Abstract
The NTRK3 gene (also known as TRKC) encodes a high affinity receptor for the neurotrophin 3′-nucleotidase (NT3), which is implicated in oligodendrocyte and myelin development. We previously found that white matter integrity in young adults related to genetic variants in genes encoding neurotrophins and their receptors. This underscores the importance of neurotrophins for white matter development. NTRK3 variants are putative risk factors for schizophrenia, bipolar disorder, and obsessive-compulsive disorder hoarding, suggesting that some NTRK3 variants may affect the brain.
To test this, we scanned 392 healthy adult twins and their siblings (mean age, 23.6 ± 2.2 years; range: 20-29 years) with 105-gradient 4-Tesla diffusion tensor imaging (DTI). We identified 18 single nucleotide polymorphisms (SNPs) in the NTRK3 gene that have been associated with neuropsychiatric disorders. We used a multi-SNP model, adjusting for family relatedness, age, and sex, to relate these variants to voxelwise fractional anisotropy (FA) – a DTI measure of white matter integrity.
FA was optimally predicted (based on the highest false discovery rate critical p), by five SNPs (rs1017412, rs2114252, rs16941261, rs3784406, and rs7176429; overall FDR critical p = 0.028). Gene effects were widespread and included the corpus callosum genu and inferior longitudinal fasciculus - regions implicated in several neuropsychiatric disorders and previously associated with other neurotrophin-related genetic variants in an overlapping sample of subjects. NTRK3 genetic variants, and neurotrophins more generally, may influence white matter integrity in brain regions implicated in neuropsychiatric disorders.
Keywords: Fractional anisotropy, diffusion tensor imaging, single nucleotide polymorphism, schizophrenia, obsessive compulsive disorder, bipolar disorder
Introduction
White matter integrity is impaired in many neuropsychiatric disorders (Thomason and Thompson, 2011) and much of its variability is due to genetic factors (Chiang et al., 2011b; Kochunov et al., 2011). Variants in genes encoding neurotrophins and their receptors have been associated with brain white matter integrity measures in young healthy adults (Braskie et al., 2012a; Braskie et al., 2012b; Chiang et al., 2011a), suggesting that neurotrophin-regulated signaling pathways may be important for white matter development.
The neurotrophic tyrosine kinase, receptor, type 3 gene (NTRK3 also known as TRKC) encodes the high affinity receptor (TrkC) for the neurotrophin 3′-nucleotidase (NT3). NT3 and TrkC have been implicated in oligodendrocyte and brain myelin development (Barres et al., 1994; Hashimoto et al., 2011; Kumar and de Vellis, 1996), suggesting that NTRK3 is a good candidate for affecting white matter integrity.
Some structurally connected networks of brain regions are known to modulate higher-order brain functions, such as executive function and complex behaviors (Fuster, 2001). Diminished integrity of these networks, through damage or a failure to develop normally, may disrupt these functions, resulting in profiles of symptoms that may involve mood, learning, perception, or memory, and may be diagnosed as mental illness (Kumar and Cook, 2002). Genetic factors may also affect brain connectivity and white matter integrity in these networks (Jahanshad et al., 2013; Thompson et al., 2010; Tost et al., 2012), possibly increasing the risk for carriers of certain genetic variants to develop mental illness.
NTRK3 genetic variants are implicated in schizophrenia, bipolar disorder (BPD), and obsessive-compulsive disorder (OCD) hoarding (Alonso et al., 2008; Athanasiu et al., 2011; Otnaess et al., 2009). These disorders tend to aggregate in families and also show some degree of comorbidity within the same individuals (Bramon and Sham, 2001; Joshi et al., 2010; LaSalle-Ricci et al., 2006; Potash et al., 2001). This evidence suggests that typical functioning of this gene may be important to mental health and atypical variants may make deviations from normal mental health more likely, regardless of which symptoms are demonstrated. This is in keeping with some prior work – including very recent work by the Psychiatric Genomics Consortium – suggesting that gene effects may extend beyond standard diagnostic boundaries (Cross-Disorder Group of the Psychiatric Genomics Consortium, 2013; Moskvina et al., 2009). All of these disorders have been associated with differences in white matter microstructure (measured as diffusion tensor imaging fractional anisotropy; DTI FA) in patients versus controls (Ellison-Wright and Bullmore, 2009; Vederine et al., 2011; White et al., 2008).
Most DTI studies of these disorders have found regionally reduced FA in patients versus controls e.g. (Friedman et al., 2008; Menzies et al., 2008; Sprooten et al., 2011), but others found regionally increased FA associated with disorder symptoms e.g. (Buchsbaum et al., 2006; den Braber et al., 2011; Hubl et al., 2004; Li et al., 2011; Mahon et al., 2009). Several voxelwise studies also found lower regional FA in those at high risk for these disorders (Bloemen et al., 2010; Chaddock et al., 2009; Clark et al., 2011; Hao et al., 2009; Hoptman et al., 2008; Menzies et al., 2008; Sprooten et al., 2011), suggesting that white matter deficits begin before overt symptoms, and may relate to genetic risk.
Some mental illness risk variants are also associated with FA variability in healthy subjects without other known risk for psychiatric conditions (Braskie et al., 2012a; Chiang et al., 2011a; Konrad et al., 2009; McIntosh et al., 2008; Winterer et al., 2008; Zuliani et al., 2011), suggesting one way that genetics may contribute to mental illness vulnerability. We identified 18 SNPs within the NTRK3 gene that also have been associated with schizophrenia, BPD, or OCD. Our subjects were young and healthy, without a known predisposition for mental illness, so we are unable to study the specific risk for mental illness. Furthermore, the variants we chose were not top risk factors for these disorders. However, as the selected SNPs were common variants that were previously related to psychiatric disorder risk, they are likely to physically affect the brain in ways we can detect, in our normally developing population. We evaluated the relationship between FA and these genetic variants using a multi-SNP model. This model evaluates the predictive effect of each SNP while adjusting for the effects of all other SNPs in the model; it also builds an aggregate model of the overall effect of a set of SNPs. In this way, the SNPs with the strongest associations can be identified, and the aggregate effect of all the SNPs considered can be evaluated. A multi-SNP model also reduces the number of individual statistical tests made, thus increasing the power to detect effects. In this model, we did not fit interactions among SNPs, as the power to pick up interactions (modulatory effects of one SNP on the effects of another) is very low. In other work, we have begun to assess some of the challenges in picking up SNP interactions (Hibar et al., 2013).
We performed our analyses in a large sample of monozygotic and dizygotic young adults twins and their singleton siblings. Twin samples (or any family-based sample, such as a pedigree) are useful for establishing the heritability of traits to determine whether they might be good candidates for examining genetic control. More precisely, the traits that are not heritable are unlikely to be good targets for more in-depth genetic analysis at the SNP level. We had already demonstrated with these twin data that DTI FA in many parts of the brain white matter is moderately to highly heritable (Chiang et al., 2011b). In the current study, we expanded on our prior work to evaluate the effect of SNPs in the NTRK3 gene on white matter FA, while controlling for age, sex, and accounting for kinship to avoid confounding effects of these covariates.
Neurotrophins and their receptors have been associated previously with white matter integrity in healthy young adults (Braskie et al., 2012a; Chiang et al., 2011a). As our selected NTRK3 SNPs were also associated with neuropsychiatric disorders, we hypothesized that they were likely to influence white matter integrity, manifested as higher or lower FA.
Materials and Methods
Subjects
High-angular resolution diffusion-weighted MRI scans, collected as part of the Queensland Twin Imaging (QTIM) Study, were performed on 468 right-handed healthy young adults of European ancestry recruited to examine genetic influences on the brain. Genome-wide genotyping was performed for all subjects, although our results here are not part of a genome-wide analysis; they arose from a candidate gene design focusing on NTRK3. From the overall available sample, we excluded 76 subjects: 8 were ancestry outliers (identified using principal components analysis of all genotyped variants to isolate those at least 6 standard deviations from HapMap CEU and TSI and the GenomeEUTWIN populations for each principal component axis), 58 had technically inadequate scans (spiking artifact, truncated field of view, or signal drop-out or distortion that prevented adequate co-registration), 3 had large ventricles that were inconsistent with good health in a young person, and 7 did not have a measured genotype for at least one of the SNPs of interest. The remaining 392 subjects largely overlapped with those in two previous studies that related DTI FA to variants in both the CLU gene (399 subjects) (Braskie et al., 2011) and the NTRK1 gene (391 subjects) (Braskie et al., 2012a). Differences in subject exclusion related entirely to different availability of genotyping for the SNPs in question. The 392 subjects included here (mean age 23.6 ± 2.2 years; range: 20-29 years) were 94 monozygotic (MZ) twins, 141 dizygotic (DZ) twins or triplets, 41 singleton siblings of other participants included in this study, and 116 individuals who had no twin or singleton sibling included in this study (total of 247 families). We used the Human610-Quad BeadChip (Illumina) to analyze DNA according to the manufacturer's protocols (Infinium HD Assay; Super Protocol Guide; Rev. A, May 2008).
All subjects provided written, informed consent. The study conformed to the National Statement on Ethical Conduct in Human Research (2007) issued by the National Health and Medical Research Council (NHMRC) of Australia. It was approved by the QIMR Human Research Ethics Committee, and by the UCLA Medical Institutional Review Board (MIRB).
Image Acquisition
Magnetic resonance imaging (MRI: on a 4-Tesla; Bruker Medspec scanner) for each subject included T1-weighted inversion recovery rapid gradient echo scans (TI/TR/TE = 700/1500/3.35 ms; flip angle = 8°; slice thickness = 0.9 mm, 256 × 256 × 256), and DTI scans (single-shot echo planar imaging with a twice-refocused spin echo sequence to reduce eddy-current induced distortions (TR/TE 6090/91.7 ms, 23 cm FOV, 128×128; 55 2-mm axial slices/0 mm gap; 1.79 mm ×1.79 mm in-plane resolution). DTI scans each had 94 diffusion-weighted images (b=1149 s/mm2) and 11 b0 images (i.e., no diffusion sensitization). Gradient directions were evenly distributed on the hemisphere.
DTI Preprocessing
As described previously (Braskie et al., 2012a), non-brain matter was automatically then manually removed from the images. The raw diffusion-weighted images were adjusted for eddy current distortions (FSL “eddy_correct”), then linearly aligned and resampled to the associated T1-weighted image (FSL's linear image registration tool -FLIRT (Jenkinson et al., 2002)). B0 images for each subject were elastically registered (using a mutual information cost function (Leow et al., 2005)) to the same subject's T1-weighted scan in common space. This registration method reduces the effects of echo planar imaging-induced susceptibility artifacts. We created FA maps using FSL software.
Template Creation and Registration
We used FA maps from 32 randomly selected, unrelated subjects (matched for sex) to create a template brain, or mean deformation target as described in detail previously (Braskie et al., 2012a; Jahanshad et al., 2010). All susceptibility-corrected FA maps were registered to the template brain using a 3D elastic warping technique with a mutual information cost function (Leow et al., 2005). Both the FA maps and the template brain were thresholded at 0.25 to exclude non-white matter. To further refine the coregistration of white matter between subject scans and the template brain, we aligned the thresholded FA maps to the thresholded template brain. We used a Gaussian kernel (7 mm full-width-half-maximum; FWHM) to spatially smooth images.
SNP and genotype information
We performed one statistical analysis that considered the association of FA with 18 SNPs in the NTRK3 gene simultaneously. These 18 SNPs included all of those in the NTRK3 gene that were both available in our genotype file and also reported previously (as of November 2012) as being associated with schizophrenia (rs999905, rs4887348) (Otnaess et al., 2009), bipolar disorder (rs991728, rs994068, rs16941261, rs2114252, rs3784405, rs3784406, rs3784410, rs4887364, rs7165979, rs9806762, rs10163123, rs11630338, rs11631112, rs12911150) (Athanasiu et al., 2011), or obsessive-compulsive hoarding (rs1017412, rs7176429) (Alonso et al., 2008).
Table 1 shows the genotype summary for each SNP we included. When only one MZ twin from each MZ twin pair was assessed, the percentage of subjects with a given genotype for any SNP was similar to when all subjects were included (≤ 1% difference). Hardy-Weinberg equilibrium (HWE) and minor allele frequencies (MAF) for each SNP are listed in Table 2. The MAF for each SNP studied in our sample was similar to that previously identified for these same SNPs in the HapMap CEPH population (i.e., Utah residents with ancestry from northern and western Europe) (Altshuler et al., 2010). The genotype distribution in our sample showed no detectable deviation from HWE. None of the SNPs of interest are in regions of DNA that code for proteins. All are intronic with the exception of rs1017412 and rs7176429, which are in the 3′ downstream region of NTRK3 (Sherry et al., 2001). The SNPs in intron 5 to 12 that were associated with bipolar disorder are also predicted to be binding sites for transcription factors Nkx2.2 and Pax6, which are involved in neural system development (Athanasiu et al., 2011). The effect on gene expression of each variant is not well characterized.
Table 1.
Variants of interest and genotypes in all included subjects.
| SNP | Minor allele | Major allele | Subjects with 0 minor alleles | Subjects with 1 minor allele | Subjects with 2 minor alleles |
|---|---|---|---|---|---|
| rs991728 | G | C | 210 | 160 | 22 |
| rs994068 | C | G | 209 | 159 | 24 |
| rs999905 | C | G | 186 | 158 | 48 |
| rs1017412 | G | A | 197 | 163 | 32 |
| rs2114252 | A | C | 206 | 161 | 25 |
| rs3784405 | C | T | 243 | 132 | 17 |
| rs3784406 | T | C | 128 | 193 | 71 |
| rs3784410 | T | C | 130 | 202 | 60 |
| rs4887348 | G | A | 211 | 149 | 32 |
| rs4887364 | C | T | 208 | 158 | 26 |
| rs7165979 | C | G | 200 | 161 | 31 |
| rs7176429 | T | G | 181 | 183 | 28 |
| rs9806762 | G | A | 161 | 187 | 44 |
| rs10163123 | G | C | 200 | 160 | 32 |
| rs11630338 | C | T | 218 | 154 | 20 |
| rs11631112 | T | G | 222 | 149 | 21 |
| rs12911150 | G | T | 236 | 142 | 14 |
| rs16941261 | C | G | 218 | 153 | 21 |
Table 2.
Minor allele frequency (MAF) and evidence for Hardy-Weinberg equilibrium: statistics here are compiled based on using one subject per family to avoid any confounding effects of kinship.
| SNP | MAF: our sample | MAF: in CEPH* | HWE χ2 | HWE p |
|---|---|---|---|---|
| rs991728 | 0.27 | 0.24 | 0.31 | 0.58 |
| rs994068 | 0.28 | 0.25 | 0.33 | 0.57 |
| rs999905 | 0.32 | 0.32 | 0.59 | 0.44 |
| rs1017412 | 0.30 | 0.28 | 0.69 | 0.41 |
| rs2114252 | 0.28 | 0.26 | 0.18 | 0.67 |
| rs3784405 | 0.22 | 0.20 | 0.29 | 0.59 |
| rs3784406 | 0.43 | 0.42 | 0.21 | 0.65 |
| rs3784410 | 0.40 | 0.42 | 1.16 | 0.28 |
| rs4887348 | 0.26 | 0.26 | 0.00 | 1.00 |
| rs4887364 | 0.28 | 0.25 | 0.06 | 0.81 |
| rs7165979 | 0.28 | 0.26 | 0.18 | 0.67 |
| rs7176429 | 0.32 | 0.28 | 1.02 | 0.31 |
| rs9806762 | 0.35 | 0.33 | 0.60 | 0.44 |
| rs10163123 | 0.28 | 0.29 | 0.42 | 0.52 |
| rs11630338 | 0.26 | 0.25 | 0.14 | 0.71 |
| rs11631112 | 0.25 | 0.24 | 0.04 | 0.84 |
| rs12911150 | 0.22 | 0.22 | 0.59 | 0.44 |
| rs16941261 | 0.25 | 0.23 | 0.40 | 0.52 |
CEPH is a reference population of Utah residents with ancestry from northern and western Europe (Altshuler et al., 2010).
To get a sense of how many subjects in the sample carry different risk genotypes, one could consider what proportion of subjects in the sample carried (1) a specific number of minor alleles, or (2) a specific number of risk alleles implicated in mental illness, or (3) a specific number of alleles that might be expected to decrease FA (although each SNP's direction of effect is not typically known in advance). The three may not coincide (i.e., sometimes the major, or more prevalent, allele is the risk allele, and mental illness risk alleles do not always result in decreased FA). However, we assessed how many subjects in our sample carried one or two mental illness risk alleles in multiple SNPs. Subjects carried between 6 and 35 “risk” alleles (mean 15.56). Each “risk” allele may or may not be associated with lower FA.
For more information about how SNPs were selected for inclusion in our model, please see the Multi-SNP Model Reduction section below.
Statistical analyses
To limit partial volume effects, statistical analyses were performed only in voxels having FA > 0.25 in the template brain, i.e., likely white matter (Figure 1). We used linear mixed-model regression at each voxel to estimate how white matter FA differed by the individual and joint associations of genotypes while controlling for age, sex, and family relatedness as described previously in detail (Kohannim et al., 2012). Family relatedness between each subject and all others was adjusted for using a symmetric N × N kinship matrix. In this matrix, a coefficient of 1 between subjects indicated that they were MZ twins; a value of 0.5 indicated that they were DZ twins or siblings; and a 0 indicated that they were not closely related. Ancestry outliers, identified using principal component analysis across genetic markers, were excluded.
Figure 1. Voxels in which statistical analyses were performed.
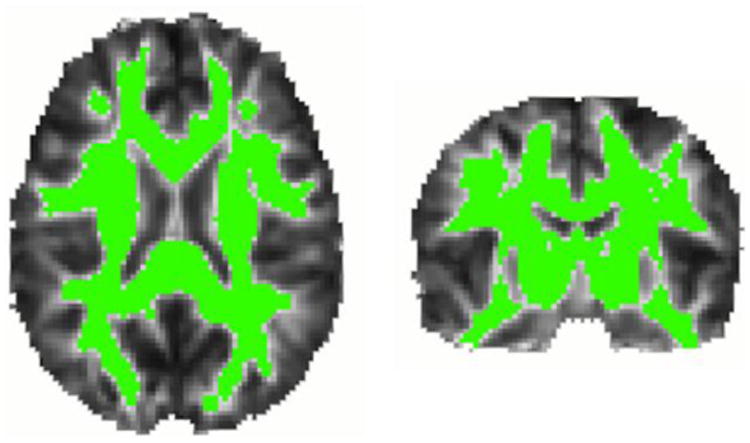
Regions highlighted in green represent voxels in which FA exceeded 0.25 in the MDT (i.e., likely white matter).
Genotype effects were determined using an additive model. The additive model determines whether having more than one copy of a certain allele in a given SNP has additional effect over having only one copy of that allele. All the SNPs are treated as if they are in a multiple regression model, but with step-wise elimination of SNPs that are not improving the fit. This model is essentially ordinary least squares, so the fit of the overall model will treat the presence of multiple SNPs in a unique individual in the same way as additive effects of SNPs across individuals. The effect of each SNP is also controlled for in evaluating the effect of all other SNPs considered, by fitting all of them at once. The statistical interactions between SNPs are not included in the model, because in general the power is extremely low to pick up modulatory effects of one SNP on another. For efforts to boost power for detecting interactions, please see (Hibar et al., 2013). The analyses were performed using Efficient Mixed-Model Association (Kang et al., 2008) (EMMA; http://mouse.cs.ucla.edu/emma/) within the R statistical package (version 2.9.2; http://www.r-project.org/). P values for the significance of individual and joint SNP associations with FA were assessed using an F-test as described previously in detail (Kohannim et al., 2012).
We employed the widely-used false discovery rate (FDR) method with a threshold of 5% to control for multiple comparisons across all voxels considered (Benjamini and Hochberg, 1995). FDR controls the expected proportion of null results that are falsely identified as significant to a set rate (in our case this rate, q = 5%, which is intended to be similar to the standard p = 0.05 frequently used to define significance). Using FDR, comparisons (voxels) having significance that is below a critical p value are considered significantly associated with the variable in question, even given the multiple comparisons. Thus, a higher critical p (closer to 0.05) may allow more voxels to be considered significant after adjustment for multiple comparisons. Actual p values in many of the significant voxels are much lower than the critical p reported.
Multi-SNP Model Reduction
We included all 18 identified SNPs in our initial model, successively removing the SNP with the weakest association to FA. This method gained us FDR-corrected significant results after removing the weakest SNP. Later statistical models were fitted to help identify which SNPs contributed most to the fit. A single multi-SNP analysis provides two main results (as with any regression having multiple predictors). First, we learn whether the included SNPs as a whole are significantly associated with FA after FDR correction across voxels, and if so, what the overall critical p value is. Second, we learn the significance for the partial contribution of each SNP in the model to FA. In other words, we learn whether each individual SNP within the model is significantly associated with FA after adjusting for the effects of every other SNP in the model and correcting for FDR across voxels. As with all multiple regressions, the significance values for each SNP in the model are only relevant if the overall model fits.
To identify the SNPs in order of their likely contribution to the model fit, we proceeded as follows. For each SNP in the model, we first identified only those voxels that, for that SNP, had a nominal association (i.e. p < 0.05) with FA. Next we obtained the mean p value across nominally associated voxels for each individual SNP in the model. SNPs with a higher proportion of voxels in which strong relationships existed between genotype and FA would be considered to have stronger associations with FA than those with mean voxelwise p values closer to 0.05. The single SNP with the highest mean p value across voxels in each multi-SNP analysis was considered the weakest association and was removed from the model before re-running. This procedure was repeated until only two SNPs were included. We then examined the critical p values for each model. The overall model with the greatest number of voxels that pass FDR correction for multiple comparisons was considered the optimal model. For a given statistical map of p values, we can threshold it to only show voxels with p values lower than a given threshold, so the higher the threshold, more voxels will be shown. In the end, a cumulative distribution function is compiled of all the p values in the map, the threshold is chosen so as to be the highest p value (i.e. the critical p value) for which the false discovery rate is controlled. We do tend to choose models with higher critical p values but it is worth noting that other criteria could be used to define the best fitting model for the anatomical data, such as the one with the lowest p value in a region of interest.
We assessed the strength of the association only in nominally significant voxels to identify those SNPs having voxels that were strongly associated with FA, even in fairly small regions. In this way, we identified relationships in which genotype strongly predicts FA in voxels of interest without preferentially selecting relationships that are more widespread throughout the brain. If all 42,804 voxels in our white matter regions were considered in our analysis, voxels with p values close to one would likely diminish small regions of high significance. However, using this threshold does weight small regions of strong significance more than larger regions with voxels having p values close to 0.05. Arguably, both very significant results and wide spread nominal results could be considered “strong.” We therefore also performed the same step-wise elimination analysis without thresholding voxels to p < 0.05 (i.e., we obtained the mean p value across all voxels in our FA > 0.25 region). We note that there are many ways to sort the nested submodels, and for trading off modeling complexity versus goodness of fit, but this step-wise method had the advantage of eliminating SNPs that contributed least to the prediction, leading to a more efficient and parsimonious model.
To evaluate whether the association between genotypes in our original optimal five SNP model and FA were robust across the entire white matter mask (i.e., FA>0.25), we additionally ran this optimal model on mean FA within that mask. We again controlled for age, sex, and kinship.
In order to determine whether our optimal multi-SNP model provided additional information over the comparison of individual SNPs with FA (outside of a multi-SNP model), we then evaluated the individual association of each SNP from the optimal model in five single SNP analyses (one separate analysis for each SNP in our optimal model). These analyses may provide different results from the partial contributions of SNPs to FA within a multi-SNP model because when evaluated individually, the relationships between SNPs are not considered. We controlled for age, sex, and family relatedness as with the multiple SNP association tool. For these tests, we used the same parameters as with the multiple SNP association. Namely, FA maps were thresholded at 0.25 to exclude non-white matter and we used an isotropic 3D Gaussian kernel (7 mm FWHM) to smooth the images spatially.
Linkage disequilibrium
Linkage disequilibrium (LD) is the non-random association of alleles at two loci – there tends to be a statistical correlation between any given SNP and other SNPs that are close to it on the genome. Therefore, a strong association between a given allele in one SNP and certain brain differences would increase the likelihood that another SNP in moderate linkage disequilibrium with the first would show a similar effect. The multiple SNP association tool used here adjusts the effect of a given SNP for the effects of all others. Thus, a SNP showing a strong association with a brain characteristic if examined alone, may manifest a weaker association if examined in concert with other SNPs in LD with the first. Figure 2 displays the LD between SNPs examined here.
Figure 2.
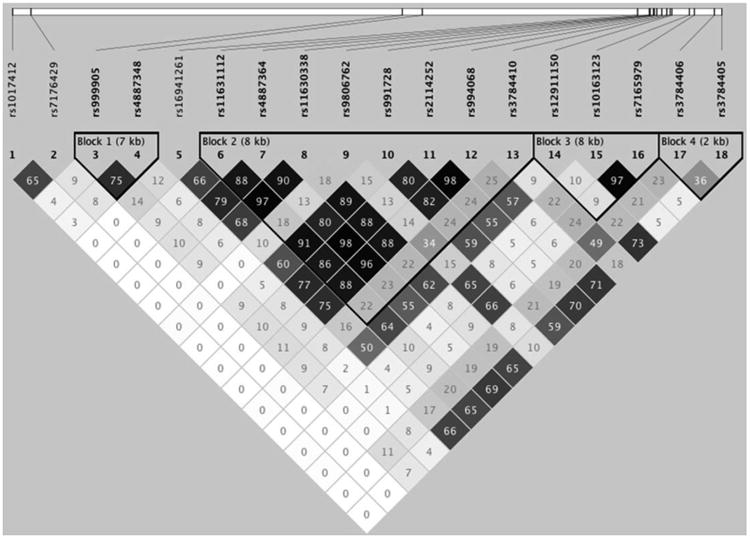
Linkage disequilibrium plot. The plot below, created in Haploview (Barrett et al., 2005), shows R2 values representing the degree of linkage disequilibrium among all SNPs considered in this study. One subject per family was selected at random and included in the analysis to avoid any confounding effects of kinship. SNPs that were significantly associated with FA in the optimal model were rs1017412 and rs2114252. Also included in the optimal model were rs16941261, rs3784406, and rs7176429.
Association of FA with top schizophrenia SNPs
To evaluate whether our results would apply to SNPs implicated in neuropsychiatric disorders in general or were more specific to our SNPs in question, we used the same sample of 392 subjects to compare DTI FA with genotypes for the top five SNPs associated with schizophrenia in November 2011 (http://www.szgene.org/TopResults.asp) (Allen et al., 2008). We included all five SNPs in the multi-SNP model, using an additive model, a smoothing kernel of 7 mm FWHM, and an FA threshold of >0.25 as we used for the NTRK3 analyses. The SNPs we included were PRSS16 rs6932590, PGBD1 rs13211507, NRGN rs12807809, NOTCH4 rs3131296, and HIST1H2BJ rs6913660.
Results
We examined the joint effect of the set of candidate SNPs on voxelwise FA in the white matter, using a partial F-test and a linear mixed-effects model to compute p values. In a stepwise fashion, we removed SNPs in the order of their effects (weakest to strongest). After removing the first SNP, rs11631112, the full model achieved significance after FDR correction. Each subsequent model also remained significant as a whole. The five-SNP model that included rs1017412, rs2114252, rs16941261, rs3784406, and rs7176429 had the most widespread association with FA in the white matter, with a FDR critical p = 0.028. Approximately 55.6% of white matter voxels (i.e., 23,806 out of the 42,804 voxels considered) survived this multiple comparison corrected threshold, showing that the model was a relatively good predictor of white matter integrity across widespread regions of the brain. In fact, when the SNPs were considered together, they were also significantly associated with mean FA across the entire white matter mask (R2 = 0.06; joint p = 0.0004). Two of the SNPs in our optimal model, rs1017412 and rs7176429, are found in the 3′ downstream region of NTRK3 (Sherry et al., 2001). Our strongest local effects (uncorrected p < 0.0001) were in the corticospinal tract, superior and inferior longitudinal fasciculus (SLF and ILF), inferior fronto-occipital fasciculus (IFO), cingulum, anterior thalamic radiation, and forceps major and minor. We also found significant effects in the posterior thalamic radiations, corpus callosum (genu, body, and splenium), anterior, posterior, and superior corona radiata, interior and exterior capsule, fornix (column and body), fornix/stria terminalis, and uncinate fasciculus. All effects were bilateral (Figure 3). The optimal model explained a mean of 5.2% of the variance in FA in those voxels that survived FDR correction.
Figure 3. FA association with the optimal five-SNP model of theNTRK3gene, displayed on a study-specific FA template.
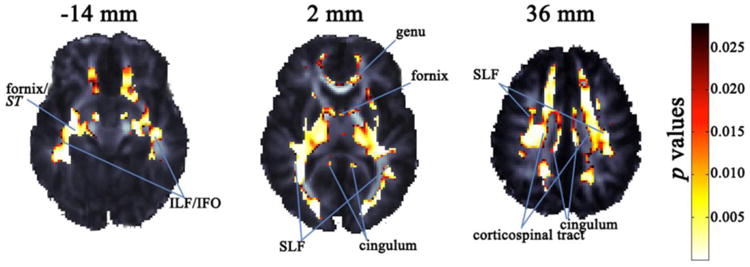
Highlighted areas are the p values indicating voxels in which the NTRK3 SNPs (rs1017412, rs2114252, rs16941261, rs3784406, and rs7176429) in our optimal model are associated with FA after adjusting for age and sex (FDR critical p value = 0.028). The left brain hemisphere is displayed on the right. Coordinates listed are for the Z direction in ICBM space. SLF and ILF denote the superior and inferior longitudinal fasciculi, IFO is the inferior fronto-occipital fasciculus, and ST is the stria terminalis.
In the optimal multiple SNP model, the combined SNPs within the model as well as the individual contributions of two of the five SNPs (rs1017412 and rs2114252) were significantly related to FA after FDR correction. For rs1017412, which was previously associated nominally with OCD hoarding (Alonso et al., 2008), the minor allele, G, was associated in our study with lower DTI FA (critical p = 0.024). Strongest effects (p <0.0001) were found in the corticospinal tract, SLF and ILF, IFO, cingulum, anterior thalamic radiation, forceps major and minor, internal and external capsules, uncinate fasciculus, and the genu of the corpus callosum. We also found significant effects in the posterior thalamic radiation, corpus callosum (body, and splenium), anterior, posterior, and superior corona radiata, and fornix/stria terminalis. Again, all effects were bilateral. Additionally, the minor allele, A, of rs2114252, which was previously associated nominally with bipolar disorder (Athanasiu et al., 2011), was associated with lower FA in our sample (critical p = 0.00026). Effects with p < 0.0001 were in the right corticospinal tract and ILF, left cingulum and anterior thalamic radiation, and bilateral SLF, IFO, and forceps major. There were no other regions in which FA was significantly associated with the genotype at rs2114252.
To assess whether our thresholding of voxels to p < 0.05 emphasized small regions of significance to the exclusion of broader regions of nominal significance in other SNPs, we also performed the same step-wise analysis, evaluating the mean p value of all voxels included in our white matter mask (FA > 0.25). Again, the optimal model included five SNPs (critical p = 0.032), including rs1017412, rs2114252, and rs7176429 from the thresholded optimal model. The new model also included schizophrenia risk variant, rs4887348, and bipolar disorder risk variant, rs3784405, which were not previously included. Again, rs1017412 (critical p = 0.028) and rs2114252 (critical p = 0.017) were significantly related to FA after FDR correction as part of the optimal model. The pattern of significance was similar to that in the optimal model obtained through thresholded step-wise elimination of SNPs except was slightly more extensive (particularly in the genu of the corpus callosum, bilateral anterior thalamic radiation, and left posterior thalamic radiation) when the non-thresholded method was used.
In an additional test, we considered the individual association of each of the five SNPs from the optimal model using a single SNP analysis (i.e., in this analysis, we no longer considered the effects of other SNPs from the optimal model), controlling for age, sex, and family relatedness. In these analyses, none of the SNPs had alleles significantly associated with DTI FA on a voxelwise basis after FDR correction.
Using the same parameters, multi-SNP tool, and subject set as used for the NTKR3 SNPs, the top five schizophrenia risk SNPs (PRSS16 rs6932590, PGBD1 rs13211507, NRGN rs12807809, NOTCH4 rs3131296, and HIST1H2BJ rs6913660) were not significantly associated with DTI FA.
Discussion
Identifying genetic variants that affect white matter integrity is of great interest for personal prediction of brain structure, to better understand disease mechanisms, and to identify therapeutic targets for psychiatric disorders involving aberrant brain connectivity. Eighteen variants in the NTRK3 gene have been implicated in neuropsychiatric disorders (Alonso et al., 2008; Athanasiu et al., 2011; Otnaess et al., 2009), suggesting that this gene may be important for neuropsychological function, as well as its known role in brain development. When evaluating the effect of NTRK3 variants on white matter integrity, we found that when considered together, five SNPs (rs1017412, rs2114252, rs16941261, rs3784406, and rs7176429) were most strongly related to voxelwise and mean DTI FA in the brain white matter of young healthy adults. None of those SNPs showed a significant effect on FA when evaluated individually, without considering the effect of the accompanying SNPs. In line with many other studies that have advocated multi-SNP or gene-based testing (Hibar et al., 2011), our results demonstrate the advantage of considering the joint effect of numerous risk SNPs when evaluating their effect on the brain. Three of the SNPs in our optimal model (rs1017412, rs2114252, and rs7176429) were also included in our optimal model when all voxels under our FA > 0.25 mask were considered (rather than only examining voxels with p < 0.05), suggesting that these SNPs had fairly broad regions of strong significance.
FA differences in neuropsychiatric patients or those at risk may differ in location from study to study, partly because some studies examine only specific regions rather than performing voxelwise analysis that allow for brain-wide comparison of results across studies. Data analysis methods may also vary, and many studies have small sample sizes. When only the largest (100 or more total subjects) voxelwise studies of adult onset schizophrenia, BPD, or OCD are considered (Buchsbaum et al., 2006; Clark et al., 2011; den Braber et al., 2011; Hao et al., 2009; Kanaan et al., 2009; Perez-Iglesias et al., 2010; Sprooten et al., 2011; Sussmann et al., 2009; Wang et al., 2011), FA was lower in affected individuals across diagnoses in the SLF (56% of studies), and the genu and ILF (44% of studies for each), suggesting that lower white matter integrity in these regions may increase the vulnerability for mental illness. In our study, two of these regions, the genu and ILF, were among the regions showing the strongest association between lower FA and NTRK3 variants. The genu, or anterior part of the corpus callosum connects the left and right brain hemispheres, particularly carrying fibers that innervate the prefrontal cortex. Lower FA in the anterior part of the corpus callosum has been associated with avolition, or lack of drive (Nakamura et al., 2012) and with poorer executive function ability (Zheng et al., 2013). The ILF connects the occipital and temporal cortex. It is an important part of the visual–limbic pathway that promotes vision-specific emotions, learning and memory. Lower FA in the ILF has been associated with decreased working memory ability (Gu et al., 2013), object recognition (Ortibus et al., 2012) and processing speed (Choi et al., 2012) and with greater depression and anxiety (Choi et al., 2012).
Genetic variation in neurotrophins and their receptors may influence white matter development in regions implicated in a range of neuropsychiatric disorders. Deeper study of genes that encode neurotrophins and their receptors may offer insights into increased vulnerability for developing these disorders. When we used the multi-SNP tool to compare DTI FA with genotypes in the top five variants currently associated with schizophrenia (Allen et al., 2008), none of those SNPs was significantly associated with FA, demonstrating that our effect may be related more to variations within neurotrophin systems than to neuropsychiatric risk per se.
Our optimal model found that NTRK3 variants significantly related to DTI FA broadly throughout the brain, including strongly in the SLF and ILF, and also in the genu of the corpus callosum, all regions in which FA is most consistently affected in those with schizophrenia, BPD, and OCD in prior large, voxelwise DTI studies of white matter integrity (Buchsbaum et al., 2006; Clark et al., 2011; den Braber et al., 2011; Hao et al., 2009; Kanaan et al., 2009; Perez-Iglesias et al., 2010; Sprooten et al., 2011; Sussmann et al., 2009; Wang et al., 2011). NTRK3 variants may modulate white matter integrity in regions implicated in a variety neuropsychiatric disorders. We examined NTRK3 SNPs that previously increased the risk for developing mental illness in order to increase the likelihood that the SNPs would affect white matter. However, our subjects were young and healthy, and not specifically selected for symptoms or family history that increase their risk for mental illness. Additionally, for our SNP with the strongest relationship to FA (rs1017412), the OCD hoarding risk allele, A, was actually associated with higher FA in our sample. For our SNP with the next strongest relationship to FA (rs2114252), the A allele, previously associated with BPD (Athanasiu et al., 2011), was associated with lower FA in our sample. It is therefore unlikely that our results herald a progression toward mental illness in this sample, and are more likely to be a result of normal development. Developmental effects still may increase the risk of mental illness in people having other factors that predispose them to the disorders (Insel, 2010; Weinberger, 1987). Our study may direct future research toward examining NTRK3 gene effects on white matter integrity in those afflicted by or at increased risk for schizophrenia, BPD, and OCD. Gaining a better understanding of which gene variants help to control brain development, particularly in regions vulnerable to disease, is an important step in understanding those maladies.
Highlights.
A multi-SNP analysis compared DTI FA with genotypes in 18 SNPs in the NTRK3 gene.
SNPs were selected for being previously associated with mental illness.
FA was best predicted by 5 SNPs (rs1017412, rs2114252, rs16941261, rs3784406, and rs7176429).
Gene effects included regions implicated in mental illness.
Effects overlapped with those of other neurotrophin-related genetic variants.
Acknowledgments
We thank the twins and siblings for their participation. In Brisbane we thank Marlene Grace and Ann Eldridge for twin recruitment, Aiman Al Najjar and other radiographers for scanning, Kerrie McAloney and Daniel Park for research support, and staff in the Molecular Epidemiology Laboratory for DNA sample processing and preparation. This study was supported by the U.S. National Institutes of Health (R01 HD050735, EB008432, EB008281, EB007813, and AG040060 to PT), and the National Health and Medical Research Council, Australia (NHMRC 486682 and 389875). MNB was funded, in part, by the NIH (P50 AG16570) and by the UCLA Easton Consortium for Biomarker and Drug Discovery in Alzheimer's Disease. OK was funded, in part, by a UCLA Dissertation Year Fellowship and by an NIH NRSA Award (F30 AG041681). GIdZ was supported by an ARC Future Fellowship (FT0991634).
Abbreviations
- NTRK3 aka TRKC
neurotrophic tyrosine kinase, receptor, type 3 gene
- NT3
neurotrophin 3′-nucleotidase
- BPD
bipolar disorder
- OCD
obsessive compulsive disorder
- DTI
diffusion tensor imaging
- FA
fractional anisotropy
- SNP
single nucleotide polymorphism
- FLIRT
FSL's linear image registration tool
- FWHM
full-width at half maximum
- LD
linkage disequilibrium
- MZ
monozygotic
- DZ
dizygotic
- HWE
Hardy-Weinberg equilibrium
- MAF
minor allele frequencies
- FDR
false discovery rate
- ILF and SLF
inferior and superior longitudinal fasciculus
- IFO
inferior fronto-occipital fasciculus
- BDNF
brain-derived neurotrophic factor
- NTRK1
neurotrophic tyrosine kinase, receptor, type 1
Footnotes
Conflict of interest: The authors declare actual or potential conflict of interest including any financial, personal or other relationships with other people or organizations.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- Allen NC, Bagade S, McQueen MB, Ioannidis JP, Kavvoura FK, Khoury MJ, Tanzi RE, Bertram L. Systematic meta-analyses and field synopsis of genetic association studies in schizophrenia: the SzGene database. Nature Genetics. 2008;40:827–834. doi: 10.1038/ng.171. [DOI] [PubMed] [Google Scholar]
- Alonso P, Gratacos M, Menchon JM, Segalas C, Gonzalez JR, Labad J, Bayes M, Real E, de Cid R, Pertusa A, Escaramis G, Vallejo J, Estivill X. Genetic susceptibility to obsessive-compulsive hoarding: the contribution of neurotrophic tyrosine kinase receptor type 3 gene. Genes, brain, and behavior. 2008;7:778–785. doi: 10.1111/j.1601-183X.2008.00418.x. [DOI] [PubMed] [Google Scholar]
- Altshuler DM, Gibbs RA, Peltonen L, Dermitzakis E, Schaffner SF, Yu F, Bonnen PE, de Bakker PI, Deloukas P, Gabriel SB, Gwilliam R, Hunt S, Inouye M, Jia X, Palotie A, Parkin M, Whittaker P, Chang K, Hawes A, Lewis LR, Ren Y, Wheeler D, Muzny DM, Barnes C, Darvishi K, Hurles M, Korn JM, Kristiansson K, Lee C, McCarrol SA, Nemesh J, Keinan A, Montgomery SB, Pollack S, Price AL, Soranzo N, Gonzaga-Jauregui C, Anttila V, Brodeur W, Daly MJ, Leslie S, McVean G, Moutsianas L, Nguyen H, Zhang Q, Ghori MJ, McGinnis R, McLaren W, Takeuchi F, Grossman SR, Shlyakhter I, Hostetter EB, Sabeti PC, Adebamowo CA, Foster MW, Gordon DR, Licinio J, Manca MC, Marshall PA, Matsuda I, Ngare D, Wang VO, Reddy D, Rotimi CN, Royal CD, Sharp RR, Zeng C, Brooks LD, McEwen JE. Integrating common and rare genetic variation in diverse human populations. Nature. 2010;467:52–58. doi: 10.1038/nature09298. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Athanasiu L, Mattingsdal M, Melle I, Inderhaug E, Lien T, Agartz I, Lorentzen S, Morken G, Andreassen OA, Djurovic S. Intron 12 in NTRK3 is associated with bipolar disorder. Psychiatry Research. 2011;185:358–362. doi: 10.1016/j.psychres.2010.05.011. [DOI] [PubMed] [Google Scholar]
- Barres BA, Raff MC, Gaese F, Bartke I, Dechant G, Barde YA. A crucial role for neurotrophin-3 in oligodendrocyte development. Nature. 1994;367:371–375. doi: 10.1038/367371a0. [DOI] [PubMed] [Google Scholar]
- Barrett JC, Fry B, Maller J, Daly MJ. Haploview: analysis and visualization of LD and haplotype maps. Bioinformatics. 2005;21:263–265. doi: 10.1093/bioinformatics/bth457. [DOI] [PubMed] [Google Scholar]
- Bloemen OJ, de Koning MB, Schmitz N, Nieman DH, Becker HE, de Haan L, Dingemans P, Linszen DH, van Amelsvoort TA. White-matter markers for psychosis in a prospective ultra-high-risk cohort. Psychological Medicine. 2010;40:1297–1304. doi: 10.1017/S0033291709991711. [DOI] [PubMed] [Google Scholar]
- Bramon E, Sham PC. The common genetic liability between schizophrenia and bipolar disorder: a review. Current Psychiatry Reports. 2001;3:332–337. doi: 10.1007/s11920-001-0030-1. [DOI] [PubMed] [Google Scholar]
- Braskie MN, Jahanshad N, Stein JL, Barysheva M, Johnson K, McMahon KL, de Zubicaray GI, Martin NG, Wright MJ, Ringman JM, Toga AW, Thompson PM. Relationship of a variant in the NTRK1 gene to white matter microstructure in young adults. The Journal of Neuroscience. 2012a;32:5964–5972. doi: 10.1523/JNEUROSCI.5561-11.2012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braskie MN, Jahanshad N, Stein JL, Barysheva M, McMahon KL, de Zubicaray GI, Martin NG, Wright MJ, Ringman JM, Toga AW, Thompson PM. Common Alzheimer's Disease Risk Variant Within the CLU Gene Affects White Matter Microstructure in Young Adults. The Journal of Neuroscience. 2011;31:6764–6770. doi: 10.1523/JNEUROSCI.5794-10.2011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braskie MN, Jahanshad N, Toga AW, McMahon KL, de Zubicaray GI, Martin NG, Wright MJ, Thompson PM. How a common variant in the growth factor receptor gene, NTRK1, affects white matter. Bioarchitecture. 2012b;2:181–184. doi: 10.4161/bioa.22190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buchsbaum MS, Friedman J, Buchsbaum BR, Chu KW, Hazlett EA, Newmark R, Schneiderman JS, Torosjan Y, Tang C, Hof PR, Stewart D, Davis KL, Gorman J. Diffusion tensor imaging in schizophrenia. Biological Psychiatry. 2006;60:1181–1187. doi: 10.1016/j.biopsych.2005.11.028. [DOI] [PubMed] [Google Scholar]
- Chaddock CA, Barker GJ, Marshall N, Schulze K, Hall MH, Fern A, Walshe M, Bramon E, Chitnis XA, Murray R, McDonald C. White matter microstructural impairments and genetic liability to familial bipolar I disorder. The British Journal of Psychiatry. 2009;194:527–534. doi: 10.1192/bjp.bp.107.047498. [DOI] [PubMed] [Google Scholar]
- Chiang MC, Barysheva M, Toga AW, Medland SE, Hansell NK, James MR, McMahon KL, de Zubicaray GI, Martin NG, Wright MJ, Thompson PM. BDNF gene effects on brain circuitry replicated in 455 twins. Neuroimage. 2011a;55:448–454. doi: 10.1016/j.neuroimage.2010.12.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chiang MC, McMahon KL, de Zubicaray GI, Martin NG, Hickie I, Toga AW, Wright MJ, Thompson PM. Genetics of white matter development: a DTI study of 705 twins and their siblings aged 12 to 29. Neuroimage. 2011b;54:2308–2317. doi: 10.1016/j.neuroimage.2010.10.015. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J, Jeong B, Polcari A, Rohan ML, Teicher MH. Reduced fractional anisotropy in the visual limbic pathway of young adults witnessing domestic violence in childhood. Neuroimage. 2012;59:1071–1079. doi: 10.1016/j.neuroimage.2011.09.033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark KA, Nuechterlein KH, Asarnow RF, Hamilton LS, Phillips OR, Hageman NS, Woods RP, Alger JR, Toga AW, Narr KL. Mean diffusivity and fractional anisotropy as indicators of disease and genetic liability to schizophrenia. Journal of Psychiatric Research. 2011;45:980–988. doi: 10.1016/j.jpsychires.2011.01.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cross-Disorder Group of the Psychiatric Genomics Consortium. Identification of risk loci with shared effects on five major psychiatric disorders: a genome-wide analysis. Lancet. 2013 doi: 10.1016/S0140-6736(12)62129-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- den Braber A, van 't Ent D, Boomsma DI, Cath DC, Veltman DJ, Thompson PM, de Geus EJ. White matter differences in monozygotic twins discordant or concordant for obsessive-compulsive symptoms: a combined diffusion tensor imaging/voxel-based morphometry study. Biological Psychiatry. 2011;70:969–977. doi: 10.1016/j.biopsych.2011.03.029. [DOI] [PubMed] [Google Scholar]
- Ellison-Wright I, Bullmore E. Meta-analysis of diffusion tensor imaging studies in schizophrenia. Schizophrenia Research. 2009;108:3–10. doi: 10.1016/j.schres.2008.11.021. [DOI] [PubMed] [Google Scholar]
- Friedman JI, Tang C, Carpenter D, Buchsbaum M, Schmeidler J, Flanagan L, Golembo S, Kanellopoulou I, Ng J, Hof PR, Harvey PD, Tsopelas ND, Stewart D, Davis KL. Diffusion tensor imaging findings in first-episode and chronic schizophrenia patients. The American Journal of Psychiatry. 2008;165:1024–1032. doi: 10.1176/appi.ajp.2008.07101640. [DOI] [PubMed] [Google Scholar]
- Fuster JM. The prefrontal cortex--an update: time is of the essence. Neuron. 2001;30:319–333. doi: 10.1016/s0896-6273(01)00285-9. [DOI] [PubMed] [Google Scholar]
- Gu L, Li J, Feng DF, Cheng ET, Li DC, Yang XQ, Wang BC. Detection of white matter lesions in the acute stage of diffuse axonal injury predicts long-term cognitive impairments: a clinical diffusion tensor imaging study. The journal of trauma and acute care surgery. 2013;74:242–247. doi: 10.1097/TA.0b013e3182684fe8. [DOI] [PubMed] [Google Scholar]
- Hao Y, Yan Q, Liu H, Xu L, Xue Z, Song X, Kaneko Y, Jiang T, Liu Z, Shan B. Schizophrenia patients and their healthy siblings share disruption of white matter integrity in the left prefrontal cortex and the hippocampus but not the anterior cingulate cortex. Schizophrenia Research. 2009;114:128–135. doi: 10.1016/j.schres.2009.07.001. [DOI] [PubMed] [Google Scholar]
- Hashimoto R, Udagawa J, Kagohashi Y, Matsumoto A, Hatta T, Otani H. Direct and indirect effects of neuropeptide Y and neurotrophin 3 on myelination in the neonatal brains. Brain Research. 2011;1373:55–66. doi: 10.1016/j.brainres.2010.12.027. [DOI] [PubMed] [Google Scholar]
- Hibar DP, Kohannim O, Stein JL, Chiang MC, Thompson PM. Multilocus genetic analysis of brain images. Frontiers in genetics. 2011;2:73. doi: 10.3389/fgene.2011.00073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hibar DP, Stein JL, Jahanshad N, Toga AW, McMahon KL, de Zubicaray GI, Montgomery GW, Martin NG, Wright MJ, Weiner MW, Thompson PM. Exhaustive search of the SNP-SNP interactome identifies replicated epistatic effects on brain volume. Medical Image Computing and Computer Assisted Intervention, annual meeting; Nagoya, Japan. 2013. pp. 22–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoptman MJ, Nierenberg J, Bertisch HC, Catalano D, Ardekani BA, Branch CA, Delisi LE. A DTI study of white matter microstructure in individuals at high genetic risk for schizophrenia. Schizophrenia Research. 2008;106:115–124. doi: 10.1016/j.schres.2008.07.023. [DOI] [PubMed] [Google Scholar]
- Hubl D, Koenig T, Strik W, Federspiel A, Kreis R, Boesch C, Maier SE, Schroth G, Lovblad K, Dierks T. Pathways that make voices: white matter changes in auditory hallucinations. Archives of General Psychiatry. 2004;61:658–668. doi: 10.1001/archpsyc.61.7.658. [DOI] [PubMed] [Google Scholar]
- Insel TR. Rethinking schizophrenia. Nature. 2010;468:187–193. doi: 10.1038/nature09552. [DOI] [PubMed] [Google Scholar]
- Jahanshad N, Lee AD, Barysheva M, McMahon KL, de Zubicaray GI, Martin NG, Wright MJ, Toga AW, Thompson PM. Genetic influences on brain asymmetry: A DTI study of 374 twins and siblings. Neuroimage. 2010;52:455–469. doi: 10.1016/j.neuroimage.2010.04.236. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jahanshad N, Rajagopalan P, Hua X, Hibar DP, Nir TM, Toga AW, Jack CR, Jr, Saykin AJ, Green RC, Weiner MW, Medland SE, Montgomery GW, Hansell NK, McMahon KL, de Zubicaray GI, Martin NG, Wright MJ, Thompson PM. Genome-wide scan of healthy human connectome discovers SPON1 gene variant influencing dementia severity. National Academy of Sciences; USA: 2013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jenkinson M, Bannister P, Brady M, Smith S. Improved optimization for the robust and accurate linear registration and motion correction of brain images. Neuroimage. 2002;17:825–841. doi: 10.1016/s1053-8119(02)91132-8. [DOI] [PubMed] [Google Scholar]
- Joshi G, Wozniak J, Petty C, Vivas F, Yorks D, Biederman J, Geller D. Clinical characteristics of comorbid obsessive-compulsive disorder and bipolar disorder in children and adolescents. Bipolar disorders. 2010;12:185–195. doi: 10.1111/j.1399-5618.2010.00795.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kanaan R, Barker G, Brammer M, Giampietro V, Shergill S, Woolley J, Picchioni M, Toulopoulou T, McGuire P. White matter microstructure in schizophrenia: effects of disorder, duration and medication. The British Journal of Psychiatry. 2009;194:236–242. doi: 10.1192/bjp.bp.108.054320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kochunov P, Glahn DC, Nichols TE, Winkler AM, Hong EL, Holcomb HH, Stein JL, Thompson PM, Curran JE, Carless MA, Olvera RL, Johnson MP, Cole SA, Kochunov V, Kent J, Blangero J. Genetic analysis of cortical thickness and fractional anisotropy of water diffusion in the brain. Frontiers in neuroscience. 2011;5:120. doi: 10.3389/fnins.2011.00120. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohannim O, Jahanshad N, Braskie MN, Stein JL, Chiang MC, Reese AH, Hibar DP, Toga AW, McMahon KL, de Zubicaray GI, Medland SE, Montgomery GW, Martin NG, Wright MJ, Thompson PM. Predicting White Matter Integrity from Multiple Common Genetic Variants. Neuropsychopharmacology. 2012;37:2012–2019. doi: 10.1038/npp.2012.49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Konrad A, Vucurevic G, Musso F, Stoeter P, Dahmen N, Winterer G. ErbB4 genotype predicts left frontotemporal structural connectivity in human brain. Neuropsychopharmacology. 2009;34:641–650. doi: 10.1038/npp.2008.112. [DOI] [PubMed] [Google Scholar]
- Kumar A, Cook IA. White matter injury, neural connectivity and the pathophysiology of psychiatric disorders. Developmental neuroscience. 2002;24:255–261. doi: 10.1159/000066746. [DOI] [PubMed] [Google Scholar]
- Kumar S, de Vellis J. Neurotrophin activates signal transduction in oligodendroglial cells: expression of functional TrkC receptor isoforms. Journal of neuroscience research. 1996;44:490–498. doi: 10.1002/(SICI)1097-4547(19960601)44:5<490::AID-JNR9>3.0.CO;2-A. [DOI] [PubMed] [Google Scholar]
- LaSalle-Ricci VH, Arnkoff DB, Glass CR, Crawley SA, Ronquillo JG, Murphy DL. The hoarding dimension of OCD: psychological comorbidity and the five-factor personality model. Behaviour research and therapy. 2006;44:1503–1512. doi: 10.1016/j.brat.2005.11.009. [DOI] [PubMed] [Google Scholar]
- Leow A, Huang SC, Geng A, Becker J, Davis S, Toga A, Thompson P. Inverse consistent mapping in 3D deformable image registration: its construction and statistical properties. Information Processing in Medical Imaging. 2005;19:493–503. doi: 10.1007/11505730_41. [DOI] [PubMed] [Google Scholar]
- Li F, Huang X, Yang Y, Li B, Wu Q, Zhang T, Lui S, Kemp GJ, Gong Q. Microstructural brain abnormalities in patients with obsessive-compulsive disorder: diffusion-tensor MR imaging study at 3.0 T. Radiology. 2011;260:216–223. doi: 10.1148/radiol.11101971. [DOI] [PubMed] [Google Scholar]
- Mahon K, Wu J, Malhotra AK, Burdick KE, DeRosse P, Ardekani BA, Szeszko PR. A voxel-based diffusion tensor imaging study of white matter in bipolar disorder. Neuropsychopharmacology. 2009;34:1590–1600. doi: 10.1038/npp.2008.216. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McIntosh AM, Moorhead TW, Job D, Lymer GK, Munoz Maniega S, McKirdy J, Sussmann JE, Baig BJ, Bastin ME, Porteous D, Evans KL, Johnstone EC, Lawrie SM, Hall J. The effects of a neuregulin 1 variant on white matter density and integrity. Molecular Psychiatry. 2008;13:1054–1059. doi: 10.1038/sj.mp.4002103. [DOI] [PubMed] [Google Scholar]
- Menzies L, Williams GB, Chamberlain SR, Ooi C, Fineberg N, Suckling J, Sahakian BJ, Robbins TW, Bullmore ET. White matter abnormalities in patients with obsessive-compulsive disorder and their first-degree relatives. The American Journal of Psychiatry. 2008;165:1308–1315. doi: 10.1176/appi.ajp.2008.07101677. [DOI] [PubMed] [Google Scholar]
- Moskvina V, Craddock N, Holmans P, Nikolov I, Pahwa JS, Green E, Owen MJ, O'Donovan MC. Gene-wide analyses of genome-wide association data sets: evidence for multiple common risk alleles for schizophrenia and bipolar disorder and for overlap in genetic risk. Molecular Psychiatry. 2009;14:252–260. doi: 10.1038/mp.2008.133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakamura K, Kawasaki Y, Takahashi T, Furuichi A, Noguchi K, Seto H, Suzuki M. Reduced white matter fractional anisotropy and clinical symptoms in schizophrenia: a voxel-based diffusion tensor imaging study. Psychiatry Research. 2012;202:233–238. doi: 10.1016/j.pscychresns.2011.09.006. [DOI] [PubMed] [Google Scholar]
- Ortibus E, Verhoeven J, Sunaert S, Casteels I, de Cock P, Lagae L. Integrity of the inferior longitudinal fasciculus and impaired object recognition in children: a diffusion tensor imaging study. Developmental medicine and child neurology. 2012;54:38–43. doi: 10.1111/j.1469-8749.2011.04147.x. [DOI] [PubMed] [Google Scholar]
- Otnaess MK, Djurovic S, Rimol LM, Kulle B, Kahler AK, Jonsson EG, Agartz I, Sundet K, Hall H, Timm S, Hansen T, Callicott JH, Melle I, Werge T, Andreassen OA. Evidence for a possible association of neurotrophin receptor (NTRK-3) gene polymorphisms with hippocampal function and schizophrenia. Neurobiology of Disease. 2009;34:518–524. doi: 10.1016/j.nbd.2009.03.011. [DOI] [PubMed] [Google Scholar]
- Perez-Iglesias R, Tordesillas-Gutierrez D, Barker GJ, McGuire PK, Roiz-Santianez R, Mata I, de Lucas EM, Quintana F, Vazquez-Barquero JL, Crespo-Facorro B. White matter defects in first episode psychosis patients: a voxelwise analysis of diffusion tensor imaging. Neuroimage. 2010;49:199–204. doi: 10.1016/j.neuroimage.2009.07.016. [DOI] [PubMed] [Google Scholar]
- Potash JB, Willour VL, Chiu YF, Simpson SG, MacKinnon DF, Pearlson GD, DePaulo JR, Jr, McInnis MG. The familial aggregation of psychotic symptoms in bipolar disorder pedigrees. The American Journal of Psychiatry. 2001;158:1258–1264. doi: 10.1176/appi.ajp.158.8.1258. [DOI] [PubMed] [Google Scholar]
- Sherry ST, Ward MH, Kholodov M, Baker J, Phan L, Smigielski EM, Sirotkin K. dbSNP: the NCBI database of genetic variation. Nucleic Acids Research. 2001;29:308–311. doi: 10.1093/nar/29.1.308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sprooten E, Sussmann JE, Clugston A, Peel A, McKirdy J, Moorhead TW, Anderson S, Shand AJ, Giles S, Bastin ME, Hall J, Johnstone EC, Lawrie SM, McIntosh AM. White matter integrity in individuals at high genetic risk of bipolar disorder. Biological Psychiatry. 2011;70:350–356. doi: 10.1016/j.biopsych.2011.01.021. [DOI] [PubMed] [Google Scholar]
- Sussmann JE, Lymer GK, McKirdy J, Moorhead TW, Munoz Maniega S, Job D, Hall J, Bastin ME, Johnstone EC, Lawrie SM, McIntosh AM. White matter abnormalities in bipolar disorder and schizophrenia detected using diffusion tensor magnetic resonance imaging. Bipolar disorders. 2009;11:11–18. doi: 10.1111/j.1399-5618.2008.00646.x. [DOI] [PubMed] [Google Scholar]
- Thomason ME, Thompson PM. Diffusion Imaging, White Matter, and Psychopathology. Annual Review of Clinical Psychology. 2011:63–85. doi: 10.1146/annurev-clinpsy-032210-104507. [DOI] [PubMed] [Google Scholar]
- Thompson PM, Martin NG, Wright MJ. Imaging genomics. Current opinion in neurology. 2010;23:368–373. doi: 10.1097/WCO.0b013e32833b764c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tost H, Bilek E, Meyer-Lindenberg A. Brain connectivity in psychiatric imaging genetics. Neuroimage. 2012;62:2250–2260. doi: 10.1016/j.neuroimage.2011.11.007. [DOI] [PubMed] [Google Scholar]
- Vederine FE, Wessa M, Leboyer M, Houenou J. A meta-analysis of whole-brain diffusion tensor imaging studies in bipolar disorder. Progress in Neuro-Psychopharmacology & Biological Psychiatry. 2011 doi: 10.1016/j.pnpbp.2011.05.009. [DOI] [PubMed] [Google Scholar]
- Wang Q, Deng W, Huang C, Li M, Ma X, Wang Y, Jiang L, Lui S, Huang X, Chua SE, Cheung C, McAlonan GM, Sham PC, Murray RM, Collier DA, Gong Q, Li T. Abnormalities in connectivity of white-matter tracts in patients with familial and non-familial schizophrenia. Psychological Medicine. 2011;41:1691–1700. doi: 10.1017/S0033291710002412. [DOI] [PubMed] [Google Scholar]
- Weinberger DR. Implications of normal brain development for the pathogenesis of schizophrenia. Archives of General Psychiatry. 1987;44:660–669. doi: 10.1001/archpsyc.1987.01800190080012. [DOI] [PubMed] [Google Scholar]
- White T, Nelson M, Lim KO. Diffusion tensor imaging in psychiatric disorders. Topics in Magnetic Resonance Imaging. 2008;19:97–109. doi: 10.1097/RMR.0b013e3181809f1e. [DOI] [PubMed] [Google Scholar]
- Winterer G, Konrad A, Vucurevic G, Musso F, Stoeter P, Dahmen N. Association of 5′ end neuregulin-1 (NRG1) gene variation with subcortical medial frontal microstructure in humans. Neuroimage. 2008;40:712–718. doi: 10.1016/j.neuroimage.2007.12.041. [DOI] [PubMed] [Google Scholar]
- Zheng Z, Shemmassian S, Wijekoon C, Kim W, Bookheimer SY, Pouratian N. DTI correlates of distinct cognitive impairments in Parkinson's disease. Human Brain Mapping. 2013 doi: 10.1002/hbm.22256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuliani R, Moorhead TW, Bastin ME, Johnstone EC, Lawrie SM, Brambilla P, O'Donovan MC, Owen MJ, Hall J, McIntosh AM. Genetic variants in the ErbB4 gene are associated with white matter integrity. Psychiatry Research. 2011;191:133–137. doi: 10.1016/j.pscychresns.2010.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]


