been derived using RNAs of different lengths and sequences. Moreover, differences may be accounted for in part by variations in sample purification and buffer composition. The SAXS-derived structure shows SL1 and SL3 directed in nearly opposite directions, with SL2 protruding perpendicularly (Fig. 3B), supporting the presence of distinct sites for known intermolecular interactions. For example, the DIS sequence in the apical SL1 loop must be exposed before genome dimerization (19, 20), and our model suggests the apo form of Psi is poised to do so. In addition, the NC domain of Gag recognizes the Psi sequence to select the vRNA for packaging (52). NC binds to the exposed G-rich bulge regions of SL1 (13) and has been shown to bind to the loops of individual SL2 and SL3 constructs with the same low-nanomolar affinity (31). The alternative fold of SL2 in our Psi model relative to that observed in a smaller construct designed for NMR studies (31) suggests that this domain may be dynamic in the context of the larger construct. This is consistent with structure-probing data (12, 13, 38, 53) and may explain why SL1 and SL3 are the primary determinants for HIV-1 genome encapsidation (53), despite similar affinities of SL2 and SL3 to NC.
Within the resolution restrictions of SAXS experiments, our model also shares structural similarities with high-resolution NMR structures of SL1 (PDB ID code 1M5L; heavy atom rmsd = 5.9 Å) (32) and SL3 (PDB ID code 1BN0; heavy atom rmsd = 4.5 Å) (33) (Fig. S8). A comparison of SL2 was not performed, because the difference in our secondary structure changes the identity of SL2 (Fig. S3; Psi Mfold 1) relative to the hairpin used for NMR structure determination (PDB ID code 1ESY) (31).
The secondary structure of the PBS/TLE domain has been extensively characterized through chemical and enzymatic probing in the free and tRNALys3-bound states (13, 23, 54–57). Despite its critical role in regulating initiation of reverse transcription, little is known about its tertiary structure. The solution structure of an A-rich stem loop in the HIV-1 MAL isolate PBS domain has been solved by NMR, revealing a tRNA anticodon-like U-turn conformation (58). Recently, probing data and computational approaches were used to develop a model of the tertiary structure of the PBS and Psi packaging domains (23). In that model, the PBS domain was not well defined and was suggested to be highly flexible, owing to the presence of a long, single-stranded bulge. Our analysis of the PBS/TLE domain suggests a less dynamic structure, giving a single, well-defined peak on SEC and high-quality SAXS data (Fig. S5). Although the single-stranded region is expected to be dynamic and to refold in the presence of tRNALys3, the PBS/TLE domain also contains well-structured helices, and the SAXS data were best fit when using the starting secondary structure shown in Fig. 1 rather than alternative secondary structures (Fig. S6).
Annealing of the 18-nt anti-PBS resulted in only minor changes to the TLE region of the PBS/TLE domain (Fig. S4C). Our analysis of the scattering curves revealed that the apo and annealed PBS/TLE constructs had nearly indistinguishable (within error) Guinier and P(r)-derived Rg values and only a slight increase in Dmax (from 118 to 123 Å). Superimposing the two structures shows that the loop region underwent a significant conformational shift, whereas the TLE and closing stem were altered only slightly (Fig. S4C). The poor fit between the envelopes and all-atom models in the single-stranded loop region of the apo and anti-PBS–annealed PBS/TLE constructs reduces confidence in the precise conformation of this region; however, the loop region of the envelope is rotated nearly 180° with respect to the TLE helix and closing stem, supporting a large conformational change on anti-PBS annealing. We hypothesize that this conformational change in the loop may contribute to a functional shift between a largely monomeric population of vRNA to dimeric vRNA, which occurs on primer annealing (30). Our model of PBS/TLE is also consistent with the proposal that during primer recruitment, tRNALys3 is released from LysRS through competition with the TLE (29). On primer annealing, the TLE remains in a tRNA-like conformation, inhibiting release of LysRS from the TLE. Further studies are needed to test this idea.
In conclusion, we report a robust method of producing reliable structural models of large RNAs through a combination of SAXS and molecular modeling. We have shown that the TAR and polyA hairpins adopt stable, coaxially stacked helices, and, similarly, the Psi domain adopts an extended conformation with SL1–SL3 available for interactions with viral/host factors. The PBS/TLE domain adopts a bent conformation, with an extended TLE that resembles a tRNA. Although anti-PBS annealing results in formation of an extended duplex with the PBS sequence, only subtle conformational changes occur in the TLE region, maintaining a tRNA-like fold. Additional conformational changes are expected to occur upon full tRNALys3 primer annealing, which involves extended interactions between tRNALys3 and the PBS region of the genome.
Methods
Experimental procedures are described in detail in SI Methods. In brief, RNA samples were in vitro transcribed and initially purified by denaturing gel electrophoresis, followed by refolding and native purification by SEC. Refolding conditions and SEC buffers were optimized to ensure homogeneity by varying the MgCl2 and NaCl concentrations. Purified monodisperse samples were analyzed at the Advanced Light Source using their mail-in system.
SAXS scattering profiles were analyzed using the PRIMUS (53) module within the ATSAS package (59). Ab initio envelope generation, averaging, and filtering were carried out using the DAMMIN module of ATSAS (59). To generate an all-atom model of the RNA tertiary structure, a three-step approach was applied, using de novo modeling to generate a starting ensemble of 10 structures, followed by ESMD and refinement through SA. An ensemble of the 10 lowest-energy structures from 50 SA refinements is reported here.
Supplementary Material
Acknowledgments
We thank Dr. R. Rambo (Lawrence Berkeley National Labs) and E. Ihms (The Ohio State University) for helpful suggestions. This work was supported by National Institutes of Health (NIH) Grant R01 AI077387 (to K.M.-F. and Dr. L. Kleiman). C.P.J. was supported by The Ohio State University Center for RNA Biology, E.D.O. by NIH Grant T32-GM008512, and W.A.C. by the Pelotonia Postdoctoral Fellowship Program. The work was conducted at the Advanced Light Source, a national user facility operated by Lawrence Berkeley National Laboratory on behalf of the US Department of Energy, Office of Basic Energy Sciences, through the Integrated Diffraction Analysis Technologies Program, supported by the Department of Energy’s Office of Biological and Environmental Research. Additional support was provided by the NIH Main Injector Neutrino Oscillation Search project (Grant R01 GM105404).
Footnotes
The authors declare no conflict of interest.
*This Direct Submission article had a prearranged editor.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1319658111/-/DCSupplemental.
References
- 1.Bieniasz PD. An overview of intracellular interactions between immunodeficiency viruses and their hosts. AIDS. 2012;26(10):1243–1254. doi: 10.1097/QAD.0b013e328353bd04. [DOI] [PubMed] [Google Scholar]
- 2.Lu K, Heng X, Summers MF. Structural determinants and mechanism of HIV-1 genome packaging. J Mol Biol. 2011;410(4):609–633. doi: 10.1016/j.jmb.2011.04.029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Clever JL, Eckstein DA, Parslow TG. Genetic dissociation of the encapsidation and reverse transcription functions in the 5′ R region of human immunodeficiency virus type 1. J Virol. 1999;73(1):101–109. doi: 10.1128/jvi.73.1.101-109.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Beerens N, Groot F, Berkhout B. Initiation of HIV-1 reverse transcription is regulated by a primer activation signal. J Biol Chem. 2001;276(33):31247–31256. doi: 10.1074/jbc.M102441200. [DOI] [PubMed] [Google Scholar]
- 5.Clever JL, Miranda D, Jr, Parslow TG. RNA structure and packaging signals in the 5′ leader region of the human immunodeficiency virus type 1 genome. J Virol. 2002;76(23):12381–12387. doi: 10.1128/JVI.76.23.12381-12387.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Kim HJ, Lee K, O’Rear JJ. A short sequence upstream of the 5′ major splice site is important for encapsidation of HIV-1 genomic RNA. Virology. 1994;198(1):336–340. doi: 10.1006/viro.1994.1037. [DOI] [PubMed] [Google Scholar]
- 7.Bannwarth S, Gatignol A. HIV-1 TAR RNA: The target of molecular interactions between the virus and its host. Curr HIV Res. 2005;3(1):61–71. doi: 10.2174/1570162052772924. [DOI] [PubMed] [Google Scholar]
- 8.Berkhout B, Vastenhouw NL, Klasens BI, Huthoff H. Structural features in the HIV-1 repeat region facilitate strand transfer during reverse transcription. RNA. 2001;7(8):1097–1114. doi: 10.1017/s1355838201002035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Beerens N, Kjems J. Circularization of the HIV-1 genome facilitates strand transfer during reverse transcription. RNA. 2010;16(6):1226–1235. doi: 10.1261/rna.2039610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Helga-Maria C, Hammarskjöld ML, Rekosh D. An intact TAR element and cytoplasmic localization are necessary for efficient packaging of human immunodeficiency virus type 1 genomic RNA. J Virol. 1999;73(5):4127–4135. doi: 10.1128/jvi.73.5.4127-4135.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Das AT, Harwig A, Vrolijk MM, Berkhout B. The TAR hairpin of human immunodeficiency virus type 1 can be deleted when not required for Tat-mediated activation of transcription. J Virol. 2007;81(14):7742–7748. doi: 10.1128/JVI.00392-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Damgaard CK, Andersen ES, Knudsen B, Gorodkin J, Kjems J. RNA interactions in the 5′ region of the HIV-1 genome. J Mol Biol. 2004;336(2):369–379. doi: 10.1016/j.jmb.2003.12.010. [DOI] [PubMed] [Google Scholar]
- 13.Wilkinson KA, et al. High-throughput SHAPE analysis reveals structures in HIV-1 genomic RNA strongly conserved across distinct biological states. PLoS Biol. 2008;6(4):e96. doi: 10.1371/journal.pbio.0060096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baudin F, et al. Functional sites in the 5′ region of human immunodeficiency virus type 1 RNA form defined structural domains. J Mol Biol. 1993;229(2):382–397. doi: 10.1006/jmbi.1993.1041. [DOI] [PubMed] [Google Scholar]
- 15.Aboul-ela F, Karn J, Varani G. Structure of HIV-1 TAR RNA in the absence of ligands reveals a novel conformation of the trinucleotide bulge. Nucleic Acids Res. 1996;24(20):3974–3981. doi: 10.1093/nar/24.20.3974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Aboul-ela F, Karn J, Varani G. The structure of the human immunodeficiency virus type-1 TAR RNA reveals principles of RNA recognition by Tat protein. J Mol Biol. 1995;253(2):313–332. doi: 10.1006/jmbi.1995.0555. [DOI] [PubMed] [Google Scholar]
- 17.Das AT, Klaver B, Klasens BI, van Wamel JL, Berkhout B. A conserved hairpin motif in the R-U5 region of the human immunodeficiency virus type 1 RNA genome is essential for replication. J Virol. 1997;71(3):2346–2356. doi: 10.1128/jvi.71.3.2346-2356.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Paillart JC, Skripkin E, Ehresmann B, Ehresmann C, Marquet R. In vitro evidence for a long-range pseudoknot in the 5′-untranslated and matrix coding regions of HIV-1 genomic RNA. J Biol Chem. 2002;277(8):5995–6004. doi: 10.1074/jbc.M108972200. [DOI] [PubMed] [Google Scholar]
- 19.Paillart JC, Marquet R, Skripkin E, Ehresmann B, Ehresmann C. Mutational analysis of the bipartite dimer linkage structure of human immunodeficiency virus type 1 genomic RNA. J Biol Chem. 1994;269(44):27486–27493. [PubMed] [Google Scholar]
- 20.Skripkin E, Paillart JC, Marquet R, Ehresmann B, Ehresmann C. Identification of the primary site of the human immunodeficiency virus type 1 RNA dimerization in vitro. Proc Natl Acad Sci USA. 1994;91(11):4945–4949. doi: 10.1073/pnas.91.11.4945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Schwartz S, Felber BK, Benko DM, Fenyö EM, Pavlakis GN. Cloning and functional analysis of multiply spliced mRNA species of human immunodeficiency virus type 1. J Virol. 1990;64(6):2519–2529. doi: 10.1128/jvi.64.6.2519-2529.1990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Yu ET, Hawkins A, Eaton J, Fabris D. MS3D structural elucidation of the HIV-1 packaging signal. Proc Natl Acad Sci USA. 2008;105(34):12248–12253. doi: 10.1073/pnas.0800509105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Stephenson JD, et al. Three-dimensional RNA structure of the major HIV-1 packaging signal region. Structure. 2013;21(6):951–962. doi: 10.1016/j.str.2013.04.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mak J, Kleiman L. Primer tRNAs for reverse transcription. J Virol. 1997;71(11):8087–8095. doi: 10.1128/jvi.71.11.8087-8095.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Huang Y, et al. Incorporation of excess wild-type and mutant tRNA(3Lys) into human immunodeficiency virus type 1. J Virol. 1994;68(12):7676–7683. doi: 10.1128/jvi.68.12.7676-7683.1994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pavon-Eternod M, Wei M, Pan T, Kleiman L. Profiling non-lysyl tRNAs in HIV-1. RNA. 2010;16(2):267–273. doi: 10.1261/rna.1928110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cen S, et al. Incorporation of lysyl-tRNA synthetase into human immunodeficiency virus type 1. J Virol. 2001;75(11):5043–5048. doi: 10.1128/JVI.75.11.5043-5048.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kleiman L, Jones CP, Musier-Forsyth K. Formation of the tRNALys packaging complex in HIV-1. FEBS Lett. 2010;584(2):359–365. doi: 10.1016/j.febslet.2009.11.038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jones CP, Saadatmand J, Kleiman L, Musier-Forsyth K. Molecular mimicry of human tRNALys anti-codon domain by HIV-1 RNA genome facilitates tRNA primer annealing. RNA. 2013;19(2):219–229. doi: 10.1261/rna.036681.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Seif E, Niu M, Kleiman L. Annealing to sequences within the primer binding site loop promotes an HIV-1 RNA conformation favoring RNA dimerization and packaging. RNA. 2013;19(10):1384–1393. doi: 10.1261/rna.038497.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Amarasinghe GK, De Guzman RN, Turner RB, Summers MF. NMR structure of stem-loop SL2 of the HIV-1 psi RNA packaging signal reveals a novel A-U-A base-triple platform. J Mol Biol. 2000;299(1):145–156. doi: 10.1006/jmbi.2000.3710. [DOI] [PubMed] [Google Scholar]
- 32.Greatorex J, Gallego J, Varani G, Lever A. Structure and stability of wild-type and mutant RNA internal loops from the SL-1 domain of the HIV-1 packaging signal. J Mol Biol. 2002;322(3):543–557. doi: 10.1016/s0022-2836(02)00776-3. [DOI] [PubMed] [Google Scholar]
- 33.Pappalardo L, Kerwood DJ, Pelczer I, Borer PN. Three-dimensional folding of an RNA hairpin required for packaging HIV-1. J Mol Biol. 1998;282(4):801–818. doi: 10.1006/jmbi.1998.2046. [DOI] [PubMed] [Google Scholar]
- 34.Rambo RP, Tainer JA. Improving small-angle X-ray scattering data for structural analyses of the RNA world. RNA. 2010;16(3):638–646. doi: 10.1261/rna.1946310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Andersen ES, et al. Role of the trans-activation response element in dimerization of HIV-1 RNA. J Biol Chem. 2004;279(21):22243–22249. doi: 10.1074/jbc.M314326200. [DOI] [PubMed] [Google Scholar]
- 36.Zuker M. Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res. 2003;31(13):3406–3415. doi: 10.1093/nar/gkg595. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Popenda M, et al. Automated 3D structure composition for large RNAs. Nucleic Acids Res. 2012;40(14):e112. doi: 10.1093/nar/gks339. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Harrison GP, Lever AM. The human immunodeficiency virus type 1 packaging signal and major splice donor region have a conserved stable secondary structure. J Virol. 1992;66(7):4144–4153. doi: 10.1128/jvi.66.7.4144-4153.1992. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bénas P, et al. The crystal structure of HIV reverse-transcription primer tRNA(Lys,3) shows a canonical anticodon loop. RNA. 2000;6(10):1347–1355. doi: 10.1017/s1355838200000911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Rambo RP, Tainer JA. Super-resolution in solution X-ray scattering and its applications to structural systems biology. Annu Rev Biophys. 2013;42:415–441. doi: 10.1146/annurev-biophys-083012-130301. [DOI] [PubMed] [Google Scholar]
- 41.Pérard J, Leyrat C, Baudin F, Drouet E, Jamin M. Structure of the full-length HCV IRES in solution. Nat Commun. 2013;4:1612. doi: 10.1038/ncomms2611. [DOI] [PubMed] [Google Scholar]
- 42.Zuo X, et al. Solution structure of the cap-independent translational enhancer and ribosome-binding element in the 3′ UTR of turnip crinkle virus. Proc Natl Acad Sci USA. 2010;107(4):1385–1390. doi: 10.1073/pnas.0908140107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kazantsev AV, et al. Solution structure of RNase P RNA. RNA. 2011;17(6):1159–1171. doi: 10.1261/rna.2563511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Burke JE, Sashital DG, Zuo X, Wang YX, Butcher SE. Structure of the yeast U2/U6 snRNA complex. RNA. 2012;18(4):673–683. doi: 10.1261/rna.031138.111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ganser-Pornillos BK, Yeager M, Pornillos O. Assembly and architecture of HIV. Adv Exp Med Biol. 2012;726:441–465. doi: 10.1007/978-1-4614-0980-9_20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Das AT, Klaver B, Berkhout B. A hairpin structure in the R region of the human immunodeficiency virus type 1 RNA genome is instrumental in polyadenylation site selection. J Virol. 1999;73(1):81–91. doi: 10.1128/jvi.73.1.81-91.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Gao L, Balakrishnan M, Roques BP, Bambara RA. Insights into the multiple roles of pausing in HIV-1 reverse transcriptase-promoted strand transfers. J Biol Chem. 2007;282(9):6222–6231. doi: 10.1074/jbc.M610056200. [DOI] [PubMed] [Google Scholar]
- 48.Klasens BI, Huthoff HT, Das AT, Jeeninga RE, Berkhout B. The effect of template RNA structure on elongation by HIV-1 reverse transcriptase. Biochim Biophys Acta. 1999;1444(3):355–370. doi: 10.1016/s0167-4781(99)00011-1. [DOI] [PubMed] [Google Scholar]
- 49.Guo J, et al. Zinc finger structures in the human immunodeficiency virus type 1 nucleocapsid protein facilitate efficient minus- and plus-strand transfer. J Virol. 2000;74(19):8980–8988. doi: 10.1128/jvi.74.19.8980-8988.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Levin JG, Mitra M, Mascarenhas A, Musier-Forsyth K. Role of HIV-1 nucleocapsid protein in HIV-1 reverse transcription. RNA Biol. 2010;7(6):754–774. doi: 10.4161/rna.7.6.14115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Al-Hashimi HM, et al. Concerted motions in HIV-1 TAR RNA may allow access to bound state conformations: RNA dynamics from NMR residual dipolar couplings. J Mol Biol. 2002;315(2):95–102. doi: 10.1006/jmbi.2001.5235. [DOI] [PubMed] [Google Scholar]
- 52.Berkowitz RD, Ohagen A, Höglund S, Goff SP. Retroviral nucleocapsid domains mediate the specific recognition of genomic viral RNAs by chimeric Gag polyproteins during RNA packaging in vivo. J Virol. 1995;69(10):6445–6456. doi: 10.1128/jvi.69.10.6445-6456.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.McBride MS, Panganiban AT. Position dependence of functional hairpins important for human immunodeficiency virus type 1 RNA encapsidation in vivo. J Virol. 1997;71(3):2050–2058. doi: 10.1128/jvi.71.3.2050-2058.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Isel C, Marquet R, Keith G, Ehresmann C, Ehresmann B. Modified nucleotides of tRNA(3Lys) modulate primer/template loop-loop interaction in the initiation complex of HIV-1 reverse transcription. J Biol Chem. 1993;268(34):25269–25272. [PubMed] [Google Scholar]
- 55.Isel C, et al. Structural basis for the specificity of the initiation of HIV-1 reverse transcription. EMBO J. 1999;18(4):1038–1048. doi: 10.1093/emboj/18.4.1038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Goldschmidt V, et al. Structural variability of the initiation complex of HIV-1 reverse transcription. J Biol Chem. 2004;279(34):35923–35931. doi: 10.1074/jbc.M404473200. [DOI] [PubMed] [Google Scholar]
- 57.Isel C, Ehresmann C, Keith G, Ehresmann B, Marquet R. Initiation of reverse transcription of HIV-1: Secondary structure of the HIV-1 RNA/tRNA(3Lys) (template/primer) J Mol Biol. 1995;247(2):236–250. doi: 10.1006/jmbi.1994.0136. [DOI] [PubMed] [Google Scholar]
- 58.Puglisi EV, Puglisi JD. HIV-1 A-rich RNA loop mimics the tRNA anticodon structure. Nat Struct Biol. 1998;5(12):1033–1036. doi: 10.1038/4141. [DOI] [PubMed] [Google Scholar]
- 59.Konarev PV, Petoukhov MV, Volkov VV, Svergun DI. ATSAS 2.1, a program package for small-angle scattering data analysis. J Appl Cryst. 2006;39:277–286. doi: 10.1107/S0021889812007662. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.