Abstract
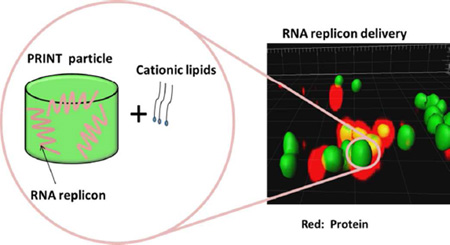
Herein we report the development of a non-viral lipid-complexed PRINT® (particle replication in non-wetting templates) protein particle system (LPP particle) for RNA replicon delivery with a view towards RNA replicon-based vaccination. Cylindrical bovine serum albumin (BSA) particles (diameter (d) 1 µm, height (h) 1 µm) loaded with RNA replicon and stabilized with a fully reversible disulfide cross-linker were fabricated using PRINT technology. Highly efficient delivery of the particles to Vero cells was achieved by complexing particles with a mixture of 1,2-dioleoyl-3-trimethylammonium-propane (DOTAP) and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) lipids. Our data suggest that: 1) this lipid-complexed protein particle is a promising system for delivery of RNA replicon-based vaccines, and 2) it is necessary to use a degradable cross-linker for successful delivery of RNA replicon via protein-based particles.
Keywords: RNA replicon, gene delivery, PRINT, protein particle, lipid
INTRODUCTION
Nucleic acids offer a unique opportunity for vaccination and have emerged as excellent candidates for the treatment of cancers and infectious diseases.1–4 Compared with traditional protein-based vaccination strategy, direct immunization with RNA or DNA has the advantages of the simplicity and purity with which they can be produced, as well as coding for a single protein of interest (i.e. an antigen) at high levels in a reproducible manner, potentially triggering immune response in both cellular and humoral branches.5–7
To protect nucleic acids from enzyme degradation and facilitate entry into cells, many delivery techniques including physical techniques such as the gene gun and electroporation,4, 8 nonviral delivery vehicles such as liposomes and cationic polymers,9–11 and viral vector-based carriers,12 have been extensively explored. However, despite decades of research efforts, commercial products for human use are still unavailable.
An RNA replicon is an important form of a nucleic acid-based vaccine and is derived from either positive- or negative-strand RNA viruses, from which the gene sequences encoding structural proteins are replaced by mRNA encoding antigens of interest as well as the RNA polymerase for RNA replicon replication and transcription.13–15 RNA replicons can be regarded as “disabled” virus vectors that are capable of amplifying within the cytoplasm of host cells for a prolonged period but are unable to produce infectious progeny.16, 17 Compared with DNA vaccines, which are also under active investigation, an RNA replicon-based vaccine has two advantages: first, it is capable of replicating exclusively in the cytoplasm of host cells, thus avoiding the requirement of nuclear entry which represents a daunting hurdle in DNA delivery. By eliminating the dependence on cellular transcription machinery and transport of nucleic acids to and from the nucleus, an RNA replicon is potentially a more efficient form of nucleic acid vaccine.18–20 Secondly, an RNA replicon has superior biosafety features, which is crucial for vaccination purposes. Compared with DNA, RNA replicon can avoid the potential integration into the genome of host cells and also prevent generation of anti-DNA antibodies, both of which may affect the host cell’s gene expression in an uncontrollable manner and thus represent incalculable risks.21 RNA replicon combines the safety characteristics of inactivated vaccines with the superior immunogenicity of live, attenuated vaccines.
Studies have shown that RNA replicon-based vaccination is highly effective for generating cellular and protective immune responses, but has been delivered mainly as naked RNA transcribed in vitro or via viral vectors. 13, 22 The practical utility of viral vectors, however, is limited by manufacturing considerations, cost-effectiveness, and potential adverse health effects.23, 24 The delivery of RNA replicon via synthetic vectors is a highly attractive approach. However, RNA replicons usually have very large sizes (~ 9 kb) compared to mRNA (~2 kb), which adds additional challenge to the development of drug delivery vehicles. There has been very limited data published on nonviral delivery of RNA replicons and it was not until recently that Geall et al. demonstrated the delivery of RNA replicons via liposome nanoparticles.25 In our previous work, we demonstrated the delivery of RNA replicon to Vero cells via PRINT protein particles with the assistance from a TransIT®-mRNA transfection reagent (TransIT).26 However, we encountered batch-to-batch reproducibility issues when TransIT was used as a transfection reagent. TransIT is also cost prohibitive especially for large-scale studies. In addition, at the optimal dosage recommended by manufacturer, TransIT can only deliver particles at very low concentrations, which represents a bottleneck for effective RNA replicon delivery for vaccine purposes (SI, issues with TransIT for transfection of PRINT protein particles). These issues with TransIT motivated us to find another reagent that can facilitate cell uptake and particle transfection. To avoid this limitation, we explored lipids as a much more efficient and effective transfection reagent for RNA replicon loaded PRINT particles, which can be potentially developed into a next generation vaccine. In this study, we also investigated in detail the loading of RNA replicon into PRINT particles, the stabilization of RNA replicon-loaded particles, as well as the role of a reversible disulfide linker in delivery of RNA replicon via protein-based particles.
EXPERIMENTAL SECTION
Materials
Bovine serum albumin and Fluorsave™ reagent were from Calbiochem. Tyramine and 1'-Carbonyldiimidazole was purchased from Sigma Aldrich. BCA protein assay reagent was from Thermo Scientific. DIC as synthesized as previously published in ref. 26. Alexa fluor 488® labeled Bovine serum albumin, Alexa Fluor® 546 goat anti-rabbit IgG (H+L) and Quant-iT ™ RNA assay kit were purchased from invitrogen. Anti-Chloramphenicol Acetyltransferase antibody was purchased from Abcam. TransIT®-mRNA transfection kit was purchased from Mirus. α-D-Lactose and glycerol were purchased from Acros. 1,2-dioleoyl-3-trimethylammonium-propane (chloride salt) (DOTAP), and 1,2-dioleoyl-sn-glycero-3-phosphoethanolamine (DOPE) were from Avanti polar lipids, Inc.
Cells and culture
Cells were maintained at 37 °C in an atmosphere containing 5% CO2. Vero cells were grown in Minimum Essential Medium (MEM; Invitrogen, Carlsbad, CA) supplemented with 5% fetal bovine serum (FBS, HyClone, Logan, UT), MEM non-essential amino acid solution (Invitrogen) and antibiotic–antimycotic (Invitrogen). A549 cells were maintained at 37 °C in an atmosphere containing 5% CO2. The cells were grown in Dulbecco’s Minimum Essential Medium (DMEM; Invitrogen, Carlsbad, CA) supplemented with 10% fetal bovine serum (FBS, HyClone, Logan, UT), and antibiotic–antimycotic (Invitrogen).
RNA replicon construction and particle preparation
Construction of CAT, Luciferase and HA RNA replicons and preparation of RNA replicon loaded BSA-based particles, preparation of RNA replicon loaded Alexa fluor 488® labeled BSA particles and particle crosslinking reaction followed the procedures described in [ 26].
RNA replicon extraction from un-crosslinked particles and DIC-crosslinked particles
For particles prior to crosslinking, 50 µL of PBS was added to dissolve 0.15 mg particles. For crosslinked particles, 50 µL of PBS containing 10 mM dithiothreitol (DTT) was added to dissolve 0.15 mg particles. A Qiazol-chloroform extraction procedure was used to extract RNA replicon from the RNA replicon-BSA mixture. The RNA pellet acquired was dissolved in 20 µL of DEPC-treated water. For un-crosslinked particles, 5 µL of sample was loaded on the gel and10 µL of sample was loaded for cross-linked particles.
Agarose gel electrophoresis
Agarose gel was prepared by dissolving agrasose in 1× NorthernMax®-Gly gel preparation and running buffer (Ambion) at 1 wt%. Typically, 5 µL of sample was mixed with 5 µL of water (or 10 µL of sample) and 10 µL of NorthernMax®-Gly load dye (Ambion) and heated at 50 °C for 10 min before loading onto the gel. The gel was then run in 1× NorthernMax®-Gly gel preparation and running buffer (Ambion) at 70 V for 35 min before being imaged by a GE ImageQuant LAS 4000 biomaolecular imager.
Preparation of lipid-complexed particles
DOTAP:DOPE (75:25) was prepared by first dissolving 1 mg of DOTAP and 1 mg of DOPE in 20 µL of trifluoroethanol (TFE) separately and mixing the two components according to the above ratio. Then water was added to achieve a concentration of 3 mg/mL. RNA replicon-loaded particles were prepared and cross-linked as previously described and were resuspended in isopropanol at 1 mg/mL. Particles were pelleted down using a minicentrifuge and isopropanol was removed. DOTAP/DOPE (75:25) solution (3 mg/mL) were added to the vial to achieve a particle concentration of 1 mg/mL and mixed by pippeting. The particle-lipid solution was incubated at room temperature for 1h. The particles were then pelleted down using a minicentrifuge and supernatant was then removed. The particles were washed three times through centrifugation with water at a particle concentraion of 1 mg/mL and finally dispersed in water with 5 wt% dextrose at 1 mg/mL.
Quantification of DOTAP/DOPE on LPP particles
The amount of DOTAP and DOPE were measured using HPLC (Agilent Technologies 1260) with a C18 rapid resolution column (Zorbax Eclipse plus, 4.6 × 100 mm, 3.5 micron) and a column temperature of 40 °C was used. A mobile phase of water and methanol on a gradient from 85% of methanol to pure methanol over 10 min followed by pure methanol for another 2 min with a flow rate of 2 mL/min was employed with a detection temperature of 50 °C on the ELSD (Agilent Technologies 1260). The DOTAP peak appeared at 8.0 minutes and the DOPE peak appeared at 9.8 minutes. The peak areas were compared to the DOTAP and DOPE standard curves to determine the concentration of lipids in the solution. The amounts of DOTAP and DOPE on the LPP particles were calculated by subtracting the amount of lipids in the supernatants and washes from the original lipid solution.
Physical Characterization of the PRINT Protein Particles
After the cross-linking reaction followed by incubation in water for 24h at room temperature, the particles were deposited on a glass slide, coated with palladium/gold and imaged by a scanning electron microscopy (Hitachi modelS-4700). The hydrodynamic diameters of the PRINT particles were measured by dynamic light scattering (Brookhaven Instruments Inc., 90Plus). For zeta potential measurements, the particles were dispersed in 1 mM potassium chloride at a concentration of 20 µg/ml and tested by a Zetasizer Nano Analyzer (Malvern Instruments Inc., Nano Zetasizer).
Analysis of luciferase and HA expression
The dosing of LPP particles and preparation of cell lysates followed procedures described in reference 26. Luciferase assay was carried out according to the manufacturer's instructions. The relative bioluminescence was calculated using following method:
Where Ar: the relative bioluminescence. Aa: the bioluminescence acquired by plate reader for samples dosed with RNA replicon or particles. Ac: the bioluminescence acquired by plate reader for untreated cells. Vero cells plated at on cover slips in 6-well dishes and grown for 24 hours. Cells were treated with LPP particles for 4h, medium was aspirated and replaced with fresh medium for 44h. Cells were then washed three times with ice cold 1×PBS and fixed with cold (−20°C) acetone/methanol (1:1 mix) for 5 minutes. Cells were washed in ice cold 1×PBS 3 times. For HA detection, cells were then incubated in primary antibody (Rabbit IgG, Immune Technology Corp, cat# IT-003-SW) for 1 hr at 37°C in the dark, cells were then washed with three times with 1XPBS and incubated in secondary Alexa Fluor® 546 goat anti-rabbit IgG (H+L) (cat #A11010, Invitrogen) for 1hr at RT in dark. For luciferase detection, cells were incubated with primary antibody conjugated with FITC (US Biological, cat# L5999-09A) for 1h at 37°C in the dark. Washed twice in PBS and mounted with Fluorsave™ reagent. Samples were then analyzed by confocal microscopy. Confocal images were acquired using a Ziess 710 laser scanning confocal imaging system (Olympus) fluorescence microscope fitted with a PlanApo 60× oil objective (Olympus). The final composite images were created using Adobe Photoshop CS (Adobe Systems, San Jose, CA).
Cytotoxcity study of RNA loaded LPP particles
Typically, 5×103 Vero cells were plated into 96 well tissue cultured treated plates 18–24 h prior to assay. The LPP particles were dosed to cells as previously described. The mixture was subsequently incubated with Vero cells for 4h at 37 °C and the non-internalized particles were removed. The cells were further incubated for another 24h at 37 °C before ATP was quantitated using the CellTiter-G10 Luminescent Cell Viability Assay (Promega) following vendor’s protocol.
RESULTS
1) Encapsulation of RNA Replicon and Stabilization of the Particles
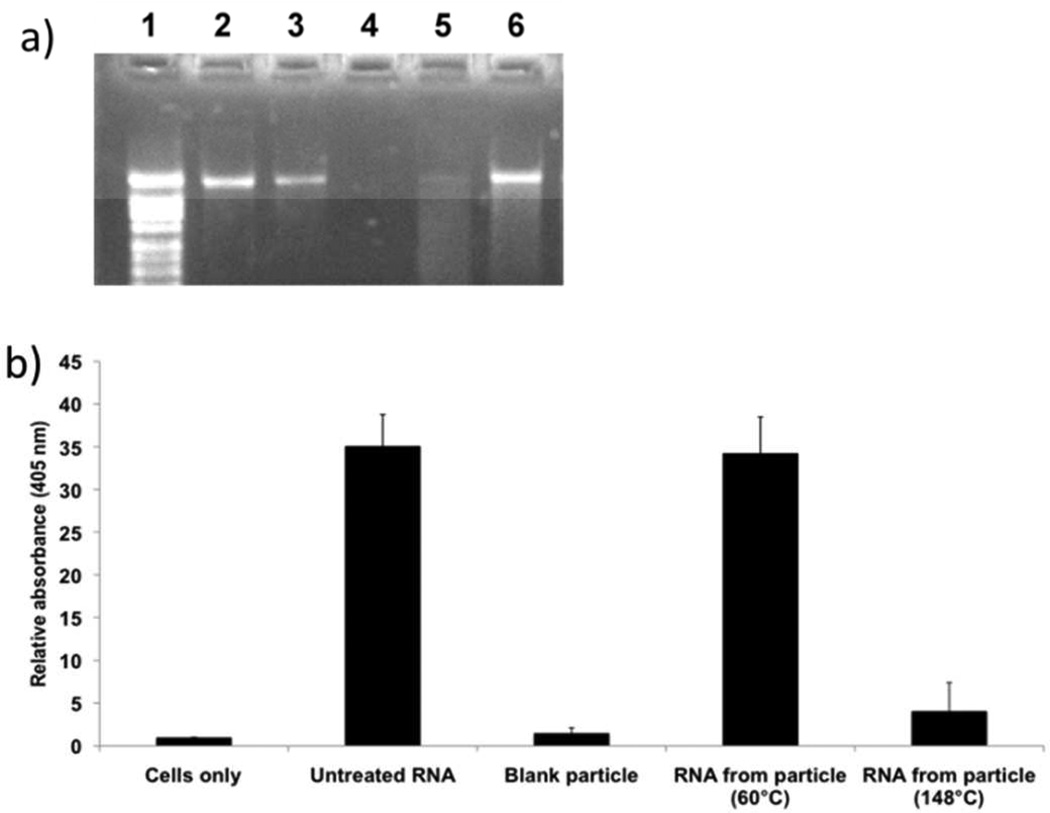
We have previously demonstrated that RNA replicon can be incorporated into cylindrical bovine serum albumin (BSA) PRINT particles (diameter (d) 1 µm, height (h) 1 µm) by mixing an RNA replicon with BSA, lactose and glycerol to form a preparticle solution that can flow into the cavities of the PRINT mold when heated (SI, fabrication of RNA replicon-incorporated PRINT particle).26 RNA replicon is a single stranded RNA with low stability. Maintaining its integrity in the process of PRINT particle fabrication is essential. We first investigated the influence of temperature used for particle fabrication on the biological activity of RNA replicon. A model RNA replicon encoding chloramphenicol acetyl transferase (CAT) was chosen in this study because CAT is a bacterial enzyme and exogenous for mammalian cells and the assays to quantify CAT activity have been well established. Typically, 1 wt% of CAT RNA replicon was formulated with 37 wt% of BSA, 37 wt% of lactose and 25 wt% of glycerol. This dry mixture was processed into cylindrical albumin particles by using two temperatures on the heated laminator roller: 60°C vs. 148°C. The particles were dissolved in phosphate buffered saline (PBS) and followed by extraction of RNA replicon from the BSA-RNA replicon mixture. It should be noted that the un-crosslinked particles used in this experiment were readily soluble in water. The integrity of the extracted RNA replicon was first evaluated using agarose gel electrophoresis, as shown in Figure 1a. Electrophoresis analysis showed that only RNA replicon encapsulated at 60°C displayed a bright tight band at the same position as the untreated RNA replicon. Below the band, there was some smearing representing the degraded RNA replicon, but the brightness of the smearing was significantly weaker than the band. By contrast, the RNA replicon encapsulated at 148 °C mainly showed smearing. The integrity of the RNA replicon was further accessed by a CAT ELISA assay after the RNA replicon was transfected into Vero cells, a kidney epithelial cell line developed from an African green monkey, using TransIT. Results showed that RNA replicon encapsulated at 148°C showed very minimal biological activity and RNA extracted from particles fabricated at 60°C produced similar protein expression levels to an untreated RNA replicon control (Figure 1b). The results indicated that for encapsulation of RNA replicon into BSA PRINT particles, lower temperature is more favorable for maintaining the integrity of the RNA cargo. When PRINT replicon particles are processed at 60°C, some of the RNA replicon was damaged, however, the amount of degraded RNA was so minimal that no difference was discernable in the CAT ELISA assay. After the harvest and purification processes using isopropanol, the particles fabricated at 60°C were determined to contain 1.5±0.1 wt% of RNA replicon in the final particle composition (Table S1). Given the size of the particles and the level of loading of RNA, this corresponds to approximately 2000 copies of the RNA replicon per particle (calculated based on a particle density of 1.0 g/mL).
Figure 1.
(a) Agarose gel of RNA replicon before and after particle crosslinking. 1: RNA ladder, 2: untreated RNA 200 ng, 3: untreated RNA 100 ng, 4: RNA replicon extracted out 37.5 µg of blank BSA particles, 5:RNA replicon extracted out of 37.5 µg of BSA particles fabricated at 148°C, 6: RNA replicon extracted out of 37.5 µg of BSA particles fabricated at 60°C. (b). Relative absorbance obtained from CAT ELISA. The absorbance from un-treated cells (cells only) were defined as 1. Error bars= mean + SD.
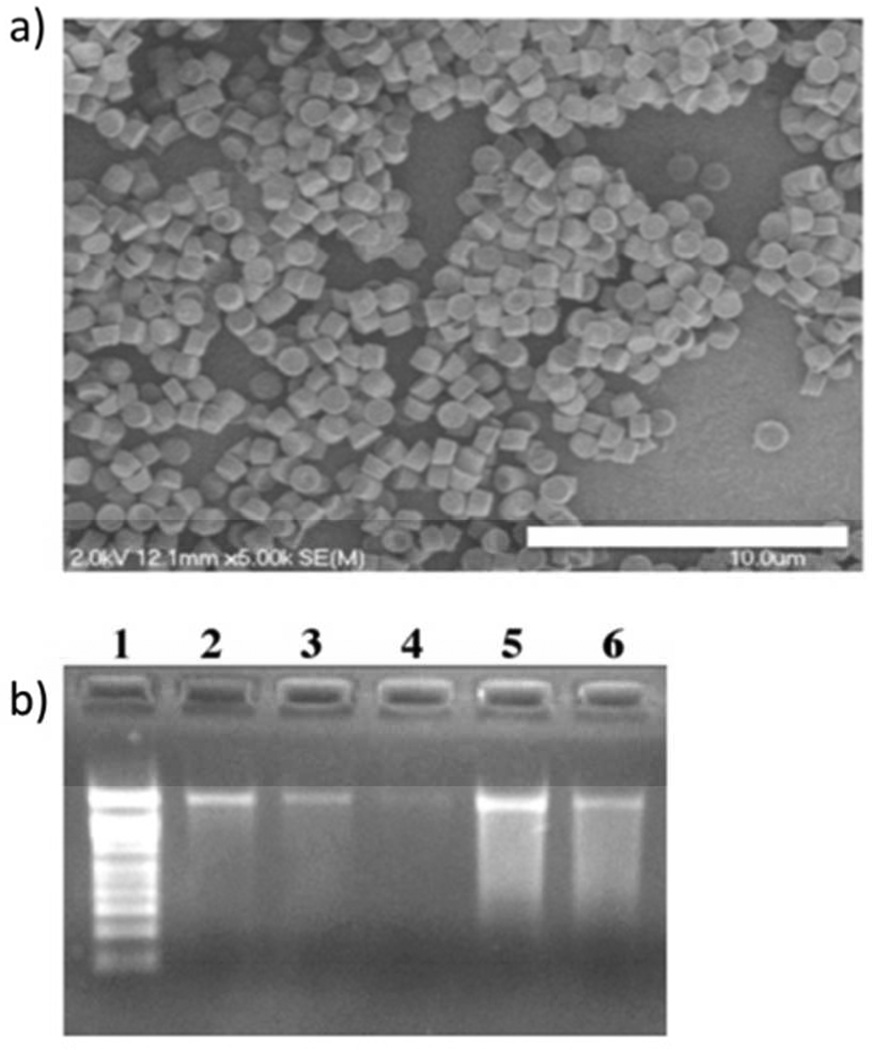
The site of action for RNA replicon is cytoplasm, which is known for its high concentration of reduced glutathione (GSH) compared to the extracellular environment (GSH intracellular concentration between 5 mM and 15 mM).27 Therefore, a disulfide-based cross-linker was chosen to cross-link the particle to act like a Trojan Horse (an image of the particles after incubation in water is shown in Figure 2a). We used dithio-bis(ethyl 1 H-imidazole-1-carboxylate) (DIC) as the cross-linker to stabilize the protein particles in aqueous solutions (SI, Figure S5). This cross-linker was developed by our group and, compared to a commercially available disulfide linker dithiobis[succinimidyl propionate] (DSP), has several advantages for protein particle stabilization for RNA replicon delivery.26 Imidazoles as the leaving groups enhanced the reaction selectivity for amines over hydroxyl groups, which is essential to maintain the integrity of RNA replicon in the cross-linking procedure. Furthermore, DIC is a “traceless” reversible cross-linker, which does not leave any molecular pendants after disulfide cleavage. A truly reversible chemistry has many benefits for the delivery of RNA replicon. It releases the amino groups in its original form and avoids the unknown immune response towards novel antigens. Additionally, even if the amine functionalities on nucleobases were cross-linked by DIC, due to the fully reversible nature of the cross-linker, the nucleobases should revert to its original state upon disulfide cleavage if DIC is used.
Figure 2.
(a) Scanning electron microscope (SEM) image of DIC-crosslinked particles containing CAT RNA replicon, image was taken after incubation with PBS, scale bar stands for 10 µm. (b) Agarose gel of RNA replicon before and after particle crosslinking: lane 1: RNA marker, 2: untreated RNA replicon 200 ng, 3: untreated RNA replicon 100 ng, 4: untreated RNA replicon 50 ng, 5: RNA replicon extracted out of 37.5 µg of BSA particles before crosslinking reaction, 6: RNA replicon extracted out of 75 µg of BSA particles after cross-linking reaction.
To assess the integrity of the RNA replicon after the cross-linking step, particles were dissolved in 10 mM dithiothreitol (DTT) solution and the RNA replicon extracted from the mixture was again evaluated by agarose gel electrophoresis (Figure 2b). The agarose electrophoresis gel results demonstrate that after the cross-linking step there is still a band representing intact RNA replicon associated with particles although there was also some smearing below the band. The RNA replicon integrity was confirmed by induced expression of CAT protein after transfection to Vero cells (SI, Characterization of cross-linked BSA particles). We further analyzed the impact of cross-linking reaction on the integrity of the RNA replicon. These results suggest that the amount of CAT protein expressed by RNA replicon treated with the cross-linker is comparable to untreated RNA replicon (SI, Figure S6). This indicates that the cross-linking reaction is RNA-friendly. More information about release of RNA from cross-linked protein particles is available in SI (Characterization of cross-linked BSA particles, Figure S7).
2) Delivery of CAT RNA Replicon-loaded Particles
Due to the isoelectric point of BSA (pI = 4.75), the cross-linked BSA particles with RNA replicons are negatively charged (ζ potential = −15.4 ± 1.0 mV). Based on the previous studies from our group and other groups, cells generally preferentially internalize positively charged particles through a non-specific electrostatic interactions between the positively charged particles and the negatively charged cell membrane. 28 DOTAP and DOPE have been widely used as transfection reagents for gene delivery and they also enjoy the advantage of being commercially available and cost-effective. 29, 30 The introduction of lipids is also expected to enhance the endosomal escape of BSA particles, which is the major roadblock for RNA replicon delivery.
A lipid mixture of DOTAP:DOPE (75:25, weight ratio) was mixed with the RNA replicon loaded BSA particles and incubated for 1h at room temperature to enable the lipids to complex with the particles. The particles were washed three times with water through centrifugation to remove excess lipids. High-performance liquid chromatography (HPLC) coupled with evaporative light scattering detection (ELSD) was used to determine the amount of lipids complexed with BSA RNA replicon particles and it was found that 0.46 ± 0.06 mg of DOTAP and 0.11 ±0.02 mg of DOPE were associated with 1 mg of particles (the actual ratio of DOTAP:DOPE=80:20). The interaction between lipids and the cross-linked particles was studied and the results indicated that hydrophobic interaction contributes to the lipid-particle complexation, and ionic interaction does not (SI, Table S2). From the results, we also suspect that lipids are not only present on the surface of the negatively-charged particles, but also penetrate to the interior of the particles.
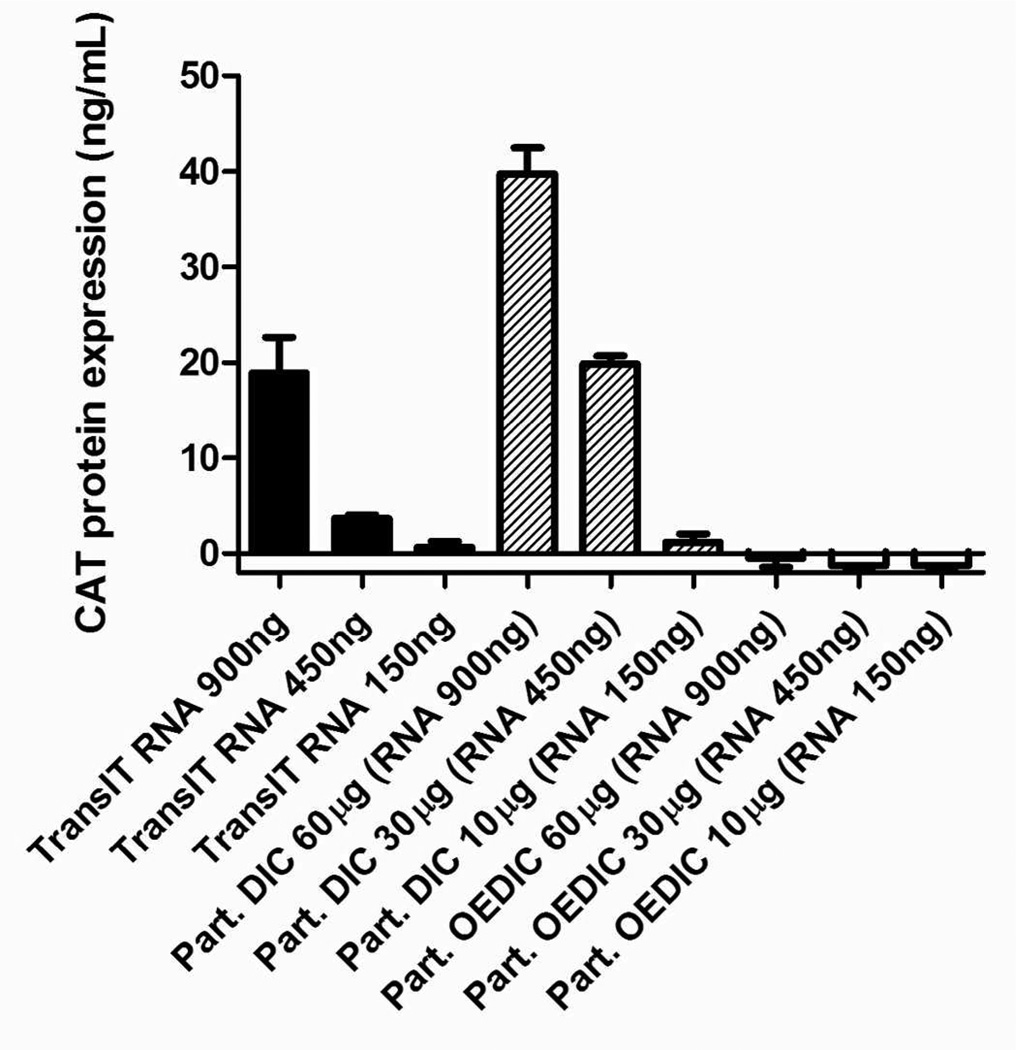
The lipid-complexed PRINT protein particles (LPP particles) achieved a positively charged surface (ζ potential = +29.3 ± 2.5 mV) and were subsequently incubated with Vero cells for 4 h at 37 °C and the non-internalized particles were removed (SI,Table S3). The cells were further incubated for another 24h at 37 °C to allow for the expression of CAT protein. The LPP particle delivery of the CAT protein was comparable to the same amount of RNA replicon directly delivered by TransIT (Figure 3). Compared to our previous work which uses TransIT to assist particle transfection ,26 LPP particles showed substantial improvement in the amount of CAT protein that can be produced by Vero cells, and achieved dose dependence up to 60 µg/mL. Utilizing TransIT as the transfection reagent for the protein particles can only effectively deliver particles at particle concentrations below 2 µg/mL. In addition, LPP particles can deliver RNA replicon in a highly reproducible manner.
Figure 3.
CAT protein concentration generated from cells. Black bars: CAT RNA replicon standards delivered by TransIT, hatched bars: DIC-crosslinked LPP particles containing CAT RNA replicon, square pattern bars: OEDIC-crosslinked LPP particles containing CAT RNA replicon. Based on quantification results, 60, 30 and 10 µg/mL of particles contain 900, 450, and 150 ng/mL of RNA respectively. Error bars= mean + SD.
To study the necessity of the disulfide cross-linker in the delivery of RNA replicon via protein particles, particles cross-linked with a non-degradable linker 2,2'-oxybis(ethane-2,1-diyl) bis(1H-imidazole-1-carboxylate) (OEDIC) under the same reaction condition as DIC was also investigated. Because both DIC and OEDIC have imidazole leaving groups and the concentrations of cross-linkers and the reaction time were held constant, therefore we expect the BSA particles cross-linked with DIC and OEDIC to have similar cross-linking density. It was observed that very minimal protein was expressed with particles cross-linked by OEDIC compared to particles cross-linked by the disulfide cross-linker DIC. This result demonstrated that 1) a disulfide linker is important to achieve cytoplasm release of RNA replicon; and 2) the intracellular protease were not responsible for the BSA particle degradation and RNA replicon release at least during the 24-h time frame of our assay. Therefore, the disulfide linker DIC can not only stabilize particles in the process of delivery, but also efficiently release RNA replicon at the ultimate site of action.
Analysis using confocal microscopy was carried out to visually confirm the generation of CAT protein. Fixed and permeabilized Vero cells treated with LPP particles were stained with a primary antibody that binds specifically to CAT protein, and further treated with dye-labeled secondary antibody. Compared to untransfected cells, cells transfected with DIC-crosslinked RNA replicon-containing particles showed intense fluorescence, indicating for high levels of expression of CAT proteins in those cells (Figure 4). A 3D image of encoded CAT protein and the LPP particles inside the Vero cells was captured and showed in Figure S8. Cells transfected with OEDIC-crosslinked RNA replicon-containing particles showed no trace of CAT protein.
Figure 4.
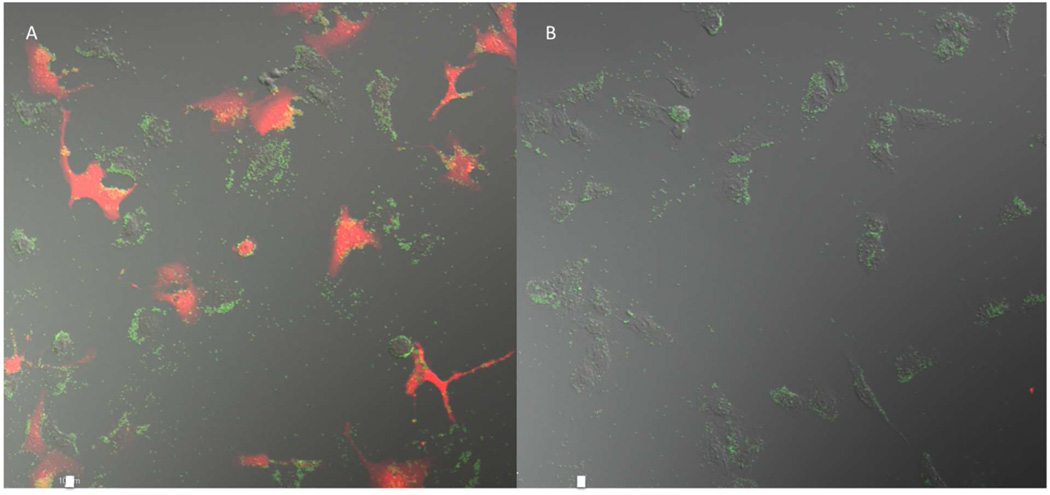
Confocal image of CAT protein. (A) BSA particles containing CAT RNA replicon cross-linked with DIC, 30 µg/mL. (B) BSA particles containing CAT RNA replicon cross-linked with OEDIC, 30 µg/mL. The red represents the CAT protein and the green represents PRINT particles. The scale bar represents 10 µm.
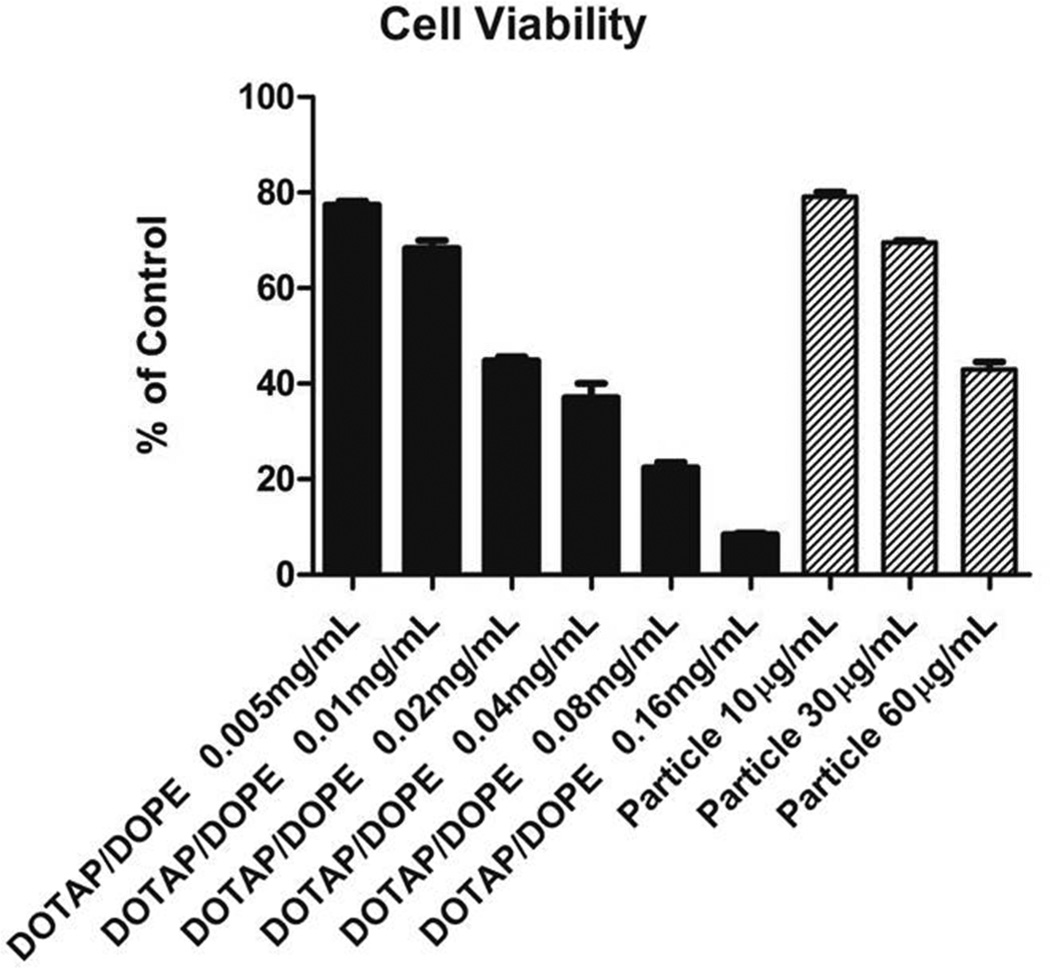
The viability of Vero cells was evaluated after the 24-h CAT protein expression (Figure 5). Cytocompatibility was maintained at lower dosing concentrations; however, the LPP particles appeared to be slightly toxic at particle concentration of 60 µg/mL. The toxicity may be attributed to the lipids which alone cause cell death at higher dosing concentrations.
Figure 5.
Viability of Vero cells dosed with DOTAP/DOPE lipids only (black bars, the ratio of DOTAP:DOPE=80:20) and LPP particles (hatched bars). Cells were dosed with samples for 4h followed by removal of particles and 24h incubation at 37 °C. Based on HPLC-ELSD quantification results, 10, 30 and 60 µg/mL of LPP particles contains 0.006, 0.018 and 0.036 mg/mL of DOTAP/DOPE lipids respectively. Error bars= mean + SD.
3) Delivery of Luciferase RNA replicon and Influenza hemagglutinin (HA) RNA replicon
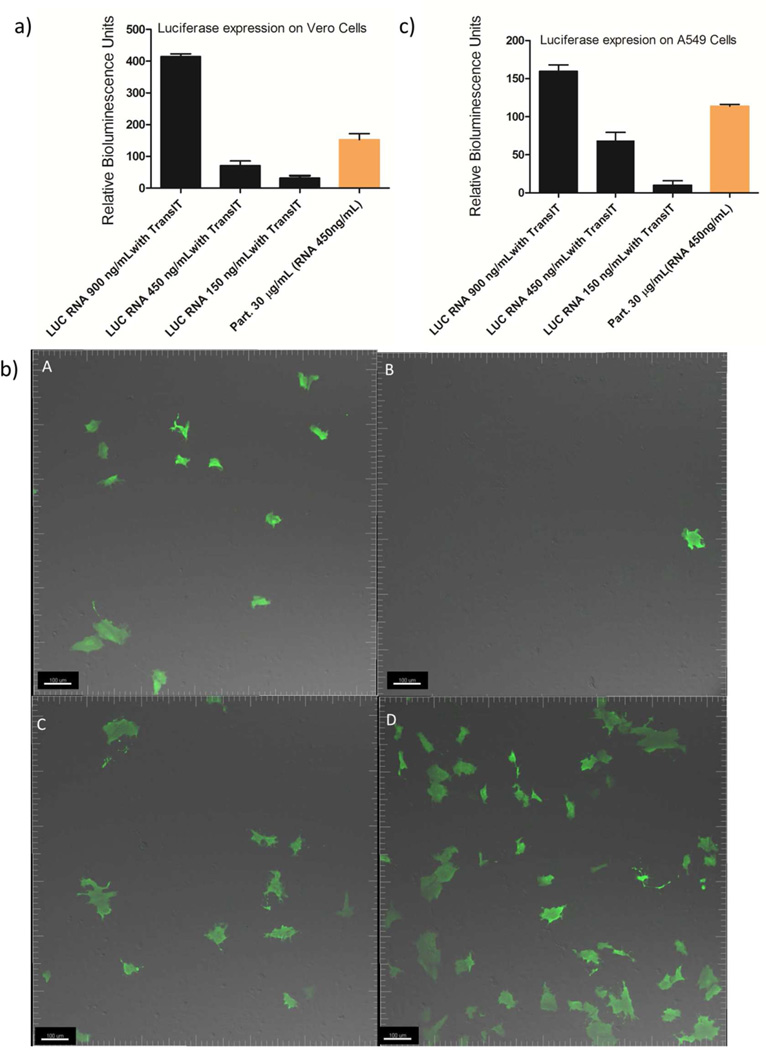
To show that RNA replicons encoding different proteins can be encapsulated and delivered within the same LPP particle, RNA replicons encoding a luciferase protein and a therapeutically relevant antigen influenza hemagglutinin (HA) were incorporated into the particles and delivered to cells via aforementioned methods. Both Luciferase and HA are exogenous for the cell lines investigated and their quantification methods have been well established. HA is a type of integral envelope proteins found on the surface of the influenza virus and is a critical antigen in eliciting protection against influenza virus challenges.31 The bioluminescence generated by luciferase RNA replicon-loaded LPP particles was comparable to the same amount of RNA replicon directly delivered by TransIT (Figure 6a). The generation of luciferase protein and the advantage of LPP particles over particles mixed with TransIT were also confirmed by confocal microscopy (Figure 6b and Figure S9). Here we further explored the necessity of using PRINT particles for delivery of RNA replicon. Compared to using DOTAP/DOPE alone for RNA replicon transfection, LPP particle-mediated delivery showed five times higher protein expression (SI, Comparison between DOTAP/DOPE only and LPP particles). In addition to Vero cells, LPP particles also delivered luciferase-encoding RNA replicon to an A549 cell line derived from adenocarcinomic human alveolar basal epithelial cells, with a lower protein expression level compared to Vero cells (Figure 6c).
Figure 6.
(a) Relative bioluminesence generated by LPP particles on Vero cells. The luminescence generated by untreated cells is defined as 1. Black: Luciferase (LUC) RNA replicon standards delivered by TransIT, orange: LPP particles (30 µg/mL) containing Luciferase RNA replicon (450 ng/mL). Error bars= mean + SD. (b) Confocal image of luciferase protein, Vero cells for all samples are over 90% confluent in 6-well plate : (A) RNA replicon (150 ng/mL) transfected with TransIT , (B) PRINT particles (10 µg/mL ) containing RNA replicon (150 ng/mL) dosed with TransIT. (C) LPP particles (10 µg/mL) containing luciferase RNA replicon (150 ng/mL), (D) LPP particles (30 µg/mL) containing luciferase RNA replicon(450 ng/mL). The green represents the luciferase protein. The scale bar represents 100 µm. (c) Relative luminesence generated by PRINT particles on A549 cells. The luminescence generated by untreated cells is defined as 1. Black: Luciferase (LUC) RNA replicon standards delivered by TransIT, orange: DIC-crosslinked BSA particles containing Luciferase RNA replicon. Error bars= mean + SD.
The ability of a cross-linked BSA particle matrix to act as a shield to protect the delicate RNA cargo from degradation especially from endogenous nucleases is an important design characteristic that we were hoping to observe. To test this hypothesis, the RNA replicon encapsulated into the LPP particles was treated with RNase A as a function of RNase A concentration for 1h at 37 °C (SI, Figure S11). The results indicated that the LPP particles successfully protected the RNA replicon from degradation by RNase A. In contrast, the naked RNA was completely degraded after incubation with RNase A of the same concentration under the same condition (SI, Figure S12).
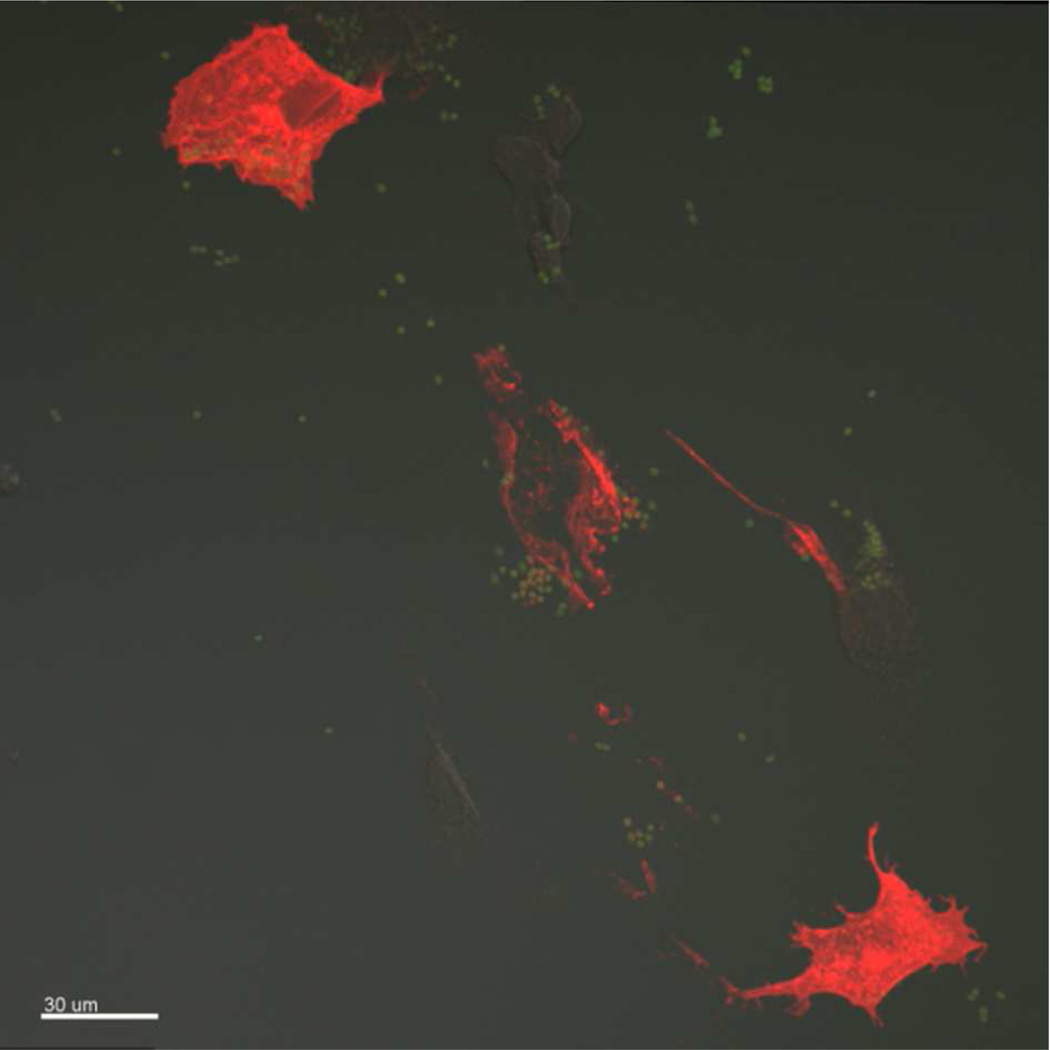
The cells treated with HA replicon-loaded LPP particles were observed to show HA expression, indicating successful delivery of HA RNA replicon to the Vero cells (Figure 7). These results suggest that PRINT is a general approach for delivering RNA replicons with straightforward incorporation of different cargos and can work in a “plug and play” manner, which is particularly important for development of new vaccines for epidemic diseases.
Figure 7.
Confocal image of HA protein. The red fluorescence represents the HA protein, the green represents particles. Particles dosed to Vero cells at 30 µg/mL. Scale bar represents 30 µm.
DISCUSSION
The PRINT technique developed in our lab is a platform that enables the generation of engineered micro- and nanoparticles having precisely controlled properties including size, shape, modulus, chemical composition and surface functionality for drug delivery applications. 26, 28, 32–35 We have demonstrated that PRINT particles can be designed to impact biodistribution,36, 37 lung deposition,38 cell internalization profiles,28, 32 and to deliver therapeutic cargos.30, 39 PRINT is also amenable to continuous roll-to-roll fabrication techniques that enable the scale-up of the particle fabrication under good manufacturing practice (GMP) compliance. Using PRINT, a variety of cargos can be incorporated through inclusion in the particle matrices in a “plug-and-play” manner. This can potentially speed up the development of new vaccines, which is particularly important in the vaccination against epidemic diseases. PRINT allows for the precise and simultaneous control over all the parameters that are essential to protect and deliver the RNA replicon and provides a platform for development of safe and effective vaccines against a wide variety of diseases.
For delivery of mRNA or RNA replicons, the most-frequently used synthetic strategies include liposomes,12, 25, 40 polyplexes,29, 41 or surface absorption onto cationic particles,11 etc. The LPP particle system is a unique approach in that the RNA replicon is encapsulated into particles chemically stabilized by a reversible linker, allowing the RNA to be selectively released in the cytoplasm of cells. This design affords robust protection for the RNA replicon against endogenous nucleases. Here by comparing with a control non-degradable linker, the reversible disulfide linker was clearly demonstrated to play a vital role in the successful delivery of RNA replicon. This result may provide important information for design of particle systems for cytoplasmic delivery of therapeutics including proteins, peptides or other nucleic acids. In this initial proof-of-concept study, we used BSA as the matrix of the particles. We are aware that humans will generate an immune response to BSA and this may result in adverse health effects upon multiple doses. In the future work, human serum albumin, synthetic polymers or polypeptides can be used as the particle matrix to reduce this risk.
CONCLUSION
Herein, a novel method for the delivery of RNA replicon via lipid-complexed PRINT protein (LPP) particles was demonstrated. This particle fabrication method, built on the PRINT technology platform, not only allows for the fabrication of particles of controlled sizes and shapes, but also was gentle enough to encapsulate RNA replicon without abolishing their biological activity. A disulfide cross-linker was essential to not only maintaining the BSA-based particles from dissolution in aqueous media, but was also essential in degrading in the intracellular reducing environment to enable release of the RNA replicon cargo. By complexing the particles with DOTAP/DOPE lipids, the particles were able to be successfully transfected to Vero cells and encoded protein was expressed via delivery of PRINT particles. The reversible disulfide linker was demonstrated to play a vital role in the successful delivery of RNA replicon. RNA replicons encoding different proteins including luciferase and HA were successfully incorporated into the PRINT particles and delivered using the same strategy. The LPP particles were also shown to protect the RNA replicon effectively against RNase A. Future work will focus on investigation of in vivo delivery efficiency and immunogenicity.
Supplementary Material
ACKNOWLEDGMENT
Funding Sources The work is supported in part by National Institutes of Health Director’s Pioneer Award (1DP1OD006432) and R01 (R01EB009565), University of North Carolina Cancer Research Fund, the Chancellor’s Eminent Professorship of Chemistry at the University of North Carolina at Chapel Hill, and a sponsored research agreement with Liquidia Technologies.
We would like to thank Jillian L. Perry (Lineberger Comprehensive Cancer Center, University of North Carolina, Chapel Hill, North Carolina 27599, United States) for help with editing.
ABBREVIATIONS
- LPP particles
Lipid-complexed PRINT protein particles
- BSA
Bovine serum albumin
- CAT
chloramphenicol acetyl transferase
- TransIT
TransIT®-mRNA transfection reagent
- DOTAP
1,2-dioleoyl-3-trimethylammonium-propane
- DOPE
1,2-dioleoyl-sn-glycero-3-phosphoethanolamine
- HA
hemagglutinin
Footnotes
ASSOCIATED CONTENT
Supporting Information. Issues with TransIT for transfection of PRINT protein particles. Fabrication and composition analysis of RNA replicon incorporated PRINT particles. Characterization of cross-linked BSA particles. Investigation of the complexation between the DOTAP/DOPE and cross-linked RNA replicon loaded protein particles. The 3D image of CAT protein and LPP particles in the Vero cells. Comparison between DOTAP/DOPE only and LPP particles. Particle protection of RNA replicon against RNase A. This material is available free of charge via the Internet at http://pubs.acs.org.”
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
REFERENCES
- 1.Tang DC, DeVit M, Johnston SA. Genetic immunization is a simple method for eliciting an immune response. Nature. 1992;356(6365):152–154. doi: 10.1038/356152a0. [DOI] [PubMed] [Google Scholar]
- 2.Weide B, Garbe C, Rammensee H-G, Pascolo S. Plasmid DNA- and messenger RNA-based anti-cancer vaccination. Immunol. Lett. 2008;115(1):33–42. doi: 10.1016/j.imlet.2007.09.012. [DOI] [PubMed] [Google Scholar]
- 3.Bringmann A, Held SAE, Heine A, Brossart P. RNA Vaccines in Cancer Treatment. J. Biomedi. Biotechnol. 2010;2010:1–13. doi: 10.1155/2010/623687. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kutzler MA, Weiner DB. DNA vaccines: ready for prime time? Nat. Rev. Genet. 2008;9(10):776–788. doi: 10.1038/nrg2432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cannon G, Weissman D. RNA based vaccines. DNA Cell Biol. 2002;21(12):953–961. doi: 10.1089/104454902762053882. [DOI] [PubMed] [Google Scholar]
- 6.Vajdy M, Srivastava I, Polo J, Donnelly J, O'Hagan D, Singh M. Mucosal adjuvants and delivery systems for protein-, DNA- and RNA-based vaccines. Immunol. Cell Biol. 2004;82(6):617–627. doi: 10.1111/j.1440-1711.2004.01288.x. [DOI] [PubMed] [Google Scholar]
- 7.Wolff JA, Malone RW, Williams P, Chong W, Acsadi G, Jani A, Felgner PL. Direct gene transfer into mouse muscle in vivo. Science. 1990;247(4949 Pt 1):1465–1468. doi: 10.1126/science.1690918. [DOI] [PubMed] [Google Scholar]
- 8.Van Tendeloo VF, Ponsaerts P, Lardon F, Nijs G, Lenjou M, Van Broeckhoven C, Van Bockstaele DR, Berneman ZN. Highly efficient gene delivery by mRNA electroporation in human hematopoietic cells: superiority to lipofection and passive pulsing of mRNA and to electroporation of plasmid cDNA for tumor antigen loading of dendritic cells. Blood. 2001;98(1):49–56. doi: 10.1182/blood.v98.1.49. [DOI] [PubMed] [Google Scholar]
- 9.Lu D, Benjamin R, Kim M, Conry RM, Curiel DT. Optimization of methods to achieve mRNA-mediated transfection of tumor cells in vitro and in vivo employing cationic liposome vectors. Cancer Gene Ther. 1994;1(4):245–252. [PubMed] [Google Scholar]
- 10.Eisele K, Gropeanu RA, Zehendner CM, Rouhanipour A, Ramanathan A, Mihov G, Koynov K, Kuhlmann CR, Vasudevan SG, Luhmann HJ, Weil T. Fine-tuning DNA/albumin polyelectrolyte interactions to produce the efficient transfection agent cBSA-147. Biomaterials. 2010;31(33):8789–8801. doi: 10.1016/j.biomaterials.2010.07.088. [DOI] [PubMed] [Google Scholar]
- 11.Su X, Fricke J, Kavanagh DG, Irvine DJ. In vitro and in vivo mRNA delivery using lipid-enveloped pH-responsive polymer nanoparticles. Mol. Pharm. 2011;8(3):774–787. doi: 10.1021/mp100390w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yamamoto A, Kormann M, Rosenecker J, Rudolph C. Current prospects for mRNA gene delivery. Eur. J. Pharm. Biopharm. 2009;71(3):484–489. doi: 10.1016/j.ejpb.2008.09.016. [DOI] [PubMed] [Google Scholar]
- 13.Zimmer G. RNA Replicons - A New Approach for Influenza Virus Immunoprophylaxis. Viruses. 2010;2(2):413–434. doi: 10.3390/v2020413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Anraku I, Harvey TJ, Linedale R, Gardner J, Harrich D, Suhrbier A, Khromykh AA. Kunjin Virus Replicon Vaccine Vectors Induce Protective CD8+ T-Cell Immunity. J. Virol. 2002;76(8):3791–3799. doi: 10.1128/JVI.76.8.3791-3799.2002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Tannis L, Gauthier A, Evelegh C, Parsons R, Nyholt D, Khromykh A, Bramson J. Semliki Forest virus and Kunjin virus RNA replicons elicit comparable cellular immunity but distinct humoral immunity. Vaccine. 2005;23(33):4189–4194. doi: 10.1016/j.vaccine.2005.04.005. [DOI] [PubMed] [Google Scholar]
- 16.Ying H, Zaks TZ, Wang RF, Irvine KR, Kammula US, Marincola FM, Leitner WW, Restifo NP. Cancer therapy using a self-replicating RNA vaccine. Nat. Med. 1999;5(7):823–827. doi: 10.1038/10548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Diken M, Kreiter S, Selmi A, Britten CM, Huber C, Türeci Ö, Sahin U. Selective uptake of naked vaccine RNA by dendritic cells is driven by macropinocytosis and abrogated upon DC maturation. Gene Ther. 2011;18(7):702–708. doi: 10.1038/gt.2011.17. [DOI] [PubMed] [Google Scholar]
- 18.Lechardeur D, Sohn KJ, Haardt M, Joshi PB, Monck M, Graham RW, Beatty B, Squire J, O'Brodovich H, Lukacs GL. Metabolic instability of plasmid DNA in the cytosol: a potential barrier to gene transfer. Gene Ther. 1999;6(4):482–497. doi: 10.1038/sj.gt.3300867. [DOI] [PubMed] [Google Scholar]
- 19.Nishimura K, Segawa H, Goto T, Morishita M, Masago A, Takahashi H, Ohmiya Y, Sakaguchi T, Asada M, Imamura T, Shimotono K, Takayama K, Yoshida T, Nakanishi M. Persistent and Stable Gene Expression by a Cytoplasmic RNA Replicon Based on a Noncytopathic Variant Sendai Virus. J. Biol. Chem. 2007;282(37):27383–27391. doi: 10.1074/jbc.M702028200. [DOI] [PubMed] [Google Scholar]
- 20.Smerdou C, Liljestrom P. Non-viral amplification systems for gene transfer: vectors based on alphaviruses. Curr. Opin. Mol. Ther. 1999;1(2):244–251. [PubMed] [Google Scholar]
- 21.Wang Z, Troilo PJ, Wang X, Griffiths TG, Pacchione SJ, Barnum AB, Harper LB, Pauley CJ, Niu Z, Denisova L, Follmer TT, Rizzuto G, Ciliberto G, Fattori E, Monica NL, Manam S, Ledwith BJ. Detection of integration of plasmid DNA into host genomic DNA following intramuscular injection and electroporation. Gene Ther. 2004;11(8):711–721. doi: 10.1038/sj.gt.3302213. [DOI] [PubMed] [Google Scholar]
- 22.Kofler RM. Mimicking live flavivirus immunization with a noninfectious RNA vaccine. Proc. Natl. Acad. Sci. USA. 2004;101(7):1951–1956. doi: 10.1073/pnas.0307145101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Grgacic EVL, Anderson DA. Virus-like particles: Passport to immune recognition. Methods. 2006;40(1):60–65. doi: 10.1016/j.ymeth.2006.07.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Ramsey JD, Vu HN, Pack DW. A top-down approach for construction of hybrid polymer-virus gene delivery vectors. J. Control. Release. 2010;144(1):39–45. doi: 10.1016/j.jconrel.2010.01.031. [DOI] [PubMed] [Google Scholar]
- 25.Geall AJ, Verma A, Otten GR, Shaw CA, Hekele A, Banerjee K, Cu Y, Beard CW, Brito LA, Krucker T, O'Hagan DT, Singh M, Mason PW, Valiante NM, Dormitzer PR, Barnett SW, Rappuoli R, Ulmer JB, Mandl CW. Nonviral delivery of self-amplifying RNA vaccines. Proc. Natl. Acad. Sci. USA. 2012;109(36):14604–14609. doi: 10.1073/pnas.1209367109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Xu J, Wang J, Luft JC, Tian S, Owens G, Jr, Pandya AA, Berglund P, Pohlhaus P, Maynor BW, Smith J, Hubby B, Napier ME, DeSimone JM. Rendering protein-based particles transiently insoluble for therapeutic applications. J. Am. Chem. Soc. 2012;134(21):8774–8777. doi: 10.1021/ja302363r. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Saito G, Swanson JA, Lee K-D. Drug delivery strategy utilizing conjugation via reversible disulfide linkages: role and site of cellular reducing activities. Adv. Drug Delivery Rev. 2003;55:199–215. doi: 10.1016/s0169-409x(02)00179-5. [DOI] [PubMed] [Google Scholar]
- 28.Gratton SEA, Ropp PA, Pohlhaus PD, Luft JC, Madden VJ, Napier ME, DeSimone JM. The effect of particle design on cellular internalization pathways. Proc. Natl. Acad. Sci. USA. 2008;105(33):11613–11618. doi: 10.1073/pnas.0801763105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bettinger T, Carlisle RC, Read ML, Ogris M, Seymour LW. Peptide-mediated RNA delivery: A novel approach for enhanced transfection of primary and post-mitotic cells. Nucleic Acids Res. 2001;29:3882–3891. doi: 10.1093/nar/29.18.3882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hasan W, Chu K, Gullapalli A, Dunn SS, Enlow EM, Luft JC, Tian S, Napier ME, Pohlhaus PD, Rolland JP, DeSimone JM. Delivery of multiple siRNAs using lipid-coated PLGA nanoparticles for treatment of prostate cancer. Nano Lett. 2012;12(1):287–292. doi: 10.1021/nl2035354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Wei CJ, Yassine HM, McTamney PM, Gall JG, Whittle JR, Boyington JC, Nabel GJ. Elicitation of broadly neutralizing influenza antibodies in animals with previous influenza exposure. Sci Transl Med. 2012;4(147):147ra114. doi: 10.1126/scitranslmed.3004273. [DOI] [PubMed] [Google Scholar]
- 32.Wang J, Tian S, Petros RA, Napier ME, Desimone JM. The complex role of multivalency in nanoparticles targeting the transferrin receptor for cancer therapies. J. Am. Chem. Soc. 2010;132(32):11306–11313. doi: 10.1021/ja1043177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Enlow EM, Luft JC, Napier ME, DeSimone JM. Potent Engineered PLGA Nanoparticles by Virtue of Exceptionally High Chemotherapeutic Loadings. Nano Lett. 2011;11(2):808–813. doi: 10.1021/nl104117p. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kelly JY, DeSimone JM. Shape-specific, monodisperse nano-molding of protein particles. J. Am. Chem. Soc. 2008;130(16):5438–5439. doi: 10.1021/ja8014428. [DOI] [PubMed] [Google Scholar]
- 35.Merkel TJ, Jones SW, Herlihy KP, Kersey FR, Shields AR, Napier M, Luft JC, Wu H, Zamboni WC, Wang AZ, Bear JE, DeSimone JM. Using mechanobiological mimicry of red blood cells to extend circulation times of hydrogel microparticles. Proc. Natl. Acad. Sci. USA. 2011;108(2):586–591. doi: 10.1073/pnas.1010013108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Merkel TJ, Chen K, Jones SW, Pandya AA, Tian S, Napier ME, Zamboni WE, Desimone JM. The effect of particle size on the biodistribution of low-modulus hydrogel PRINT particles. J. Control. Release. 2012;162(1):37–44. doi: 10.1016/j.jconrel.2012.06.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Perry JL, Reuter KG, Kai MP, Herlihy KP, Jones SW, Luft JC, Napier M, Bear JE, DeSimone JM. PEGylated PRINT nanoparticles: the impact of PEG density on protein binding, macrophage association, biodistribution, and pharmacokinetics. Nano Lett. 2012;12(10):5304–5310. doi: 10.1021/nl302638g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Garcia A, Mack P, Williams S, Fromen C, Shen T, Tully J, Pillai J, Kuehl P, Napier M, Desimone JM, Maynor BW. Microfabricated engineered particle systems for respiratory drug delivery and other pharmaceutical applications. J. Drug. Deliv. 2012;2012:941243. doi: 10.1155/2012/941243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dunn SS, Tian S, Blake S, Wang J, Galloway AL, Murphy A, Pohlhaus PD, Rolland JP, Napier ME, DeSimone JM. Reductively responsive siRNA-conjugated hydrogel nanoparticles for gene silencing. J. Am. Chem. Soc. 2012;134(17):7423–7430. doi: 10.1021/ja300174v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Malone RW, Felgner PL, Verma IM. Cationic liposome-mediated RNA transfection. Proc. Natl. Acad. Sci. USA. 1989;86(16):6077–6081. doi: 10.1073/pnas.86.16.6077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Read ML, Singh S, Ahmed Z, Stevenson M, Briggs SS, Oupicky D, Barrett LB, Spice R, Kendall M, Berry M, Preece JA, Logan A, Seymour LW. A versatile reducible polycation-based system for efficient delivery of a broad range of nucleic acids. Nucleic Acids Res. 2005;33(9):e86. doi: 10.1093/nar/gni085. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.