Significance
Comprehending the responses of organisms to pollutants by a systems-based approach allows characterization of molecular events and the cellular pathways that have been perturbed. However, mapping only adverse outcomes of a toxicant in an organism falls short of describing the defense response that is mounted to maintain homeostasis and resistance to the toxic insult. Our study provides the understanding of molecular mechanisms of algae in response to silver, which in turn indicates how the algae might behave in a silver contamination scenario. We have used complementary information obtained from the transcriptome, proteome, and physiology to gain mechanistic insights into the responses of Chlamydomonas reinhardtii. We show here the importance of stress and adaptive responses, especially at sublethal concentrations of pollutant.
Keywords: toxicity response, adaptive pathway, algae, systems biology, adverse outcome pathway
Abstract
Understanding mechanistic and cellular events underlying a toxicological outcome allows the prediction of impact of environmental stressors to organisms living in different habitats. A systems-based approach aids in characterizing molecular events, and thereby the cellular pathways that have been perturbed. However, mapping only adverse outcomes of a toxicant falls short of describing the stress or adaptive response that is mounted to maintain homeostasis on perturbations and may confer resistance to the toxic insult. Silver is a potential threat to aquatic organisms because of the increasing use of silver-based nanomaterials, which release free silver ions. The effects of silver were investigated at the transcriptome, proteome, and cellular levels of Chlamydomonas reinhardtii. The cells instigate a fast transcriptome and proteome response, including perturbations in copper transport system and detoxification mechanisms. Silver causes an initial toxic insult, which leads to a plummeting of ATP and photosynthesis and damage because of oxidative stress. In response, the cells mount a defense response to combat oxidative stress and to eliminate silver via efflux transporters. From the analysis of the perturbations of the cell’s functions, we derived a detailed mechanistic understanding of temporal dynamics of toxicity and adaptive response pathways for C. reinhardtii exposed to silver.
Assessing the effects of stressors, such as metal ions, on organisms in the environment usually involves the quantification of impact on parameters, such as growth, reproduction, or survival. These parameters, however, reveal neither subtle effects that precede organism level changes nor adaptive responses that allow the organism to recover. Advances in “omics” technologies increasingly facilitate the comprehensive analysis of stressor effects in organisms living in different habitats at different subcellular levels (1, 2), including abundance of RNA transcripts (transcriptome) (2) and expression of proteins (proteome). The genome-wide transcriptome and proteome changes should aid in determining the molecular pathways underlying the response of the organism to a stressor. The “adverse outcome pathway” concept has recently been introduced as a conceptual framework to link multiple levels of biological organization, bridging a direct molecular initiating event and an adverse outcome at a biological level of organization relevant to risk assessment (3). Although an adverse outcome pathway allows us to obtain comprehensive knowledge of the effects of a toxicant, the adaptive response that is mounted to maintain homeostasis in the organism on perturbations and may confer resistance or adaptation to the toxic insult is at least as relevant because it determines the outcome of exposure to a toxicant (4). Therefore, the toxicity and adaptive response pathways together, along with the physiological state, need to be explored to gain insight into the recovering capacity of the organisms. In our present study we have elucidated toxicity and adaptive response pathways by identifying and linking the perturbations across the transcriptome, proteome, and physiological phenotype in the green algae Chlamydomonas reinhardtii on exposure to silver.
Silver toxicity to aquatic organisms, historically, has been a concern because of the effluents of photo-processing and mining industries (5). The toxicity is related to silver speciation with only free silver ions (Ag+) being highly toxic. Silver ions readily complex with high affinity to ligands, such as sulfide, chloride, dissolved organic carbon, and biomolecules (6).
Silver is considered an important contaminant that has high environmental impact because of the effects on health of the ecosystem (7) and bioaccumulation (8, 9). A recent trend is the increased use of silver as silver nanoparticles in consumer products (www.nanotechproject.org/inventories). This widespread use potentially increases the release of Ag+ into the aquatic environment (10). Despite complexation of Ag+, which can render the ions nonbioavailable, even low nanomolar concentrations of Ag in surface waters are of concern because not only are they highly toxic, but also tend to bioconcentrate in organisms similar to essential metals (11).
Studies have shown that nanomolar concentrations of Ag+ are toxic to organisms, such as the freshwater crustacean Daphnia magna (12), the freshwater fish rainbow trout (Oncorhynchus mykiss) (13), protozoans (8), and the unicellular algae C. reinhardtii (14). In C. reinhardtii, the uptake of Ag+ is fast (15) and is assumed to be via copper transporters (16). Moreover, the toxicity of silver nanoparticles is attributed to free Ag+ (17).
Although several studies have explored the mechanisms of silver toxicity, mostly in fish, there are not many insights into the molecular perturbations that lead to the outcome of either toxicity or adaptation to silver. Our previous studies have shown that Ag+ causes inhibition of photosynthesis and induction of reactive oxygen species (ROS) in C. reinhardtii (18). In this study we exploited the technologies of microarray and multidimensional protein identification (MudPIT) to analyze the transcriptome and proteome, respectively, to characterize the molecular changes. Such an integrated approach elucidated the effects on the network of biological pathways and, thereby, both the toxicity and adaptive pathways, which regulate the response of C. reinhardtii to Ag+.
Results and Discussion
Global Changes on the Transcriptome and Proteome Levels.
C. reinhardtii instigated a strong response at the transcriptome level to Ag+ exposure. Multigroup analysis of the transcriptome by two-way ANOVA showed that ∼8,200 transcripts were significantly regulated across all time points (SI Appendix, Table S1). The microarray data are submitted to the Gene Expression Omnibus database (GEO accession no. GSE48677). Two-group analysis of each exposure condition compared with the control demonstrates that most of the regulation of the transcriptome is at the early time points of 15 min and 1 h. Beyond the 1-h time point, the transcriptome of the exposed algae was not different from the respective control algae (SI Appendix, Fig. S1). This result indicates that already after 5 h of exposure the cell had organized its response at the transcriptome level. This finding is especially true for algae exposed to 10 nM Ag+, where the transcriptome resembled that of the control already after 1 h of exposure. With use of the publicly available annotation database for C. reinhardtii (19), ∼3,125 transcripts could be assigned to functional groups. Although a large number of transcripts were commonly regulated between the different time points and concentrations, there were also some which were unique to the exposure conditions (SI Appendix, Fig. S2).
By MudPIT analysis for time points of 1 and 5 h (SI Appendix, Fig. S3), we identified ∼4,000 proteins with at least two discrete peptide hits at an FDR of 0.5%. The proteins identified in this study represent 29% of the Chlamydomonas database (20). The statistical analysis of the spectral counts (21) revealed significant differences in protein abundance between control and Ag+-exposed algae. Of the 4,000 identified proteins, around 1,000 were significantly differentially expressed on exposure to Ag+ at 1 and 5 h. This result is in contrast to the transcriptome response, where no changes relative to the control were observed at 5 h and beyond.
MapMan Ontology term enrichment (22) of the differentially regulated proteins showed that biological pathways with significant enrichment common at the transcriptome and proteome levels were those involved in photosynthesis, the light reaction centers, tetrapyrrole synthesis, mitochondrial electron transport chain, amino acid metabolism, lipid metabolism, glycolysis, protein targeting, glutathione and ascorbate reduction-oxidation (redox) processes, and cell wall synthesis (SI Appendix, Fig S4). However, protein processing was enriched only at the proteome level (SI Appendix, Fig. S5). The proteins that were exclusively enriched at the level of the transcriptome were those of vesicle transport, signaling of proteins, and MAP kinases (SI Appendix, Fig. S6). These results indicate that not all pathways regulated at the transcriptome are reflected at the proteome level. One reason for the discrepancy is that, although mRNA abundance is a good indication for the presence of a protein, it does not always explain the variation in protein quantities. Only about 40% of the variation of protein amounts can be explained by mRNA abundance and 60% has been suggested to be mostly controlled at the level of translation (23). Another explanation is that MudPIT discriminates more against low copy-number proteins than microarray does and, thus, possibly produces some discrepancies.
Uptake and Transport of Silver.
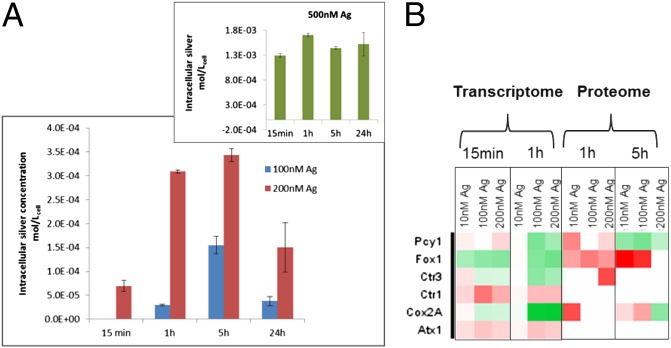
One factor determining the outcome of exposure to Ag+ is the intracellular concentration. Intracellular concentrations of silver increased up to 5 h of exposure (Fig. 1A), after which a decrease was observed. It is remarkable that after 5-h exposure duration, C. reinhardtii exposed to 200 nM Ag+ reached micromolar concentrations of intracellular silver (3.5 × 10−4 mol/Lcell-1). The intracellular concentration confirms that Ag+ in C. reinhaardtii is taken up fast and the bioconcentration factor is high (15). The accumulation of silver suggests that Ag+ likely is complexed with proteins or other ligands. The Ag+ is a soft Lewis acid and, like copper ions (Cu+), has affinity to coordinate with the S and N groups. Earlier studies hypothesize that the uptake of Ag+ into the cells occur via the Cu+ transport systems (16). Indeed, our transcriptome and proteome analysis indicate that several Cu+ transporters are regulated (Fig. 1B). One transporter is Ctr1, which facilitates the transport across the plasma membrane, supporting its role in the transport of Ag+. Moreover, Ag+ induced a regulation of Atx1, a copper metallo-chaperone in the cytoplasm that transports Cu+ to those proteins that require copper to be functional. Transporters, such as P-type ATPases present on organelle membranes, were also regulated. These results suggest that the intracellular distribution of Ag+ to the cellular compartments is mediated via Cu+ chaperones. The findings also indicate that, similar to that in insects (24), intracellular silver interferes with transport and homeostasis of copper in the algae.
Fig. 1.
Uptake and transport of silver in C. reinhardtii. Intracellular concentrations of silver (A) on exposure to 100 and 200 nM, and (Inset) to 500-nM exposure. No intracellular silver was quantifiable in C. reinhardtii exposed to 10 nM silver and is therefore not shown in the graph. Heat map of Cu transporters (B) in C. reinhardtii exposed to silver for varying durations with each box representing a protein at the transcriptome and proteome level, green being down-regulated and red up-regulated. The SD is shown in the figures. Atx1, antioxidant 1, copper chaperone; Cox2A, subunit 2A of cytochrome oxidase; Ctr1 and -3, copper transporter 1 and 3; Fox1, Ferroxidase; Pcy1, Plastocyanin. For the molecular responses, algae exposed to 500 nM Ag+ were not analyzed (see Methods).
We observed a decrease in the intracellular silver concentrations at 24 h for 100 nM and 200 nM Ag+ but not for the 500 nM Ag+ exposure (Fig. 1A, Inset). The decrease at lower exposure concentrations suggests that there is an efflux of Ag+ from the cells. At the transcriptome level, we observed an up-regulation of a P-type ATPase similar to CopATPase of Enterococcus hirae (25), indicating that the efflux of Ag+ occurs via a Cu-efflux system. Moreover, at later time points of exposure, free Ag+ in the medium may decrease because of the release of ligands from the algae, which is expected to lower the bioavailability through complexation, as shown for Cu+ (26) and Ag+ (6). In contrast, on exposure to 500 nM Ag+, cells appeared to be unable to actively eliminate the metal because they were severely damaged (see below).
Effects of Silver on Cellular Processes at Transcriptome, Proteome, and Physiological Levels.
Photosynthesis.
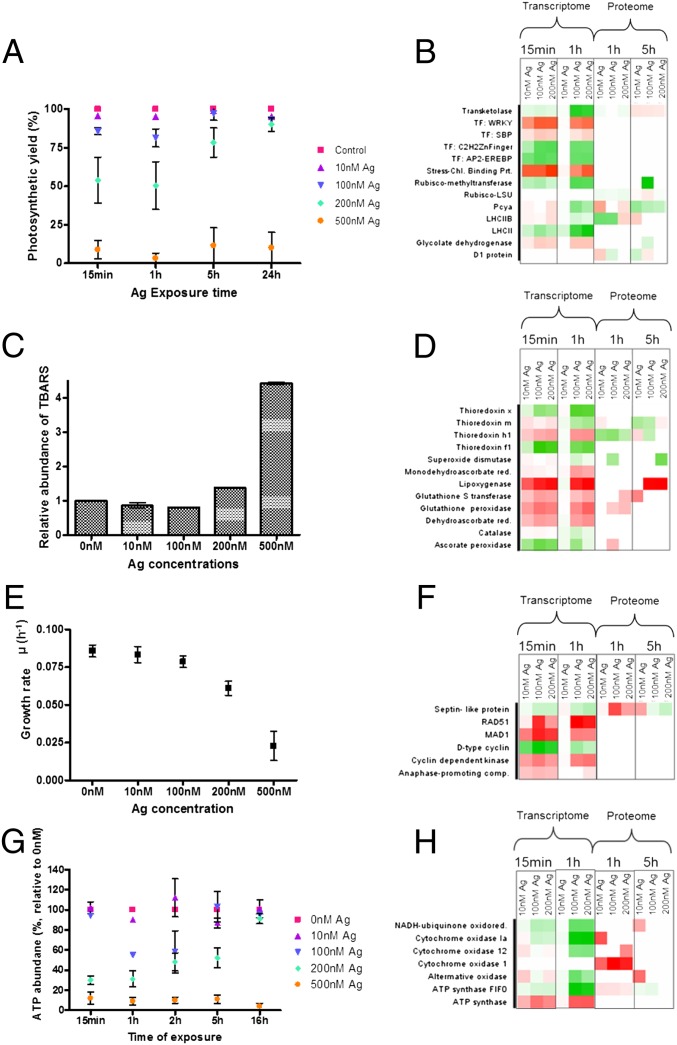
Exposure to Ag+ caused the photosynthetic yield of photosystem II (PSII) to be adversely affected (Fig. 2A). The inhibition of PSII yield was quick, observed within 15 min of exposure, and followed a clear dose–response up to the time point of 5 h. Within 1 h, 200 nM Ag+ reduced the photosynthetic yield by nearly 50%. However, after this time the yield returned to control levels. The inhibition of PSII can be explained by the sensitivity of the photosynthetic machinery to Ag+, which binds to thiols of functional proteins, displacing trace metals—such as Cu+—in the process, making the proteins nonfunctional (27). The recovery of the PSII yield after 5 h coincides with the decreased intracellular Ag+ concentrations and suggests that the cell has mounted a detoxifying response. However, at higher exposure concentrations (500 nM Ag+) a recovery in the PSII yield with time was not observed. Apparently, the algae could not balance the toxicity with the detoxifying response. To understand the molecular mechanisms that lead to the inhibition and recovery of the photosynthetic machinery, we analyzed its transcriptome and proteome (Fig. 2B).
Fig. 2.
Regulation of functional pathways in C. reinhardtii at physiological (A, C, E, G) and molecular (B, D, F, H) levels. Photosynthesis (A, B); lipid peroxidation compared with the control (C) and oxidative stress response (D); growth (E, F); ATP content (G) and synthesis (H). In the heat maps each square represents a protein, with green being down-regulated and red up-regulated. For the molecular responses, algae exposed to 500 nM Ag+ were not analyzed (see Methods).
Both at the transcriptome and proteome levels, photosynthesis is one of the functionally enriched groups with a large number of identifiers (SI Appendix, Figs. S1 and S7). Several of the light-harvesting complex (LHC) proteins were regulated upon exposure to Ag+. These proteins, especially the LHC proteins B and M, are sensitive to stress, such as high light (28), and a regulation is indicative for the recovery of the photosynthetic machinery to the normal state. Serine-threonine protein kinase, which phosphorylates the LHC proteins, was also regulated, indicating that the functioning of LHC and, thereby, the state transition from PSII to PSI is affected. Plastocyanin, which is an electron carrier between PSII and PSI, and cytochrome oxidase were regulated both at the transcriptome and proteome level. These proteins are particularly relevant because they require Cu+ for their activity. Thus, when Ag+ replaces Cu+ it can cause disturbance or inactivation of the photosynthetic electron transport (29). This disturbance results in the generation of ROS. We also observed a regulation of the D1 protein, at the proteome level, which indicates the presence of increased ROS (30). Thus, although Ag+ has been shown earlier to induce ROS in C. reinhardtii (18), we show here the biological pathways that lead to increased production of ROS. The ROS produced in the chloroplast compartment from disturbances in the photosynthetic machinery can also pass into the cytoplasm and activate the MAPK cascade, which in turn regulates specific stress response transcription factors (31). Upon Ag+ exposure, transcription factors belonging to the families of WRKY, AP2-EREBP (ethylene response element binding protein), and Cys2/His2-type finger, which have been shown to play a role in response to temperature and water stress (32), were regulated (Fig. 2B).
In addition to the damage on the PS, Ag+ exposure also caused changes in the closely associated Calvin cycle. The key enzymes, such as rubisco, triose-phosphate isomerase, and transketolase were regulated. This regulation indicates that to cope with inhibition of photosynthesis, the carbon fixation is possibly also regulated, leading to changes in the starch and fatty acid synthesis and glycolysis, as observed by the regulation of proteins involved. Earlier studies have linked the regulation of carbon fixation to the production of ROS caused by the disruption of the electron transport chain (33). From the analysis it can be concluded that the disturbance of the PS machinery by Ag+, and thereby the regulation of the proteins involved, has an impact on several other biological functions.
Antioxidant response.
The presence of ROS is one of the reasons for oxidative stress; they can cause damage to membranes by peroxidation of lipids. We measured the presence of malon-dialdehyde (MDA), one of the products of lipid peroxidation, in the presence of Ag+. Lipid peroxidation was observed only in C. reinhardtii exposed to 500 nM Ag+ with nearly four-times more MDA than the control, indicating that damage caused by increased ROS production is a key component of Ag+ toxicity (Fig. 2C). The absence of oxidative damage as measured by MDA amounts at lower Ag+ concentrations, however, does not reflect absence of oxidative stress. In fact, the transcriptome and proteome data indicate a defense response against ROS, directing the cell to a detoxifying reaction and repair.
The responses that scavenge ROS, such as synthesis of the antioxidants glutathione and thioredoxins, and of enzymes that include glutathione peroxidase (GPX-H or GPX5), GST (GSTS1), catalase, superoxide dismutase, and ascorbate peroxidase, were regulated (Fig. 2D) in algae even when no lipid peroxidation was quantifiable. The proteins GPX-H (GPX5) and GSTS1, which are specifically induced in the presence of singlet oxygen (34), were highly up-regulated both at the transcriptome and proteome. These enzymes contribute to the cell acclimatizing to oxidative stress. At low concentrations of Ag+, lipoxygenase, which is involved in the oxidation of membrane lipids, was regulated. Additionally, monodehydroascorbate reductase and dehydroascorbate reductase, which maintain the pool of the reducing agents, glutathione and ascorbate, were regulated; they have been shown before to enhance tolerance to abiotic stress, like heat, and to confer tolerance to metal induced stress in higher plants (35). The thioredoxin (Trx) proteins, Trxh1, x, m, and f1, which have been described to be induced by H2O2 and DNA damage (36), and which facilitate the reduction of cysteine double bonds of proteins, were also regulated. The transcriptome and proteome therefore indicate that, despite the fact that lipid peroxidation was not observed at the phenotype level at lower concentrations, Ag+ induces an oxidative stress response, likely triggering detoxification, repair, and adaptation.
Growth rate.
Upon exposure to Ag+, the growth rate of C. reinhardtii was significantly reduced in a dose-dependent manner. Whereas at the highest concentration (500 nM) of Ag+ growth was completely inhibited, the growth rates were notably reduced at concentrations <500 nM (Fig. 2E). This finding is in contrast to the effect on photosynthesis, where the algae exposed to less than 500 nM Ag+ recovered by 5 h. Several proteins involved in growth and cell division were regulated both at transcriptome and proteome levels (Fig. 2F). These proteins included: the cyclin-dependent kinase regulating cell cycle; a septin-like protein that is a scaffold recruiting protein and essential for cytokinesis; dynein 1b, required for centrosome assembly; and the anaphase-promoting complex, which is crucial for the transition from the metaphase to the anaphase of the cell cycle. Proteins required for DNA double-strand repair, such as RAD51, and those preventing anaphase of the cell cycle, such as MAD1, were also differentially regulated. Most of these proteins were represented only at the transcriptome level and not at the proteome level (Fig. 2F). The reason for the discrepancy is that proteins involved in cell proliferation are unstable and have short half-lives (23). From the transcriptome analysis we can conclude that there is an effect at the metaphase of the cell division and this is reflected in the dose-dependent reduction of growth.
Energy.
Energy content of the cells in the form of ATP is a sensitive endpoint in C. reinhardtii for determining the toxicity of chemicals (37). The ATP content responded to Ag+ exposure and decreased significantly in C. reinhardtii at concentrations greater than 100 nM (Fig. 2G). The chloroplast and mitochondria are the primary sites of ATP production, and the inhibition of the function of these organelles as seen by the regulation of key proteins might explain the decrease in ATP. Moreover, detoxifying mechanisms, such as active efflux of Ag+, also requires ATP, impacting the total content within the cell. In mitochondria, the electrochemical proton gradient produced by the electron transport chain is used to generate ATP. The transcriptome and proteome showed a regulation of several of the proteins involved (SI Appendix, Fig. S8). The NADH-ubiquinone oxidoreductase, the cytochrome oxidases, and alternate oxidase, all of which are involved in transport of electrons, were significantly regulated (Fig. 2H). Also regulated were several subunits of ATP-synthase, which is involved in the synthesis of ATP. The NADH-dehydrogenases, which were regulated, have cysteine residues, which have high affinity for Ag+. Moreover, in bacteria the inactivation of NADH-dehydrogenase as a result of the replacement of Cu+ by Ag+ has been shown to cause the formation of an inactive complex (38), resulting in inefficient pumping of protons across the membrane and loss of the proton gradient. Thereby an increased production of ROS occurs, which has been linked to the antibacterial properties of Ag+ (39). Our data provide evidence that Ag+ produces the same effects in the mitochondria in C. reinhardtii.
Lipid metabolism and autophagy.
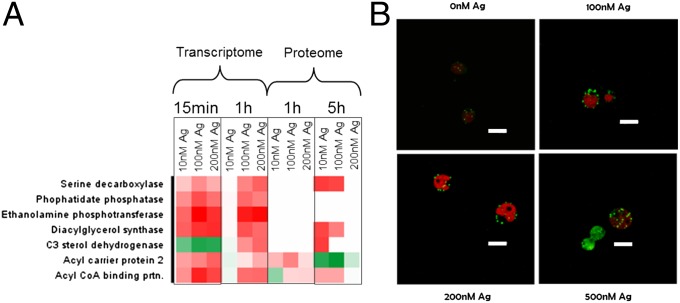
The transcriptome and proteome responses indicated that lipid metabolism is one of the pathways regulated upon exposure to Ag+. Several proteins involved in lipid biosynthesis, such as the diacyl glycerol acyl transferase (type 2) and phospho-lipid diacyl glycerol synthase, key enzymes for the synthesis of diacyl glycerol, were regulated (Fig. 3A). Diacyl glycerol is the precursor of triacyl glycerol (TAG), which is a component of the membrane lipids and is normally stored in lipid bodies (40). The number of lipid bodies in the algae indeed increased in a dose-dependent manner upon exposure to Ag+ (Fig. 3B and SI Appendix, Fig. S9). Interestingly, the increase in lipid bodies was also seen at concentrations of Ag+, where no effects on photosynthesis or growth were observed, suggesting that lipid body accumulation is a particularly sensitive response. During normal growth, C. reinhardtii has only few lipid bodies. However, several stress conditions, such as nitrogen or sulfur deprivation, promote the accumulation of TAG-containing lipid bodies (41). The accumulation of lipid bodies is also related to the triggering of the sophisticated autophagy program, which includes destruction of cytoplasmic and chloroplastic ribosomes, degradation of the cytochrome b6-f complex, regulation of the proteasome, and plummeting of carbon fixation (42). In addition to the lipid biosynthesis proteins, the 5′-AMP-activated protein kinase (AMPK) and autophagy-related proteins 3 and 7, involved in the regulation of the autophagy program, were regulated on exposure to Ag+ (SI Appendix, Fig. S10). These results indicate that Ag+ induces lipid bodies either directly as a response to lipid membrane damage, acting as storage for precursors of lipid membranes, or indirectly as a response to the autophagy process, which requires new membranes lipids (42).
Fig. 3.
Molecular and physiological changes of lipid synthesis in C. reinhardtii exposed to silver for 1 h. Regulation of proteins (A) and lipid bodies containing precursors of lipids stained green with Nile red (B). The chloroplasts autoflourescence is seen as red. (Scale bars in B, 10 µm.) For the molecular responses, algae exposed to 500 nM Ag+ were not analyzed (see Methods).
Targeting and transport of proteins.
Protein targeting to various organelles was significantly regulated in response to Ag+ exposure (SI Appendix, Fig. S11). Most of the changes were, however, only observed at the transcriptome and not at the proteome level. Markedly, mRNA transcripts of proteins involved in transport of other proteins and molecules to the nucleus, endoplasmic reticulum (ER), and plasma membrane were up-regulated. Genes involved in the secretory pathways, such as Sec71 (snare complex protein) and Sec61, required for the vesicular transport between the ER and the Golgi complex, were up-regulated. In contrast, transcripts of proteins involved in protein export to the chloroplast and mitochondria were down-regulated. These include the chloroplast proteins Tic20, a component of the chloroplast protein import apparatus, SRP54 (signal recognition particle 54), and SecY, a preprotein translocase. In the mitochondria, several translocases were also regulated. From these results it appears that mitochondria and chloroplast, both organelles of prokaryotic origin, are affected by Ag+, as seen not only from their reduced functionality, but also by reduced transport of essential proteins to these organelles.
Temporal dynamics of the regulation.
The expression of the proteins at the transcriptome level display temporal dynamics that also differ with concentration, as observed from Euclidean clustering (SI Appendix, Fig. S12). At the lowest exposure concentration (i.e., 10 nM Ag+), differential expression was observed only at 15 min. At the later time points of 1, 5, and 16 h, the transcriptome of 10 nM Ag+-exposed algae resembled that of the control algae. This finding indicates that the early response at the transcriptome level is sufficient to allow the cells to recover from the toxicity, if any, caused by 10 nM Ag+. Among the regulated transcripts, of interest are those regulated uniquely, such as the alternative oxidase 1 (Aox1) and carbonic anhydrase, which were up-regulated at 10 nM at 15 min but down-regulated at all other time points and concentrations. Alternative oxidase is a key protein in mitochondrial electron transport and helps to control the excess production of ROS under stress conditions (43). In addition, carbonic anhydrase, involved in carbon fixation, has been shown to play a key role in detoxification processes by activation of the lysosomal system in toxicant exposed organisms (44). The regulation of the proteins that play a key role in detoxification suggests that these proteins are involved in the recovery from Ag+-induced stress at low concentrations. At the higher concentrations of 100 nM and 200 nM Ag+, the transcriptome is similar to that of the unexposed control sample after 5 h of exposure. This result indicates that the defense responses require a longer duration to allow recovery from the stress at higher concentrations of Ag+.
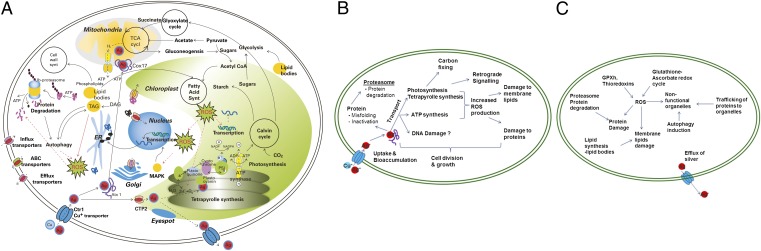
To conclude, the transcriptome, proteome, and physiological profiles provided detailed insights into the mechanisms of toxicity, detoxification, and repair at different biological levels in C. reinhardtii on exposure to Ag+ (Fig. 4). From the detailed analysis of the perturbations of the cell’s functional networks, we can derive toxicity and adaptive pathways for Ag+ in C. reinhardtii. The initiating event is the binding and translocation across the cell membrane of Ag+ by copper transporters. In the cells, Ag+ is distributed via the Cu+ chaperones and this concomitantly elicits several effects. Ag+ binds to thiol groups of proteins causing mis-folding and damage, replaces Cu+ in key proteins in ATP- and photosynthesis, and regulates the expression of proteins leading to inhibition of ATP- and photosynthesis. Interestingly, the functioning of mitochondria and chloroplast, both organelles of prokaryotic origin, is specifically affected. The damage is mainly because of increased ROS production, which causes lipid peroxidation. As a defense mechanism against the oxidative stress, the algae mount an antioxidant response. At lower concentration of Ag+ the antioxidant response is seemingly adequate for the recovery. At higher concentrations, in addition to the antioxidant response, efflux removes Ag+ as seen by decreasing intracellular concentrations, confirming a detoxification process. Our analyses, which consider both the concentration and time-dependent responses, allowed us to identify the dynamics of detoxification and recovery from Ag+ toxicity. Despite the recovery of the algae within 24 h at low-exposure concentrations, the high intracellular silver concentrations raise concerns about long-term effects of Ag+ exposure in the algae. On continuous exposures for long durations, it is probable that the adaptive responses of C. reinhardtii are overwhelmed even at low concentrations, leading to toxicity. Moreover, because algae are primary producers, bio-accumulated silver may affect other organisms when introduced into the food chain. At this point we demonstrate the necessity to take into account the adaptive responses to estimate how toxic the contaminant is. However, a comprehensive understanding of responses of nonmodel organisms, especially in the environment, is still a challenge. Nonetheless, it is probable that other primary producers do respond similarly to silver.
Fig. 4.
The toxicity and adaptive response pathways, as derived from linking transcriptome and proteome responses to physiological effects. (A) Schematic representation of biological pathways in C. reinhardtii affected by Ag+. (B) Schematic representation of the toxicity pathway. (C) Schematic representation of the adaptive-response pathway.
Methods
Medium and Growth Conditions.
C. reinhardtii (CC-125, wild-type mt+137c) were grown in Talaquil (45) medium (pH 7.5) (SI Appendix).
Silver Exposure Conditions.
For the exposure experiments, algae in the log-phase of growth were used. C. reinhardtii were added to a final cell density of 2.5 × 105 cells/mL to Talaquil medium, which was pre-equilibrated with silver nitrate. For durations of 15 min, 1 h, 5 h, and 16 h, algae were exposed to silver concentrations of 10 nM, 100 nM, 200 nM, and 500 nM, respectively. The control algae were exposed to no silver. The nominal ion concentrations of silver estimated using the free software VMinTeq (www2.lwr.kth.se/English/OurSoftware/vminteq) showed that 95% of silver occurs as free silver ions (Ag+) and is bioavailable at the start of the experiment. At later time points, the amounts of Ag+ may be different, if bound to biomolecule ligands potentially released from the algae into the medium. After exposure, various physiological responses, such as the growth rate, photosynthetic yield, total cellular ATP, and lipid body content were determined (SI Appendix). All exposure experiments were done in biological triplicates.
Bioaccumulation.
To ensure quantification of intracellular silver, cysteine, a ligand which binds to Ag+ with high affinity, was used to remove the extracellular Ag+ from algal cells. C. reinhardtii cultures exposed to Ag+ were treated with 1 mM cysteine and the algae were collected on nitrocellulose filters (Sartorius). The samples were digested and diluted (SI Appendix) and analyzed by high-resolution inductively coupled plasma-MS analysis (Thermo Finnigan).
Transcriptome Analysis.
Total RNA was isolated from C. reinhardtii cultures using the RNeasy Kit (Qiagen) RNA integrity was determined using an Agilent BioAnalyzer 2100 and only those samples with an RNA integrity number higher than 7 were used for the microarray. The RNA isolated from algae exposed to 500 nM Ag+ was of insufficient quality for analysis by microarrays as reflected by the RNA integrity number. Therefore, for the transcriptome and proteome analysis only algae exposed to concentrations of silver at 10, 100, and 200 nM were analyzed.
Microarray Design and Experiment.
A C. reinhardtii whole-genome gene-expression microarray (Genotypic Technology) was designed using the JGI V4 Chlamydomonas best-transcripts database. The array (AMADID # 025588) is in the format 4 × 44 k (Agilent Technologies) and covers 15,150 nuclear encoded transcripts with one probe per transcript in triplicates. The labeling of RNA and hybridization was done according to the manufacturer’s protocol (Agilent Technologies) (SI Appendix).
Proteome Analysis.
Protein extraction and tryptic digestion.
Algae exposed for 1 and 5 h to varying concentrations of Ag+ were centrifuged down at 1,200 × g and total protein was extracted. Trypsin digestion of protein to peptides, two dimension liquid chromatography separation, mass spectrometry, and analysis were done as described previously with slight modifications (46) (SI Appendix).
Supplementary Material
Acknowledgments
The authors thank Bettina Wagner for the help with Chlamydomonas reinhardtii cultures; René Schönenberger for the help with multidimensional protein identification; and David Kistler for the inductively coupled plasma-MS analysis. This work was supported in part by Eawag and the National Research Programm 64 of Swiss National Research Foundation.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. H.H. is a guest editor invited by the Editorial Board.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, www.ncbi.nlm.nih.gov/geo (accession no. GSE48677).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1319388111/-/DCSupplemental.
References
- 1.Van Aggelen G, et al. Integrating omic technologies into aquatic ecological risk assessment and environmental monitoring: Hurdles, achievements, and future outlook. Environ Health Perspect. 2010;118(1):1–5. doi: 10.1289/ehp.0900985. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schirmer K, Fischer BB, Madureira DJ, Pillai S. Transcriptomics in ecotoxicology. Anal Bioanal Chem. 2010;397(3):917–923. doi: 10.1007/s00216-010-3662-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ankley GT, et al. Adverse outcome pathways: A conceptual framework to support ecotoxicology research and risk assessment. Environ Toxicol Chem. 2010;29(3):730–741. doi: 10.1002/etc.34. [DOI] [PubMed] [Google Scholar]
- 4.Jennings P. Stress response pathways, toxicity pathways and adverse outcome pathways. Arch Toxicol. 2013;87(1):13–14. doi: 10.1007/s00204-012-0974-4. [DOI] [PubMed] [Google Scholar]
- 5.Purcell TW, Peters JJ. Sources of silver in the environment. Environ Toxicol Chem. 1998;17(4):539–546. [Google Scholar]
- 6.Stevenson LM, et al. Environmental feedbacks and engineered nanoparticles: Mitigation of silver nanoparticle toxicity to Chlamydomonas reinhardtii by algal-produced organic compounds. PLoS ONE. 2013;8(9):e74456. doi: 10.1371/journal.pone.0074456. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Hogstrand C, Wood CM. Toward a better understanding of the bioavailability, physiology and toxicity of silver in fish: Implications for water quality criteria. Environ Toxicol Chem. 1998;17(4):547–561. [Google Scholar]
- 8.Ratte HT. Bioaccumulation and toxicity of silver compounds: A review. Environ Toxicol Chem. 1999;18(1):89–108. [Google Scholar]
- 9.Bianchini A, Grosell M, Gregory SM, Wood CM. Acute silver toxicity in aquatic animals is a function of sodium uptake rate. Environ Sci Technol. 2002;36(8):1763–1766. doi: 10.1021/es011028t. [DOI] [PubMed] [Google Scholar]
- 10.Benn TM, Westerhoff P. Nanoparticle silver released into water from commercially available sock fabrics. Environ Sci Technol. 2008;42(11):4133–4139. doi: 10.1021/es7032718. [DOI] [PubMed] [Google Scholar]
- 11.Veltman K, Huijbregts MAJ, Van Kolck M, Wang WX, Hendriks AJ. Metal bioaccumulation in aquatic species: Quantification of uptake and elimination rate constants using physicochemical properties of metals and physiological characteristics of species. Environ Sci Technol. 2008;42(3):852–858. doi: 10.1021/es071331f. [DOI] [PubMed] [Google Scholar]
- 12.Bianchini A, Wood CM. Mechanism of acute silver toxicity in Daphnia magna. Environmental toxicology and chemistry / SETAC. 2003;22(6):1361–1367. [PubMed] [Google Scholar]
- 13.Morgan IJ, Henry RP, Wood CM. The mechanism of acute silver nitrate toxicity in freshwater rainbow trout (Oncorhynchus mykiss) is inhibition of gill Na+ and Cl− transport. Aquat Toxicol. 1997;38(1–3):145–163. [Google Scholar]
- 14.Lee DY, Fortin C, Campbell PGC. Contrasting effects of chloride on the toxicity of silver to two green algae, Pseudokirchneriella subcapitata and Chlamydomonas reinhardtii. Aquat Toxicol. 2005;75(2):127–135. doi: 10.1016/j.aquatox.2005.06.011. [DOI] [PubMed] [Google Scholar]
- 15.Piccapietra F, Allué CG, Sigg L, Behra R. Intracellular silver accumulation in Chlamydomonas reinhardtii upon exposure to carbonate coated silver nanoparticles and silver nitrate. Environ Sci Technol. 2012;46(13):7390–7397. doi: 10.1021/es300734m. [DOI] [PubMed] [Google Scholar]
- 16.Fortin C, Campbell PGC. Silver uptake by the green alga Chlamydomonas reinhardtii in relation to chemical speciation: Influence of chloride. Environ Toxicol Chem. 2000;19(11):2769–2778. [Google Scholar]
- 17.Navarro E, et al. Toxicity of silver nanoparticles to Chlamydomonas reinhardtii. Environ Sci Technol. 2008;42(23):8959–8964. doi: 10.1021/es801785m. [DOI] [PubMed] [Google Scholar]
- 18.Szivak I, Behra R, Sigg L. Metal-induced reactive oxygen species production in Chlamydomonas reinhardtii (Chlorophyceae) J Phycol. 2009;45(2):427–435. doi: 10.1111/j.1529-8817.2009.00663.x. [DOI] [PubMed] [Google Scholar]
- 19.May P, Christian JO, Kempa S, Walther D. ChlamyCyc: An integrative systems biology database and web-portal for Chlamydomonas reinhardtii. BMC Genomics. 2009;10:209. doi: 10.1186/1471-2164-10-209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wienkoop S, et al. Targeted proteomics for Chlamydomonas reinhardtii combined with rapid subcellular protein fractionation, metabolomics and metabolic flux analyses. Mol Biosyst. 2010;6(6):1018–1031. doi: 10.1039/b920913a. [DOI] [PubMed] [Google Scholar]
- 21.Zhang B, et al. Detecting differential and correlated protein expression in label-free shotgun proteomics. J Proteome Res. 2006;5(11):2909–2918. doi: 10.1021/pr0600273. [DOI] [PubMed] [Google Scholar]
- 22.Lopez D, Casero D, Cokus SJ, Merchant SS, Pellegrini M. Algal Functional Annotation Tool: A Web-based analysis suite to functionally interpret large gene lists using integrated annotation and expression data. BMC Bioinformatics. 2011;12:282. doi: 10.1186/1471-2105-12-282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Schwanhäusser B, et al. Global quantification of mammalian gene expression control. Nature. 2011;473(7347):337–342. doi: 10.1038/nature10098. [DOI] [PubMed] [Google Scholar]
- 24.Armstrong N, Ramamoorthy M, Lyon D, Jones K, Duttaroy A. Mechanism of silver nanoparticles action on insect pigmentation reveals intervention of copper homeostasis. PLoS ONE. 2013;8(1):e53186. doi: 10.1371/journal.pone.0053186. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Solioz M, Odermatt A. Copper and silver transport by CopB-ATPase in membrane vesicles of Enterococcus hirae. J Biol Chem. 1995;270(16):9217–9221. doi: 10.1074/jbc.270.16.9217. [DOI] [PubMed] [Google Scholar]
- 26.Soldo D, Hari R, Sigg L, Behra R. Tolerance of Oocystis nephrocytioides to copper: intracellular distribution and extracellular complexation of copper. Aquat Toxicol. 2005;71(4):307–317. doi: 10.1016/j.aquatox.2004.11.011. [DOI] [PubMed] [Google Scholar]
- 27.Xu FF, Imlay JA. Silver(I), mercury(II), cadmium(II), and zinc(II) target exposed enzymic iron-sulfur clusters when they toxify Escherichia coli. Appl Environ Microbiol. 2012;78(10):3614–3621. doi: 10.1128/AEM.07368-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McKim SM, Durnford DG. Translational regulation of light-harvesting complex expression during photoacclimation to high-light in Chlamydomonas reinhardtii. Plant Physiol Biochem. 2006;44(11–12):857–865. doi: 10.1016/j.plaphy.2006.10.018. [DOI] [PubMed] [Google Scholar]
- 29.Sujak A. Interaction between cadmium, zinc and silver-substituted plastocyanin and cytochrome b(6)f complex—Heavy metals toxicity towards photosynthetic apparatus. Acta Physiol Plant. 2005;27(1):61–69. [Google Scholar]
- 30.Nishiyama Y, Allakhverdiev SI, Yamamoto H, Hayashi H, Murata N. Singlet oxygen inhibits the repair of photosystem II by suppressing the translation elongation of the D1 protein in Synechocystis sp. PCC 6803. Biochemistry. 2004;43(35):11321–11330. doi: 10.1021/bi036178q. [DOI] [PubMed] [Google Scholar]
- 31.Kovtun Y, Chiu WL, Tena G, Sheen J. Functional analysis of oxidative stress-activated mitogen-activated protein kinase cascade in plants. Proc Natl Acad Sci USA. 2000;97(6):2940–2945. doi: 10.1073/pnas.97.6.2940. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Qiu YP, Yu DQ. Over-expression of the stress-induced OsWRKY45 enhances disease resistance and drought tolerance in Arabidopsis. Environ Exp Bot. 2009;65(1):35–47. [Google Scholar]
- 33.Marri L, et al. Prompt and easy activation by specific thioredoxins of Calvin cycle enzymes of Arabidopsis thaliana associated in the GAPDH/CP12/PRK supramolecular complex. Mol Plant. 2009;2(2):259–269. doi: 10.1093/mp/ssn061. [DOI] [PubMed] [Google Scholar]
- 34.Fischer BB, Eggen RIL, Trebst A, Krieger-Liszkay A. The glutathione peroxidase homologous gene Gpxh in Chlamydomonas reinhardtii is upregulated by singlet oxygen produced in photosystem II. Planta. 2006;223(3):583–590. doi: 10.1007/s00425-005-0108-9. [DOI] [PubMed] [Google Scholar]
- 35.Cuypers A, Vangronsveld J, Clijsters H. Biphasic effect of copper on the ascorbate-glutathione pathway in primary leaves of Phaseolus vulgaris seedlings during the early stages of metal assimilation. Physiol Plant. 2000;110(4):512–517. [Google Scholar]
- 36.Sarkar N, Lemaire S, Wu-Scharf D, Issakidis-Bourguet E, Cerutti H. Functional specialization of Chlamydomonas reinhardtii cytosolic thioredoxin h1 in the response to alkylation-induced DNA damage. Eukaryot Cell. 2005;4(2):262–273. doi: 10.1128/EC.4.2.262-273.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nestler H, et al. Multiple-endpoint assay provides a detailed mechanistic view of responses to herbicide exposure in Chlamydomonas reinhardtii. Aquat Toxicol. 2012;110–111:214–224. doi: 10.1016/j.aquatox.2012.01.014. [DOI] [PubMed] [Google Scholar]
- 38.Friedrich T. The NADH:ubiquinone oxidoreductase (complex I) from Escherichia coli. Biochim Biophys Acta. 1998;1364(2):134–146. doi: 10.1016/s0005-2728(98)00024-3. [DOI] [PubMed] [Google Scholar]
- 39.Park HJ, et al. Silver-ion-mediated reactive oxygen species generation affecting bactericidal activity. Water Res. 2009;43(4):1027–1032. doi: 10.1016/j.watres.2008.12.002. [DOI] [PubMed] [Google Scholar]
- 40.Li Y, et al. Chlamydomonas starchless mutant defective in ADP-glucose pyrophosphorylase hyper-accumulates triacylglycerol. Metab Eng. 2010;12(4):387–391. doi: 10.1016/j.ymben.2010.02.002. [DOI] [PubMed] [Google Scholar]
- 41.Miller R, et al. Changes in transcript abundance in Chlamydomonas reinhardtii following nitrogen deprivation predict diversion of metabolism. Plant Physiol. 2010;154(4):1737–1752. doi: 10.1104/pp.110.165159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang ZT, Ullrich N, Joo S, Waffenschmidt S, Goodenough U. Algal lipid bodies: Stress induction, purification, and biochemical characterization in wild-type and starchless Chlamydomonas reinhardtii. Eukaryot Cell. 2009;8(12):1856–1868. doi: 10.1128/EC.00272-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Molen TA, Rosso D, Piercy S, Maxwell DP. Characterization of the alternative oxidase of Chlamydomonas reinhardtii in response to oxidative stress and a shift in nitrogen source. Physiol Plant. 2006;127(1):74–86. [Google Scholar]
- 44.Caricato R, Lionetto MG, Dondero F, Viarengo A, Schettino T. Carbonic anhydrase activity in Mytilus galloprovincialis digestive gland: Sensitivity to heavy metal exposure. Comp Biochem Physiol C Toxicol Pharmacol. 2010;152(3):241–247. doi: 10.1016/j.cbpc.2010.04.011. [DOI] [PubMed] [Google Scholar]
- 45.Le Faucheur S, Behra R, Sigg L. Phytochelatin induction, cadmium accumulation, and algal sensitivity to free cadmium ion in Scenedesmus vacuolatus. Environ Toxicol Chem. 2005;24(7):1731–1737. doi: 10.1897/04-394r.1. [DOI] [PubMed] [Google Scholar]
- 46.Nestler H, Groh KJ, Schönenberger R, Eggen RI, Suter MJ. Linking proteome responses with physiological and biochemical effects in herbicide-exposed Chlamydomonas reinhardtii. J Proteomics. 2012;75(17):5370–5385. doi: 10.1016/j.jprot.2012.06.017. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.