Significance
TubZ is a tubulin superfamily member that forms filaments necessary for faithful segregation of certain low copy-number bacterial plasmids. Here, we observe that filament formation of the Bacillus thuringiensis TubZ forms completely different filaments depending on the ability of GTP to hydrolyze. Using GTPγS or hydrolysis-blocked mutants results in two-stranded filaments, and polymerization with GTP yields a four-stranded morphology. Through a combination of high-resolution cryo-electron microscopy, light scattering, and mutational analyses, we propose that the two-stranded filament is a prehydrolysis intermediate on-pathway to the posthydrolysis four-stranded form. The density maps also suggest that the C-terminal region not visible in prior crystal structures makes critical interactions in the two-stranded filaments, explaining why even short truncations block polymerization.
Keywords: bacterial cytoskeleton, plasmid segregation
Abstract
Cytoskeletal filaments form diverse superstructures that are highly adapted for specific functions. The recently discovered TubZ subfamily of tubulins is involved in type III plasmid partitioning systems, facilitating faithful segregation of low copy-number plasmids during bacterial cell division. One such protein, TubZ-Bt, is found on the large pBtoxis plasmid in Bacillus thuringiensis, and interacts via its extended C terminus with a DNA adaptor protein TubR. Here, we use cryo-electron microscopy to determine the structure of TubZ-Bt filaments and light scattering to explore their mechanism of polymerization. Surprisingly, we find that the helical filament architecture is remarkably sensitive to nucleotide state, changing from two-stranded to four-stranded depending on the ability of TubZ-Bt to hydrolyze GTP. We present pseudoatomic models of both the two- and four-protofilament forms based on cryo-electron microscopy reconstructions (10.8 Å and 6.9 Å, respectively) of filaments formed under different nucleotide states. These data lead to a model in which the two-stranded filament is a necessary intermediate along the pathway to formation of the four-stranded filament. Such nucleotide-directed structural polymorphism is to our knowledge an unprecedented mechanism for the formation of polar filaments.
The tubulin family of cytoskeletal proteins plays important roles in both eukaryotic and prokaryotic cells. αβ-Tubulin dimers form microtubules in eukaryotic cells that are necessary for cell division and intracellular transport. The most common prokaryotic tubulin, FtsZ, plays an essential role in cytokinesis and is found ubiquitously in bacteria and also in many archaea. Additionally, a diverse set of less-conserved tubulin family members have been identified including a variety of monomeric eukaryotic tubulins (γ-, δ-, ε-, ζ-, and η-) (1), the αβ-tubulin–like heterodimer BtubA/B (2), the prokaryotic extrachromosomal TubZs involved in plasmid segregation (3–5), and the recently discovered bacteriophage encoded tubulins, PhuZ (6, 7).
Among eukaryotic αβ-tubulins, sequence identity is quite high (75–85%), as it is also among FtsZs (40–50%) (8). However, sequence identity between eukaryotic and prokaryotic family members is quite low (10–20%), and new tubulins are often discovered using only the limited number of highly conserved residues involved in nucleotide binding and hydrolysis. Despite this sequence diversity, the core structures of individual tubulin subunits are extraordinarily well conserved, and this structural conservation extends to the longitudinal interactions between monomers (9). In contrast, such striking structural conservation stops at the protofilament level: αβ-tubulin forms tubes of varying protofilament number (10), FtsZ forms a variety of straight and curved protofilament structures (11), and BtubA/B forms five protofilament structures (12). The lack of conservation of residues involved in lateral interactions has allowed the evolution of diverse higher-order filament structures, the organization and dynamics of which are precisely tuned to their cellular function.
The TubZ tubulin family, discovered recently on several Bacillus virulence plasmids (4, 5) and in a Clostridial bacteriophage (3), is a group of proteins involved in bacterial plasmid partitioning (par) systems. At their core, par systems contain a polymer-forming NTPase, a DNA-binding protein, and a centromeric binding site on the DNA (13, 14). Together, these components ensure that low copy-number plasmids are efficiently segregated to both daughter cells during cell division. The TubZ found on the pBtoxis plasmid from Bacillus thurigiensis (TubZ-Bt) has been shown to treadmill (growing at one end and shrinking at the other end), and these dynamics are important for proper plasmid segregation (15). Additionally TubZ-Bt assembly has been monitored in vitro by light scattering (16) and crystal structures have been solved in the presence of GDP, GTPγS, and in the apo state (17, 18).
TubZ-Bt acts in concert with the TubR helix-turn-helix DNA binding protein (18, 19), and binds to a centromeric region of the plasmid DNA, tubC, which contains seven 12-bp pseudorepeats. The TubRC complex binds in an unknown manner to the C terminus of TubZ (residues ∼407–484). Additionally, a new protein located downstream of tubZRC, named TubY, was recently implicated in the function of the Clostridium botulinum bacteriophage c-st TubZ. TubY stabilizes the assembly of TubZ filaments alone, but depolymerizes TubZ assembled in the presences of the TubRC complex. Other TubZ systems, including TubZ-Bt, have been found to have a similar protein located upstream of the operon (3), but it is unknown whether this putative TubY plays a role in pBtoxis TubZRC plasmid segregation.
Here we show that the filament morphology of TubZ-Bt is linked to a nucleotide state. Untagged TubZ-Bt forms almost exclusively four-stranded helical filaments in vitro in the presence of GTP, whereas the previously observed (16–18) two-stranded filaments only accumulate under conditions where GTP hydrolysis is blocked. We present ∼11 Å and ∼7 Å resolution cryo-electron microscopy (cryo-EM) reconstructions of the helical structure of TubZ-Bt grown in the presence of GTP or GTPγS, respectively. We can unambiguously fit the TubZ-Bt crystal structure into both of these filament morphologies to form detailed pseudoatomic models. We observed that removing the extreme C terminus of the protein compromises or abrogates filament assembly, although the majority of these residues are unresolved in the reconstructions. These structures, coupled with additional biochemical and mutational studies, lead to a model in which TubZ-Bt forms an unstable two-stranded intermediate on pathway to the formation of highly stable four-stranded filaments that is only reached upon GTP hydrolysis.
Results
TubZ Filament Morphology Is Affected by Nucleotide State.
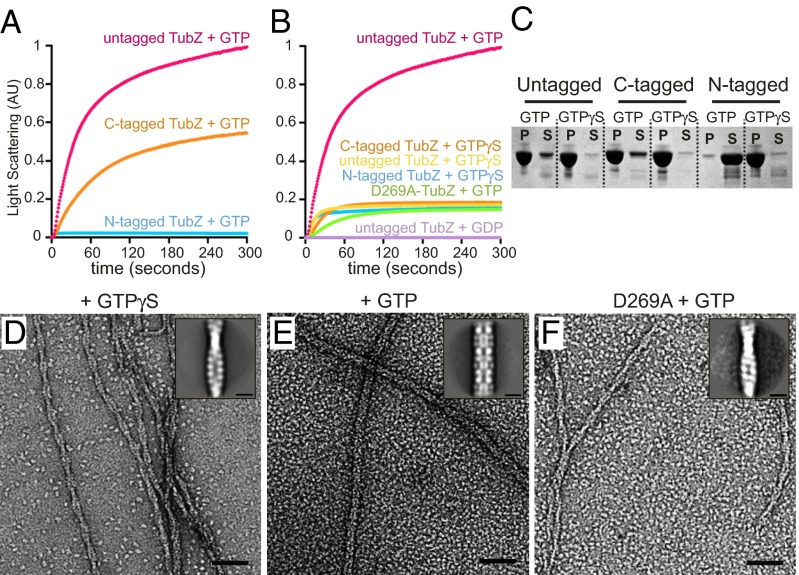
During early experiments to understand the kinetics of TubZ-Bt, we observed that an N-terminally tagged TubZ-Bt, with six residues before the start methionine after tobacco etch virus protease cleavage of a His-tag, exhibited extremely poor polymerization in the presence of GTP compared with a C-terminally tagged construct. Previous studies had used an N-terminally tagged construct for polymerization studies (16) and a C-terminally tagged construct for structural studies (17). Because the extreme C terminus has been shown to be important for binding to TubRC complexes, we cloned, expressed, and purified an untagged version of wild-type TubZ. Quantitatively comparing the polymerization ability of these different proteins using 90° light scattering in the presence of GTP revealed that C-terminally tagged protein exhibited strong polymerization, whereas the N-terminally tagged protein showed almost no polymerization (Fig. 1A). In contrast, untagged TubZ showed an even more robust polymerization than the C-terminally tagged construct, reaching a nearly doubled plateau value (Fig. 1A). Because this seemed at odds with previously published data, we next compared the polymerization kinetics of all three constructs (N-tagged, C-tagged, untagged) in the presence of the nonhydrolyzable GTP analog GTPγS by 90° light scattering. We observed that although all constructs show similar rapid polymerization with GTPγS (Fig. 1B), they plateau at a much lower scattering value than the assembly of untagged TubZ with GTP. GDP alone did not appear to polymerize TubZ to any detectable degree (Fig. 1B).
Fig. 1.
Changes in TubZ-Bt filament morphology and behavior in the presence of terminal tags. (A and B) Monitoring polymerization of 3 μM TubZ by light scattering. (A) N-terminally tagged TubZ in the presence of GTP, C-terminally His-tagged TubZ in the presence of GTP, and untagged TubZ in the presence of GTP. (B) N- or C-terminally His-tagged TubZ in the presence of GTPγS, and untagged TubZ in the presence of GTPγS, GTP, or GDP. C-terminally His-tagged D269A-TubZ (hydrolysis dead) in the presence of GTP. (C) SDS/PAGE of pelleting assays of N- or C-terminally tagged, untagged TubZ in the presence of GTP or GTPγS. P, pellet; S, supernatant. (D) Micrograph of negative-stain EM of untagged TubZ assembled with GTPγS. (Inset) Average of boxed two-stranded filaments. (E) Negative-stain EM of untagged TubZ assembled with GTP, inset is average of boxed filaments. (F) Negative-stain EM of C-terminally His-tagged D269A-TubZ assembled with GTP. (Inset) Average of boxed filaments. (Scale bars, 50 nm; Insets, 10 nm.)
To test the possibility that the observed lower plateaus with GTPγS were caused by unfavorable interactions with the nonhydrolysable analog, the catalytic aspartate on the T7 loop of TubZ was mutated (D269A) to abolish GTP hydrolysis, allowing us to directly assess the impact of hydrolysis. This mutant contains a C-terminal His-tag as the fully untagged D269A mutant proved refractory to purification. Notably, TubZ-D269A in the presence of GTP showed a polymerization curve comparable to that of GTPγS, with a similarly low plateau value (Fig. 1B).
Two possibilities exist to explain the striking differences in observed light-scattering plateau values. One possibility is that without hydrolysis, polymerization is somehow stunted: fewer polymers are formed, leading to a commensurate loss of signal. A second possibility is that the two filaments have different morphologies, with the GTP filament scattering more strongly (they are wider or longer) than the GTPγS/D269A TubZ filaments. The first possibility was tested by pelleting filaments polymerized using either GTP or GTPγS. Surprisingly, we observed that a greater percentage of TubZ polymerizes in GTPγS than in GTP, clearly showing that the lower plateau in light scattering cannot be a consequence of decreased polymer formation (Fig. 1C). To test the second possibility, TubZ filaments formed in the presence of either GTP or GTPγS were examined by negative-stain EM. Surprisingly, untagged TubZ polymerized with GTP predominantly formed thick filaments, ∼120 Å in diameter (Fig. 1E), whereas polymerization with GTPγS led to the thinner two-stranded filaments (Fig. 1 D and F) observed by previous groups (16, 17), although short thicker regions were occasionally observed. The ratio of filament types observed for TubZ-D269A in the presence of GTP or GTPγS was comparable to that in the WT-GTPγS sample, indicating that GTP hydrolysis must play a fundamental role in determining TubZ-Bt filament morphology (Fig. 1 D–F and Fig. S1 A–F), and that the two-stranded species is not an artifact of using a nucleotide analog.
TubZ-Bt Filaments Are Able to Convert Between Filament Forms.
Two-stranded filaments dominate TubZ preparations containing GTPγS, but thicker segments are also present (17). Whether two-stranded filaments emerge from the thicker filaments or vice versa is unclear. We boxed out these branching sites for analysis, and it appears that the two-stranded filaments emerging from thicker filaments have a similar morphology to unattached two-stranded filaments. Thick filaments predominate when assembling TubZ in the presence of GTP (Fig. 1E). However, if the two-stranded form were an intermediate to the thicker filaments, it might be possible to stabilize two-stranded filaments in the presence of GTP by either adding glycerol or forming filaments directly on carbon-coated grids rather than in solution before visualization, helping trap early intermediates. Using these strategies we were able to visualize two-stranded filaments leading in and out of thicker filaments, suggesting that the two-stranded filaments are transient intermediates in the formation of the thick filaments (Fig. S2).
Helical Reconstruction of TubZ Four-Stranded Filaments by Cryo-EM.
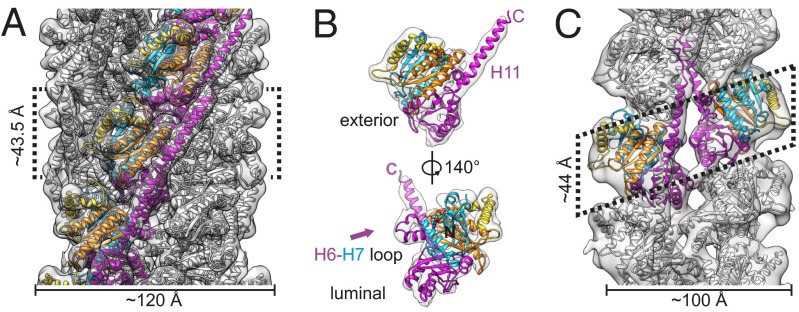
To further understand the structure of both filament types, we performed cryo-EM reconstructions (examples of collected data in Fig. S3 A–C) using iterative helical real-space refinement (20, 21). The wild-type TubZ-GTP filaments were reconstructed to a resolution of 6.9 Å by the 0.143 Fourier shell correlation (FSC) criteria using “gold-standard” refinement (22) (Fig. S4A). The TubZ-GTP filaments consist of four parallel protofilaments with fourfold planar symmetry (Fig. 2). The fiber has an outer diameter of ∼120 Å and an inner diameter of ∼19 Å, showing no strand–strand contacts across the lumen. The helical symmetry has a rise of ∼43.5 Å and an azimuthal angle of ∼31.8°. Crystal structures of TubZ fit quite well into the density of the reconstruction (Fig. 2 B and C, and Movie S1). This result indicates that that no large intramonomer conformational changes occur upon polymerization, suggesting the difference in four- and two-stranded morphologies is dictated largely by intermonomer conformational changes. Subtle adjustments to the structure of the loop between H6 and H7, and H11 (Fig. 2B and Fig. S4C) were made using Molecular Dynamics Flexible Fitting (MDFF) (23). An alignment between the individual chains of the filamentous crystal structures highlighted this range of structural conformations, and we observed that our modified structure falls well within the conformational range of known structures (Fig. S4C). Because of the weak density in the EM reconstruction for the T3 loop, it was difficult to fit this region accurately. Comparison of our density at a low contour level with the two conformations observed in the crystal structure shows that the conformation is likely in an intermediate conformation (Fig. S4E). We also sought to characterize the quality of our pseudoatomic model by comparing the FSC between it and the experimental map (Fig. S4A, red line), indicating an FSC of 0.5 at 7.6 Å, whereas the starting pseudoatomic model before MDFF has an FSC value of 0.5 at 8.77 Å.
Fig. 2.
Morphology of TubZ-GTP four-stranded filaments and two-stranded filaments. (A) Cryo-EM TubZ-GTP filament reconstruction. Filament has been low-pass filtered to 7 Å, a B-factor of ∼−309 applied, and high-pass filtered to 30 Å. A pseudoatomic model is fit, using a structure determined using MDFF. Residues 0–80 and 236–257 are colored in blue, 81–129 in gold, 130–212 in orange, and 213–235 and 258–414 in magenta. (B) Cut-out of one monomer of TubZ from the four-stranded filament. (C) Cryo-EM TubZ-GTPγS filament reconstruction. Filament has been low-pass filtered to 11 Å, a B-factor of ∼−452 applied, and high-pass filtered to 35 Å. A pseudoatomic model is fit using PDB ID code 2XKA chain F. In all, unconnected density has been hidden for improved visualization.
The important and generally well-conserved protofilament longitudinal interactions can be clearly observed and are comparable to those observed in the filamentous crystal structures. The rotation between longitudinal pairs in the four-stranded filament is most consistent with the rotation of those of the GDP crystal structure (31.8° vs. 30°, respectively) (17). One feature highlighted by Aylett et al. was a motion of helix H2 upon coordination of Mg2+ in the TubZ nucleotide-binding pocket (17). The authors observed that helix H2 takes on an “in” conformation with aspartate 64 coordinating with the Mg2+ in the binding pocket, whereas H2 is in an “out” conformation with the Mg2+ absent. In our EM structure, the density of the helix is clearly in the “out” position, indicating that Mg2+ is likely missing from the binding site, despite saturating amounts (mM) in the polymerization buffer, supporting the expectation that GTP would be hydrolyzed in the four-stranded filaments corresponding to a GDP state (Fig. S4D). Consequently, we will refer to this four-stranded filament grown from GTP as the GDP-state.
We were able to directly visualize lateral contacts between adjacent protofilaments. The lateral contacts form two charged surfaces (Fig. S4B) in one layer. Helices 6, 9, and 10, in addition to the loop between H6 and H7, forming one side of the interface, and residues on loops S9–S10, H1-S2 forming the other (Fig. S5A). The N-terminal helix H0 is also found in this interface, explaining the sensitivity of the four-stranded form to N-terminal purification tags, whereas the C terminus is on the outside surface of the filament (Fig. 2). In addition, the residues on H9 and H10 of the 0 monomer are potentially interacting with the residues between H2 and S3 on the adjacent layer (+1 monomer) (Fig. S4B).
Helical Reconstruction of TubZ Two-Stranded Filaments by Cryo-EM.
To understand the relationship between the two filament forms, we also performed a helical reconstruction of the TubZ-GTPγS filaments (examples of collected data in Fig. S3 D–F). To optimize formation of the two-stranded filaments, an N-terminally tagged construct was used in addition to GTPγS. The filaments were reconstructed to a resolution of 10.8 Å by the 0.143 FSC criteria (Fig. S6A). TubZ-GTPγS filaments refine to a helical rise of 22 Å and an azimuthal angle of 191.8°. Unlike the TubZ-GDP state, the two protofilaments are offset from each other by a half a monomer length (Fig. 2). We were able to unambigiously fit the TubZ crystal structure into the EM density (Movie S2), and a comparison of the FSC between a psuedo-atomic model using PDB ID code 2XKA chain F and the experimental map (Fig. S6A, red line), indicates an FSC of 0.5 at 14.4 Å. As in the four-stranded filament, we see that the longitudinal interactions are similar to those previously observed. The distance between longitudinally interacting monomers on the same strand is 44 Å, as dictated by the helical symmetry. (For clarity, we will refer to the monomers along the same strand as 0, +1, and so forth.).The long C-terminal helix of TubZ H11, and the N-terminal helix H0 are both exposed to solution on nonluminal, nonexternal opposite flat sides of the filament, which we designate the H4-H5 and N (for N terminal) interfaces, respectively (labeled in Fig. S6D). Surprisingly, there is a limited amount of density at the luminal junction of the two protofilaments. As with the four-stranded interface, this two-stranded interface is laced with charged residues (Fig. S5B). The positioning of the cross-strand monomers suggests interactions between H9 and H10 on one monomer and H6 and H11 of the opposite monomer (Fig. S6B). At this resolution it is hard to pinpoint definitively which residues are interacting, because the residue to residue distance varies depending on which rotamer is present.
Comparison of Two- and Four-Stranded Filaments.
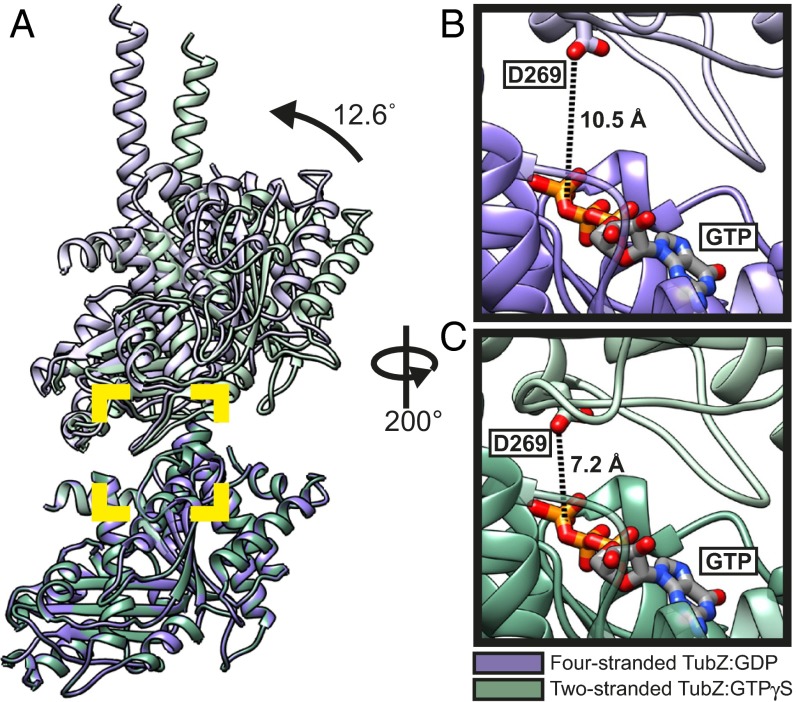
We compared the two filament forms to gain insight into the mechanism of filament assembly and the role of GTP binding and hydrolysis in the process. Although longitudinal interactions are generally well conserved, our EM structures reveal small but important differences between the longitudinal arrangements in the two- and four-stranded polymers (Fig. 3). The two-stranded filament has a significantly smaller intermonomer twist angle (∼23.8°) versus the four-stranded filament (∼31.8°). This finding is similar to—but more exaggerated than—the differences observed between the protofilament crystal forms of GTPγS (∼25.7°) and GDP (30°) (17), more precisely representing the dictates of the respective filament forms. In contrast, the longitudinal rises along the protofilament axes are nearly identical, only differing by ∼0.5 Å. The difference in twist angle can be explained by looking at an alignment of the bottom monomer of a longitudinal pair fit into our two maps (Fig. 3), which reveals a hinge-like opening motion transitioning between the GTPγS and GDP filament forms. The closed conformation of the GTPγS interaction allows for a tighter twist angle. The two interfaces that form the GTPase pocket open ∼12° between the two- and four-stranded structures, which moves the catalytic aspartate (residue 269) in the T7 loop of the +1 monomer further away from the location of the γ-phosphate in the GTP binding site of the +0 monomer (Fig. 3 B and C), suggesting that the two-stranded filaments are in a hydrolysis-competent state. In contrast, the distance of D269 to the predicted location of the γ-phosphate in the four-stranded filament would be considerably larger (∼10 Å vs. ∼7 Å). This finding suggests that the interaction with the γ-phosphate on GTP holds the longitudinal pair in a “closed” conformation, capable of forming the intermediary two-stranded morphology. After hydrolysis, this interaction is broken, and the longitudinal pairs “open,” forming the four-stranded filament structure. This observation, together with the position of H2, again supports that the four-stranded filament is in a posthydrolysis GDP state, and the two-stranded filament is in prehydrolysis state, consistent with results showing that the TubZ filaments grown in GTP contain mostly GDP (16).
Fig. 3.
Comparison of longitudinal fits of two- and four-stranded atomic models of TubZ-Bt. Structure used for representations in all panels is PDB ID code 2XKA chain F, with residues from the T3 and H6/H7 loops, as well as some residues at the C terminus of H11 removed. (A) Comparison of longitudinal fits of the two-stranded (in green) and four-stranded (in purple) atomic models, aligned using the bottom chain of the longitudinal pair. The angle change between the two models was determined using the difference in angle between planes of residues 1-383 of the top and bottom chains of each pair. These planes and their angles were generated and measured using Chimera (26). B and C represent a rotated zoom into the area marked by the yellow square of the longitudinal interface of the four- and two-stranded models, respectively. The distance measurement shown is the shortest distance between Aspartate 269 on the T7 loop, and the O3 oxygen of the β-phosphate of GTP.
Location and Importance of Extended C-Terminal Tail.
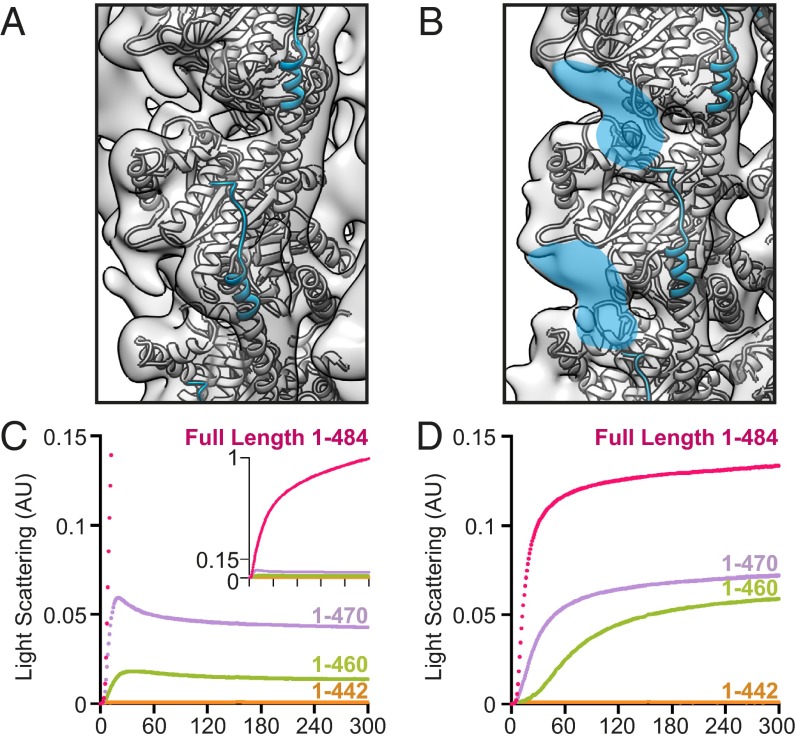
An important feature missing in the crystal structures is the extended C-terminal tail of TubZ-Bt, with the electron density only extending to residue 421 of 484 residues. This tail is necessary for interaction with TubRC complexes (18) and is highly charged. We anticipated the TubZ reconstructions would provide extra information about the structure and possible interactions of the C-terminal tail. However, density in the four-stranded reconstruction ends around residue 414, and little density beyond that expected for the crystal structure is visible anywhere in the map, even at a low contour level (Figs. 2B and 4A, and Fig. S7A).
Fig. 4.
The C-terminal tail of TubZ is important for assembly of both TubZ filament morphologies. (A) Helix H11 is highlighted in blue, with last residues in density (414) shown in spheres. The z axis has been rotated ∼19.5° for ease of comparison. (B) Residues 414–421 of Helix H11 are highlighted in blue. Extra density leading from the C-terminal tail is highlighted in blue. Structure used for representations in A and B is PDB ID code 2XKA chain F, with reconstructions low-pass filtered to 11 Å. (C and D) Monitoring polymerization of tail truncations of 3 μM TubZ by 90° light scattering. (C) Full-length (magenta), 1-470 (purple), 1-460 (green), and 1-442 (orange) untagged TubZ assembled in the presence of GTP. (D) Full-length (magenta), 1-470 (purple), 1-460 (green), and 1-442 (orange) untagged TubZ assembled in the presence of GTPγS.
In contrast, several prominent density features that cannot be accounted for by the crystal structure are seen in the two-stranded reconstruction. Unlike the four-stranded structure, density for H11 is visible at least up to the limit seen in the crystal structure (421). Notably, extra density (especially in the difference map, Fig. S7B) seems to extend beyond the end of the H11 emerging from the +0 monomer, but making contact with the +1 monomer. There is also additional density on the +2 monomer along a single protofilament (Fig. 4B and Fig. S7B). Because of the resolution of the reconstruction and the predicted lack of structure in this region, we were unable to convincingly model this area, but propose that both of these major regions of extra density arise from a continuation of the C-terminal tail of the +0 monomer. This finding suggests that C-terminal tail interactions along the protofilament could be important in the formation of the two-stranded filament.
To investigate the functional importance of the C-terminal tail for polymerization, several tagless truncations of varying lengths (1-442, 1-460, and 1-470) were cloned, expressed, and purified. These mutants were first characterized by light scattering to confirm their activity. To our surprise, even a 14-residue truncation (1-470) seriously compromised the polymerization kinetics of TubZ, both in the presence of GTP and GTPγS (Fig. 4 C and D). Removing another 10 residues (1-460) further impairs polymerization, whereas removal of 42 residues (1-442) completely abolishes observable polymerization of either form at concentrations comparable to full-length protein. By negative-stain EM, the filament morphologies of truncated constructs 1-470 and 1-460 are identical to that seen with full-length filament (Fig. S1 G–L), however at substantially lower abundance.
The Two-Stranded TubZ Filament Is on Pathway to the Four-Stranded Filament.
We observed that C-terminal truncations comparably affected TubZ polymerization efficiency in the presence of GTP or GTPγS, whereas the expectation from the degree of C-terminal contacts made in the four- and two-stranded filaments would suggest a much larger affect on the formation of the two-stranded filament. This finding, along with the structural differences described above, lead to a model in which a two-stranded, prehydrolysis intermediate is a necessary prerequisite to formation of the mature four-stranded posthydrolysis filament. The existence of such a transient two-stranded intermediate is also supported by our visualizations of the two-stranded filament branching into and out of four-stranded filaments. Given that a requirement for such a structurally different intermediate in tubulin family assembly is unprecedented, we pursued evidence for this transition through mutations.
Because we had observed that adding residues on the N terminus disrupted formation of the four-stranded TubZ filament, we designed a set of point mutations based on our structure that should preferentially compromise the four-stranded filament. To have a robust effect, three lysines on a loop adjacent to the N terminus in the four-stranded filament were mutated to alanines [K224A/K227A/K230A (3K)]. As observed by negative-stain EM, this mutation prevents polymerization in the presence of GTP. Notably, in the presence of GTPγS, mutant TubZ polymerizes similarly to wild-type protein, forming two-stranded filaments. In addition, single protofilaments can also be seen on grids of TubZ-3K, implying that these residues might be weakly involved in stabilizing the two-stranded filament via luminal interactions (Fig. S8). These observations further support the proposal of the two-stranded filaments being on pathway to the four-stranded form.
Discussion
B. thurigiensis TubZ Forms Distinct Two- and Four-Stranded Species.
Our results show that TubZ-Bt forms four-stranded filaments in vitro, in the presence of the native substrate GTP, and assumes a predominantly two-stranded conformation in the presence of the nonhydrolyzable nucleotide analog GTPγS. We propose that both conformations are biologically relevant, with the two-stranded form being a transient intermediate on the pathway to the four-stranded state. Previous reports have focused on the two-stranded form, either because of their use of GTPγS (17) or their use of N-terminally tagged protein (16, 18). We show here that both of these factors have an unexpectedly profound effect on filament morphology, either by disruption of critical lateral interfaces or by affecting nucleotide turnover and filament kinetics.
Beyond these overall observations, we determined cryo-EM reconstructions of both filament forms at resolutions of 6.9 Å and 10.8 Å (four- and two-stranded forms, respectively), allowing the crystal structure of TubZ-Bt to be unambiguously positioned within our experimental density maps. The resultant pseudoatomic resolution molecular models are notably different from previously published work using negative-stain reconstructions (17); this discrepancy is likely a result of the higher resolution data available when using cryo-EM, and the lack of features on TubZ-Bt making accurate fitting at low resolution problematic. Our model for the two-stranded filament is rotated roughly 90° from the previous fitting, with H11 and the N terminus located on opposite surfaces on the side of the lumen of the filament. In an effort to confirm the prior model, the authors sought to determine the location of the C terminus using a C-terminal GFP tag. Although the location of the GFP in their negative-stain map was consistent with H11 being on the outside of the filament, the position of GFP is unfortunately a very weak constraint because there are ∼60 intervening residues. Indeed, our two-stranded map suggests that a significant portion of the C terminus binds along the filament and ends up on the external surface, whereas part of H11 is oriented toward the luminal interface of the filament (Figs. S6B and S7B).
Effects of Nucleotide State on TubZ-Bt Filament Morphology.
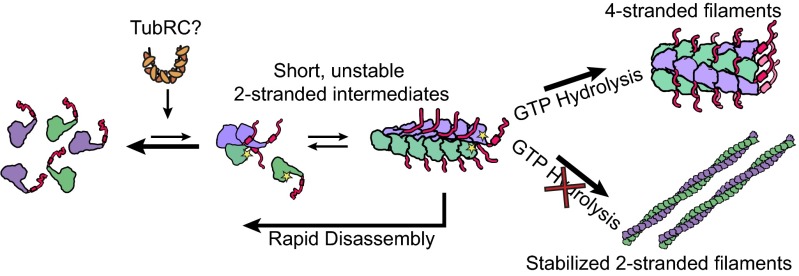
We observed that TubZ-Bt filaments have the ability to transition between an unstable two-stranded filament and a four-stranded filament. Four observations suggest that the two-stranded filament is an on-pathway intermediate to the formation of the more stable four-stranded filament: First, blocking the ability of TubZ-Bt to hydrolyze GTP by either mutagenesis or chemical means stabilizes the two-stranded filament and reduces the population of four-stranded filaments. Second, in the presence of GTP the population of two-stranded filaments can be significantly increased by using conditions that would either be expected to trap initial events or stabilize transient species. Third, mutations that disrupt the four-stranded lateral interface but do not impact GTP hydrolysis lead to a loss of polymer in GTP but not in GTPγS, indicating that the two-stranded filaments are unstable after GTP hydrolysis. Finally, the four-stranded filament structure appears to be in a posthydrolysis conformation containing GDP, whereas the two-stranded filament is in a prehydrolysis conformation. The lack of filament growth in the presence of GDP leads to the supposition that there is an obligate structural intermediate that is in a hydrolysis-competent conformation, and allows for formation of the four-stranded filament. This dramatic reconfiguration of filament morphology is an unprecedented mechanism of filament formation (Fig. 5).
Fig. 5.
Model of the polymerization mechanism of TubZ. TubZ monomers assemble into short unstable two-stranded intermediates, and convert into four-stranded mature filaments in the presence of GTP. If nucleotide hydrolysis in blocked either mutationally or chemically, the two stranded filaments are stabilized and able to form long filaments. Formation of four-stranded filaments is diminished, as the conversion from two- to four-strands is slower than the formation of the two-stranded filament.
In eukaryotic tubulins both intra- and intermonomer conformational changes have been observed between unassembled αβ-dimers and αβ-dimers in the microtubule lattice. This change happens via a piston-like motion of H6 and H7 (24); no such conformational switch has been observed in the crystal structures of TubZ-Bt, nor do we have evidence of it happening in our EM structures. Instead, it appears that the conformational change underlying the different TubZ-Bt filament morphologies is driven by an opening of the nucleotide cleft and consequent changes in protofilament twist, implying that the TubZ-Bt:GTPγS and TubZ-Bt:GDP structures represent pre- and posthydrolysis states, respectively. This motion appears to be based solely on changes in the intermonomer interactions, rather than an intramonomer rearrangement of TubZ-Bt, and represents a unique mechanism to couple nucleotide state to filament morphology.
Dramatic rearrangements would have to occur for long stretches of two-stranded filaments to become four-stranded. Changes in longitudinal twist upon GTP hydrolysis must directly disrupt interactions that stabilize the staggered two-stranded state and provide new opportunities for stabilizing four-stranded planar interactions. Whether the preexisting two strands (Fig. S9A) initially reform as a planar adjacent pair (Fig. S9B), or first separate completely to become diametrically opposed protofilaments (Fig. S9C) within the four-stranded filament, is unclear. Either way, additional monomers would be necessary to stabilize the final form, adding either in between opposed strands or on to the reformed planar pairs.
Given the infrequent visualization of two-stranded filaments of any length under normal conditions, it seems likely that the two-stranded filaments are short in both length and duration. Thus, only a small number of “closed” protofilament interactions would need to decouple upon hydrolysis as the “opening” of longitudinal interactions occurs. Such a model would suggest the possibility of a short two-stranded cap at the growing end of the filament. Determining how the transformation occurs, and looking for a specialized cap structure, would shed light on the underlying polymer dynamics by which TubZ-Bt is able to segregate plasmids.
TubZ-Bt Polymerization Is Dependent on the C-Terminal Tail.
It is well known that the C-terminal tail of tubulin family members plays an important role in the binding of accessory proteins. We have shown that even small truncations of the C-terminal tail induce a dramatic loss of polymerization of the TubZ-Bt filament. Similarly, removal of the C-terminal tail of TubZ-Bc leads to a loss of polymerization (25). Intriguingly, there is no observed density for ∼70 residues of this tail in the four-stranded structure, but there is extra density in the two-stranded intermediate that potentially represents at least a portion of the C-terminal tail. It seems most likely that the residues in this density are the residues immediately following those seen in the crystal structures, although parts of the C terminus may loop out before returning to bind to the +2 subunit. In the context of our model of TubZ polymerization, this finding suggests that the C-terminal tail may be involved in filament nucleation via longitudinal interactions in the two-stranded filament. We propose that in the closed two-stranded GTP state, the residues C-terminal to H11 are able to form important stabilizing interactions along the protofilaments, between 0, +1, and +2 subunits. Upon hydrolysis, the loss of the D269/γ-phosphate interaction causes cleft opening, straining the C-terminal interaction, and allowing for formation of the more stable four-stranded filament.
It has been observed that the TubRC complex nucleates TubZ filaments (19). The TubRC complex has been shown to bind to the C-terminal tail, and it appears that residues 408–484 have an important effect, although the most dramatic loss in binding comes from the loss of the same 14 residues that have most dramatic affect on TubZ polymerization (18). An intriguing speculation is that the TubRC complex is involved in modulating the interaction of the C-terminal tail with the plus-end subunit as the filament grows. An important future direction is to obtain structural understanding of the interaction between the TubRC complex and the TubZ filament. Does TubR bind on the side of TubZ filaments and act like a tram for the transport of DNA, or does it form a formin-like ring that either caps or slides along the TubZ filament to facilitate proper location of the pBtoxis plasmid during cell division? Does it facilitate conversion between two- and four-stranded polymers?
TubZ-Bt Filaments in the Cell.
Previous whole-cell tomography data suggest that TubZ-Bt may be two-stranded in vivo. These data were collected in Escherichia coli cells, with TubZ-Bt overexpressed and in the absence of the TubRC complex (17). Unfortunately, we have not yet been able to observe the filament morphology of TubZ-Bt in native cells and in the presence of pBtoxis. We note, however, that we have not been able to identify conditions that eliminate the preferential formation of four-stranded filaments of wild-type TubZ-Bt in the presence of GTP, even when including TubRC.
If TubZ does in fact preferentially form a four-stranded filament in the cell, what would be the purpose of this unique two-stranded intermediate? Whether it helps the fine-tuning of filament dynamics alone or in concert with TubRC and the pBtoxis plasmid or provides a means to better couple TubRC to just the growing end of filaments are two possibilities. A tomographic reconstruction of TubZ-Bt in cells, similar to recent work on BtubA/B filaments (12) would help resolve structural questions that might remain.
Materials and Methods
Detailed methods are provided in SI Materials and Methods. Briefly, TubZ proteins were expressed in E. coli and purified. TubZ polymerization was monitored by 90° light scattering using an in-house designed stopped-flow system monitored at 530 nm. Negative-stain electron microscopy was carried out using 0.75% uranyl formate, and imaged on FEI Tecnai T12. Cryo-EM was carried out using sample applied to holey carbon grids (C-flat, Protochips) frozen in liquid ethane using an FEI Vitrobot. Images of frozen sample were collected on a FEI Tecnai F20 operating at 200 kV with an 8k × 8k TemCam-F816 camera (TVIPS). Data were processed using SPIDER, EMAN1.9 and in-house CTF correction software. Reconstructions were performed using Iterative Helical Real Space Refinement. Molecular modeling of four-stranded pseudoatomic model was performed using MDFF.
Supplementary Material
Acknowledgments
We thank M. Erb and J. Pogliano for the gift of DNA encoding TubZ-Bt, and many discussions; J. Kraemer, S. Li, M. Moritz, R. Choy, D. Southworth, J. Polka, and C. Rivera for helpful discussions; E. Zehr for providing important assistance to using Molecular Dynamics Flexible Fitting software; Dr. Justin Kollman for electron microscopy advice and support throughout this project; and M. Braunfeld, C. Waddling, and P. Wassam for providing assistance with electron microscope and computing facilities. This work was supported by both the National Institutes of Health (GM031627) and the Howard Hughes Medical Institute.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The atomic coordinates and structure factors have been deposited in the Protein Data Bank, www.pdb.org [PDB ID codes 3J4T (Pseudo-Atomic Model) and 3J4S (Pseudo-Atomic Model)] and the Electron Microscopy Data Bank (accession nos. EMDB-5762 and EMDB-5763).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1318339111/-/DCSupplemental.
References
- 1.Dutcher SK. Long-lost relatives reappear: Identification of new members of the tubulin superfamily. Curr Opin Microbiol. 2003;6(6):634–640. doi: 10.1016/j.mib.2003.10.016. [DOI] [PubMed] [Google Scholar]
- 2.Jenkins C, et al. Genes for the cytoskeletal protein tubulin in the bacterial genus Prosthecobacter. Proc Natl Acad Sci USA. 2002;99(26):17049–17054. doi: 10.1073/pnas.012516899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Oliva MA, Martin-Galiano AJ, Sakaguchi Y, Andreu JM. Tubulin homolog TubZ in a phage-encoded partition system. Proc Natl Acad Sci USA. 2012;109(20):7711–7716. doi: 10.1073/pnas.1121546109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tang M, Bideshi DK, Park HW, Federici BA. Minireplicon from pBtoxis of Bacillus thuringiensis subsp. israelensis. Appl Environ Microbiol. 2006;72(11):6948–6954. doi: 10.1128/AEM.00976-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Tinsley E, Khan SA. A novel FtsZ-like protein is involved in replication of the anthrax toxin-encoding pXO1 plasmid in Bacillus anthracis. J Bacteriol. 2006;188(8):2829–2835. doi: 10.1128/JB.188.8.2829-2835.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Aylett CH, Izoré T, Amos LA, Löwe J. Structure of the tubulin/FtsZ-like protein TubZ from Pseudomonas bacteriophage ΦKZ. J Mol Biol. 2013;425(12):2164–2173. doi: 10.1016/j.jmb.2013.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kraemer JA, et al. A phage tubulin assembles dynamic filaments by an atypical mechanism to center viral DNA within the host cell. Cell. 2012;149(7):1488–1499. doi: 10.1016/j.cell.2012.04.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Erickson HP. Evolution of the cytoskeleton. Bioessays. 2007;29(7):668–677. doi: 10.1002/bies.20601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Nogales E, Downing KH, Amos LA, Löwe J. Tubulin and FtsZ form a distinct family of GTPases. Nat Struct Biol. 1998;5(6):451–458. doi: 10.1038/nsb0698-451. [DOI] [PubMed] [Google Scholar]
- 10.Sui H, Downing KH. Structural basis of interprotofilament interaction and lateral deformation of microtubules. Structure. 2010;18(8):1022–1031. doi: 10.1016/j.str.2010.05.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Erickson HP, Taylor DW, Taylor KA, Bramhill D. Bacterial cell division protein FtsZ assembles into protofilament sheets and minirings, structural homologs of tubulin polymers. Proc Natl Acad Sci USA. 1996;93(1):519–523. doi: 10.1073/pnas.93.1.519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Pilhofer M, Ladinsky MS, McDowall AW, Petroni G, Jensen GJ. Microtubules in bacteria: Ancient tubulins build a five-protofilament homolog of the eukaryotic cytoskeleton. PLoS Biol. 2011;9(12):e1001213. doi: 10.1371/journal.pbio.1001213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ebersbach G, Gerdes K. Plasmid segregation mechanisms. Annu Rev Genet. 2005;39:453–479. doi: 10.1146/annurev.genet.38.072902.091252. [DOI] [PubMed] [Google Scholar]
- 14.Salje J. Plasmid segregation: How to survive as an extra piece of DNA. Crit Rev Biochem Mol Biol. 2010;45(4):296–317. doi: 10.3109/10409238.2010.494657. [DOI] [PubMed] [Google Scholar]
- 15.Larsen RA, et al. Treadmilling of a prokaryotic tubulin-like protein, TubZ, required for plasmid stability in Bacillus thuringiensis. Genes Dev. 2007;21(11):1340–1352. doi: 10.1101/gad.1546107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Chen Y, Erickson HP. In vitro assembly studies of FtsZ/tubulin-like proteins (TubZ) from Bacillus plasmids: Evidence for a capping mechanism. J Biol Chem. 2008;283(13):8102–8109. doi: 10.1074/jbc.M709163200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Aylett CH, Wang Q, Michie KA, Amos LA, Löwe J. Filament structure of bacterial tubulin homologue TubZ. Proc Natl Acad Sci USA. 2010;107(46):19766–19771. doi: 10.1073/pnas.1010176107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Ni L, Xu W, Kumaraswami M, Schumacher MA. Plasmid protein TubR uses a distinct mode of HTH-DNA binding and recruits the prokaryotic tubulin homolog TubZ to effect DNA partition. Proc Natl Acad Sci USA. 2010;107(26):11763–11768. doi: 10.1073/pnas.1003817107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Aylett CH, Löwe J. Superstructure of the centromeric complex of TubZRC plasmid partitioning systems. Proc Natl Acad Sci USA. 2012;109(41):16522–16527. doi: 10.1073/pnas.1210899109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Egelman EH. The iterative helical real space reconstruction method: Surmounting the problems posed by real polymers. J Struct Biol. 2007;157(1):83–94. doi: 10.1016/j.jsb.2006.05.015. [DOI] [PubMed] [Google Scholar]
- 21.Kollman JM, Polka JK, Zelter A, Davis TN, Agard DA. Microtubule nucleating gamma-TuSC assembles structures with 13-fold microtubule-like symmetry. Nature. 2010;466(7308):879–882. doi: 10.1038/nature09207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Scheres SH, Chen S. Prevention of overfitting in cryo-EM structure determination. Nat Methods. 2012;9(9):853–854. doi: 10.1038/nmeth.2115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Trabuco LG, Villa E, Mitra K, Frank J, Schulten K. Flexible fitting of atomic structures into electron microscopy maps using molecular dynamics. Structure. 2008;16(5):673–683. doi: 10.1016/j.str.2008.03.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ravelli RB, et al. Insight into tubulin regulation from a complex with colchicine and a stathmin-like domain. Nature. 2004;428(6979):198–202. doi: 10.1038/nature02393. [DOI] [PubMed] [Google Scholar]
- 25.Hoshino S, Hayashi I. Filament formation of the FtsZ/tubulin-like protein TubZ from the Bacillus cereus pXO1 plasmid. J Biol Chem. 2012;287(38):32103–32112. doi: 10.1074/jbc.M112.373803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Pettersen EF, et al. UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem. 2004;25(13):1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.