Significance
Sinorhizobium meliloti and its legume hosts establish a symbiosis in which bacterial fixed nitrogen is exchanged for plant carbon compounds. We study this symbiosis because it is agriculturally and ecologically important and to identify mechanisms used in host–microbe interactions. S. meliloti is internalized in specialized host nodule cells that then use small, cysteine-rich peptides to drive their differentiation into polyploid cells that fix nitrogen. We found that a representative host peptide blocks cell division, in part by eliciting significant changes in the expression of genes that regulate the cell cycle and mediate cell division. We also found that the peptide activated pathways conserved in related pathogens. Our study provides insights into how host peptides cause differentiation of S. meliloti during symbiosis.
Keywords: rhizobia-legume, host–microbe interactions
Abstract
The α-proteobacterium Sinorhizobium meliloti establishes a chronic intracellular infection during the symbiosis with its legume hosts. Within specialized host cells, S. meliloti differentiates into highly polyploid, enlarged nitrogen-fixing bacteroids. This differentiation is driven by host cells through the production of defensin-like peptides called “nodule-specific cysteine-rich” (NCR) peptides. Recent research has shown that synthesized NCR peptides exhibit antimicrobial activity at high concentrations but cause bacterial endoreduplication at sublethal concentrations. We leveraged synchronized S. meliloti populations to determine how treatment with a sublethal NCR peptide affects the cell cycle and physiology of bacteria at the molecular level. We found that at sublethal levels a representative NCR peptide specifically blocks cell division and antagonizes Z-ring function. Gene-expression profiling revealed that the cell division block was produced, in part, through the substantial transcriptional response elicited by sublethal NCR treatment that affected ∼15% of the genome. Expression of critical cell-cycle regulators, including ctrA, and cell division genes, including genes required for Z-ring function, were greatly attenuated in NCR-treated cells. In addition, our experiments identified important symbiosis functions and stress responses that are induced by sublethal levels of NCR peptides and other antimicrobial peptides. Several of these stress-response pathways also are found in related α-proteobacterial pathogens and might be used by S. meliloti to sense host cues during infection. Our data suggest a model in which, in addition to provoking stress responses, NCR peptides target intracellular regulatory pathways to drive S. meliloti endoreduplication and differentiation during symbiosis.
Interspecies signaling is critical throughout the process that establishes the symbiosis between Medicago plant species and the nitrogen-fixing bacterium Sinorhizobium meliloti (1). Secretion of flavonoid molecules by plant roots signals the presence of a potential host to S. meliloti. In response, S. meliloti secretes lipochitooligosaccharide nodulation (Nod) factors, which upon perception cause the root hairs to grow into curls and entrap S. meliloti cells. Nod factor perception also leads to cellular divisions within the root that establish an indeterminate meristem by which a nodule is produced. Production of Nod factors and exopolysaccharides by entrapped S. meliloti elicits the formation of a host-derived structure termed the “infection thread” in which the bacteria multiply. The infection thread elongates and guides growing S. meliloti cells to specialized host nodule cells that internalize bacteria through an endocytic-like process. Intracellular bacteria then undergo terminal differentiation to become nitrogen-fixing bacteroids, a process during which they become enlarged, highly polyploid, and undergo membrane modifications (2).
The terminal differentiation of intracellular S. meliloti is controlled by a family of small host peptides called nodule-specific cysteine-rich (NCR) peptides (3, 4). The genomes of Medicago plants code for more than 600 NCR peptides, which are of various lengths and charge and bear four or six cysteine residues that likely form disulfide-bond pairs (5–8). Recent research has established that NCR peptides are both required and sufficient to drive differentiation of intracellular S. meliloti bacteroids (3, 4). Remarkably, in vitro treatment of S. meliloti with sublethal levels of individual cationic NCR peptides also increases bacterial size and ploidy, indicating that at least some of the symbiotically important effects of NCR peptides can be observed ex planta (4, 9). NCR peptides have been postulated to mediate the increase in S. meliloti ploidy by altering its cell cycle (4), which is a highly regulated process involving sequential changes in gene expression (10). However, the molecular mechanisms by which NCR peptides manipulate the cell cycle and promote differentiation are poorly understood.
In addition to provoking bacteroid differentiation, NCR peptides exhibit antimicrobial activity (4, 9, 11). NCR peptides are similar to plant defensins, which are antimicrobial peptides (AMPs) that function in the innate immune response (4, 12, 13). Investigations of a small number of synthesized NCR peptides have revealed that at high concentrations some of these peptides display potent antimicrobial activity against S. meliloti as well as other bacterial species (4, 9, 11). Work by Gail Ferguson and colleagues (9) provided strong evidence that NCR peptides are also antimicrobial in planta; mutation of the S. meliloti bacA gene, which codes for an inner membrane protein (14), increases the sensitivity of bacteria to the cytotoxic effects of NCR peptides and prevents the establishment of a functional symbiosis. Evidence suggests that the antimicrobial activity of NCR peptides is caused, at least in part, by the disruption of bacterial lipid membrane integrity and increased cell permeabilization (4, 9, 11).
The relationship between the effects of sublethal and lethal doses of NCR peptides remains unclear. Surprisingly, in addition to increasing in size and ploidy, viable S. meliloti cells treated with a sublethal dose of NCR peptides readily take up stains that typically indicate membrane permeabilization and cellular death (4, 9). This paradoxical observation strongly suggests that at sublethal levels NCR peptides cause significant but nonlethal stress on treated bacteria. Physiological responses mounted by S. meliloti in response to sublethal levels of NCR peptides could be important in symbiosis.
In this study, we took advantage of our recent insights into the control of the S. meliloti cell cycle (10) to explore how S. meliloti responds to sublethal levels of an NCR peptide at the molecular level. We used the cationic NCR247 peptide for this study because its biochemical properties are well understood and its biological activities are similar to the activities exhibited by other cationic NCR peptides (4, 9, 11). We discovered that a sublethal level of NCR247 specifically blocks cell division and antagonizes Z-ring function, in part by eliciting a transcriptional response in which expression of master cell-cycle regulators and critical cell-division genes is greatly attenuated. We also identified important symbiosis genes that are induced by NCR247-mediated stress and AMP-activated signaling pathways that are conserved in related pathogenic α-proteobacteria. Our data suggest a model in which, in addition to provoking stress responses, NCR peptides target intracellular regulatory pathways and processes to cause endoreduplication and differentiation of S. meliloti during symbiosis.
Results
NCR247 Peptide Specifically and Robustly Blocks Cell Division in S. meliloti.
Like most α-proteobacteria, S. meliloti initiates DNA replication once during the cell cycle so that under normal conditions cells only carry one or two copies of the genome (15). In principle, NCR peptides could cause polyploidy in S. meliloti by either of two general mechanisms. In one mechanism that is reminiscent of Escherichia coli and Bacillus subtilis multifork DNA replication (16), NCR-peptide treatment could induce multiple rounds of DNA replication initiation during a single S phase and thus cause extra copies of the genome to accumulate. An alternative mechanism is that NCR-peptide treatment could block cell division but allow the cell to skip the division (M) phase of the cell cycle so that DNA accumulates at the usual rate of replication over a series of cell cycles.
To differentiate between these possible mechanisms, we tested how a sublethal dose of the NCR247 peptide affects the cell cycle of S. meliloti. We generated synchronized cultures arrested in G1 phase of the cell cycle as described in ref. 10 and resuspended these cells in modified minimal medium that supports synchronized growth and NCR247 peptide activity (Fig. 1A). After a period of incubation, arrested S. meliloti cells initiated DNA replication synchronously and progressed through the cell cycle in unison. By 120 min, replication was completed, and cells had entered the predivisional or G2 phase. After 180 min, most cells in the synchronized culture had divided to yield two daughter cells (Fig. 1A).
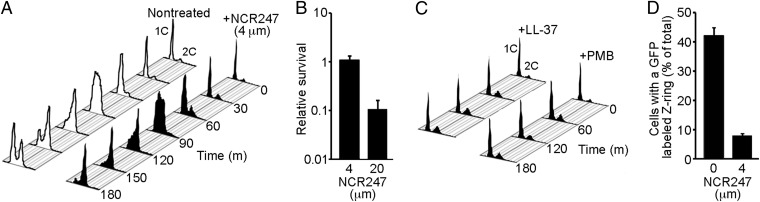
Fig. 1.
Sublethal levels of NCR247 peptide specifically and robustly block S. meliloti cell division. (A) DNA content per cell during synchronized growth of S. meliloti cultures treated with or without 4 μM of NCR247. Cultures were treated at time 0. Cells have either one copy (1C) or two copies (2C) of their genome. (B) Relative survival of S. meliloti treated with 4 μM or 20 μM of NCR247 for 180 min. Relative survival was calculated by dividing the number of cfus after NCR247 treatment by the number of cfus at time 0. Error bars indicate SD. (C) DNA content per cell in synchronized S. meliloti cultures treated with a sublethal inhibitory concentration of LL-37 (1 μΜ) or polymyxin B (PMB) (1.25 μg/mL). (D) Relative abundance of cells with an FtsZ-GFP–labeled Z-ring in synchronized cultures treated with or without 4 μM of NCR247. Cultures were viewed at 120 min after treatment. Isopropyl β-d-1-thiogalactopyranoside (0.125 mM) was added at time 0 to induce FtsZ-GFP expression. Error bars indicate SD.
Treatment of synchronized S. meliloti cultures at the G1 phase with a sublethal concentration of the cationic NCR247 peptide did not alter the initiation or progression of DNA replication (Fig. 1 A and B). However, we found that NCR247 treatment robustly blocked cell division (Fig. 1A). Although morphological signs of septation were observed in nontreated G2-phase cells, we observed little to no evidence of septation in cells treated with NCR247 (Fig. S1 A and B). This block in cell division was reversible, because plating of serial dilutions to assess relative survival after peptide treatment revealed no loss of viability (Fig. 1B). The response of S. meliloti to NCR247 treatment was highly specific, because sublethal levels of other well-studied cationic antimicrobial peptides (LL-37 and polymyxin B) arrested the cell cycle immediately upon treatment so that the cells remained in G1 (Fig. 1C). Thus, NCR247 is unique in its action in that it specifically and robustly blocks S. meliloti cell division.
NCR247-Peptide Treatment Disrupts Z-Ring Function and Leads to Down-Regulation of Master Cell-Cycle Regulators gcrA and ctrA.
Research from other systems suggests two general possibilities for how NCR247-peptide treatment could block cell division. One possibility is that NCR247 simply inhibits the formation, stability, or function of the Z-ring or other components of the cell-division machinery, as found for other small proteins that regulate bacterial cell division (17, 18). A second possibility is that NCR247-peptide treatment blocks cell division by eliciting responses that disrupt cell-cycle regulatory pathways, as has been found to occur in Caulobacter crescentus cells in response to certain stresses (19, 20).
To test for the first possibility, we used an inducible FtsZ-GFP translational fusion protein (21) to visualize the Z-ring in cells in synchronous cultures at the G2 phase. In nontreated synchronized cultures, ∼45% of cells exhibited a Z-ring labeled with FtsZ-GFP. The modest frequency of cells with a fluorescent Z-ring likely results from the short period of FtsZ-GFP induction and the low concentration of the inducer used in this experiment. We found that the frequency of cells with a labeled Z-ring was significantly lower in synchronized cultures treated with a sublethal level of NCR247 than in nontreated cultures (Fig. 1D and Fig. S1C). Instead of FtsZ-GFP being localized in the Z-ring at the midcell, NCR247-treated cells mostly exhibited diffuse localization of FtsZ-GFP (Fig. S1C). These observations indicate that Z-ring formation or stability is disrupted in synchronized NCR247-treated cells.
To test the second possibility, that sublethal NCR247 treatment perturbs cell-cycle regulation, we measured the expression level of the master cell-cycle–regulator genes dnaA, gcrA, and ctrA in synchronized, NCR247-treated S. meliloti cultures. Each of these regulators controls a distinct step in the C. crescentus cell cycle; DnaA initiates DNA replication and activates gcrA expression, GcrA promotes cell-cycle progression and activates ctrA expression, and CtrA regulates cell division (22). Strikingly, we found that the expression level of dnaA, gcrA, and ctrA was significantly perturbed by NCR247 treatment as compared with nontreated cultures (Fig. 2). The effect on dnaA and ctrA expression was particularly pronounced. In nontreated cultures, dnaA expression was maintained at a constant level during the cell cycle, and ctrA expression rose sharply during the early replication phase and reached maximum levels in predivisional cells (Fig. 2). In NCR247-treated cultures, dnaA expression levels dropped to approximately half the normal level, and the cell-cycle–dependent increase in ctrA expression was attenuated significantly (Fig. 2). Thus, NCR247 treatment of S. meliloti significantly disrupts the normal expression pattern of master cell-cycle regulators.
Fig. 2.
Treatment with NCR247 peptide disrupts the expression of master cell-cycle regulators. Shown is the fold change in dnaA, gcrA, and ctrA expression in nontreated and NCR247-treated synchronized cultures relative to time 0. Expression of dnaA, gcrA, and ctrA is relative to smc00128. Error bars indicate the 95% confidence interval (CI).
NCR247 Perturbs Expression of Cell-Cycle–Regulated Genes Involved in Motility, Cell Division, and Cell-Cycle Regulation.
We hypothesized that the changes in gene expression during the cell cycle in NCR247-treated cells contributed significantly to the block in cell division for two reasons. First, NCR247-peptide treatment affected the expression of cell-cycle regulators early in the S phase, well before the establishment of a Z-ring at the midcell in the G2 phase. Second, low levels of ctrA expression in C. crescentus can cause defects in cell division (23, 24).
We tested our hypothesis by using microarrays to profile and compare cell-cycle gene expression during synchronous growth of nontreated cultures or cultures treated with a sublethal dose of NCR247 (Fig. 3A). Surprisingly, we found that expression of 902 genes, ∼15% of the genome, was significantly altered by NCR247 treatment (a twofold or greater change, P ≤ 0.01) (Fig. 3A and Dataset S1). Of the differentially regulated genes, 45% were up-regulated, and 55% were down-regulated in NCR247-treated cells. The frequency of differentially expressed genes increased progressively during the treatment of synchronized cultures, with the greatest number being observed at 90 and 120 min (between mid-S and G2 phases of the cell cycle) after NCR247 treatment (Fig. 3A and Fig. S2). To verify the microarray data, we used quantitative PCR to compare the expression levels of eight genes in control and NCR247-treated cultures and found similar gene-expression patterns as determined by the microarray experiments (Fig. S3).
Fig. 3.
Massive transcriptional response to the NCR247 peptide by S. meliloti contributes to the block in cell division. (A) Hierarchical clustered expression profiles for 902 genes in nontreated and NCR247-treated cultures during synchronous growth. Shown are log2 expression levels relative to time 0 for each gene. The scale for expression level is located at the figure bottom. (B) Profiles of the log2 expression level for 462 cell-cycle–regulated genes in nontreated and NCR247-treated cultures during synchronous growth at the indicated times. (C) Profiles of the log2 expression level for 64 genes putatively regulated by CtrA in nontreated and NCR247-treated cultures during synchronous growth at the indicated times.
De Nisco et al. (10) identified 462 genes whose expression is significantly regulated during the S. meliloti cell cycle. Remarkably, we found that a sublethal dose of NCR247 significantly altered the expression of 217 of these genes, nearly half of the cell-cycle–regulated genes (Fig. 3B and Dataset S1). Most of the genes affected in NCR247-treated cultures were down-regulated in the replication and predivisional phases (60–120 min), a period when these genes normally increase sharply in expression and reach maximal levels in the cell cycle (Fig. 3B; compare nontreated and NCR247-treated heat maps). Genes required for cell polarity (podJ1), cell division (ftsk1, ftsk2, ftsI, divIC-like, and xerC) and cell-cycle regulation (ccrM, divK, divJ, pleD, and sciP) were significantly down-regulated in NCR247-treated cells (Dataset S1). In C. crescentus, the bifunctional FtsK protein and FtsI transpeptidase are required for cell division (25, 26), and FtsK also is required for Z-ring stabilization at the midcell (25). If FtsK function is conserved in S. meliloti, the down-regulation of ftsK1 and ftsK2 might contribute to the low abundance of cells with a Z-ring in NCR247-treated cultures. These data clearly demonstrate that treatment with the NCR247 peptide causes a significant change in cell-cycle–regulated transcription in S. meliloti.
In S. meliloti, CtrA is predicted to control directly 64 cell-cycle–regulated genes involved in motility, cell division, and cell-cycle regulation (10). Because ctrA expression was attenuated in NCR247-treated cells (Fig. 2), we analyzed the expression profile of the predicted CtrA regulon in NCR247-treated cultures. We found that NCR247 treatment affected the expression of most of the predicted CtrA-regulated genes (∼77%), nearly all of which were down-regulated in the replication and predivisional phase (Fig. 3C and Dataset S1). Thus, the expression profile of the predicted CtrA regulon in NCR247-treated cultures is consistent with the low ctrA expression levels at these cell-cycle phases (Fig. 2). Within the predicted CtrA regulon are genes encoding important signaling proteins (divJ and pleC), cell polarity proteins (podJ1), and transcriptional regulators, including the transcription factor rem that likely is involved in activating most flagellar biosynthesis and chemotaxis genes during the cell cycle (10). Thus, transcriptional down-regulation of ctrA in response to NCR247 treatment contributes significantly to the expression profile of cell-cycle–regulated genes. Collectively, these data strongly support the hypothesis that the transcriptional response elicited by treatment with NCR247 peptide greatly contributes to the block in cell division.
NCR247-Peptide Treatment Affects the Expression of Genes and Functions Important in Symbiosis.
In addition to affecting cell-cycle–regulated gene expression, sublethal levels of NCR247 altered the expression of symbiotically important functions, including chemotaxis and motility and biosynthesis of exopolysaccharide and cyclic glucan (Dataset S1). These changes in expression likely represent physiological adaptations by S. meliloti to stress produced by sublethal levels of NCR247. To gain a better understanding of these physiological adaptations, we compared the profile of differentially expressed genes in NCR247-treated cultures with the profiles from cultures grown under heat, acid, osmotic, iron, carbon, phosphate, and nitrogen stress (Table S1 and Dataset S1). Significant overlap was found between the genes differentially expressed in peptide-treated cells and in most other stresses. These overlapping genes likely constitute a general stress response activated by sublethal doses of NCR247 (Dataset S1). This analysis also identified 384 S. meliloti genes whose expression is altered specifically by a sublethal dose of NCR247 (Dataset S1). Repressed genes in this list include many hypothetical genes, nitrate metabolism genes (napA and napD-F), and nitric oxide metabolism genes (norC–E). Genes specifically induced by NCR247 include genes involved in glycogen metabolism (glgA1, glgC, and glgX1), potassium transport (kdpA–C), global stress responses (fecI and rpoH1), and hypothetical genes. The effect on rpoH1 expression is particularly notable, because RpoH1 is required for bacteroid differentiation and survival in host legumes that produce cationic NCR peptides (27, 28). RpoH1 regulates a suite of genes during heat and pH stress (29, 30), and we found that a subset of RpoH1-dependent genes was induced in NCR247-treated cells (Dataset S1).
NCR247 and Other Cationic Peptides Activate Three Regulons Important in Host–Microbe Interactions.
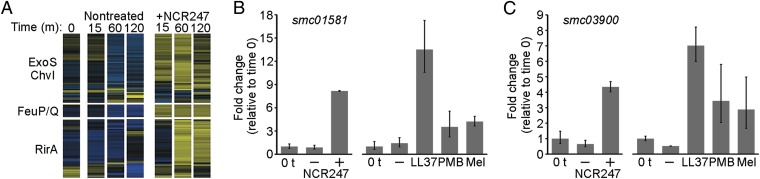
Hierarchical clustering of the differentially expressed genes revealed a subset of 153 genes that was strongly induced within 30 min of peptide treatment (Fig. 3A and Dataset S1, clusters 6 and 7). Nearly two-thirds of these genes are known or predicted to code for proteins localized in the membrane or periplasm (Dataset S1). We discovered that many of these genes fell into three defined regulons controlled by ExoS-ChvI, FeuP-FeuQ, and RirA (Fig. 4A) (31–33). ExoS-ChvI and FeuP-FeuQ are two-component systems that control exopolysaccharide and cyclic glucan production, respectively, in addition to other genes. RirA is a transcriptional repressor that regulates iron acquisition and metabolism genes. ExoS-ChvI and FeuP-FeuQ signaling is required for S. meliloti symbiosis (32, 34), and work on related pathogenic α-proteobacteria has determined that all three regulators have critical functions during host infection (35–37).
Fig. 4.
NCR247 and other cationic AMPs strongly induce the expression of multiple regulons conserved in α-proteobacteria. (A) Profiles of the log2 expression level for genes regulated by ExoS-ChvI, FeuP-FeuQ, and RirA in nontreated and NCR247-treated cultures. Shown are log2 expression levels at 0, 15, 60, and 120 min after NCR247 treatment (see scale in Fig. 3). (B and C) Fold change in smc01581 (B) and smc03900 (C) expression relative to time 0 in cultures after 15 min of treatment with no peptide or with a sublethal inhibitory concentration of NCR247, LL-37 (1 μΜ), melittin (0.5 μΜ), or PMB (1.25 μg/mL). The expression of the control gene smc00128 was used to compare smc01581 and smc03900 expression levels. ExoS-ChvI regulates smc01581, and FeuP-FeuQ regulates smc03900. Error bars indicate the 95% CI.
The immediate and strong induction of the ExoS-ChvI, FeuP-FeuQ, and RirA regulons suggested that these regulators in S. meliloti control primary physiological responses to cationic peptide exposure. To test this possibility, we assayed the expression of direct targets of ExoS-ChvI (smc01581), FeuP-FeuQ (smc03900), and RirA (smc01747) in S. meliloti cells treated with a sublethal concentration of NCR247 and other cationic peptides (LL-37, melittin, or polymyxin B). We found that treatment of S. meliloti with NCR247 or any other tested cationic peptides significantly and similarly induced the expression of ExoS-ChvI and FeuP-FeuQ target genes (Fig. 4 B and C). Treatment of S. meliloti with sublethal levels of NCR247 and LL-37 also induced the RirA target gene (Fig. S4). The similar effects of NCR247 and other cationic peptides on gene expression were in sharp contrast to the different effects they had on the S. meliloti cell cycle (Fig. 1C). Thus, sublethal levels of cationic peptides activate ExoS-ChvI–, FeuP-FeuQ–, and RirA-regulated genes in S. meliloti.
Discussion
Our findings provide important mechanistic insights into how NCR peptides exert their crucial symbiotic role. After S. meliloti has penetrated into the nodules it elicited on its legume host and has been released into the plant cytoplasm inside host membrane compartments, NCR peptides drive the intracellular bacteria to differentiate terminally into bacteroids. Differentiation into bacteroids is a process essential for symbiosis that includes endoreduplication, cellular enlargement, and membrane changes. We found that the cationic peptide NCR247 robustly blocks S. meliloti cell division without affecting DNA replication initiation or S-phase progression. Cells treated with NCR247 peptide failed to construct a Z-ring at the midcell and exhibited little or no morphological signs of septation. Moreover, through gene-expression profiling, we found that sublethal levels of the NCR247 peptide cause a massive remodeling of the bacterial transcriptome that includes highly significant perturbations of the expression of master cell-cycle regulators and many cell-cycle–regulated genes. The timing and direction of these changes in gene expression and the fact that many genes critical for cell division were affected indicate that this transcriptional response elicited by treatment with the NCR247 peptide contributes significantly to the blocking of cell division. Our data suggest that during symbiosis NCR peptides drive intracellular S. meliloti to endoreduplicate and enlarge, at least in part by altering cell-cycle gene expression to block cell division.
Progression through the S. meliloti cell cycle involves sequential changes in gene expression, a pattern that is caused by the action of master cell-cycle regulators (10). The altered cell-cycle gene-expression profile in NCR247-treated cultures likely was caused by the perturbed expression of dnaA, gcrA, and ctrA. The NCR247-mediated effect on ctrA expression and its known role in cell division are consistent with the model in which NCR247 alters cell-cycle gene expression to cause endoreduplication. However, at face value, the effect of NCR247 treatment on dnaA expression does not conform to this model, because DnaA likely is needed for the repeated rounds of DNA replication initiation during endoreduplication in vivo. In C. crescentus, DnaA activity is regulated at both the transcriptional and posttranscriptional level (22). Thus, it is possible that changes to posttranscriptional regulatory pathways that affect DnaA levels or activity in NCR247-treated cells could compensate for lower dnaA transcript levels (see below).
How sublethal levels of NCR247 affect the expression of master cell-cycle regulators is unclear. Recent research suggests that the regulatory mechanisms acting on ctrA are conserved between C. crescentus and S. meliloti (10, 38, 39). In C. crescentus and S. meliloti, ctrA expression is regulated by two distinct promoter sequences (38, 40). During the C. crescentus cell cycle, GcrA initially binds one promoter to activate ctrA transcription, and phosphorylated CtrA binds the other promoter to up-regulate ctrA expression further (41). The attenuated pattern of ctrA expression in NCR247-treated S. meliloti cells during the cell cycle could be caused by reduced GcrA activity or by defects in the CtrA autoregulation pathway. As in C. crescentus, the transcriptional regulatory activity of S. meliloti CtrA is thought to be dependent on phosphorylation and a phosphoregulatory pathway (42). In addition, S. meliloti CtrA likely is regulated proteolytically, as it is in C. crescentus (43). Investigating how sublethal levels of NCR247 affect master cell-cycle regulators will provide insights into the biological activities of NCR peptides and the regulatory mechanisms controlling the cell cycle during symbiosis.
S. meliloti cells treated with a sublethal dose of cationic NCR peptides exhibit alterations in membrane permeability (4, 9), suggesting that treated bacteria experience significant, nonlethal stress. The global transcriptional remodeling we observed in NCR247-treated cells strongly supports this idea. Notably, many of the genes induced immediately after sublethal NCR247 treatment code for membrane or periplasmic proteins. These proteins regulate the transport of ions, minerals, siderophores, and polysaccharides, suggesting that the increased regulation of levels of these ions and molecules is important during cellular adaptation to NCR247 peptide-mediated stress. Interestingly, we also observed that treatment of S. meliloti with a sublethal dose of NCR247 activated the expression of rpoH1 and several RpoH1-regulated genes, similar to the effects seen when S. meliloti cells were treated with a lethal dose of NCR247 (11). These data suggest that, even at sublethal levels, NCR247-peptide treatment activates a cytoplasmic stress response in S. meliloti cells. This finding is particularly intriguing because it could explain why S. meliloti rpoH1 mutants die soon after being internalized within plant cells that express NCR peptides (27, 28). Collectively, our data strongly suggest that cationic NCR peptides exert their effects by targeting the S. meliloti cell envelope, periplasm, and cytoplasm and suggest mechanisms through which S. meliloti adapts to NCR peptide-mediated stress in vitro and during symbiosis.
The very rapid activation of the ExoS-ChvI and FeuP-FeuQ regulons in response to sublethal NCR247 treatment is particularly interesting because numerous bacterial pathogens use two-component systems to sense AMP activity. These two-component systems are comprised of a membrane-embedded histidine kinase sensor (ExoS and FeuP) and a cytoplasmic response regulator (ChvI and FeuQ) (32, 33). Both ExoS-ChvI and FeuP-FeuQ are critical for S. meliloti symbiosis. Bacteria with mutations that severely perturb ExoS-ChvI or FeuP-FeuQ signaling produce insufficient levels of exopolysaccharides or cyclic glucans, respectively, and fail to stimulate infection thread growth properly (32, 44). Genetic studies on these signaling systems also indicate that they have additional important functions in later stages of the symbiosis (32, 45, 46). In pathogenic bacterial species, two-component systems have evolved to sense AMP activity directly, along with other signals, and activation of these systems leads to changes in gene expression that increase AMP resistance (47). Whether the S. meliloti ExoS-ChvI and FeuP-FeuQ also sense AMPs directly and regulate important AMP-resistance functions is not yet known, but the ortholog of ExoS-ChvI in the animal pathogen Brucella abortus is required for AMP resistance (37). An intriguing possibility suggested by our results is that ExoS-ChvI and FeuP-FeuQ may use nodule cationic AMPs as signals to induce functions such as exopolysaccharides and cyclic glucans that are needed for infection. Further research is needed to investigate the link between cationic AMPs and these signaling systems.
Nodules of Medicago plants express hundreds of NCR peptides that vary in length, charge, number of cysteine residues, and spatiotemporal expression (4, 5, 7, 8, 48). It has been suggested that this chemical and structural diversity impacts peptide activity, as found in other families of small disulfide-linked peptides that exhibit rich chemical and structural diversity (4, 49). Indeed, experiments on modified versions of the NCR247 peptide indicated that the presence and order of disulfide bonds between cysteine residues significantly influences peptide antimicrobial activity (50). It has been proposed that NCR peptides (i) alter the S. meliloti cell cycle and metabolism to achieve a more efficient symbiosis (4), (ii) sanction non–nitrogen-fixing rhizobia (4), (iii) kill senescent bacteroids so that their nutrients can be absorbed by the plant (4, 48), and (iv) serve as signals to the plant (48). In addition, the cytotoxicity of NCR peptides could help ensure that other bacterial species are unable to survive inside the nodule (12). Nodules also produce other small non-NCR peptides, including antimicrobial peptides that are part of the innate immune system (51) and a small family of glycine-rich peptides that also exhibit chemical and structural diversity (5). Our study establishes a platform for further interrogation of the biological activities of nodule peptides.
Methods
Bacterial Strains and Growth Conditions.
All experiments were done on synchronized cultures of strain Sm1021 (10). Treatment and culture conditions are described in SI Methods. The time of treatment was designated time 0.
Gene-Expression Profiling and Heatmap Generation.
RNA isolation, cDNA synthesis, and quantitative PCR are described in SI Methods. The microarray and labeling procedures are as described in ref. 10. Only one channel was used for hybridization. Three biological replicates were hybridized for each time point, except for the 15-min time point, which was represented by two biological replicates. Microarray data normalization and analysis are described in SI Methods. Heatmaps were generated as described in SI Methods. Hierarchical clustering was used to identify clusters of differentially expressed genes that have similar expression profiles. Gene-expression profiles in S. meliloti cells exposed to NCR247 and other stresses were compared as described in SI Methods.
Supplementary Material
Acknowledgments
We thank Stuart Levine, Andreas Haag, and members of the G.C.W. laboratory for helpful discussions and Chris Brennan for help on experiments. We also thank the late Gail P. Ferguson for her supportive enthusiasm and generosity in the early stages of this project. This work was supported by National Institutes of Health Grants GM31010 (to G.C.W.), P30 ES002109 to the MIT Center for Environmental Health Sciences, and P30-CA14051 to the Cancer Center Support (core facilities). G.C.W. is an American Cancer Society Professor.
Footnotes
The authors declare no conflict of interest.
Data deposition: The microarray data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE54244).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1400450111/-/DCSupplemental.
References
- 1.Gibson KE, Kobayashi H, Walker GC. Molecular determinants of a symbiotic chronic infection. Annu Rev Genet. 2008;42:413–441. doi: 10.1146/annurev.genet.42.110807.091427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mergaert P, et al. Eukaryotic control on bacterial cell cycle and differentiation in the Rhizobium-legume symbiosis. Proc Natl Acad Sci USA. 2006;103(13):5230–5235. doi: 10.1073/pnas.0600912103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wang D, et al. A nodule-specific protein secretory pathway required for nitrogen-fixing symbiosis. Science. 2010;327(5969):1126–1129. doi: 10.1126/science.1184096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Van de Velde W, et al. Plant peptides govern terminal differentiation of bacteria in symbiosis. Science. 2010;327(5969):1122–1126. doi: 10.1126/science.1184057. [DOI] [PubMed] [Google Scholar]
- 5.Kondorosi E, Mergaert P, Kereszt A. A paradigm for endosymbiotic life: Cell differentiation of Rhizobium bacteria provoked by host plant factors. Annu Rev Microbiol. 2013;67:611–628. doi: 10.1146/annurev-micro-092412-155630. [DOI] [PubMed] [Google Scholar]
- 6.Young ND, et al. The Medicago genome provides insight into the evolution of rhizobial symbioses. Nature. 2011;480(7378):520–524. doi: 10.1038/nature10625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mergaert P, et al. A novel family in Medicago truncatula consisting of more than 300 nodule-specific genes coding for small, secreted polypeptides with conserved cysteine motifs. Plant Physiol. 2003;132(1):161–173. doi: 10.1104/pp.102.018192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fedorova M, et al. Genome-wide identification of nodule-specific transcripts in the model legume Medicago truncatula. Plant Physiol. 2002;130(2):519–537. doi: 10.1104/pp.006833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haag AF, et al. Protection of Sinorhizobium against host cysteine-rich antimicrobial peptides is critical for symbiosis. PLoS Biol. 2011;9(10):e1001169. doi: 10.1371/journal.pbio.1001169. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.De Nisco NJ, Abo RP, Wu CM, Penterman J, Walker GC. Global analysis of cell cycle gene expression of the legume symbiont Sinorhizobium meliloti. Proc Natl Acad Sci USA. 2014;111:3217–3224. doi: 10.1073/pnas.1400421111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tiricz H, et al. Antimicrobial nodule-specific cysteine-rich peptides induce membrane depolarization-associated changes in the transcriptome of Sinorhizobium meliloti. Appl Environ Microbiol. 2013;79(21):6737–6746. doi: 10.1128/AEM.01791-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Graham MA, Silverstein KA, Cannon SB, VandenBosch KA. Computational identification and characterization of novel genes from legumes. Plant Physiol. 2004;135(3):1179–1197. doi: 10.1104/pp.104.037531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Silverstein KA, Graham MA, VandenBosch KA. Novel paralogous gene families with potential function in legume nodules and seeds. Curr Opin Plant Biol. 2006;9(2):142–146. doi: 10.1016/j.pbi.2006.01.002. [DOI] [PubMed] [Google Scholar]
- 14.Glazebrook J, Ichige A, Walker GC. A Rhizobium meliloti homolog of the Escherichia coli peptide-antibiotic transport protein SbmA is essential for bacteroid development. Genes Dev. 1993;7(8):1485–1497. doi: 10.1101/gad.7.8.1485. [DOI] [PubMed] [Google Scholar]
- 15.Hallez R, Bellefontaine AF, Letesson JJ, De Bolle X. Morphological and functional asymmetry in alpha-proteobacteria. Trends Microbiol. 2004;12(8):361–365. doi: 10.1016/j.tim.2004.06.002. [DOI] [PubMed] [Google Scholar]
- 16.Katayama T, Ozaki S, Keyamura K, Fujimitsu K. Regulation of the replication cycle: Conserved and diverse regulatory systems for DnaA and oriC. Nat Rev Microbiol. 2010;8(3):163–170. doi: 10.1038/nrmicro2314. [DOI] [PubMed] [Google Scholar]
- 17.Modell JW, Hopkins AC, Laub MT. A DNA damage checkpoint in Caulobacter crescentus inhibits cell division through a direct interaction with FtsW. Genes Dev. 2011;25(12):1328–1343. doi: 10.1101/gad.2038911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Handler AA, Lim JE, Losick R. Peptide inhibitor of cytokinesis during sporulation in Bacillus subtilis. Mol Microbiol. 2008;68(3):588–599. doi: 10.1111/j.1365-2958.2008.06173.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Wortinger M, Sackett MJ, Brun YV. CtrA mediates a DNA replication checkpoint that prevents cell division in Caulobacter crescentus. EMBO J. 2000;19(17):4503–4512. doi: 10.1093/emboj/19.17.4503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jonas K, Liu J, Chien P, Laub MT. Proteotoxic stress induces a cell-cycle arrest by stimulating Lon to degrade the replication initiator DnaA. Cell. 2013;154(3):623–636. doi: 10.1016/j.cell.2013.06.034. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Ma X, et al. Interactions between heterologous FtsA and FtsZ proteins at the FtsZ ring. J Bacteriol. 1997;179(21):6788–6797. doi: 10.1128/jb.179.21.6788-6797.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Collier J. Regulation of chromosomal replication in Caulobacter crescentus. Plasmid. 2012;67(2):76–87. doi: 10.1016/j.plasmid.2011.12.007. [DOI] [PubMed] [Google Scholar]
- 23.Curtis PD, Klein D, Brun YV. Effect of a ctrA promoter mutation, causing a reduction in CtrA abundance, on the cell cycle and development of Caulobacter crescentus. BMC Microbiol. 2013 doi: 10.1186/1471-2180-13-166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Schredl AT, Perez Mora YG, Herrera A, Cuajungco MP, Murray SR. The Caulobacter crescentus ctrA P1 promoter is essential for the coordination of cell cycle events that prevent the overinitiation of DNA replication. Microbiology. 2012;158(Pt 10):2492–2503. doi: 10.1099/mic.0.055285-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang SC, West L, Shapiro L. The bifunctional FtsK protein mediates chromosome partitioning and cell division in Caulobacter. J Bacteriol. 2006;188(4):1497–1508. doi: 10.1128/JB.188.4.1497-1508.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Costa T, Priyadarshini R, Jacobs-Wagner C. Localization of PBP3 in Caulobacter crescentus is highly dynamic and largely relies on its functional transpeptidase domain. Mol Microbiol. 2008;70(3):634–651. doi: 10.1111/j.1365-2958.2008.06432.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Mitsui H, Sato T, Sato Y, Ito N, Minamisawa K. Sinorhizobium meliloti RpoH1 is required for effective nitrogen-fixing symbiosis with alfalfa. Mol Genet Genomics. 2004;271(4):416–425. doi: 10.1007/s00438-004-0992-x. [DOI] [PubMed] [Google Scholar]
- 28.Oke V, Rushing BG, Fisher EJ, Moghadam-Tabrizi M, Long SR. Identification of the heat-shock sigma factor RpoH and a second RpoH-like protein in Sinorhizobium meliloti. Microbiology. 2001;147(Pt 9):2399–2408. doi: 10.1099/00221287-147-9-2399. [DOI] [PubMed] [Google Scholar]
- 29.Barnett MJ, Bittner AN, Toman CJ, Oke V, Long SR. Dual RpoH sigma factors and transcriptional plasticity in a symbiotic bacterium. J Bacteriol. 2012;194(18):4983–4994. doi: 10.1128/JB.00449-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.de Lucena DK, Pühler A, Weidner S. The role of sigma factor RpoH1 in the pH stress response of Sinorhizobium meliloti. BMC Microbiol. 2010 doi: 10.1186/1471-2180-10-265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rodionov DA, Gelfand MS, Todd JD, Curson AR, Johnston AW. Computational reconstruction of iron- and manganese-responsive transcriptional networks in alpha-proteobacteria. PLOS Comput Biol. 2006;2(12):e163. doi: 10.1371/journal.pcbi.0020163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Griffitts JS, et al. A Sinorhizobium meliloti osmosensory two-component system required for cyclic glucan export and symbiosis. Mol Microbiol. 2008;69(2):479–490. doi: 10.1111/j.1365-2958.2008.06304.x. [DOI] [PubMed] [Google Scholar]
- 33.Chen EJ, Fisher RF, Perovich VM, Sabio EA, Long SR. Identification of direct transcriptional target genes of ExoS/ChvI two-component signaling in Sinorhizobium meliloti. J Bacteriol. 2009;191(22):6833–6842. doi: 10.1128/JB.00734-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bélanger L, Dimmick KA, Fleming JS, Charles TC. Null mutations in Sinorhizobium meliloti exoS and chvI demonstrate the importance of this two-component regulatory system for symbiosis. Mol Microbiol. 2009;74(5):1223–1237. doi: 10.1111/j.1365-2958.2009.06931.x. [DOI] [PubMed] [Google Scholar]
- 35.Lestrate P, et al. Attenuated signature-tagged mutagenesis mutants of Brucella melitensis identified during the acute phase of infection in mice. Infect Immun. 2003;71(12):7053–7060. doi: 10.1128/IAI.71.12.7053-7060.2003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ngok-Ngam P, et al. Roles of Agrobacterium tumefaciens RirA in iron regulation, oxidative stress response, and virulence. J Bacteriol. 2009;191(7):2083–2090. doi: 10.1128/JB.01380-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Manterola L, et al. The lipopolysaccharide of Brucella abortus BvrS/BvrR mutants contains lipid A modifications and has higher affinity for bactericidal cationic peptides. J Bacteriol. 2005;187(16):5631–5639. doi: 10.1128/JB.187.16.5631-5639.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Schlüter JP, et al. Global mapping of transcription start sites and promoter motifs in the symbiotic α-proteobacterium Sinorhizobium meliloti 1021. BMC Genomics. 2013 doi: 10.1186/1471-2164-14-156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Brilli M, et al. The diversity and evolution of cell cycle regulation in alpha-proteobacteria: A comparative genomic analysis. BMC Syst Biol. 2010 doi: 10.1186/1752-0509-4-52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Barnett MJ, Hung DY, Reisenauer A, Shapiro L, Long SR. A homolog of the CtrA cell cycle regulator is present and essential in Sinorhizobium meliloti. J Bacteriol. 2001;183(10):3204–3210. doi: 10.1128/JB.183.10.3204-3210.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Domian IJ, Reisenauer A, Shapiro L. Feedback control of a master bacterial cell-cycle regulator. Proc Natl Acad Sci USA. 1999;96(12):6648–6653. doi: 10.1073/pnas.96.12.6648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Pini F, et al. The DivJ, CbrA and PleC system controls DivK phosphorylation and symbiosis in Sinorhizobium meliloti. Mol Microbiol. 2013;90(1):54–71. doi: 10.1111/mmi.12347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kobayashi H, De Nisco NJ, Chien P, Simmons LA, Walker GC. Sinorhizobium meliloti CpdR1 is critical for co-ordinating cell cycle progression and the symbiotic chronic infection. Mol Microbiol. 2009;73(4):586–600. doi: 10.1111/j.1365-2958.2009.06794.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Cheng HP, Walker GC. Succinoglycan is required for initiation and elongation of infection threads during nodulation of alfalfa by Rhizobium meliloti. J Bacteriol. 1998;180(19):5183–5191. doi: 10.1128/jb.180.19.5183-5191.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Wells DH, Chen EJ, Fisher RF, Long SR. ExoR is genetically coupled to the ExoS-ChvI two-component system and located in the periplasm of Sinorhizobium meliloti. Mol Microbiol. 2007;64(3):647–664. doi: 10.1111/j.1365-2958.2007.05680.x. [DOI] [PubMed] [Google Scholar]
- 46.Pobigaylo N, Szymczak S, Nattkemper TW, Becker A. Identification of genes relevant to symbiosis and competitiveness in Sinorhizobium meliloti using signature-tagged mutants. Mol Plant Microbe Interact. 2008;21(2):219–231. doi: 10.1094/MPMI-21-2-0219. [DOI] [PubMed] [Google Scholar]
- 47.Otto M. Bacterial sensing of antimicrobial peptides. Contrib Microbiol. 2009;16:136–149. doi: 10.1159/000219377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Nallu S, et al. Regulatory patterns of a large family of defensin-like genes expressed in nodules of Medicago truncatula. PLoS ONE. 2013;8(4):e60355. doi: 10.1371/journal.pone.0060355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Olivera BM, Teichert RW. Diversity of the neurotoxic Conus peptides: A model for concerted pharmacological discovery. Mol Interv. 2007;7(5):251–260. doi: 10.1124/mi.7.5.7. [DOI] [PubMed] [Google Scholar]
- 50.Haag AF, et al. Role of cysteine residues and disulfide bonds in the activity of a legume root nodule-specific, cysteine-rich peptide. J Biol Chem. 2012;287(14):10791–10798. doi: 10.1074/jbc.M111.311316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tesfaye M, et al. Spatio-temporal expression patterns of Arabidopsis thaliana and Medicago truncatula defensin-like genes. PLoS ONE. 2013;8(3):e58992. doi: 10.1371/journal.pone.0058992. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.