Significance
The innate immune response is initiated by the recognition of conserved microbial signatures via membrane-resident receptor complex. The dimerization and phosphorylation of Arabidopsis protein kinase complex FLS2/BAK1/BIK1 is essential to initiate and transduce immune signaling to bacterial flagellin. BIK1, a classic serine/threonine kinase, was found here to be autophosphorylated and transphosphorylated by BAK1 at multiple tyrosine residues to relay plant immune signaling. The essential function of tyrosine kinase activity of BIK1 in plants echoes the function of nonreceptor tyrosine kinases that transduce receptor tyrosine kinase signaling via dimerization and phosphorylation in metazoans. Thus, despite lack of classical tyrosine kinases, tyrosine phosphorylation is also an important regulatory mechanism to control membrane-resident receptor signaling in plants.
Abstract
The sessile plants have evolved a large number of receptor-like kinases (RLKs) and receptor-like cytoplasmic kinases (RLCKs) to modulate diverse biological processes, including plant innate immunity. Phosphorylation of the RLK/RLCK complex constitutes an essential step to initiate immune signaling. Two Arabidopsis plasma membrane-resident RLKs, flagellin-sensing 2 and brassinosteroid insensitive 1-associated kinase 1 (BAK1), interact with RLCK Botrytis-induced kinase 1 (BIK1) to initiate plant immune responses to bacterial flagellin. BAK1 directly phosphorylates BIK1 and positively regulates plant immunity. Classically defined as a serine/threonine kinase, BIK1 is shown here to possess tyrosine kinase activity with mass spectrometry, immunoblot, and genetic analyses. BIK1 is autophosphorylated at multiple tyrosine (Y) residues in addition to serine/threonine residues. Importantly, BAK1 is able to phosphorylate BIK1 at both tyrosine and serine/threonine residues. BIK1Y150 is likely catalytically important as the mutation blocks both tyrosine and serine/threonine kinase activity, whereas Y243 and Y250 are more specifically involved in tyrosine phosphorylation. The BIK1 tyrosine phosphorylation plays a crucial role in BIK1-mediated plant innate immunity as the transgenic plants carrying BIK1Y150F, Y243F, or Y250F (the mutation of tyrosine to phenylalanine) failed to complement the bik1 mutant deficiency in immunity. Our data indicate that plant RLCK BIK1 is a nonreceptor dual-specificity kinase and both tyrosine and serine/threonine kinase activities are required for its functions in plant immune signaling. Together with the previous finding of BAK1 to be autophosphorylated at tyrosine residues, our results unveiled the tyrosine phosphorylation cascade as a common regulatory mechanism that controls membrane-resident receptor signaling in plants and metazoans.
Lacking an adaptive immune system and specialized immune cells, sessile plants largely rely on the innate immune system to fend off potential infections (1, 2). The first layer of innate immunity is activated by sensing of the conserved microbial signatures, termed pathogen- or microbe-associated molecular patterns (PAMPs or MAMPs) by plasma membrane (PM)-resident pattern recognition receptors (PRRs) (3, 4). Evidence indicates that PRRs also detect the endogenous molecules derived from damaged cells, termed damage-associated molecular patterns (DAMPs) (3). PRRs are often encoded by receptor-like kinases (RLKs) or receptor-like proteins (RLPs) in plants, and mediate pattern-triggered immunity (PTI) that attributes partly to host resistance against a broad spectrum of microbial infections (5, 6).
Arabidopsis flagellin-sensing 2 (FLS2), one of the best-characterized PRRs in plants, encodes a leucine-rich repeat (LRR)-RLK that recognizes bacterial flagellin or its active peptide derivative flg22 (7). Upon flg22 perception, FLS2 instantaneously complexes with another LRR-RLK brassinosteroid (BR) insensitive 1-associated kinase 1 (BAK1) (8, 9). Botrytis-induced kinase 1 (BIK1), a PM-localized receptor-like cytoplasmic kinase (RLCK), is rapidly phosphorylated upon flg22 perception in an FLS2- and BAK1-dependent manner (10, 11). BIK1 functions as a kinase substrate of BAK1 and forms a complex with FLS2 and BAK1 in transducing flagellin signaling (10). BR-signaling kinase 1 (BSK1), another RLCK and originally identified as a substrate of the brassinosteroid receptor BRI1 (BR insensitive 1), associates with FLS2 and positively regulates PTI signaling (12). Stomatal cytokinesis-defective 1 (SCD1), an FLS2-associated protein identified from proteomics analysis, is required for certain aspects of flg22-mediated responses (13). Activation of MAP kinases (MAPKs) and calcium-dependent protein kinases (CDPKs) functions independently or synergistically downstream of FLS2 and BAK1 receptor complex to activate the expression of flg22-responsive genes (14, 15).
BAK1 is functionally required for responses triggered by multiple MAMPs in Arabidopsis and tobacco (8, 9, 16). In addition, BAK1 plays an important role in mediating plant growth hormone BR signaling (17, 18). BAK1 heterodimerizes with several RLKs including BRI1, elongation factor-Tu receptor (EFR), and Arabidopsis DAMP peptide 1 receptor (AtPEPR1) in addition to FLS2 (8, 9, 17–20). BAK1 positively regulates plant immunity and BR signaling likely through its transphosphorylation with corresponding RLK receptors. Consistent with BIK1 as a kinase substrate of BAK1, BIK1 also complexes with various RLKs, including BRI1, FLS2, EFR, and AtPEPRs (10, 11, 21, 22). However, unlike BAK1, BIK1 positively regulates plant immunity, yet negatively regulates BR signaling (22). The differential phosphorylation of BIK1 by BAK1 and BRI1 may count for the distinct functions of BIK1 in different signaling pathways.
Autophosphorylation and transphosphorylation have been demonstrated in many RLKs and RLCKs to regulate diverse signaling pathways. Sequential transphosphorylation between BRI1 and BAK1 is essential to fully activate BR signaling (23). In general, plant RLKs and RLCKs are classified as serine/threonine kinases (24). It has been shown recently that BRI1 and BAK1 possess tyrosine kinase activity in addition to serine/threonine kinase activity (25, 26). In this study, we have performed extensive mass spectrometry (MS) analysis of BIK1 autophosphorylation and transphosphorylation by BAK1. Consistent with previous mutational analyses (10, 27), several genetically defined serine/threonine residues that are important for BIK1 functions were identified from our MS assay. In particular, T237 is an essential site for BIK1 autophosphorylation and transphosphorylation by BAK1. Surprisingly, MS analysis identified three BIK1 tyrosine phosphorylation sites Y23, Y234, and Y250. In addition, Y243 and Y250 were also revealed as transphosphorylation sites by BAK1. Mutational analysis suggested that BIK1Y150 is likely catalytically important as the mutation blocks both tyrosine and serine/threonine kinase activity, whereas Y243 and Y250 are more specifically involved in tyrosine phosphorylation. Transgenic complementation assays indicate that Y150, Y243, and Y250 are crucial for BIK1 functions in plant defense. Thus, plant RLCK BIK1 is a dual-specificity kinase, and both tyrosine and serine/threonine kinase activities are essential for its function in Arabidopsis innate immunity. Our studies further suggest the complexity of phosphorylation events in plant RLK/RLCK-mediated signaling.
Results
BIK1 Directly Interacts with BAK1 in Vivo and in Vitro.
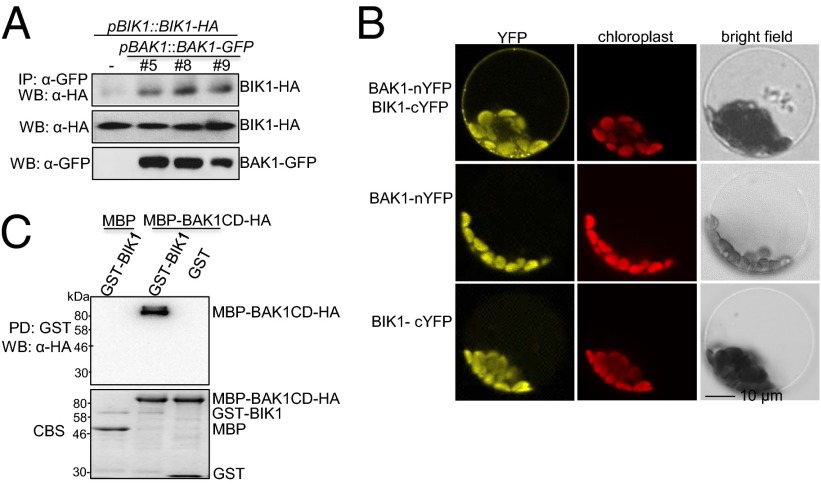
We previously reported that flg22-induced BIK1 phosphorylation depends on FLS2 and BAK1 (10). Furthermore, we found that BIK1 associated with BAK1 when transiently coexpressed in Arabidopsis protoplasts (10). To further investigate the role of BIK1 in FLS2/BAK1 receptor complex, we determined the association of BIK1 and BAK1 both in vivo and in vitro (Fig. 1). We transformed the HA epitope-tagged BIK1 under the control of its native promoter (pBIK1::BIK1-HA) into pBAK1::BAK1-GFP transgenic plants and performed the in vivo coimmunoprecipitation (Co-IP) assay in intact plants. BAK1-GFP immunoprecipitated BIK1-HA, as detected by Western blot with an α-HA antibody upon α-GFP antibody immunoprecipitation (Fig. 1A). Consistently, bimolecular fluorescence complementation (BiFC) assay indicated that BIK1 associated with BAK1 as indicated by the yellow fluorescence protein (YFP) signal primarily on the plasma membrane when coexpressing of BIK1 fused to the carboxyl-terminal half of YFP (BIK1-cYFP) and BAK1 fused to the amino-terminal half of YFP (BAK1-nYFP) in protoplasts (Fig. 1B). Neither of the individual constructs emitted YFP signal in protoplasts (Fig. 1B). To test whether BAK1 directly interacts with BIK1 through the cytosolic kinase domain, we performed an in vitro pull-down assay with GST-BIK1 fusion proteins immobilized on glutathione Sepharose beads as bait against BAK1 cytosolic domain (BAK1CD) fused to maltose-binding protein (MBP) with an HA epitope tag. As shown in Fig. 1C, MBP-BAK1CD could be pulled down by GST-BIK1, but not GST. Similarly, GST-BIK1 could be pulled down by MBP-BAK1CD amylose-agarose beads (Fig. S1). Taken together, the data demonstrate that BIK1 forms a complex with BAK1 by direct interaction with the BAK1 cytosolic kinase domain.
Fig. 1.
BIK1 interacts with BAK1 in vivo and in vitro. (A) BIK1 associates with BAK1 in transgenic plants. Total proteins from seedlings carrying pBIK1::BIK1-HA/pBAK1::BAK1-GFP or pBIK1::BIK1-HA were immunoprecipitated with an α-GFP antibody (IP: α-GFP) and analyzed with Western blot using an α-HA-HRP antibody (WB: α-HA) shown (Top). The expression of BIK1-HA and BAK1-GFP is shown in the Middle and Bottom, respectively. (B) BIK1 interacts with BAK1 in BiFC assay. The increased fluorescence achieved by coexpression of BAK1-nYFP and BIK1-cYFP is visible as the halo around the periphery of the plasma membrane that is only observed when both constructs are expressed. (C) BIK1 interacts with BAK1CD with in vitro pull-down assay. GST or GST-BIK1 immobilized on glutathione Sepharose beads was incubated with MBP or MBP-BAK1CD proteins. The beads were collected for Western blot with an α-HA-HRP antibody. The proteins were shown by Coomassie blue staining (CBS). The above experiments were repeated three times with similar results.
Transphosphorylation in FLS2/BAK1/BIK1 Complex.
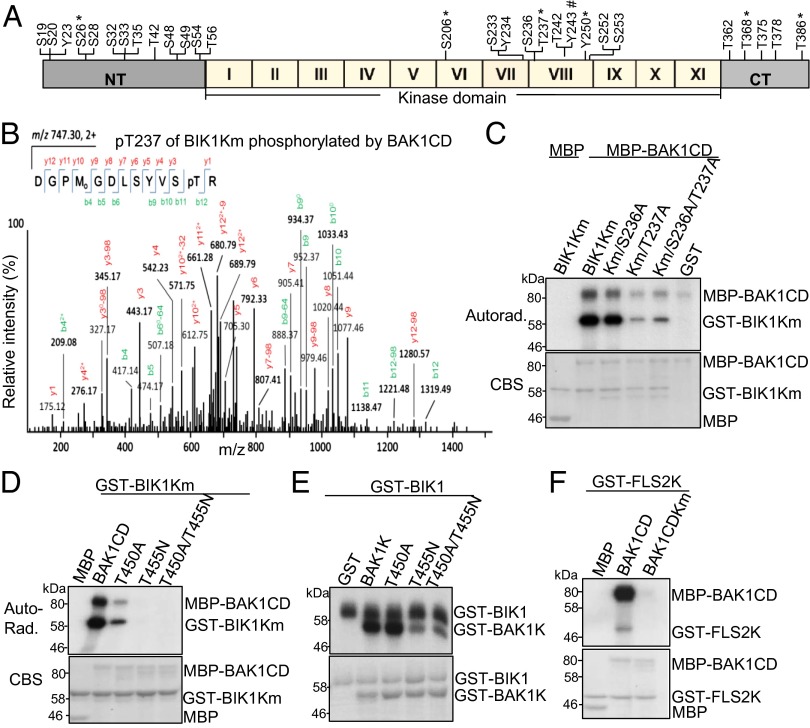
BIK1 is predicted to encode a serine/threonine-protein kinase with a typical kinase domain containing 11 motifs (I–XI), relatively short amino-terminal and carboxyl-terminal (CT) domains (Fig. 2A) (28). Site-directed mutagenesis has suggested that several serine/threonine residues are important for its kinase activity and biological functions (10, 27). However, the biochemical evidence of these phosphorylation sites is still lacking. To systemically examine BIK1 autophosphorylation and transphosphorylation mediated by BAK1, we performed a series of analyses of recombinant GST-BIK1 tryptic peptides by liquid chromatography-tandem mass spectrometry (LC-MS/MS) after in vitro BIK1 autophosphorylation or BAK1 transphosphorylation reactions. Fourteen serine (S) residues and 10 threonine (T) residues were identified in BIK1 protein after autophosphorylation reaction (Fig. 2A). Among these 24 residues, 12 (S19, S20, S26, S28, S32, S33, T35, T42, S48, S49, S54, and T56) are in the N terminus, 7 (S206, S233, S236, T237, T242, S252, and S253) are in the kinase domain, and 5 (T362, T368, T375, T378, and T386) are in the C terminus. We did not identify any phosphorylation residues with GST-BIK1 kinase inactive mutant (Km) protein, which carries a mutation in the putative ATP binding site. We further identified the BIK1 phosphorylation sites mediated by BAK1 with GST-BIK1Km as a substrate and MBP-BAK1CD as a kinase. Five serine or threonine sites (S26, S206, T237, T368, and T386) in BIK1 were phosphorylated by BAK1 (Fig. 2A). Notably, all five serine/threonine sites were also BIK1 autophosphorylation sites.
Fig. 2.
Transphosphorylation in FLS2/BAK1/BIK1 complex. (A) Schematic structure of BIK1. The position of phosphorylated amid acid detected from MS analysis of BIK1 phosphorylation is labeled. * indicates the residues identified from both BIK1 autophosphorylation and transphosphorylation by BAK1. # indicates the residues identified from transphosphorylation by BAK1. NT, N-terminal domain; I–XI, 11 kinase subdomains; CT, C-terminal domain. (B) BIK1 T237 is phosphorylated by BAK1 with MS analysis. Sequence of a doubly charged peptide ion at m/z 747.30 matches DGPMGDLSYVSpTR of BIK1. (C) BIK1 T237 is an essential phosphorylation site by BAK1 in vitro. The in vitro kinase assay was performed using MBP-BAK1CD as a kinase and BIK1Km variants as the substrates. Phosphorylation was analyzed by autoradiography (Upper), and the protein loading was shown by CBS (Lower). (D) BAK1 T450 and T455 are required for autophosphorylation and transphosphorylation of BIK1 in vitro. BIK1Km fusion proteins were used as the substrates for BAK1CD variants. (E) T455 is one of the phosphorylation sites of BAK1 by BIK1 in vitro. BAK1K variants were used as the substrates for BIK1 in an in vitro kinase assay. (F) BAK1 phosphorylates FLS2 in vitro. GST-FLS2K fusion proteins were used as the substrates for BAK1CD variants. The above kinase assays were repeated four times with similar results. The MS analysis was repeated twice.
In agreement with our previous mutagenesis studies that T237 is an important phosphorylation site of BIK1 in response to flg22 treatment (10), MS analyses revealed that T237 was phosphorylated in both reactions of autophosphorylation and transphosphorylation by BAK1 (Fig. 2 A and B). To further confirm its importance, we mutagenized T237 to alanine (T237A) in GST-BIK1Km protein and tested its ability to be phosphorylated by MBP-BAK1CD. As shown in Fig. 2C, BAK1CD directly phosphorylated BIK1Km in vitro in the presence of [32P]-γ-ATP, whereas the phosphorylation level of BIK1KmT237A by BAK1CD was largely reduced, suggesting that T237 is a major phosphorylation site for BAK1-mediated transphosphorylation on BIK1. In contrast, mutation of the adjacent S236 residue (S236A) in BIK1Km or BIK1KmT237A had little effect on its phosphorylation by BAK1CD (Fig. 2C). Taken together, the data suggest that BIK1T237 residue is an important and major phosphorylation site in flg22-mediated signaling.
Notably, T237 of BIK1 is equivalent to T450 of BAK1, based on the amino acid sequence alignment between the kinase domains of BIK1 and BAK1 (Fig. S2). MS analyses identified both T450 and T455 of BAK1 as major sites for autophosphorylation and transphosphorylation by BRI1 (23). T455 is also a highly conserved site in RLK/RLCK family members (Fig. S2) (23). To investigate whether these two sites are required for BAK1 to transphosphorylate BIK1, we generated MBP fusion proteins of BAK1CDT450A and BAK1CDT455N. As shown in Fig. 2D, compared with WT BAK1CD, the kinase activity and transphosphorylation of BAK1CDT450A or BAK1CDT455N to BIK1Km were significantly reduced or completely eliminated. The similar result was obtained for BAK1CDT450A/T455N double mutant (Fig. 2D). We previously reported that BIK1 was able to transphosphorylate BAK1 with the BAK1 kinase domain (BAK1K) as a substrate, which does not possess autophosphorylation activity due to the lack of jutax-membrane domain (10). Interestingly, BAK1KT455N, but not BAK1KT450A, dramatically reduced the ability to be phosphorylated by BIK1 (Fig. 2E), suggesting that T455 of BAK1 is an important phosphorylation site by BIK1. We also found that MBP-BAK1CD is able to phosphorylate GST-FLS2K (Fig. 2F). The data suggest the entangled phosphorylation events in FLS2/BAK1/BIK1 complex.
Specific Tyrosine Phosphorylation of BIK1.
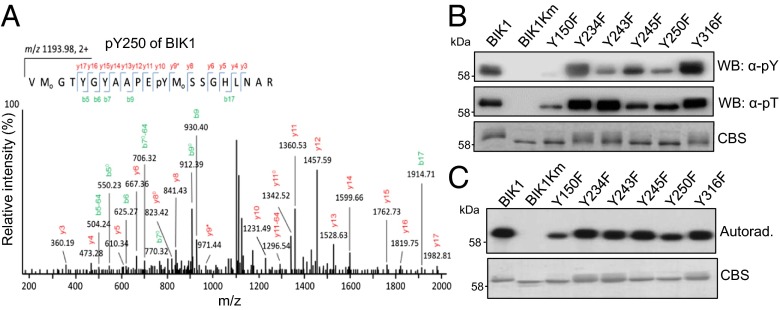
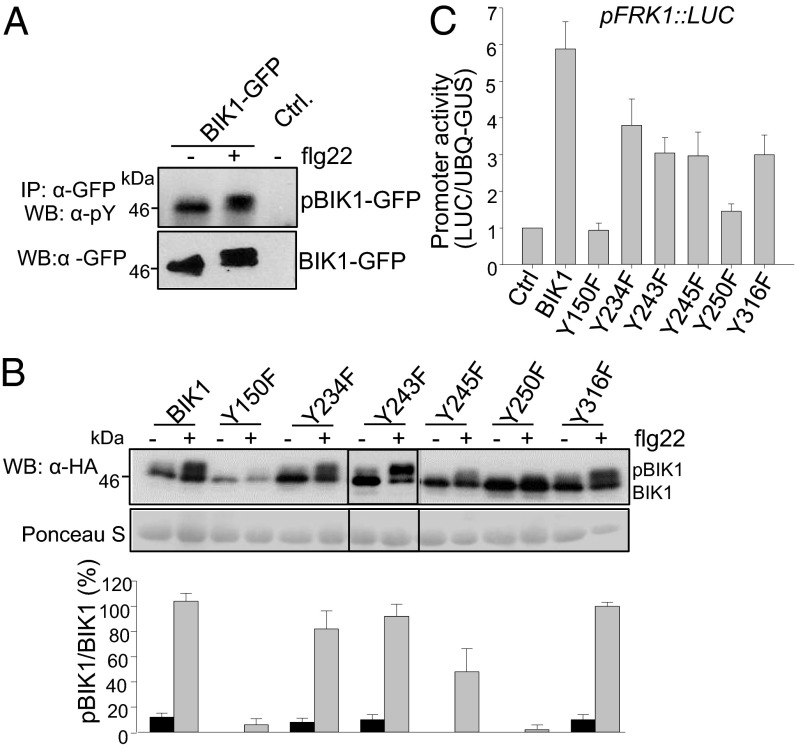
During the course of analyzing comprehensive MS data, we repetitively identified three tyrosine phosphorylation sites (Y23, Y234, and Y250) in BIK1 autophosphorylation reactions (Figs. 2A and 3A and Fig. S3 A and B). This is a rather surprising finding as plant RLCKs have been classified as serine/threonine protein kinases (24). To further confirm our MS data, we performed in vitro BIK1 phosphorylation assay and detected tyrosine phosphorylation with a specific α-phosphotyrosine antibody (α-pY Ab), which has been used to characterize tyrosine phosphorylation of BRI1 and BAK1 (25, 26). As shown in Fig. 3B, the in vitro expressed BIK1, but not BIK1Km, could cross-react strongly to α-pY Ab, indicating that BIK1 possesses tyrosine kinase activity. Thus, BIK1 is a dual-specificity kinase with both serine/threonine and tyrosine activities. We individually substituted two tyrosine residues identified by MS analysis (Y234 and Y250) and two other tyrosine residues (Y243 and Y245) in the BIK1 activation domain with phenylalanine (F) to test their involvement in BIK1 tyrosine phosphorylation (Fig. 3B and Fig. S3C). The alignment of the BIK1 kinase domain with several Arabidopsis RLKs/RLCKs and human interleukin-1 receptor-associated kinase 1 (IRAK1) revealed that Y150 and Y250 of BIK1 are highly conserved in all these RLKs/RLCKs (Fig. S2). These two corresponding residues in BRI1 (BRI1Y956 and BRI1Y1057) are essential for BRI1 kinase activity (25). In addition, BIK1Y316 is conserved in PBL1 and BAK1 (Fig. S2). Thus, we also created BIK1Y150F and BIK1Y316F for tyrosine activity assays. Compared with WT BIK1, BIK1Y150F, BIK1Y243F, and BIK1Y250F mutant proteins exhibited dramatically reduced or compromised cross-reactivity to α-pY Ab, whereas BIK1Y234F, BIK1Y245F, and BIK1Y316F retained WT tyrosine kinase activity (Fig. 3B). Notably, BIK1Y150F mutant also dramatically reduced threonine and/or serine kinase activity as detected with an α-phosphothreonine antibody (α-pT) (Fig. 3B) or autoradiograph with [32P]-γ-ATP (Fig. 3C). Apparently, Y150 is essential for BIK1 catalytic activity. Significantly, BIK1Y243F and BIK1Y250F had no or little effect on threonine and/or serine kinase activity (Fig. 3 B and C). Thus, Y243 and Y250 are important tyrosine phosphorylation sites of BIK1.
Fig. 3.
Specific tyrosine phosphorylation of BIK1. (A) BIK1 Y250 is autophosphorylated with MS analysis. (B) BIK1 is autophosphorylated on tyrosine residues in vitro. GST-BIK1 and its variants were used in the in vitro phosphorylation assay and BIK1 phosphorylation was detected by immunoblotting with an α-pY Ab (Top) and an α-phosphotheronine (α-pT) antibody (Middle). The protein loading was shown by CBS (Bottom). (C) In vitro BIK1 autophosphorylation detected by [32P]-γ-ATP (Upper). The protein loading was shown by CBS (Lower).
BAK1-Mediated BIK1 Tyrosine Phosphorylation.
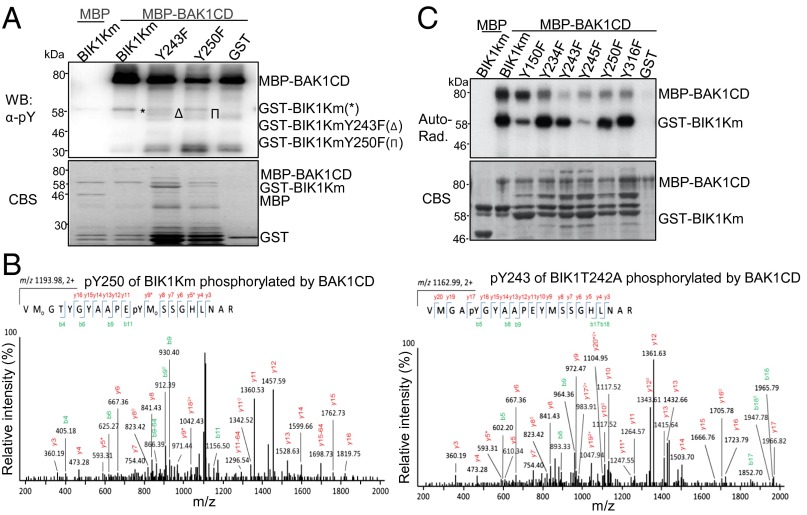
BIK1 is a substrate of BAK1 and BAK1 possesses tyrosine kinase activity (10, 26). Thus, we determined whether BIK1 could serve as a tyrosine kinase substrate of BAK1 with α-pY Ab. As shown in Fig. 4A, BIK1Km could be phosphorylated at tyrosine residues by MBP-BAK1CD as indicated by α-pY Ab. Consistent with a previous report (26), BAK1CD exhibited strong tyrosine autophosphorylation activity (Fig. 4A). Importantly, Y243 and Y250 were identified as phosphorylation sites of BIK1 by BAK1 from MS analyses (Fig. 4B). The data further unveiled the importance of BIK1Y243 and BIK1Y250 autophosphorylation and/or BAK1 transphosphorylation. Consistently, the tyrosine phosphorylation of BIK1KmY243F and BIK1KmY250F by BAK1CD was reduced compared with that of BIK1Km (Fig. 4A), suggesting the important role in mediating tyrosine phosphorylation by BAK1. The overall phosphorylation of BIK1KmY243F and BIK1KmY250F by BAK1 as detected by autoradiograph of [32P]-γ-ATP remained comparable to that of BIK1Km (Fig. 4C). Thus, BIK1 is a tyrosine kinase substrate of BAK1, and Y243 and Y250 are two important BIK1 sites that are phosphorylated by BAK1.
Fig. 4.
BAK1-mediated tyrosine phosphorylation on BIK1. (A) BAK1 phosphorylates BIK1 on tyrosine residues in vitro. BIK1Km variants were used as the substrates for MBP-BAK1CD. The α-pY Ab was used to detect tyrosine phosphorylation (Upper) and the protein loading was shown by CBS (Lower). (B) BIK1 Y250 (Left) and Y243 (Right) are phosphorylated by BAK1 with MS analysis. BIK1T242A and BIK1Km were used for MS assays. (C) In vitro phosphorylation of BIK1Km variants by BAK1. BIK1Km variants were used as the substrates for MBP-BAK1CD. The phosphorylation was shown by autoradiography (Upper) and the protein loading was shown by CBS (Lower). The above experiments were repeated three times with similar results. MS analysis was repeated twice.
Tyrosine Residues Are Involved in flg22-Induced BIK1 Phosphorylation.
BIK1 is quickly phosphorylated upon flg22 perception as shown with a mobility shift by Western blot (10). We examined flg22-mediated BIK1 tyrosine phosphorylation in vivo with an α-pY Ab after immunoprecipitation of protoplast-expressed BIK1-GFP (Fig. 5A). The mobility shift of BIK1-GFP detected by an α-GFP antibody indicates flg22-induced BIK1 phosphorylation (Fig. 5A). Importantly, the α-pY Ab showed cross-reactivity to the immunoprecipitated BIK1 either with or without flg22 treatment, providing the evidence that BIK1 tyrosine phosphorylation occurs in vivo (Fig. 5A). Notably, both the shifted and unshifted BIK1 bands could be detected by α-pY Ab, suggesting that BIK1 has a basal level of tyrosine phosphorylation in the absence of flg22 treatment.
Fig. 5.
BIK1 tyrosine phosphorylation in flg22 signaling. (A) In vivo tyrosine phosphorylation of BIK1. BIK1-GFP or the empty vector control (Ctrl) was expressed in WT protoplasts for 8 h followed by 1μM flg22 treatment for 10 min. BIK1 tyrosine phosphorylation was detected by immunoblotting with α-pY Ab (Upper) and with an α-GFP antibody for protein expression (Lower). (B) Requirement of specific tyrosine residues in flg22-induced BIK1 phosphorylation. BIK1 variants were expressed in WT protoplasts for 8 h followed by 1 μM flg22 treatment for 10 min and subjected to immunoblotting with an α-HA antibody. The flg22-mediated BIK1 phosphorylation is indicated by the mobility shift (Top) and the protein loading is shown by Ponceau S staining of the membrane (Middle). The intensity of the shifted and unshifted bands was quantified by ImageJ software and % of their ratio is shown (Bottom). (C) Activation of pFRK1::LUC by BIK1 variants. The pFRK1::LUC was cotransfected with BIK1, BIK1 variants or a vector control in protoplasts. UBQ10-GUS was included as a transfection control and the luciferase activity was normalized with GUS activity. The above experiments were repeated three to four times with similar results.
To investigate the role of specific tyrosine residues of BIK1 in vivo, we created the above mentioned tyrosine mutants in protoplast expression vector and tested their effect on flg22-induced BIK1 phosphorylation. Consistent with its requirement in catalytic activity, BIK1Y150F dramatically reduced the ratio of shifted band versus unshifted band upon flg22 treatment (Fig. 5B). Significantly, BIK1Y250F also lost the flg22-induced mobility shift compared with WT BIK1 (Fig. 5B). In addition, the mobility shift of BIK1Y245F was also partially compromised upon flg22 treatment (Fig. 5B). However, BIK1Y234, BIK1Y243, and BIK1Y316 seem to be dispensable for flg22-induced BIK1 mobility shift (Fig. 5B). This result reconciles our MS and biochemical analysis in which Y250 is an important BIK1 autophosphorylation and BAK1-mediated transphosphorylation site, suggesting its essential role in flg22-mediated signaling. Overexpression of BIK1 in protoplasts could constitutively activate the pFRK1::LUC reporter gene, a marker gene in PTI signaling (10). Consistently, expression of BIK1Y150F or BIK1Y250F in protoplasts was no longer able to activate pFRK1::LUC, whereas BIK1Y234F, BIK1Y243F, BIK1Y245F, or BIK1Y316F partially compromised the activation of pFRK1::LUC (Fig. 5C). These results establish that Y150 and Y250 are two essential sites of BIK1 in flg22-triggered phosphorylation and signaling. However, BIK1Y23F mutant retained WT tyrosine kinase activity (Fig. S3D) and did not impair flg22-induced BIK1 mobility shift (Fig. S3E) and the activation of pFRK1::LUC (Fig. S3F), suggesting that mutation of Y23 does not affect its function in flg22 signaling.
Multiple Tyrosine Residues Are Required for BIK1-Mediated Plant Immunity.
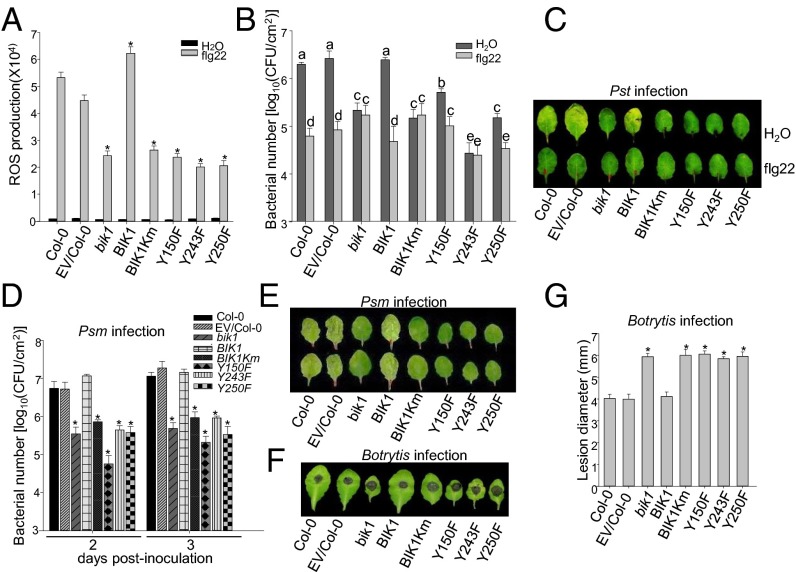
To further elucidate the functional significance of specific tyrosine residues of BIK1, we complemented the bik1 mutant plants with HA epitope-tagged WT BIK1 or various mutants, including BIK1Km, Y150F, Y243F, and Y250F under the control of its native promoter. Multiple lines of each construct were obtained and two lines with comparable protein expression level as WT BIK1 for each construct were chosen for further assays. The bik1 mutant compromises various flg22-triggered immune responses, including flg22-induced ROS production, and resistance to Pseudomonas syringae pv. tomato DC3000 (Pst) infection (10, 11, 28). The WT BIK1 construct completely restored, whereas BIK1Km, BIK1Y150F, BIK1Y243F, or BIK1Y250F still retained the compromised flg22-induced ROS production in the bik1 mutant compared with Col-0 WT or empty vector transgenic plants in Col-0 background (Fig. 6A and Fig. S4A). The bik1 mutant is more resistant to Pst infection, but is unable to mediate flg22-induced resistance. As shown in Fig. 6 B and C, the expression of WT BIK1, but not BIK1Km, BIK1Y150F, BIK1Y243F, or BIK1Y250F in the bik1 mutant plant was able to restore WT level resistance to Pst infection and flg22-mediated resistance to Pst infection (Fig. 6 B and C). Similar to enhanced resistance to Pst infection, the bik1 mutant is more resistant to P. syringae maculicola (Psm) infection compared with WT plants, indicated by in planta bacterial multiplication 2- and 3-d postinoculation (dpi) (Fig. 6D) and disease symptom development (Fig. 6E). The bik1 mutant plants expressing WT BIK1 exhibited a similar susceptibility to Psm infection as WT plants, whereas the bik1 mutant plants expressing BIK1Km, BIK1Y150F, BIK1Y243F, or BIK1Y250F showed a similar level of resistance with the bik1 mutant, 2 and 3 dpi (Fig. 6 D and E and Fig. S4B). The bik1 mutant is more susceptible to Botrytis cinerea infection (28). The bik1 transgenic plants with BIK1Km or tyrosine substitution mutants were as susceptible as the bik1 mutant compared with WT plants as measured by symptom development (Fig. 6F and Fig. S4C) and lesion diameter (Fig. 6G and Fig. S4C) after B. cinerea infection. Taken together, our genetic analysis indicate that Y150, Y243, and Y250 are important for BIK1 functions in plant innate immunity and BIK1 tyrosine phosphorylation constitutes an essential step in PTI signaling. In addition to the compromised immune responses, the bik1 mutant exhibits certain growth defects, in particular at the later developmental stage with early flowering, and twisted and curling rosette leaves (28). The WT BIK1 complementation plants rescued these growth defects in the bik1 mutants (Fig. S5). However, the transgenic plants carrying either BIK1Km, BIK1Y150F, BIK1Y243F, or BIK1Y250F resembled the bik1 mutant with curling rosette leaves at later developmental stage and early flowering phenotypes (Fig. S5). These observations indicate that the kinase activity and the tyrosine residues (Y150, Y243, and Y250) are also required for BIK1 functions in growth and development.
Fig. 6.
Y150, Y243, and Y250 are required for BIK1 functions in plant immunity. (A) flg22-triggered oxidative burst. ROS production from leaf discs of 5-wk-old plants was presented as total photon counts during 30 min of 100 nM flg22 treatment. Values presented are mean ± SE (n = 36) and significant difference is shown as *P < 0.05, established by a one-way ANOVA compared with data from WT plants. (B) flg22-mediated restriction of bacterial growth. Four-week old plants were pretreated with H2O or 100 nM flg22 for 24 h and followed by hand inoculation of Pst at 5 × 105 cfu/mL. Bacterial growth was measured at 2 dpi. The data are shown as mean ± SE of three repeats and the different letters indicate a significant difference with P < 0.05 compared with data from WT plants. (C) The disease symptom of Pst infection. The similar experiments were performed as in B with the picture taken at 3 dpi. (D) Bacterial growth of Psm infection. Plants were hand inoculated with Psm at 5 × 105 cfu/mL and the bacterial growth was measured at 2 and 3 dpi. The data are shown as mean ± SE of three repeats and significant difference is shown as *P < 0.05, compared with data from WT plants. (E) Disease symptom of Psm infection. The similar experiments were performed as in D with the picture taken at 3 dpi. (F) Disease symptom of B. cinerea infection. Leaves from 5-wk-old plants were deposited with 10 μL of B. cinerea strain BO5 at a concentration of 2.5 × 105 spores per mL. Disease symptom was recorded at 2 dpi. (G) Lesion development of B. cinerea infection. Similar assays were performed as in F and the lesion diameter was measured at 2 dpi. The data are shown as mean ± SE of at least 30 leaves and significant difference is shown at *P < 0.05, compared with data from WT plants. The above experiments were repeated two times with similar results. The BIK1 complementation transgenic plants were pBIK1::BIK1Y150F-HA line 2-1, pBIK1::BIK1Y243F-HA line 1-3, and pBIK1::BIK1Y250F-HA line A-7.
Discussion
Plant RLKs are architecturally related to metazoan receptor tyrosine kinases (RTKs). However, unlike RTKs, which are generally tyrosine protein kinases, plant RLKs/RLCKs belong to the RLK/Pelle/IRAK protein kinase family, which is classified as serine/threonine kinases (24). Recently, two Arabidopsis LRR-RLKs BRI1 and BAK1 have been shown to possess tyrosine kinase activity (25, 26). Tyrosine phosphorylation of BRI1 kinase inhibitor 1 (BKI1) by BRI1 upon BR perception releases BKI1 into the cytosol and allows the recruitment of BAK1 to BRI1 (29). BAK1 is autophosphorylated at Y610, which is important for BR signaling and some aspects of plant defense (26). Here we show that BIK1, an important component in plant PTI signaling, is autophosphorylated and transphosphorylated by BAK1 at several tyrosine residues in addition to serine/threonine residues. BAK1 physically interacts with BIK1 in vitro and in vivo, and directly phosphorylates BIK1 at multiple serine/threonine/tyrosine residues. BIK1T237 is essential for its autophosphorylation and phosphorylation by BAK1. BIK1 is also able to reciprocally phosphorylate BAK1, and BAK1T455 is likely a phosphorylation site by BIK1. BIK1Y23, BIK1Y234, and BIK1Y250 were identified as autophosphorylation sites with MS analysis. BIK1Y243 and BIK1Y250 were also phosphorylated by BAK1. Mutational and transgenic analyses indicate the importance of Y150, Y243, and Y250 in BIK1 tyrosine phosphorylation and functions in plant immunity. The data revealed that plant RLCK BIK1 functions as a dual-specificity protein kinase in plant immunity and supported the notion that tyrosine phosphorylation is likely a common regulatory mechanism that controls plasma membrane-resident receptor signaling in plants and metazoans.
Extensive mutagenesis analyses have identified many serine/threonine sites important for BIK1 functions (27). Several serine and threonine residues, including S33, T35, T42 in BIK1 N terminus, are important for BIK1 autophosphorylation and phosphorylation of an artificial substrate MBP (27). These several sites were also identified as BIK1 autophosphorylation sites in our MS analysis. In addition, we also identified 9 other serine/threonine residues in the N terminus of BIK1 autophosphorylation, suggesting the important regulatory role of N terminus in BIK1 kinase activity. In supporting this, S33 is important for BIK1-mediated flg22-induced resistance to B. cinerea and Pst infection (27). In addition, the four sites identified by MS analyses, S233, S236, T237, and T242, lie within the activation loop of BIK1 kinase domain. T237A mutation blocks flg22-induced BIK1 mobility shift and BIK1-mediated FRK1 promoter induction (10). The bik1 mutant plants carrying S236A, T237A, or T242A could not restore the compromised flg22-induced resistance to B. cinerea and Pst infections (27). Significantly, MS analysis also revealed T237 as a BIK1 transphosphorylation site by BAK1, which further reconciles the importance of this site in transducing BAK1- and BIK1-mediated signaling.
BIK1 is also phosphorylated upon ethylene (28) treatment and required for responses to ethylene (27). Recent study indicates that BIK1 regulates ethylene signaling through interaction with PEPR1, a LRR-RLK perceiving Arabidopsis endogenous peptide Pep1 (21). In contrast to its positive roles in plant immunity and ET signaling, BIK1 is a negative regulator in BR signaling. BIK1 complexes with BRI1 and is directly phosphorylated by BRI1 in transducing BR signaling (22). BRI1 also possesses tyrosine phosphorylation activity (25). It will be interesting to test whether BRI1 phosphorylates BIK1 at certain tyrosine residues, such as Y250. It is of importance to determine whether BIK1Y250 is also required for its function in BR and ET signaling. Notably, BAK1Y610 is indispensable for plant resistance to nonpathogenic Pst hrpA mutant, but is dispensable for flg22-mediated seedling growth inhibition (26), suggesting that BAK1Y610 is required for some but not all aspects of BAK1 functions. The similar scenario could exist in BIK1 tyrosine phosphorylation.
Experimental Procedures
Plant Growth Condition and Generation of Transgenic Plants.
The procedure to generate pBIK1::BIK1Y150F-HA, pBIK1::BIK1Y243F-HA, pBIK1::BIK1Y250F-HA, or BIK1km transgenic plants in the bik1 mutant background and Arabidopsis growth condition is described in SI Experimental Procedures.
Plasmid Constructs, Protoplast Transfection, and in Vivo Tyrosine Phosphorylation.
Arabidopsis BIK1 full-length BAK1CD, BAK1K, and FLS2CD constructs were reported previously (10). The BIK1 and BAK1 mutants were generated with site-directed mutagenesis kits and the primers are listed in SI Experimental Procedures. For BiFC assay, the details are described in SI Experimental Procedures. The protoplast isolation and transfection were reported previously (10), and the details of the reporter assay are described in SI Experimental Procedures.
In Vitro Pull-Down, Seedling Co-IP Assay, and MS Analysis.
Fusion protein purification and pull-down assays followed the standard protocol. For seedling Co-IP, 5 g 10-d-old seedlings were used. The in vitro phosphorylation assay for MS analysis was performed in a 10-μL reaction containing 20 mM Tris⋅HCl, pH 7.5, 10 mM MgCl2, 100 mM NaCl, 3 mM CaCl2, 1 mM DTT, and 0.1 mM ATP. The details are described in SI Materials and Methods.
Supplementary Material
Acknowledgments
We thank Dr. J. Li for various Arabidopsis mutants and transgenic plants, members of the laboratories of L.S. and P.H. for critical comments and suggestions for the experiments, and the Arabidopsis Biological Resource Center (ABRC). The work was supported by National Institutes of Health (NIH) Grants R01GM092893 (to P.H.) and R01GM097247 and the Robert A. Welch Foundation (A-1795) (to L.S.). B.L. was partially supported by Dr. Daohong Jiang’s laboratory at Huazhong Agricultural University, China.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1318817111/-/DCSupplemental.
References
- 1.Chisholm ST, Coaker G, Day B, Staskawicz BJ. Host-microbe interactions: Shaping the evolution of the plant immune response. Cell. 2006;124(4):803–814. doi: 10.1016/j.cell.2006.02.008. [DOI] [PubMed] [Google Scholar]
- 2.Jones JDG, Dangl JL. The plant immune system. Nature. 2006;444(7117):323–329. doi: 10.1038/nature05286. [DOI] [PubMed] [Google Scholar]
- 3.Boller T, Felix G. A renaissance of elicitors: Perception of microbe-associated molecular patterns and danger signals by pattern-recognition receptors. Annu Rev Plant Biol. 2009;60:379–406. doi: 10.1146/annurev.arplant.57.032905.105346. [DOI] [PubMed] [Google Scholar]
- 4.Schwessinger B, Ronald PC. Plant innate immunity: Perception of conserved microbial signatures. Annu Rev Plant Biol. 2012;63:451–482. doi: 10.1146/annurev-arplant-042811-105518. [DOI] [PubMed] [Google Scholar]
- 5.Monaghan J, Zipfel C. Plant pattern recognition receptor complexes at the plasma membrane. Curr Opin Plant Biol. 2012;15(4):349–357. doi: 10.1016/j.pbi.2012.05.006. [DOI] [PubMed] [Google Scholar]
- 6.Antolín-Llovera M, Ried MK, Binder A, Parniske M. Receptor kinase signaling pathways in plant-microbe interactions. Annu Rev Phytopathol. 2012;50(50):451–473. doi: 10.1146/annurev-phyto-081211-173002. [DOI] [PubMed] [Google Scholar]
- 7.Gómez-Gómez L, Boller T. FLS2: An LRR receptor-like kinase involved in the perception of the bacterial elicitor flagellin in Arabidopsis. Mol Cell. 2000;5(6):1003–1011. doi: 10.1016/s1097-2765(00)80265-8. [DOI] [PubMed] [Google Scholar]
- 8.Chinchilla D, et al. A flagellin-induced complex of the receptor FLS2 and BAK1 initiates plant defence. Nature. 2007;448(7152):497–500. doi: 10.1038/nature05999. [DOI] [PubMed] [Google Scholar]
- 9.Heese A, et al. The receptor-like kinase SERK3/BAK1 is a central regulator of innate immunity in plants. Proc Natl Acad Sci USA. 2007;104(29):12217–12222. doi: 10.1073/pnas.0705306104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lu D, et al. A receptor-like cytoplasmic kinase, BIK1, associates with a flagellin receptor complex to initiate plant innate immunity. Proc Natl Acad Sci USA. 2010;107(1):496–501. doi: 10.1073/pnas.0909705107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhang J, et al. Receptor-like cytoplasmic kinases integrate signaling from multiple plant immune receptors and are targeted by a Pseudomonas syringae effector. Cell Host Microbe. 2010;7(4):290–301. doi: 10.1016/j.chom.2010.03.007. [DOI] [PubMed] [Google Scholar]
- 12.Shi H, et al. BR-SIGNALING KINASE1 physically associates with FLAGELLIN SENSING2 and regulates plant innate immunity in Arabidopsis. Plant Cell. 2013;25(3):1143–1157. doi: 10.1105/tpc.112.107904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Korasick DA, et al. Novel functions of Stomatal Cytokinesis-Defective 1 (SCD1) in innate immune responses against bacteria. J Biol Chem. 2010;285(30):23342–23350. doi: 10.1074/jbc.M109.090787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Asai T, et al. MAP kinase signalling cascade in Arabidopsis innate immunity. Nature. 2002;415(6875):977–983. doi: 10.1038/415977a. [DOI] [PubMed] [Google Scholar]
- 15.Boudsocq M, et al. Differential innate immune signalling via Ca(2+) sensor protein kinases. Nature. 2010;464(7287):418–422. doi: 10.1038/nature08794. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shan LB, et al. Bacterial effectors target the common signaling partner BAK1 to disrupt multiple MAMP receptor-signaling complexes and impede plant immunity. Cell Host Microbe. 2008;4(1):17–27. doi: 10.1016/j.chom.2008.05.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Li J, et al. BAK1, an Arabidopsis LRR receptor-like protein kinase, interacts with BRI1 and modulates brassinosteroid signaling. Cell. 2002;110(2):213–222. doi: 10.1016/s0092-8674(02)00812-7. [DOI] [PubMed] [Google Scholar]
- 18.Nam KH, Li J. BRI1/BAK1, a receptor kinase pair mediating brassinosteroid signaling. Cell. 2002;110(2):203–212. doi: 10.1016/s0092-8674(02)00814-0. [DOI] [PubMed] [Google Scholar]
- 19.Postel S, et al. The multifunctional leucine-rich repeat receptor kinase BAK1 is implicated in Arabidopsis development and immunity. Eur J Cell Biol. 2010;89(2-3):169–174. doi: 10.1016/j.ejcb.2009.11.001. [DOI] [PubMed] [Google Scholar]
- 20.Roux M, et al. The Arabidopsis leucine-rich repeat receptor-like kinases BAK1/SERK3 and BKK1/SERK4 are required for innate immunity to hemibiotrophic and biotrophic pathogens. Plant Cell. 2011;23(6):2440–2455. doi: 10.1105/tpc.111.084301. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Liu Z, et al. BIK1 interacts with PEPRs to mediate ethylene-induced immunity. Proc Natl Acad Sci USA. 2013;110(15):6205–6210. doi: 10.1073/pnas.1215543110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lin W, et al. Inverse modulation of plant immune and brassinosteroid signaling pathways by the receptor-like cytoplasmic kinase BIK1. Proc Natl Acad Sci USA. 2013;110(29):12114–12119. doi: 10.1073/pnas.1302154110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang XF, et al. Sequential transphosphorylation of the BRI1/BAK1 receptor kinase complex impacts early events in brassinosteroid signaling. Dev Cell. 2008;15(2):220–235. doi: 10.1016/j.devcel.2008.06.011. [DOI] [PubMed] [Google Scholar]
- 24.Shiu SH, Bleecker AB. Receptor-like kinases from Arabidopsis form a monophyletic gene family related to animal receptor kinases. Proc Natl Acad Sci USA. 2001;98(19):10763–10768. doi: 10.1073/pnas.181141598. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Oh MH, et al. Tyrosine phosphorylation of the BRI1 receptor kinase emerges as a component of brassinosteroid signaling in Arabidopsis. Proc Natl Acad Sci USA. 2009;106(2):658–663. doi: 10.1073/pnas.0810249106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Oh MH, et al. Autophosphorylation of Tyr-610 in the receptor kinase BAK1 plays a role in brassinosteroid signaling and basal defense gene expression. Proc Natl Acad Sci USA. 2010;107(41):17827–17832. doi: 10.1073/pnas.0915064107. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Laluk K, et al. Biochemical and genetic requirements for function of the immune response regulator BOTRYTIS-INDUCED KINASE1 in plant growth, ethylene signaling, and PAMP-triggered immunity in Arabidopsis. Plant Cell. 2011;23(8):2831–2849. doi: 10.1105/tpc.111.087122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Veronese P, et al. The membrane-anchored BOTRYTIS-INDUCED KINASE1 plays distinct roles in Arabidopsis resistance to necrotrophic and biotrophic pathogens. Plant Cell. 2006;18(1):257–273. doi: 10.1105/tpc.105.035576. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Jaillais Y, et al. Tyrosine phosphorylation controls brassinosteroid receptor activation by triggering membrane release of its kinase inhibitor. Genes Dev. 2011;25(3):232–237. doi: 10.1101/gad.2001911. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.