Significance
Some genotypes of parasites can infect some genotypes of hosts but not others, whereas hosts also vary in susceptibility to a given parasite genotype. Variation in genes important for defenses against parasites could produce this specificity. Here, we find that variation in gene expression depended on both the genotype of the host and the genotype of the parasite. Moreover, we found that bumblebees that were exposed to infectious genotypes of a trypanosome parasite had low gene expression of immune genes but upregulation of genes that control expression. A poorly infecting parasite genotype, however, induced expression of immune genes. These results suggest that variation in the regulation of gene expression may also contribute to producing genotype-by-genotype specificity.
Keywords: coevolution, manipulation, Red Queen
Abstract
In many systems, host–parasite evolutionary dynamics have led to the emergence and maintenance of diverse parasite and host genotypes within the same population. Genotypes vary in key attributes: Parasite genotypes vary in ability to infect, host genotypes vary in susceptibility, and infection outcome is frequently the result of both parties’ genotypic identities. These host–parasite genotype-by-genotype (GH × GP) interactions influence evolutionary and ecological dynamics in important ways. Interactions can be produced through genetic variation; however, here, we assess the role of variable gene expression as an additional source of GH × GP interactions. The bumblebee Bombus terrestris and its trypanosome gut parasite Crithidia bombi are a model system for host–parasite matching. Full-transcriptome sequencing of the bumblebee host revealed that different parasite genotypes indeed induce fundamentally different host expression responses and host genotypes vary in their responses to the infecting parasite genotype. It appears that broadly and successfully infecting parasite genotypes lead to reduced host immune gene expression relative to unexposed bees but induce the expression of genes responsible for controlling gene expression. Contrastingly, a poorly infecting parasite genotype induced the expression of immunologically important genes, including antimicrobial peptides. A targeted expression assay confirmed the transcriptome results and also revealed strong host genotype effects. In all, the expression of a number of genes depends on the host genotype and the parasite genotype and the interaction between both host and parasite genotypes. These results suggest that alongside sequence variation in coding immunological genes, variation that controls immune gene expression can also produce patterns of host–parasite specificity.
Infection success of a parasite generally depends on the genotype of both the host and the parasite (1–5). The nature of this host–parasite genotype-by-genotype (GH × GP) interaction is important not only because it affects how characteristics of hosts and parasites evolve but because this genotypic level of specificity between hosts and parasites can also foster population-wide genetic diversity. Arguably, how genetic diversity is maintained is one of the most important questions in evolutionary ecological research. One process that has come into close focus is negative frequency-dependent selection of genotypes as a result of fluctuating, antagonistic host–parasite coevolution, captured in the Red Queen scenario (6, 7). Under certain conditions, such fluctuations will select for genetic exchange (sexual reproduction and recombination) in both hosts (6, 7) and parasites (8), and so produce and maintain genetic variation (9).
The coevolution of hosts and parasites is thought to be responsible for the rapid evolution of immune defense-related genes in various host taxa (10–14), and these genes can determine susceptibility of hosts to particular parasites (15–17). Although some genes are evolving extremely rapidly, particularly those genes involved in antiviral responses (13, 18), many other immunological genes seem to remain comparatively static (10, 11). The reasons for this static pattern can be manifold; for example, the same genes may be constrained by their function in other essential tasks [e.g., embryonic development in the case of members of the Toll pathway (19–21)]. However, an additional, nonexclusive hypothesis deserves attention: Variation in expression of otherwise invariable genes could play a major role in producing host resistance and, in particular, generate host–parasite specificity as is seen by GH × GP interactions. Existing GH × GP patterns are modulated by environmental conditions (22, 23), suggesting that immunity is a plastic trait and expression of defense mechanisms changes according to context. This environmental interaction with immunity is perhaps unsurprising, because variable contexts will shift the relative immunological costs (24). Some authors have suggested that gene expression in host–parasite interactions is likely to evolve toward greater immunological surveillance and reduced parasite conspicuousness (25); however, on a finer scale, gene expression differences among hosts may provide variation in resistance (26) without necessarily being based on differences in the coding sequence of key immunological genes. Such specific variation in how genes are expressed could therefore produce specificity in the outcome when various host and parasite genotypes interact. Interestingly, the expression of defense genes shows elevated levels of additive genetic variance in Drosophila species, which can indicate balancing selection (27) and can result from frequency-dependent selection as in Red Queen host–parasite dynamics. Indeed, host gene expression responses vary with host genotype in several systems [e.g., Drosophila melanogaster (28), Apis mellifera (29), Bombus terrestris (30–32), and Mus musculus (32)]. Similarly, some studies have described variation in host expression depending on parasite genotype (33–35). It is important to note that variation in gene expression is itself based on genetic variation in regulatory genes, but the exact regulatory pathways are poorly known in most systems. If regulatory variation is crucial, hosts may vary in their standing or induced immunological expression profiles independent of variation in the coding sequence of these genes. While phenotypic variation in parasite infectivity or host resistance is well described, little is known about how distinct host genotypes respond to distinct parasite genotypes.
The common European bumblebee, B. terrestris, has become a model of host–parasite interactions, particularly with its prevalent trypanosome gut parasite, Crithidia bombi (3, 36). C. bombi is genetically highly diverse (37–40) and infects a high proportion of B. terrestris in the wild (31, 41, 42). Different parasite genotypes have different infection success in different colonies of this host (43). Because B. terrestris mates singly, and because of the hymenopteran haplodiploid sex determination system, the relatedness of workers within one colony is high (r = 0.75). Thus, all workers originating from a colony can be considered as having a single genotypic background. Not only do genotypes of hosts and parasites vary in their abilities to resist or infect, respectively, but there is a statistical interaction between host and parasite genotypes (GH × GP) that determines the outcome of infection (3, 4), and this, in turn, is moderated by external variables, such as food availability (22).
Resistance, as measured by infection success or intensity, and the expressed immune response of bumblebees differ with their genotype (30, 31). In addition, some of these responses also differ according to the genotype of the parasite (34, 35). These targeted candidate gene approaches have established that the genotype of both the host and the parasite can influence the expression of immunologically important host genes. However, these targeted approaches can only consider known immune genes, usually derived from model insect species (e.g., D. melanogaster). As such, this approach may miss important immunological patterns if B. terrestris and model species differ in their immune repertoires. Recent full-genome sequencing of other insects, such as honeybees and pea aphids, has found that the immune repertoire and organization can differ considerably from the well-studied Dipteran models (44, 45). Furthermore, gut immunity, which is relevant for C. bombi and other parasites that reside in the gut, is generally not yet well understood. Some progress has been made concerning gut immunity of model insects used in other contexts (46), but little is known from host species serving as models in evolutionary ecology studies of host–parasite interactions, such as bumblebees. Here, we address gene expression variation that could underlie the GH × GP interactions of the B. terrestris–C. bombi system. We first infected four host colonies (GH) with three clonal parasite genotypes (GP) to assess the specificity of infection intensity and success. Infections can be readily quantified by the number of parasite cells shed in the feces. We then used RNA sequencing on dissected guts to assess how B. terrestris workers from these colonies, or genetic backgrounds, respond upon exposure to different genotypes of their ubiquitous trypanosome parasite C. bombi.
Results
Infection.
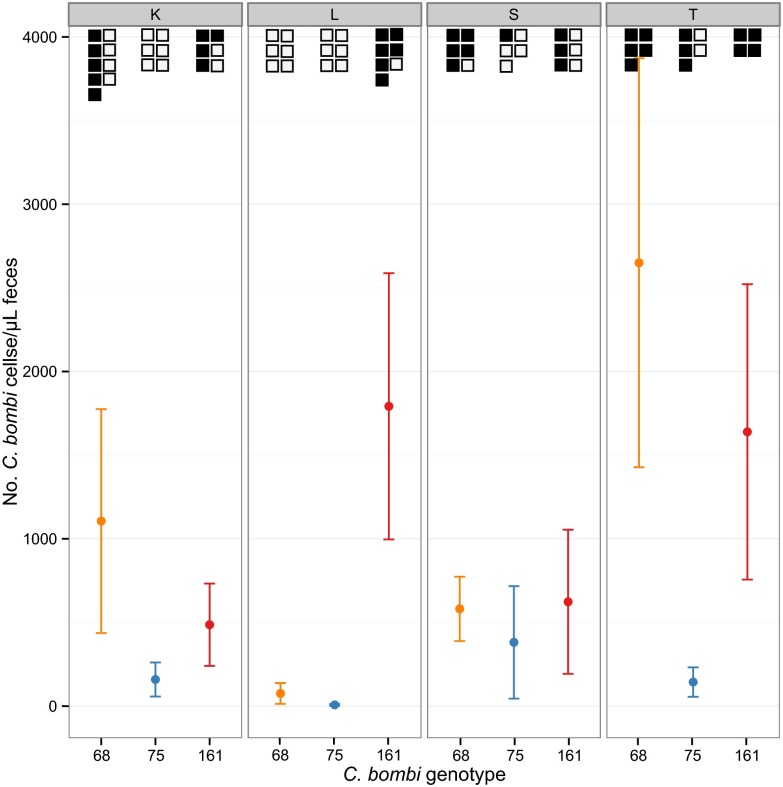
We found that the interaction of the host genotype and the parasite genotype (i.e., “who infects whom”) determined both infection intensity (measured as parasite cells per microliter of feces; Fig. 1) and infection success (presence/absence of visible C. bombi cells in the feces; squares in Fig. 1). Both parasite and host genotype alone also significantly influenced infection success. Parasite genotype no. 75 was the least likely to infect a bee and produced the lowest infection intensity. Parasite genotypes nos. 161 and 68 had higher infection success and infection intensity.
Fig. 1.
Mean number of C. bombi cells ± SE per microliter of feces according to C. bombi genotype and host genotype (colonies K, L, S, and T). The upper squares represent the number of experimentally exposed individuals checked for infection, with squares being filled if subsequent visual inspection of their feces revealed an established infection. In each panel, the x axis lists the parasite genotypes (nos. 68, 75, and 161; and are colored orange, blue, and red, respectively). Both infection intensity and whether or not an individual becomes infected varied according to the interaction between host and parasite genotypes (GH × GP intensity: F6,59 = 2.52, P = 0.031; infection: χ26,59 = 14.10, P = 0.029). Both parasite and host genotype alone significantly influenced infection success (GP: χ22,65 = 22.01, P < 0.001, GH: χ23,67 = 11.04, P = 0.012).
RNA Sequencing.
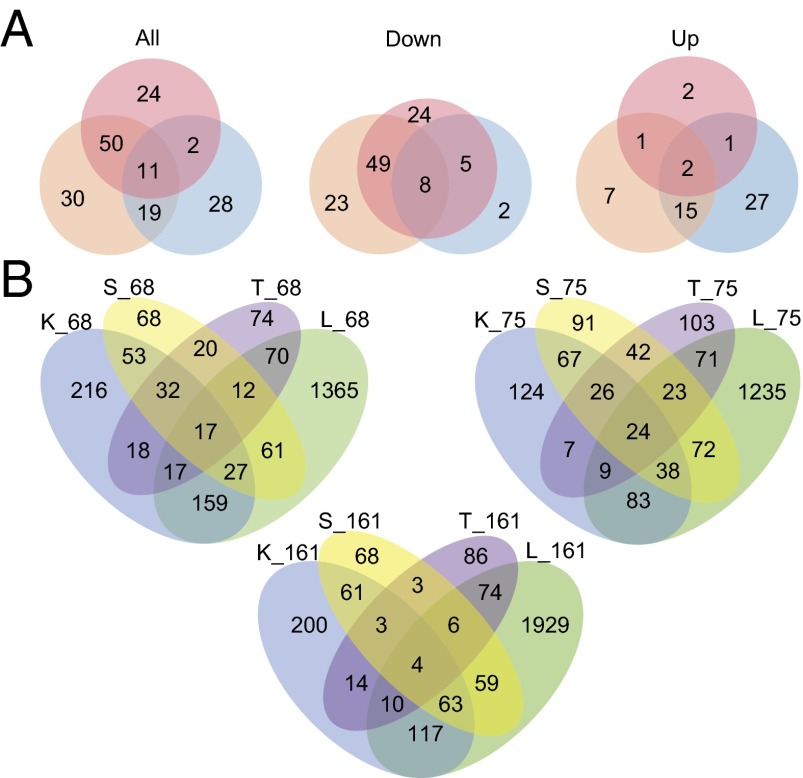
The sequencing produced over 204 million reads after quality control measures (12.3–13.3 million reads per group); across groups, 80.5–82.8% of these reads mapped to the official B. terrestris gene set. Within each parasite genotype exposure, we identified between 60 and 110 differentially expressed transcripts (Fig. 2A) of at least twofold change after applying a false discovery rate (Benjamini–Hochberg) of 5%. Bees exposed to the more infectious parasite genotypes (nos. 68 and 161) had more down-regulated than up-regulated genes (Fig. 2A). Bees exposed to the less infective genotype (no. 75) showed the opposite pattern. This was robust to the cutoff value (Fig. S1). The differentially expressed host genes were mostly distinct to the genotype of the parasite (Fig. 2A). As a preliminary exploration of a possible GH × GP interaction, we also compared the expression patterns with each parasite genotype across colonies. However, there were few overlapping differentially expressed genes across host genotypes infected with the same genotype of parasite (Fig. 2B) even when we took a liberal cutoff of P < 0.05. Host colonies differed dramatically in their response to a given parasite genotype.
Fig. 2.
(A) Venn diagrams of the number of differentially expressed genes upon exposure to three genotypes of C. bombi relative to unexposed workers [no. 68 (orange), no. 75 (blue), and no. 161 (red)] across all host colonies at a false discovery rate of 0.05. (B) Venn diagrams of the number of differentially expressed genes (P < 0.05) when bees with different genotypes (colonies K, L, S, and T) are given the same genotype of parasite (nos. 68, 75, and 161) relative to unexposed workers from the same host genotype. For example, “S_161” indicates colony “S” exposed to parasite genotype “161.”
Gene Ontology.
Of the 8,084 genes that transcripts mapped to after quality control within the edgeR analysis, 3,975 of them had gene ontology (GO) terms that could be inferred. The different parasite genotypes resulted in the over- or underrepresentation of a number of functional groups (Tables S1–S3). Notably, exposure to the poorly infecting parasite genotype (no. 75) is characterized by increased expression of a number of possible immune-related genes, including two bacterial response GO terms, hemocyte proliferation and a number of iron binding and transport categories. In contrast, the other two, more infectious, parasite genotypes induced increased expression of genes related to transcription factor binding, activity, and regulation.
Quantitative PCR Tests of GH × GP Expression.
Exposure to C. bombi, grouping all parasite genotypes together, induced host genotype-specific responses for six surveyed genes [maelstrom, serpin (SPN) 3/4a, spaetzle 4, exonuclease, d-arabinose-1-dehydrogenase, and cytochrome p450 315a; Fig. S2] and approached significance for another four genes (apidermin 2, ATP-binding cassette subfamily G member 1, esterase FE4, and limkain-b1; Table S4). Colonies differed in expression of many genes, including the antimicrobial peptide defensin (Fig. S2). The genes that varied across host genotypes also differed in our subsequent analysis, and these data are presented in Fig. 3 and Fig. S3. A summary of these statistical results can be found in Table S4.
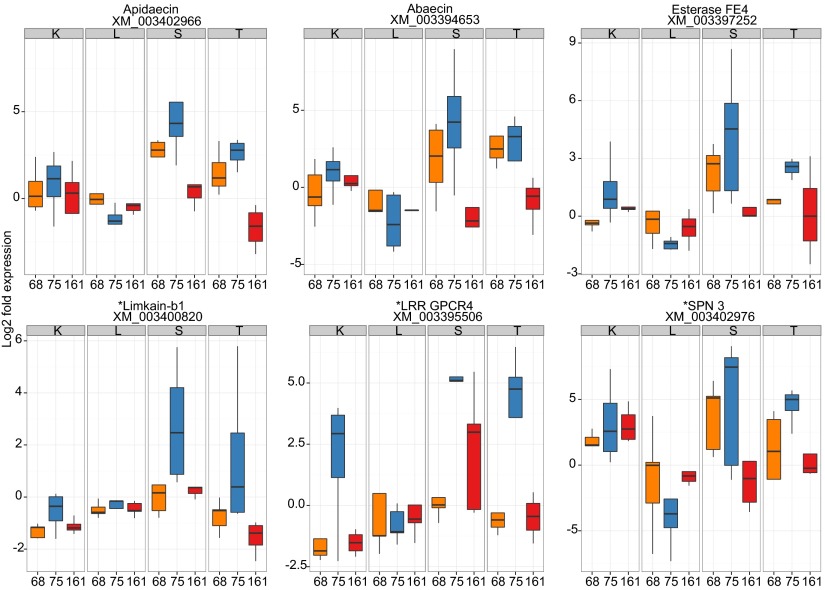
Fig. 3.
Logfold change in expression of genes based on qPCR, where there is a significant interaction of host genotype and parasite genotype [GH × GP interaction for apidaecin, abaecin, and esterase FE4 (F6,42 = 2.73, F6,42 = 2.32, and F6,42 = 2.75, respectively; P < 0.05)] determining expression; where the interaction approached significance [limkain-b1, LRR GPCR4, and SPN 3 (F6,42 = 2.09, F6,42 = 1.95, and F6,42 = 2.25, respectively; P < 0.1)], expression of both limkain-b1 and LRR GPCR4 also varied based on GH (F3,42 = 3.15, P < 0.05; F3,42 = 5.67, P < 0.01) and GP (F2,42 = 7.51, F2,42 = 5.75; P < 0.01). In each panel, the x axis lists the parasite genotypes (nos. 68, 75, and 161; bars are color-coded orange, blue, and red, respectively), whereas boxes labeled with letters refer to colonies (K, L, S, and T). The respective gene is indicated at the top of each panel, together with its GenBank accession number.
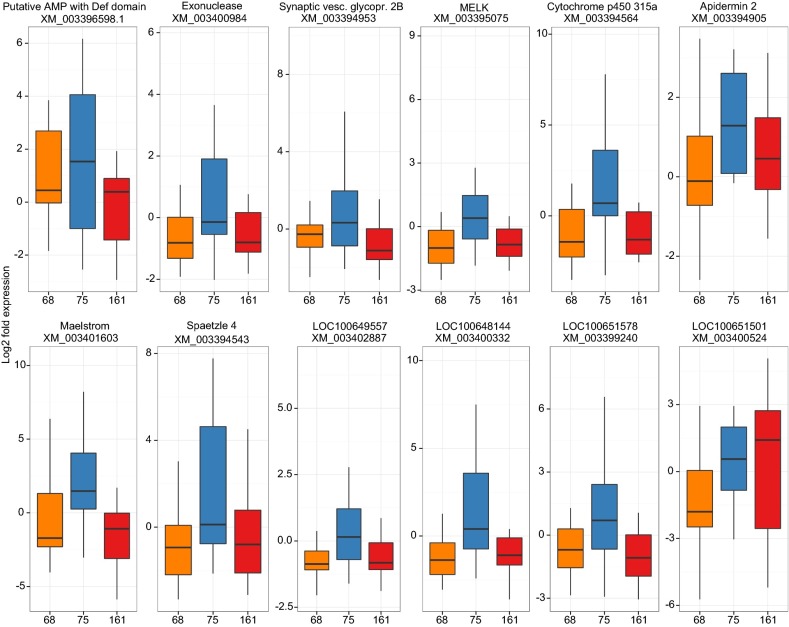
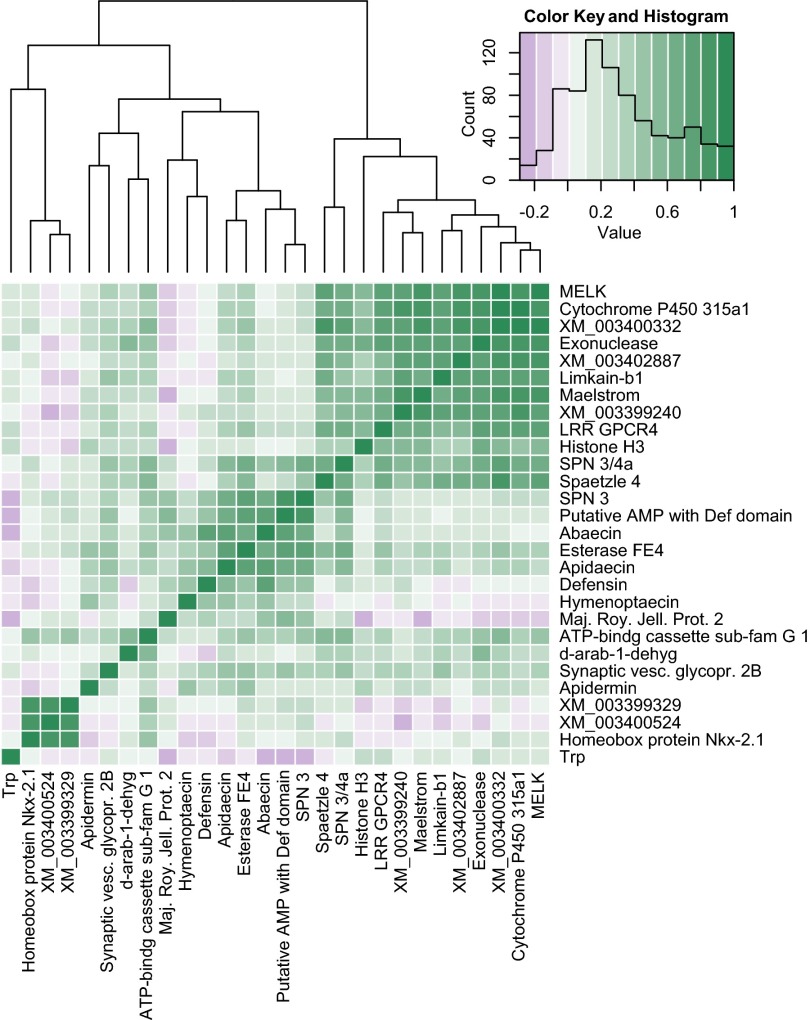
Bees from different colonies responded differently to parasite genotypes (GH × GP interaction; Fig. 3) for three of our surveyed genes, including the key antimicrobial peptides abaecin and apidaecin, as well as esterase FE4. The serine protease inhibitor SPN 3, limkain-b1, and leucine-rich repeat-containing G protein-coupled receptor 4 (LRR GPCR4) also showed a trend toward a GH × GP interaction, but these only approached significance. The C. bombi genotype differentially influenced the expression of 14 genes where the expression did not also vary across host genotypes [maternal embryonic leucine zipper kinase (MELK); spaetzle 4; synaptic vesicle glycoprotein 2B; maelstrom; cytochrome P450 315a1; limkain-b1; exonuclease; LRR GPCR4; an uncharacterized protein with a defensin-like domain LOC100644966; apidermin 2; and uncharacterized proteins LOC100651501, LOC100651578, LOC100648144, and LOC100649557; Fig. 4]. In nearly all cases, exposure to the less infectious parasite genotype no. 75 led to increased expression of these genes, whereas exposure to the most infectious genotype no. 161 led to lower expression, sometimes even below the expression levels of unexposed bees. To a lesser extent, exposure to genotype no. 68 also led to low expression of these genes. The different host genotypes also differed in the expression of 13 genes irrespective of parasite genotype (Fig. S3). Gene expression clusters into putative functional groups (Fig. 5). The expression of all antimicrobial peptides is strongly correlated with each another, and there is a large clade of genes with positively correlated expression that are involved in gene regulation (MELK, exonuclease, limkain-b1, maelstrom, and histone H3). The two serine protease inhibitors (SPN 3 and SPN 3/4a) are also coexpressed and appear to be linked to the expression of spaetzle 4.
Fig. 4.
Logfold change in expression of genes that differ by parasite genotype (GP: F2,42 = 3.30*, F2,42 = 8.44***, F2,42 = 7.00**, F2,42 = 10.28***, F2,42 = 10.47***, F2,42 = 3.64*, F2,42 = 12.87***, F2,42 = 6.41**, F2,42 = 5.88**, F2,42 = 8.11**, F2,42 = 11.61***, F2,42 = 3.99*; *P < 0.05, **P < 0.01, ***P < 0.001) irrespective of host genotype. In each panel, the x axis lists the parasite genotypes (nos. 68, 75, and 161; bars are color-coded orange, blue, and red, respectively). Genes and their accession numbers are indicated at the top of each panel.
Fig. 5.
Relationship of the expression among genes according to the strength of correlation among all genes. Clustering is produced based on Euclidean distances. ATP-bindg cassette sub-fam G 1, ATP-binding cassette subfamily G1; d-arab-1-dehyg, d-arabinose-1-dehydrogenase; Def, defensin; Maj. Roy. Jell. Prot, major royal jelly protein; Synaptic vesc. glycopr. 2B, synaptic vesicle glycoprotein 2B; Trp, transient receptor potential protein.
Discussion
We found that exposure to different genotypes of a single parasite species, C. bombi, induce strikingly different gene expression responses in their bumblebee hosts, as measured in a full-gut transcriptome. Very few differentially expressed genes were shared across all three parasite genotype exposures (Fig. 2A). Our quantitative PCR (qPCR) validation confirmed strong effects of both the genotype of the parasite and the genotype of the host on the expression of a number of candidate genes chosen from the transcriptome data. Furthermore, the expression of some of these genes is dependent on the statistical interaction of host and parasite genotypes (GH × GP). These significant interactions mirror the outcome of infection phenotypes (3, 4), suggesting that gene expression differences could underlie the genotype-by-genotype specificity in this host–parasite system.
These results suggest that the control of expression patterns of infection-relevant genes may be just as likely to produce phenotypic genotype-by-genotype patterns as sequence variation in these genes. Here, we found that there are some striking differences in how parasite genotypes influence host expression even within an admittedly modest number of host and parasite genotypes. In particular, the most infective parasite genotypes, which also attained the highest infection intensities (nos. 68 and 161), induced lower expression of the differentially expressed genes than the poorly infecting genotype (no. 75). There are three possible explanations for this reduction in expression upon exposure to infectious parasites. First, infection may cause universally reduced expression as a consequence of sickness per se. Second, effective parasites may avoid detection, as well as host responses, by evading the immune system. Finally, highly infectious genotypes may be able to suppress the immune response of their hosts actively and so achieve success. We tentatively favor this final hypothesis for the following reasons. Sickness effects on gene expression are unlikely to be pronounced 18 h after exposure to this parasite. At this point, there are few parasites in the gut relative to titers later in infection progression (47, 48). Furthermore, workers infected with this parasite exhibit only minimal damage from infection (49). Together, this suggests that exposure to C. bombi, within this time frame, would not induce a wholesale change in expression due to diminished host condition. Immune evasion is an additional plausible explanation for differences in gene expression profiles; however, the expression of important immunological genes, such as AMPs, on exposure to these infectious genotypes was, in many cases, even lower than in control bees [negative values in Figs. 3 and 4, and similar in kind to the patterns found in other studies (26, 50)]. The most protected host genotype, colony S, which consistently controlled parasite numbers across all parasite genotypes (Fig. 1), was less likely to have suppressed expression of these differentially expressed genes (Figs. S2 and S3). We also found that the enriched GO terms in the bees exposed to the two more infectious parasite genotypes (no. 68 and 161) include a number of categories involved in regulating gene expression, whereas the poorly infecting genotype (no. 75) produced differential expression of a number of terms that may be important in preventing the establishment and growth of this parasite in the host. Although this pattern of regulatory expression is suggestive of immune manipulation rather than immune evasion, it is possible that evasion or a combination of both evasion and suppression is involved in C. bombi infection dynamics.
Manipulation of host responses by parasites is surprisingly common (26, 50–52). Bacterial infection in insects can suppress immune expression, and it can particularly suppress AMP expression (50, 53 and reviewed in ref. 54). AMP expression is a key determinant of infection success for C. bombi (55) and other trypanosomes, such as Trypanosoma brucei (56), in their insect hosts. Leishmania, another genus of trypanosome more closely related to C. bombi, infection reduces host immune expression in both the vertebrate (57) and its insect vector hosts (58, 59). In mammals, this is achieved by manipulation of signaling through MAP kinases (57), and in the sandfly vector, it appears to result in reduced expression of caspar (58), a gene controlling the immune deficiency (IMD) pathway, and suppressed expression of the AMP defensin (59). A similar process occurs in other systems. North American house finches are infected by a zoonotic bacterium Mycoplasma galliseptum. Bonneaud et al. (26, 60) described a distinct difference between birds from a western population that had remained disease-free and an eastern population that had evolved resistance to this parasite over the course of the disease outbreak. Birds that came from the eastern population had a lower infection load and less parasite-induced immune suppression than those from the naive western population, or in samples from the eastern population early in the outbreak. These two populations experienced different pressures, and host genotype frequency changed. Similarly, the protozoan pathogen Toxoplasma gondii suppresses the host immune response, and the capacity to do so varies by T. gondii strain (61). Interestingly, these studies describe variation in expression of innate components of the vertebrate immune system. The innate immune pathways are highly conserved and are also shared with insects. These results in vertebrates could be interpreted as being similar to our host genotype variation in expression upon exposure to C. bombi and variation in host immune expression in response to different genotypes of C. bombi. Thus, it seems plausible that C. bombi could manipulate the control of immunological expression, producing conditions amenable to its establishment and proliferation within the gut. The overrepresentation of differentially expressed genes involved with transcriptional regulation in the two infectious parasite genotypes (Tables S1–S3) may provide some clues about how C. bombi is able to influence the host immune response. The majority of all differentially expressed genes in those bees, given the effective parasite genotypes, were down-regulated relative to controls, including those genes that are involved in gene regulation, and were confirmed by qPCR (e.g., exonuclease, limkain-b1, MELK, maelstrom; Figs. 3 and 4).
If we examine two example colonies with different susceptibility profiles, we also see some striking differences. Colony S is broadly resistant to all three of these parasite genotypes. Although some individuals were infected (topmost panel in Fig. 1), the titer of parasites was kept low, even for the highly infectious parasite strains. In contrast, colony T experienced both high infection success and high parasite load. A common pattern is that genes that varied across host genotypes or depended on both host and parasite genotypes (GH × GP interaction) were more highly expressed in colony S than in colony T (Fig. 3 and Figs. S2 and S3). This is particularly true for the cuticular protein apidermin 2. Apidermin 2 is primarily expressed in flexible cuticle, such as the trachea and gut in honeybees (62). This group of proteins is thought to be involved in cuticular development, and any role in parasite defense is unknown. The interaction between B. terrestris and C. bombi at a structural level in the gut remains unclear, but we hypothesize that C. bombi attaches to the gut to avoid being flushed through the digestive tract. Expression of apidermin 2 may represent a repair mechanism to respond to damage caused by C. bombi to the gut, or perhaps a direct defense. Apidermin 2 is predicted to be secreted in Apis mellifera and is highly hydrophobic. Hydrophobicity is a common feature of antimicrobial peptides and a direct method of action (63). Apidermin 2 in B. terrestris contains 77.5% hydrophobic amino acid residues, which is slightly higher than in A. mellifera, and contains a signal peptide from positions 1–16, resulting in a mature peptide of 64 aa, which is a size similar to known antimicrobial peptides. A number of other genes were more highly expressed in the well-protected host genotype S, including the antimicrobial peptide apidaecin, exonuclease, SPN 3/4a, d-arabinose-1-dehydrogenase, and limkain-b1 (Fig. 3 and Fig. S3).
In addition to the potential defensive role of apidermin 2, we identify what appears to be a novel antimicrobial peptide (LOC100644966) and contains key AMP characteristics, including a signal peptide from positions 1–25, with a cleavage site between position 25 and 26, which results in a 74-aa mature peptide with a defensin-like domain. The expression of this gene is strongly correlated with the expression of the other AMPs: abaecin, apidaecin, defensin, and hymenoptaecin (Fig. 5). The characteristics of this gene and its coordinated expression with other AMPs suggest that it may also function to kill invasive microorganisms, although further in vitro tests would be required to confirm antimicrobial activity.
Both serine protease inhibitors (SPN 3 and SPN 3/4a) showed a trend toward specific expression according to parasite and host genotypes. Both of these genes appear to be specific to Bombus. A particularly interesting aspect of SPN 3 is that it contains an arthropod-specific Pacifacin domain. In locusts, serine protease inhibitors with this domain have been implicated in the regulation of the important immune prophenol oxidase response (64).
Irrespective of the particular genes that were differentially expressed upon exposure to this parasite, the overarching pattern illustrates how different expression patterns can be influenced by the genotype of both the host and the parasite. This is particularly important because it provides evidence for an additional mechanism of host–parasite matching. Altering expression in response to different parasite genotypes may be especially advantageous in systems where there is high genetic diversity within the parasite population, as is the case here. C. bombi is very common and extraordinarily diverse (37–40). Although a high proportion of individuals are infected in the field and are also commonly coinfected with multiple genotypes of the parasite, the same genotype of parasite is almost never recovered twice. Given the high incidence of infection and the low probability of exposure to the same genotype of parasite, an expression response that differs according to parasite genotype may be better suited than changes to the coding sequence of the defense genes themselves.
Genotype-by-genotype interactions can result from variation in coding regions (65, 66 and reviewed in ref. 67), but our results suggest that similar patterns may derive from variation in the expression of infection-relevant genes. The very fact that environmental variation can mediate the GH × GP infection outcome in the B. terrestris–C. bombi system (22) suggests that flexible expression profiles, and not inherently fixed genetic differences, may, in some part, underlie these patterns. Previous targeted work found that antimicrobial peptide expression varies according to the combination of host and parasite genotype (35). Our results also recover this pattern in AMP expression. Parasite genotype (33, 61) and host genotype (28, 29, 32) influence host gene expression in other systems and in the B. terrestris–C. bombi system (30, 31, 34, 35). Here, we found not only that gene expression varies according to host and parasite genotype combination but that differences in expression map to infection success and intensity.
Our results suggest that we may now be in a position to predict infection outcome in this system based on expression differences shortly after exposure. We predict that genotypes of the parasite that induces strong expression of antimicrobial peptides [e.g., genotype A (34), strain II (35)] will fail to establish, whereas genotypes that do not [e.g., genotype B (34), strain IV (35)] will successfully invade and establish in their host. The control of the expression of these genes or the susceptibility of hosts to parasite manipulation will, of course, be under genetic control (i.e., regulatory elements). Further assessment of variation in regulatory elements in B. terrestris may illustrate how genetic variation outside of the directly interacting host genes may produce GH × GP variation in this system.
Materials and Methods
We exposed 7-d-old workers to 10,000 C. bombi cells of one of three genotypes (no. 68, 75, or 161) in 10 μL of 50% (vol/vol) sugar water or a sham inoculum without the parasite cells. We used second-generation bees to avoid the known effect of maternal immunological history (68, 69). Eighteen hours after exposure, we anesthetized half of the bees on ice and removed their guts. We sequenced RNA with an Illumina HiSeq 2000 sequencing system for pools of three individuals per treatment per colony. We checked the remaining bees for infection by visual checks and qPCR quantification of parasite cells in the feces after 7 d. We analyzed the transcript counts as dependent on parasite genotype across host genotypes and within each host genotype. The results from this second analysis can be viewed as a heuristic assessment of the patterns of infection with different parasite genotypes within a host genotype. We assigned GO terms by homology using Blast2GO (70). We then analyzed the over- or underrepresentation of GO categories among significantly differentially expressed transcripts. We used qPCR to validate the expression of 29 target genes (Table S4). More complete materials and methods can be found in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Miguel Jales, Daniel Heinzmann, and Elke Karaus for their technical support. Data were generated at the Genetic Diversity Centre of Eidgenössiche Technische Hochschule Zürich. We thank the Bumblebee Genome Consortium (http://hymenopteragenome.org/beebase/) for providing genomic resources that were used for this study. The transcriptome was sequenced at the Beijing Genomics Institute. This work was supported by European Research Council Advanced Grant 268853 RESIST (to P.S.-H.).
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE55035).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1318628111/-/DCSupplemental.
References
- 1.Lambrechts L, Halbert J, Durand P, Gouagna LC, Koella JC. Host genotype by parasite genotype interactions underlying the resistance of anopheline mosquitoes to Plasmodium falciparum. Malar J. 2005;4(1):3. doi: 10.1186/1475-2875-4-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Carius HJ, Little TJ, Ebert D. Genetic variation in a host-parasite association: Potential for coevolution and frequency-dependent selection. Evolution. 2001;55(6):1136–1145. doi: 10.1111/j.0014-3820.2001.tb00633.x. [DOI] [PubMed] [Google Scholar]
- 3.Schmid-Hempel P. On the evolutionary ecology of host-parasite interactions: Addressing the question with regard to bumblebees and their parasites. Naturwissenschaften. 2001;88(4):147–158. doi: 10.1007/s001140100222. [DOI] [PubMed] [Google Scholar]
- 4.Mallon EB, Loosli R, Schmid-Hempel P. Specific versus nonspecific immune defense in the bumblebee, Bombus terrestris L. Evolution. 2003;57(6):1444–1447. doi: 10.1111/j.0014-3820.2003.tb00351.x. [DOI] [PubMed] [Google Scholar]
- 5.Schulenburg H, Ewbank JJ. Diversity and specificity in the interaction between Caenorhabditis elegans and the pathogen Serratia marcescens. BMC Evol Biol. 2004;4:49. doi: 10.1186/1471-2148-4-49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hamilton WD. Sex versus non-sex versus parasite. Oikos. 1980;35(2):282–290. [Google Scholar]
- 7.Jaenike J. An hypothesis to account for the maintenance of sex within populations. Evol Theory. 1978;3:191–194. [Google Scholar]
- 8.Schulte RD, Makus C, Schulenburg H. Host-parasite coevolution favours parasite genetic diversity and horizontal gene transfer. J Evol Biol. 2013;26(8):1836–1840. doi: 10.1111/jeb.12174. [DOI] [PubMed] [Google Scholar]
- 9.Hamilton WD. Pathogens as causes of genetic diversity in their host populations. In: Anderson R, May R, editors. Population Biology of Infectious Diseases. Berlin: Springer; 1982. pp. 269–296. [Google Scholar]
- 10.Sackton TB, et al. Dynamic evolution of the innate immune system in Drosophila. Nat Genet. 2007;39(12):1461–1468. doi: 10.1038/ng.2007.60. [DOI] [PubMed] [Google Scholar]
- 11.Waterhouse RM, et al. Evolutionary dynamics of immune-related genes and pathways in disease-vector mosquitoes. Science. 2007;316(5832):1738–1743. doi: 10.1126/science.1139862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Viljakainen L, et al. Rapid evolution of immune proteins in social insects. Mol Biol Evol. 2009;26(8):1791–1801. doi: 10.1093/molbev/msp086. [DOI] [PubMed] [Google Scholar]
- 13.Lazzaro B, Clark A. Rapid evolution of innate immune response genes. In: Singh R, Xu J, Kulathinal R, editors. Rapidly Evolving Genes and Genetic Systems. Oxford: Oxford Univ Press; 2012. pp. 203–222. [Google Scholar]
- 14.Tiffin P, Moeller DA. Molecular evolution of plant immune system genes. Trends Genet. 2006;22(12):662–670. doi: 10.1016/j.tig.2006.09.011. [DOI] [PubMed] [Google Scholar]
- 15.Lazzaro B, Sackton T, Clark A. Genetic variation in Drosophila melanogaster resistance to infection: A comparison across bacteria. Genetics. 2006;174(3):1539–1554. doi: 10.1534/genetics.105.054593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Rottschaefer SM, et al. Exceptional diversity, maintenance of polymorphism, and recent directional selection on the APL1 malaria resistance genes of Anopheles gambiae. PLoS Biol. 2011;9(3):e1000600. doi: 10.1371/journal.pbio.1000600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Harris C, et al. Polymorphisms in Anopheles gambiae immune genes associated with natural resistance to Plasmodium falciparum. PLoS Pathog. 2010;6(9):e1001112. doi: 10.1371/journal.ppat.1001112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Obbard DJ, Jiggins FM, Halligan DL, Little TJ. Natural selection drives extremely rapid evolution in antiviral RNAi genes. Curr Biol. 2006;16(6):580–585. doi: 10.1016/j.cub.2006.01.065. [DOI] [PubMed] [Google Scholar]
- 19.Belvin MP, Anderson KV. A conserved signaling pathway: The Drosophila toll-dorsal pathway. Annu Rev Cell Dev Biol. 1996;12:393–416. doi: 10.1146/annurev.cellbio.12.1.393. [DOI] [PubMed] [Google Scholar]
- 20.Lemaitre B, Nicolas E, Michaut L, Reichhart JM, Hoffmann JA. The dorsoventral regulatory gene cassette spätzle/Toll/cactus controls the potent antifungal response in Drosophila adults. Cell. 1996;86(6):973–983. doi: 10.1016/s0092-8674(00)80172-5. [DOI] [PubMed] [Google Scholar]
- 21.Medzhitov R. Toll-like receptors and innate immunity. Nat Rev Immunol. 2001;1(2):135–145. doi: 10.1038/35100529. [DOI] [PubMed] [Google Scholar]
- 22.Sadd BM. Food-environment mediates the outcome of specific interactions between a bumblebee and its trypanosome parasite. Evolution. 2011;65(10):2995–3001. doi: 10.1111/j.1558-5646.2011.01345.x. [DOI] [PubMed] [Google Scholar]
- 23.Lazzaro BP, Little TJ. Immunity in a variable world. Philos Trans R Soc Lond B Biol Sci. 2009;364(1513):15–26. doi: 10.1098/rstb.2008.0141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Råberg L, Grahn M, Hasselquist D, Svensson E. On the adaptive significance of stress-induced immunosuppression. Proc Biol Sci. 1998;265(1406):1637–1641. doi: 10.1098/rspb.1998.0482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Nuismer SL, Otto SP. Host-parasite interactions and the evolution of gene expression. PLoS Biol. 2005;3(7):e203. doi: 10.1371/journal.pbio.0030203. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bonneaud C, et al. Rapid evolution of disease resistance is accompanied by functional changes in gene expression in a wild bird. Proc Natl Acad Sci USA. 2011;108(19):7866–7871. doi: 10.1073/pnas.1018580108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wayne ML, et al. Expression of defense genes in Drosophila evolves under a different selective regime from expression of other genes. Evolution. 2011;65(4):1068–1078. doi: 10.1111/j.1558-5646.2010.01197.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sackton TB, Lazzaro BP, Clark AG. Genotype and gene expression associations with immune function in Drosophila. PLoS Genet. 2010;6(1):e1000797. doi: 10.1371/journal.pgen.1000797. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Navajas M, et al. Differential gene expression of the honey bee Apis mellifera associated with Varroa destructor infection. BMC Genomics. 2008;9:301. doi: 10.1186/1471-2164-9-301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Schlüns H, Sadd BM, Schmid-Hempel P, Crozier RH. Infection with the trypanosome Crithidia bombi and expression of immune-related genes in the bumblebee Bombus terrestris. Dev Comp Immunol. 2010;34(7):705–709. doi: 10.1016/j.dci.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 31.Brunner FS, Schmid-Hempel P, Barribeau SM. Immune gene expression in Bombus terrestris: Signatures of infection despite strong variation among populations, colonies, and sister workers. PLoS ONE. 2013;8(7):e68181. doi: 10.1371/journal.pone.0068181. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lovegrove FE, et al. Simultaneous host and parasite expression profiling identifies tissue-specific transcriptional programs associated with susceptibility or resistance to experimental cerebral malaria. BMC Genomics. 2006;7:295. doi: 10.1186/1471-2164-7-295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bozdech Z, et al. The transcriptome of Plasmodium vivax reveals divergence and diversity of transcriptional regulation in malaria parasites. Proc Natl Acad Sci USA. 2008;105(42):16290–16295. doi: 10.1073/pnas.0807404105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Barribeau SM, Schmid-Hempel P. Qualitatively different immune response of the bumblebee host, Bombus terrestris, to infection by different genotypes of the trypanosome gut parasite, Crithidia bombi. Infect Genet Evol. 2013;20:249–256. doi: 10.1016/j.meegid.2013.09.014. [DOI] [PubMed] [Google Scholar]
- 35.Riddell CE, Adams S, Schmid-Hempel P, Mallon EB. Differential expression of immune defences is associated with specific host-parasite interactions in insects. PLoS ONE. 2009;4(10):e7621. doi: 10.1371/journal.pone.0007621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sadd BM, Barribeau SM. Heterogeneity in infection outcome: Lessons from a bumblebee-trypanosome system. Parasite Immunol. 2013;35(11):339–349. doi: 10.1111/pim.12043. [DOI] [PubMed] [Google Scholar]
- 37.Schmid-Hempel P, Reber Funk C. The distribution of genotypes of the trypanosome parasite, Crithidia bombi, in populations of its host, Bombus terrestris. Parasitology. 2004;129(Pt 2):147–158. doi: 10.1017/s0031182004005542. [DOI] [PubMed] [Google Scholar]
- 38.Salathé RM, Schmid-Hempel P. The genotypic structure of a multi-host bumblebee parasite suggests a role for ecological niche overlap. PLoS ONE. 2011;6(8):e22054. doi: 10.1371/journal.pone.0022054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Tognazzo M, Schmid-Hempel R, Schmid-Hempel P. Probing mixed-genotype infections II: High multiplicity in natural infections of the trypanosomatid, Crithidia bombi, in its host, Bombus spp. PLoS ONE. 2012;7(11):e49137. doi: 10.1371/journal.pone.0049137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ruiz-González MX, et al. Dynamic transmission, host quality, and population structure in a multihost parasite of bumblebees. Evolution. 2012;66(10):3053–3066. doi: 10.1111/j.1558-5646.2012.01655.x. [DOI] [PubMed] [Google Scholar]
- 41.Shykoff JA, Schmid-Hempel P. Incidence and effects of four parasites in natural populations of bumble bees in Switzerland. Apidologie. 1991;22:117–125. [Google Scholar]
- 42.Imhoof B, Schmid-Hempel P. Colony success of the bumble bee, Bombus terrestris, in relation to infections by two protozoan parasites, Crithidia bombi and Nosema bombi. Insectes Soc. 1999;46(3):233–238. [Google Scholar]
- 43.Schmid-Hempel P, Puhr K, Kruger N, Reber C, Schmid-Hempel R. Dynamic and genetic consequences of variation in horizontal transmission for a microparasitic infection. Evolution. 1999;53(2):426–434. doi: 10.1111/j.1558-5646.1999.tb03778.x. [DOI] [PubMed] [Google Scholar]
- 44.Gerardo NM, et al. Immunity and other defenses in pea aphids, Acyrthosiphon pisum. Genome Biol. 2010;11(2):R21. doi: 10.1186/gb-2010-11-2-r21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Evans JD, et al. Immune pathways and defence mechanisms in honey bees Apis mellifera. Insect Mol Biol. 2006;15(5):645–656. doi: 10.1111/j.1365-2583.2006.00682.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Lemaitre B. The drosophila gut: A new paradigm for epithelial immune response. Cytokine. 2012;59(3):494. [Google Scholar]
- 47.Otterstatter MC, Thomson JD. Within-host dynamics of an intestinal pathogen of bumble bees. Parasitology. 2006;133(Pt 6):749–761. doi: 10.1017/S003118200600120X. [DOI] [PubMed] [Google Scholar]
- 48.Schmid-Hempel P, Schmid-Hempel R. Transmission of a pathogen in Bombus terrestris, with a note on division of labour in social insects. Behav Ecol Sociobiol. 1993;33(5):319–327. [Google Scholar]
- 49.Brown MJF, Loosli R, Schmid-Hempel P. Condition-dependent expression of virulence in a trypanosome infecting bumblebees. Oikos. 2000;91(3):421–427. [Google Scholar]
- 50.Apidianakis Y, et al. Profiling early infection responses: Pseudomonas aeruginosa eludes host defenses by suppressing antimicrobial peptide gene expression. Proc Natl Acad Sci USA. 2005;102(7):2573–2578. doi: 10.1073/pnas.0409588102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Schmid-Hempel P. Parasite immune evasion: A momentous molecular war. Trends Ecol Evol. 2008;23(6):318–326. doi: 10.1016/j.tree.2008.02.011. [DOI] [PubMed] [Google Scholar]
- 52.Lefèvre T, et al. Invasion of the body snatchers: The diversity and evolution of manipulative strategies in host-parasite interactions. Adv Parasitol. 2009;68:45–83. doi: 10.1016/S0065-308X(08)00603-9. [DOI] [PubMed] [Google Scholar]
- 53.Ji D, Kim Y. An entomopathogenic bacterium, Xenorhabdus nematophila, inhibits the expression of an antibacterial peptide, cecropin, of the beet armyworm, Spodoptera exigua. J Insect Physiol. 2004;50(6):489–496. doi: 10.1016/j.jinsphys.2004.03.005. [DOI] [PubMed] [Google Scholar]
- 54.Vallet-Gely I, Lemaitre B, Boccard F. Bacterial strategies to overcome insect defences. Nat Rev Microbiol. 2008;6(4):302–313. doi: 10.1038/nrmicro1870. [DOI] [PubMed] [Google Scholar]
- 55.Deshwal S, Mallon EB. Antimicrobial peptides play a functional role in bumblebee anti-trypanosome defense. Dev Comp Immunol. 2014;42(2):240–243. doi: 10.1016/j.dci.2013.09.004. [DOI] [PubMed] [Google Scholar]
- 56.Hu C, Aksoy S. Innate immune responses regulate trypanosome parasite infection of the tsetse fly Glossina morsitans morsitans. Mol Microbiol. 2006;60(5):1194–1204. doi: 10.1111/j.1365-2958.2006.05180.x. [DOI] [PubMed] [Google Scholar]
- 57.Olivier M, Gregory DJ, Forget G. Subversion mechanisms by which Leishmania parasites can escape the host immune response: A signaling point of view. Clin Microbiol Rev. 2005;18(2):293–305. doi: 10.1128/CMR.18.2.293-305.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Telleria EL, et al. Caspar-like gene depletion reduces Leishmania infection in sand fly host Lutzomyia longipalpis. J Biol Chem. 2012;287(16):12985–12993. doi: 10.1074/jbc.M111.331561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Telleria EL, et al. Bacterial feeding, Leishmania infection and distinct infection routes induce differential defensin expression in Lutzomyia longipalpis. Parasit Vectors. 2013;6:12. doi: 10.1186/1756-3305-6-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Bonneaud C, Balenger SL, Zhang JW, Edwards SV, Hill GE. Innate immunity and the evolution of resistance to an emerging infectious disease in a wild bird. Mol Ecol. 2012;21(11):2628–2639. doi: 10.1111/j.1365-294X.2012.05551.x. [DOI] [PubMed] [Google Scholar]
- 61.Saeij JPJ, et al. Toxoplasma co-opts host gene expression by injection of a polymorphic kinase homologue. Nature. 2007;445(7125):324–327. doi: 10.1038/nature05395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Kucharski R, Maleszka J, Maleszka R. Novel cuticular proteins revealed by the honey bee genome. Insect Biochem Mol Biol. 2007;37(2):128–134. doi: 10.1016/j.ibmb.2006.10.009. [DOI] [PubMed] [Google Scholar]
- 63.Chen YX, et al. Role of peptide hydrophobicity in the mechanism of action of alpha-helical antimicrobial peptides. Antimicrob Agents Chemother. 2007;51(4):1398–1406. doi: 10.1128/AAC.00925-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Simonet G, Claeys I, Broeck JV. Structural and functional properties of a novel serine protease inhibiting peptide family in arthropods. Comp Biochem Physiol B Biochem Mol Biol. 2002;132(1):247–255. doi: 10.1016/s1096-4959(01)00530-9. [DOI] [PubMed] [Google Scholar]
- 65.Poirie M, et al. Drosophila resistance genes to parasitoids: Chromosomal location and linkage analysis. Proc Biol Sci. 2000;267(1451):1417–1421. doi: 10.1098/rspb.2000.1158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Luijckx P, Fienberg H, Duneau D, Ebert D. A matching-allele model explains host resistance to parasites. Curr Biol. 2013;23(12):1085–1088. doi: 10.1016/j.cub.2013.04.064. [DOI] [PubMed] [Google Scholar]
- 67.Schmid-Hempel P. Evolutionary Parasitology: The Integrated Study of Infections, Immunology, Ecology, and Genetics. Oxford: Oxford Univ Press; 2011. [Google Scholar]
- 68.Sadd BM, Schmid-Hempel P. Facultative but persistent trans-generational immunity via the mother’s eggs in bumblebees. Curr Biol. 2007;17(24):R1046–R1047. doi: 10.1016/j.cub.2007.11.007. [DOI] [PubMed] [Google Scholar]
- 69.Sadd BM, Schmid-Hempel P. A distinct infection cost associated with trans-generational priming of antibacterial immunity in bumble-bees. Biol Lett. 2009;5(6):798–801. doi: 10.1098/rsbl.2009.0458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Conesa A, et al. Blast2GO: A universal tool for annotation, visualization and analysis in functional genomics research. Bioinformatics. 2005;21(18):3674–3676. doi: 10.1093/bioinformatics/bti610. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.