Abstract
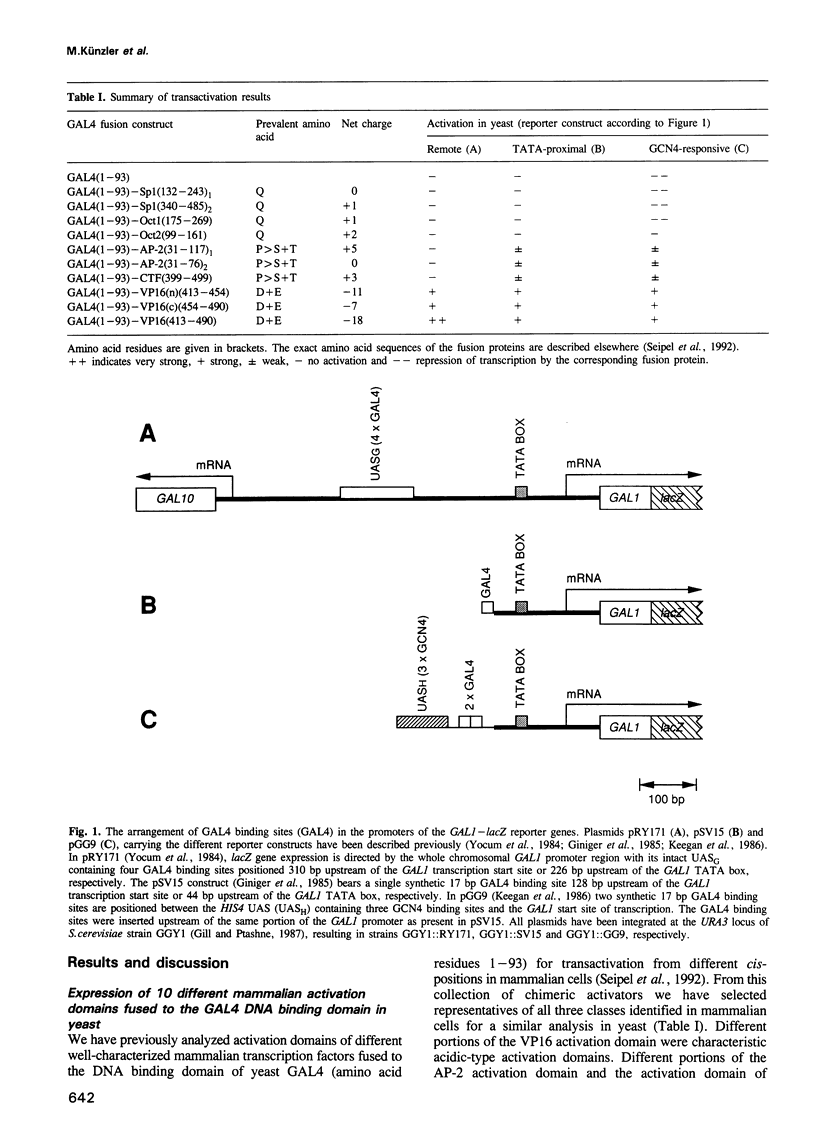
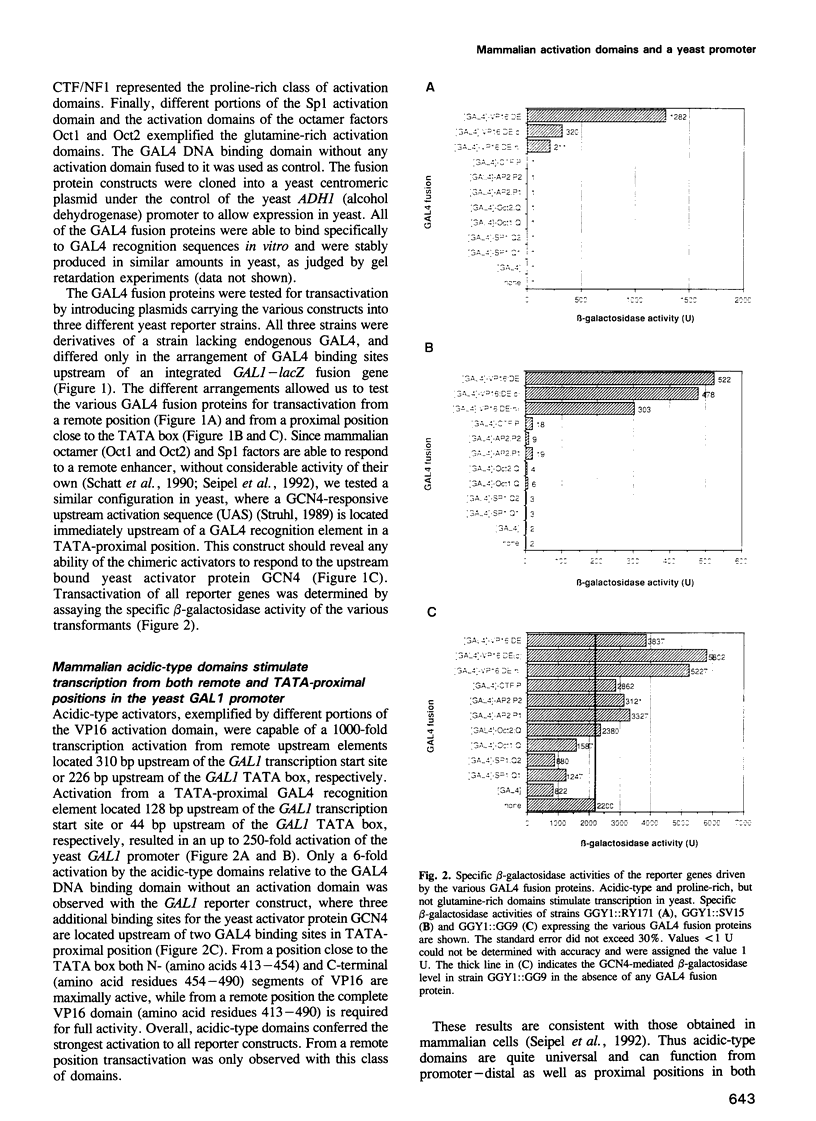
We have fused representatives of three structurally and functionally distinct classes of mammalian transcription activation domains for RNA polymerase II to the yeast GAL4 DNA binding domain. All fusion proteins were stable when expressed in yeast and were tested for their ability to activate transcription from various positions in the yeast GAL1 promoter. Activation domains functional from remote as well as TATA-proximal positions in mammalian cells, e.g. the acidic-type domain of VP16, also stimulate transcription in yeast from various promoter positions. Proline-rich domains, as e.g. in AP-2 and CTF/NF1, with considerable promoter activity and low enhancer activity in mammalian cells stimulate transcription in yeast only from a position close to the TATA box. The glutamine-rich domains of Oct1, Oct2 and Sp1, which activate transcription in mammalian cells from close to the TATA box in response to a remote enhancer, are inactive in the yeast GAL1 promoter. This finding might reflect some basic difference between the organization of yeast and mammalian promoters.
Full text
PDF


Images in this article
Selected References
These references are in PubMed. This may not be the complete list of references from this article.
- Arndt K. T., Styles C., Fink G. R. Multiple global regulators control HIS4 transcription in yeast. Science. 1987 Aug 21;237(4817):874–880. doi: 10.1126/science.3303332. [DOI] [PubMed] [Google Scholar]
- Braus G., Mösch H. U., Vogel K., Hinnen A., Hütter R. Interpathway regulation of the TRP4 gene of yeast. EMBO J. 1989 Mar;8(3):939–945. doi: 10.1002/j.1460-2075.1989.tb03455.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cochran M. D., Weissmann C. Modular structure of the beta-globin and the TK promoters. EMBO J. 1984 Nov;3(11):2453–2459. doi: 10.1002/j.1460-2075.1984.tb02155.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courey A. J., Tjian R. Analysis of Sp1 in vivo reveals multiple transcriptional domains, including a novel glutamine-rich activation motif. Cell. 1988 Dec 2;55(5):887–898. doi: 10.1016/0092-8674(88)90144-4. [DOI] [PubMed] [Google Scholar]
- Fink G. R. Pseudogenes in yeast? Cell. 1987 Apr 10;49(1):5–6. doi: 10.1016/0092-8674(87)90746-x. [DOI] [PubMed] [Google Scholar]
- Gill G., Ptashne M. Mutants of GAL4 protein altered in an activation function. Cell. 1987 Oct 9;51(1):121–126. doi: 10.1016/0092-8674(87)90016-x. [DOI] [PubMed] [Google Scholar]
- Giniger E., Varnum S. M., Ptashne M. Specific DNA binding of GAL4, a positive regulatory protein of yeast. Cell. 1985 Apr;40(4):767–774. doi: 10.1016/0092-8674(85)90336-8. [DOI] [PubMed] [Google Scholar]
- Guarente L., Bermingham-McDonogh O. Conservation and evolution of transcriptional mechanisms in eukaryotes. Trends Genet. 1992 Jan;8(1):27–32. doi: 10.1016/0168-9525(92)90021-u. [DOI] [PubMed] [Google Scholar]
- Hinnebusch A. G. Mechanisms of gene regulation in the general control of amino acid biosynthesis in Saccharomyces cerevisiae. Microbiol Rev. 1988 Jun;52(2):248–273. doi: 10.1128/mr.52.2.248-273.1988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ito H., Fukuda Y., Murata K., Kimura A. Transformation of intact yeast cells treated with alkali cations. J Bacteriol. 1983 Jan;153(1):163–168. doi: 10.1128/jb.153.1.163-168.1983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keegan L., Gill G., Ptashne M. Separation of DNA binding from the transcription-activating function of a eukaryotic regulatory protein. Science. 1986 Feb 14;231(4739):699–704. doi: 10.1126/science.3080805. [DOI] [PubMed] [Google Scholar]
- Kuenzler M., Balmelli T., Egli C. M., Paravicini G., Braus G. H. Cloning, primary structure, and regulation of the HIS7 gene encoding a bifunctional glutamine amidotransferase: cyclase from Saccharomyces cerevisiae. J Bacteriol. 1993 Sep;175(17):5548–5558. doi: 10.1128/jb.175.17.5548-5558.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lech K., Anderson K., Brent R. DNA-bound Fos proteins activate transcription in yeast. Cell. 1988 Jan 29;52(2):179–184. doi: 10.1016/0092-8674(88)90506-5. [DOI] [PubMed] [Google Scholar]
- Metzger D., White J. H., Chambon P. The human oestrogen receptor functions in yeast. Nature. 1988 Jul 7;334(6177):31–36. doi: 10.1038/334031a0. [DOI] [PubMed] [Google Scholar]
- Miozzari G., Niederberger P., Hütter R. Tryptophan biosynthesis in Saccharomyces cerevisiae: control of the flux through the pathway. J Bacteriol. 1978 Apr;134(1):48–59. doi: 10.1128/jb.134.1.48-59.1978. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mitchell P. J., Tjian R. Transcriptional regulation in mammalian cells by sequence-specific DNA binding proteins. Science. 1989 Jul 28;245(4916):371–378. doi: 10.1126/science.2667136. [DOI] [PubMed] [Google Scholar]
- Müller-Immerglück M. M., Schaffner W., Matthias P. Transcription factor Oct-2A contains functionally redundant activating domains and works selectively from a promoter but not from a remote enhancer position in non-lymphoid (HeLa) cells. EMBO J. 1990 May;9(5):1625–1634. doi: 10.1002/j.1460-2075.1990.tb08282.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Müller M. M., Gerster T., Schaffner W. Enhancer sequences and the regulation of gene transcription. Eur J Biochem. 1988 Oct 1;176(3):485–495. doi: 10.1111/j.1432-1033.1988.tb14306.x. [DOI] [PubMed] [Google Scholar]
- Oliver S. G., van der Aart Q. J., Agostoni-Carbone M. L., Aigle M., Alberghina L., Alexandraki D., Antoine G., Anwar R., Ballesta J. P., Benit P. The complete DNA sequence of yeast chromosome III. Nature. 1992 May 7;357(6373):38–46. doi: 10.1038/357038a0. [DOI] [PubMed] [Google Scholar]
- Passmore S., Maine G. T., Elble R., Christ C., Tye B. K. Saccharomyces cerevisiae protein involved in plasmid maintenance is necessary for mating of MAT alpha cells. J Mol Biol. 1988 Dec 5;204(3):593–606. doi: 10.1016/0022-2836(88)90358-0. [DOI] [PubMed] [Google Scholar]
- Pfeifer K., Kim K. S., Kogan S., Guarente L. Functional dissection and sequence of yeast HAP1 activator. Cell. 1989 Jan 27;56(2):291–301. doi: 10.1016/0092-8674(89)90903-3. [DOI] [PubMed] [Google Scholar]
- Pinkham J. L., Olesen J. T., Guarente L. P. Sequence and nuclear localization of the Saccharomyces cerevisiae HAP2 protein, a transcriptional activator. Mol Cell Biol. 1987 Feb;7(2):578–585. doi: 10.1128/mcb.7.2.578. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ptashne M. How eukaryotic transcriptional activators work. Nature. 1988 Oct 20;335(6192):683–689. doi: 10.1038/335683a0. [DOI] [PubMed] [Google Scholar]
- Sadowski I., Ma J., Triezenberg S., Ptashne M. GAL4-VP16 is an unusually potent transcriptional activator. Nature. 1988 Oct 6;335(6190):563–564. doi: 10.1038/335563a0. [DOI] [PubMed] [Google Scholar]
- Schatt M. D., Rusconi S., Schaffner W. A single DNA-binding transcription factor is sufficient for activation from a distant enhancer and/or from a promoter position. EMBO J. 1990 Feb;9(2):481–487. doi: 10.1002/j.1460-2075.1990.tb08134.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schena M., Yamamoto K. R. Mammalian glucocorticoid receptor derivatives enhance transcription in yeast. Science. 1988 Aug 19;241(4868):965–967. doi: 10.1126/science.3043665. [DOI] [PubMed] [Google Scholar]
- Seipel K., Georgiev O., Schaffner W. Different activation domains stimulate transcription from remote ('enhancer') and proximal ('promoter') positions. EMBO J. 1992 Dec;11(13):4961–4968. doi: 10.1002/j.1460-2075.1992.tb05603.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sengstag C., Hinnen A. The sequence of the Saccharomyces cerevisiae gene PHO2 codes for a regulatory protein with unusual aminoacid composition. Nucleic Acids Res. 1987 Jan 12;15(1):233–246. doi: 10.1093/nar/15.1.233. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl K. Molecular mechanisms of transcriptional regulation in yeast. Annu Rev Biochem. 1989;58:1051–1077. doi: 10.1146/annurev.bi.58.070189.005155. [DOI] [PubMed] [Google Scholar]
- Struhl K. The JUN oncoprotein, a vertebrate transcription factor, activates transcription in yeast. Nature. 1988 Apr 14;332(6165):649–650. doi: 10.1038/332649a0. [DOI] [PubMed] [Google Scholar]
- Suzuki Y., Nogi Y., Abe A., Fukasawa T. GAL11 protein, an auxiliary transcription activator for genes encoding galactose-metabolizing enzymes in Saccharomyces cerevisiae. Mol Cell Biol. 1988 Nov;8(11):4991–4999. doi: 10.1128/mcb.8.11.4991. [DOI] [PMC free article] [PubMed] [Google Scholar]
- White M. J., Hirsch J. P., Henry S. A. The OPI1 gene of Saccharomyces cerevisiae, a negative regulator of phospholipid biosynthesis, encodes a protein containing polyglutamine tracts and a leucine zipper. J Biol Chem. 1991 Jan 15;266(2):863–872. [PubMed] [Google Scholar]
- Yocum R. R., Hanley S., West R., Jr, Ptashne M. Use of lacZ fusions to delimit regulatory elements of the inducible divergent GAL1-GAL10 promoter in Saccharomyces cerevisiae. Mol Cell Biol. 1984 Oct;4(10):1985–1998. doi: 10.1128/mcb.4.10.1985. [DOI] [PMC free article] [PubMed] [Google Scholar]



